Abstract
Investigating human oligodendrogenesis and the interaction of oligodendrocytes with neurons and astrocytes would accelerate our understanding of the mechanisms underlying white matter disorders. However, this is challenging due to limited accessibility of functional human brain tissue. Here, we developed a novel differentiation method of human induced pluripotent stem cells (hiPS cells) to generate three-dimensional (3D) neural spheroids that contain oligodendrocytes as well as neurons and astrocytes, called human oligodendrocyte spheroids (hOLS). We demonstrate that oligodendrocyte-lineage cells derived in hOLS transition through developmental stages similar to primary human oligodendrocytes and that the migration of oligodendrocyte-lineage cells and their susceptibility to lysolecithin exposure can be captured by live imaging. Moreover, their morphology changes as they mature over time in vitro and start myelinating neurons. We anticipate that this method can be used to study oligodendrocyte development, myelination, and interactions with other major cell types in the central nervous system.
INTRODUCTION
Oligodendrocytes play key roles in brain development including myelinating and electrically insulating neuronal axons for impulse propagation, as well as providing trophic and metabolic support for neurons1–4. These functions are coordinated by communication between oligodendrocytes and neighboring astrocytes and neurons5–7, which occurs both through physical interactions and through secreted factors5,8–11. During neural development, oligodendrocyte-lineage cells progress from mobile, bipolar oligodendrocyte progenitor cells (OPCs) to stationary, highly branched mature oligodendrocytes. The loss of oligodendrocytes or alterations in their ability to migrate, myelinate, or communicate with other cell types can lead to diseases such as multiple sclerosis and vanishing white matter disease12,13.
While methods have been developed to generate oligodendrocytes from human pluripotent stem (hPS) cells14–18, these models cannot be maintained long term in vitro and lack the diversity of mature cell types and the cytoarchitecture that oligodendrocytes encounter in vivo. These features of two-dimensional culture make the study of cellular interactions difficult as they occur in a three-dimensional (3D) environment in vivo. The advent of organoid methodologies has allowed the generation of self-organized cellular models with complex cytoarchitecture in vitro, and these cultures can be maintained over extended periods of time19. Here, we develop an approach to differentiate human induced pluripotent (hiPS) cells into 3D neural spheroids to model the development of human oligodendrocyte-lineage cells alongside neurons and astrocytes. Our method produces human neurons, astrocytes, and oligodendrocytes that co-develop both spatially and temporally. The oligodendrocytes transition through developmental stages and are transcriptionally similar to primary oligodendrocytes. Oligodendrocyte-lineage cells mature morphologically and electrophysiologically over time and ultimately myelinate nearby axons.
RESULTS
Generation and Characterization of human oligodendrocyte spheroids (hOLS)
To generate human oligodendrocyte spheroids (hOLS) that contain oligodendrocytes, astrocytes, and neurons, we leveraged approaches to derive region-specific 3D cultures that we previously developed20,21. We enzymatically dissociated hiPS cells into a single cell suspension (n = 7 hiPS cell lines derived from 7 control subjects; Supplementary Table 1) and aggregated them into spheroids using microwells (Fig. 1a). After dislodging the spheroids from the microwells 18–24 hours later, we exposed them to dual SMAD inhibitors dorsomorphin (DM) and SB-431542 followed by epidermal growth factor (EGF) and fibroblast growth factor 2 (FGF2). Smoothened agonist (SAG) and IWP-2 were added to activate the SHH pathway (day 12–24) and inhibit the WNT pathway (day 4–24), respectively. From day 25 to day 36, we added platelet derived growth factor (PDGF), hepatocyte growth factor (HGF), insulin-like growth factor 1 (IGF1), neurotrophin 3 (NT-3), brain-derived neurotrophic factor (BDNF), insulin, triiodo-L-thyronine (T3), biotin, and a cyclic AMP analog (cAMP) to promote oligodendrocyte progenitor cell (OPC) survival and proliferation14. At day 37, insulin, biotin, T3, cAMP, and L-ascorbic acid (AA) were added to the media to promote oligodendrocyte maturation. From day 25 to day 36 of in vitro culture, hOLS showed high expression of the ectoderm marker SOX2, but not the mesoderm or endoderm markers BRACH and SOX17 (Supplementary Fig. 1a; n = 4 samples from hOLS derived from 4 hiPS cell lines). At day 37, hOLS expressed the forebrain markers FOXG1, SIX3, NKX2–1, OTX2, PAX6, and LHX2 at levels comparable to or higher than our previously described method to generate human cortical spheroids (hCS)20,22, but not midbrain (LMX1B), hypothalamus (RAX) or spinal cord (HOXB4) related genes (Supplementary Fig. 1b; n = 5 samples from hOLS derived from 5 hiPS cell lines).
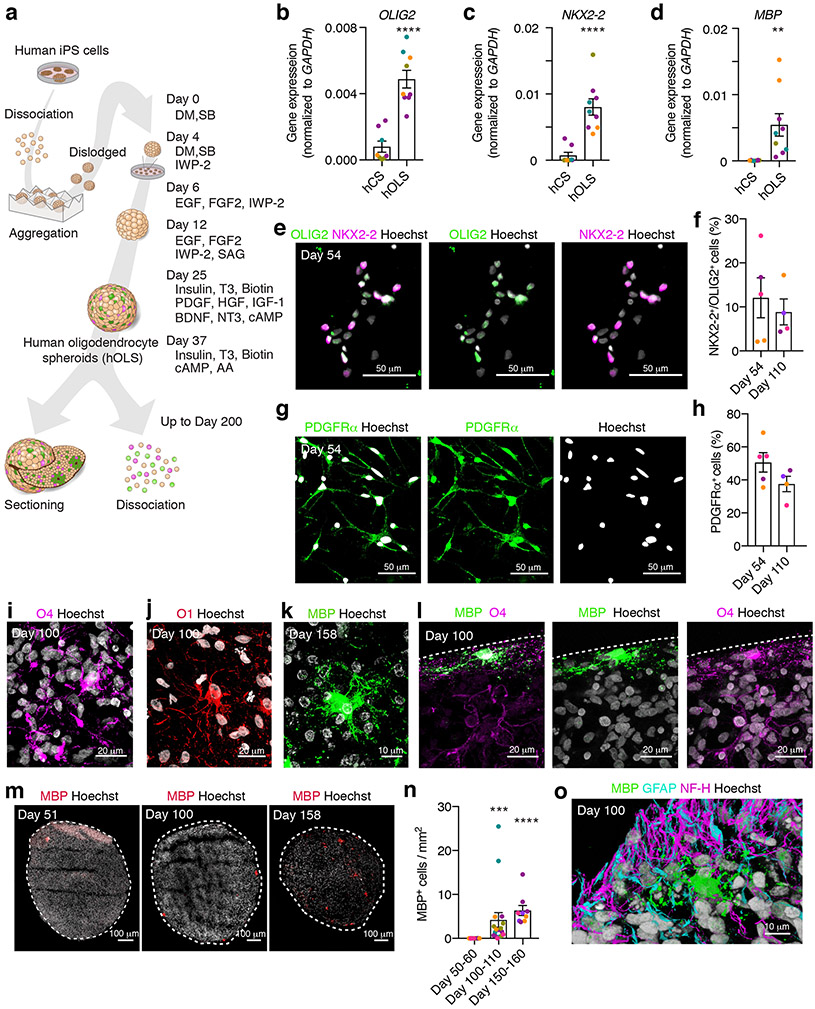
FIGURE 1. Characterization of hOLS derived from hiPS cell lines.
a, Schematic for generating hOLS. Human iPS cells are enzymatically dissociated and aggregated in microwells to form spheroids. DM, dorsomorphin; SB, SB-431542; SAG, smoothened agonist; HGF, hepatocyte growth factor; AA, ascorbic acid to the end of the description of 1a.
b–d, Relative gene expression (normalized to GAPDH) as determined by qPCR at day 100 of in vitro culture in hCS and hOLS of (b) OLIG2 (two-tailed Mann-Whitney test, ****P< 0.0001), (c) NKX2–2 (two-tailed Mann-Whitney test, ****P<0.0001) and (d) MBP (two-tailed t-test, t = 2.97, df=15, ***P=0.009). In b–d, for hCS n= 8 and for hOLS n= 9 RNA samples from spheroids derived from 4 hiPS cell lines in 1–4 differentiation experiments; see Supplementary Table 1. Lines are shown in different colors; each point represents 2–4 hOLS pooled from one differentiation experiment.
e–h, Day 54 immunostaining and quantification of OLIG2 and NKX2–2 double positive cells (e, f) (two-tailed t-test, t=0.55, df=7, P= 0.59), and of PDGFRα (g, h) (two-tailed t-test, t=1.68, df=7, P= 0.13) out of Hoechst in dissociated hOLS at day 54 and day 110 (n= 5 samples each consisting of 4–6 hOLS derived from 3 hiPS cell lines; hiPS cell lines shown in different colors; see Supplementary Table 1).
i–l, Immunostaining of (i) O4, (j) O1, (k) MBP, (l) MBP and O4 in cryosections. Immunostainings were repeated on hOLS from 6 independent inductions for O4, 3 independent inductions for O1, 5 independent inductions for MBP, and 3 independent inductions for MBP and O4 together with similar results.
m, n, Immunostaining (m) and quantification (n) of MBP+ cells over time in whole hOLS cryosection (day 50–60: n=12 hOLS from 5 hiPS cell lines; day 100–110: n=17 hOLS from 5 hiPS cell lines; day 150–160: n=9 hOLS from 4 hiPS cell lines, each point represents one hOLS; see Supplementary Table 1; Kruskal-Wallis test, P< 0.0001 with Dunn’s multiple comparison test day 50–60 versus day 100–110, ***P=0.0005 and day 50–60 versus day 100–110, ****P<0.0001).
o, Immunostaining of MBP, GFAP and NF-H in hOLS cryosections. Immunostainings were repeated on hOLS from 3 independent inductions with similar results.
Data are mean ± s.e.m.
Scale bars, 10 μm (k, o), 20 μm (i, j, l), and 50 μm (e, g), and 100 μm (m).
At day 100 of in vitro differentiation, we found a significant increase in gene expression of OLIG2, NKX2–2, and MBP in hOLS as determined by qPCR compared to hCS20, suggesting an enrichment of oligodendrocyte-lineage cells (n = 9 samples from hOLS and n = 8 samples from hCS derived from 4 hiPS cell lines; P< 0.001; Fig. 1b–d). We next performed immunofluorescent labeling on cryosections (Supplementary Fig. 1c) obtained at day 51 and dissociated cultures obtained from day 54 and day 110 hOLS to test for the presence of NKX2–2+/OLIG2+ double positive OPCs and PDGFRα+ OPCs (Fig. 1e–h). Approximately 12% of cells at day 54 and 9% of cells at day 110 were NKX2–2+/OLIG2+ double positive (Fig. 1f; n= 4–5 samples from hOLS derived from 3 hiPS cell lines) and approximately 51% of cells at day 54 and 35% of cells at day 110 were PDGFRα+ (Fig. 1h; n= 4–5 samples from hOLS derived from 3 hiPS cell lines). To determine whether OPCs were maturing into oligodendrocytes over time, we immunostained cryosections obtained from hOLS at day 100–160 of in vitro differentiation. We observed O4+, O1+, and MBP+ cells, indicating a range of oligodendrocyte stages from pre-oligodendrocytes to mature, late stage oligodendrocytes (Fig. 1i–k). Interestingly, we found both O4+ cells that were bipolar and did not express MBP, as well as O4+ cells that were highly branched and overlapped with MBP (Fig. 1l). To determine whether the abundance of mature oligodendrocytes increased in hOLS over time, we quantified the density of MBP+ cells in whole cryosections between days 50 and 160 of differentiation. We observed an increase in the density of MBP+ cells and that most MBP+ cells were located in the outer third of each section (Fig. 1m, n; Supplementary Fig. 1d; n= 9–17 hOLS from 6 hiPS cell lines; P< 0.0001).
A goal of this method was to produce oligodendrocytes in close proximity to neurons and astrocytes, so we performed immunostainings in day 100 hOLS cryosections for MBP, GFAP, and Neurofilament-H (200 kD) (NF-H). We observed both neurons and glial-lineage cells in close proximity to each other (Fig. 1o). Moreover, the relative gene expression of the neural marker RBFOX3 as determined by qPCR at day 100 was comparable between hCS and hOLS (Supplementary Fig. 1e; P> 0.05), while the glial marker GFAP was expressed at a higher level in hOLS (Supplementary Fig. 1e; P< 0.01). Quantifications in dissociated cultures revealed that approximately 45% of cells at day 54 and 20% of cells at day 110 were MAP2+, and 8% of cells at day 54 and 21% of cells at day 110 were GFAP+ (Supplementary Fig 1f–i; n= 4–5 samples from hOLS from 3 hiPS cell lines). To verify neurotransmitter identity in these cultures, we next used qPCR and found that hOLS expressed more of the GABAergic related GAD1 gene (P< 0.01) and less of the glutamatergic-transporter-encoding SLC17A7 gene (P<0.001) (also known as VGLUT1) than hCS (Supplementary Fig. 1j, k).
Single-cell RNA-seq of hOLS-derived oligodendrocytes
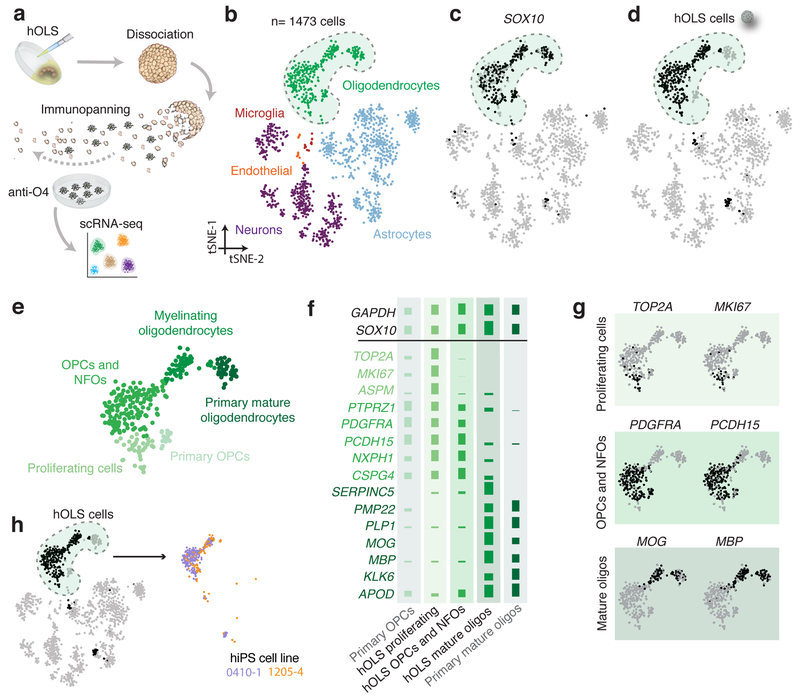
To comprehensively characterize oligodendrocyte-lineage cells in hOLS, we isolated O4+ cells from day 127 hOLS by immunopanning and performed deep single-cell RNA sequencing (Fig. 2a). We sequenced 295 cells derived from two hiPS cell lines and compared their profiles to cells isolated from primary human fetal cortex, primary human adult cortex, and hCS (Supplementary Fig. 2a). Clustering of all cells using the t-distributed stochastic neighbor embedding (tSNE)23 approach revealed distinct populations of SOX10+ oligodendrocytes, STMN2+ neurons, SOX9+ astrocytes, CX3CR1+ microglia, and FLT1+ endothelial cells (Fig. 2b, c; Supplementary Fig. 2b). The O4+ cells derived from hOLS clustered most closely to OPCs and mature oligodendrocytes from the primary human adult cortical tissue within the SOX10+ cluster (Fig. 2d, e). On closer inspection, the oligodendrocyte cluster contained populations of proliferating cells, OPCs and newly formed oligodendrocytes (NFOs), and myelinating oligodendrocytes derived from hOLS that had similar patterns of marker expression as primary OPCs and primary mature oligodendrocytes (Fig. 2f, g; Supplementary Fig. 2c; also see Supplementary Fig. 2a for examples of genes differentially expressed between primary and hOLS samples). Expression of oligodendrocyte stage-specific markers was confirmed in cells from each cluster by qPCR (Supplementary Fig. 2d). Moreover, we found O4+ cells in the three oligodendrocyte subclusters in hOLS from two hiPS cell lines and a high transcriptomic consistency across lines (Pearson’s r= 0.96, log normalized gene expression) (Fig.2h; Supplementary Fig. 2e,f).
FIGURE 2. Transcriptional comparison of hOLS oligodendrocyte-lineage cells to primary tissue cells.
a, Schematic showing the isolation of O4+ cells from hOLS.
b, tSNE clustering single cell RNA-seq data from hOLS (n = 295 cells), primary human brain tissue and hCS (n= 1473 cells total; colored by cell type).
c, Gene expression of oligodendrocyte-lineage related SOX10 in single cells.
d, O4+ hOLS-derived single cells.
e, Oligodendrocyte cluster from tSNE map in (b) with three distinct k-means subclusters of hOLS.
f, Mean expression of oligodendrocyte lineage-specific genes in hOLS as well as primary OPCs and mature oligodendrocytes isolated from adult human brain tissue (log2 data normalized across rows).
g, Single cell gene expression of subcluster-specific markers in the oligodendrocyte-lineage cluster.
h, O4+ single cells derived from hOLS indicated by hiPS cell line.
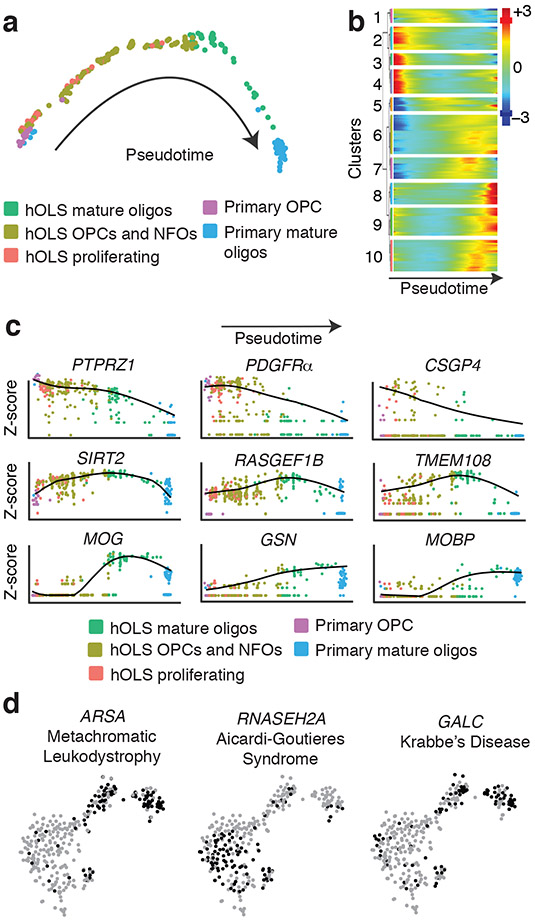
To further assess the developmental progression of oligodendrocyte-lineage cells in hOLS, we performed Monocle analysis, which utilizes an unsupervised algorithm to reveal single-cell gene expression kinetics over time and orders cells through a biological process24. This analysis revealed a spectrum of oligodendrocyte-lineage stages in hOLS ranging from dividing cells expressing MKI67 and TOP2A, which closely resemble primary OPCs, to mature cells expressing MOG and MBP, which closely resemble primary mature oligodendrocytes (Fig. 3a). This analysis further identified several temporal patterns of gene expression, including some genes that were highly expressed early and decreased over pseudotime, some that peaked mid-pseudotime, and others that were more highly expressed at later pseudotime points (Fig. 3b). Early pseudotime genes expressed in dividing oligodendrocytes included PTPRZ1, PDGFRA, and CSGP4; mid-pseudotime genes expressed in immature oligodendrocytes included SIRT2, RASGEF1B, and TMEM108; late pseudotime genes expressed in mature oligodendrocytes included MOG, GSN, and MOBP25 (Fig. 3c; Supplementary Fig. 3b).
FIGURE 3. Developmental trajectory and expression of disease-related genes in hOLS.
a, Monocle pseudotime analysis of all oligodendrocyte-lineage cells colored by tissue of origin. Most mature time points are shown on right.
b, Hierarchical clustered heat map depicting gene modules whose expression patterns covary across pseudotime (z-scores normalized by row).
c, Expression of oligodendrocyte lineage markers across pseudotime (colored by tissue of origin; log2 data normalized by gene).
d, Singe cell gene expression pattern of disease-implicated genes in the oligodendrocyte lineage cluster from (Fig. 2b).
Anticipating that hOLS may be useful for disease modeling, we analyzed the single-cell expression pattern of genes associated with oligodendrocyte-related disorders (Fig. 2e and 3d and Supplementary Fig. 3c). We found that a gene associated with Aicardi-Goutieres Syndrome (RNASEH2A)26 was expressed primarily in OPCs and NFOs, and that genes associated with metachromatic leukodystrophy (ARSA)27, and Krabbe Disease (GALC)28 were expressed in mature oligodendrocytes. These findings suggest that having access to multiple stages of oligodendrocyte development may be important for disease modeling.
Maturation of oligodendrocytes in hOLS
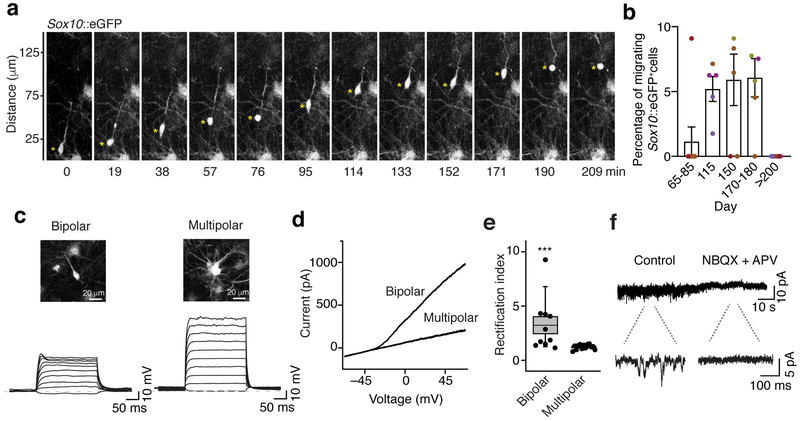
Prompted by the finding that oligodendrocyte lineage cells mature transcriptionally in hOLS, we next examined their morphology and functional properties. During development, bipolar OPCs migrate from their region of origin throughout the cortex where they ultimately cease migrating and extend multiple processes that interact with nearby neurons29. To determine whether hOLS-derived OPCs migrate in hOLS as they do in vivo, we used temperature-controlled live imaging with a previously described Sox10-MCS5::eGFP reporter30 at in vitro differentiation stages ranging from day 65 up to day 275 in culture (Fig. 4a). We found that the propensity of Sox10-MCS5::eGFP+ cells to migrate peaked between day 110 and 180 of in vitro culture, but all Sox10-MCS5::eGFP+ cells were stationary by day 235 (Fig. 4b; n = 5–8 hOLS from 5 hiPS cell lines; P< 0.01).
FIGURE 4. Oligodendrocyte maturation in hOLS.
a, Example of time lapse imaging of a migrating Sox10-MCS5::eGFP+ cell in hOLS.
b, Percentage of migrating Sox10-MCS5::eGFP+ cells over differentiation time in vitro. (day 65–85: n=8 hOLS from 3 hiPS cell lines; day 115: n=5 hOLS from 3 hiPS cell lines; day 150: n=6 hOLS from 3 hiPS cell lines; day 170–180: n=6 hOLS from 3 hiPS cell lines; day >200: n=8 hOLS from 4 hiPS cell lines; see Supplementary Table 1). Lines are shown in different colors; Kruskal-Wallis test, P=0.003). Data are mean ± s.e.m. asterisk indicates the soma of a migrating cell
c, Slice recordings in hOLS showing membrane voltage responses in bipolar and multipolar cells following intracellular current injections.
d, Example of an I-V curve showing an outward rectifying current in a bipolar cell but not in a multipolar cell. For c and d, recordings were repeated in 12 bipolar cells and 13 multipolar cells from 3 independent inductions with similar results.
e, Quantification of the rectification index in bipolar and multipolar Sox10-MCS5::eGFP+ cells (n=13 multipolar cells, n=12 bipolar cells, two-tailed Mann-Whitney test, ***P= 0.0009). Dots represent individual cells, box edges represent s.e.m., the middle horizontal lines within the box represent the mean, and whiskers represent the 10th and 90th percentile of the population.
f, Voltage clamp recording from a representative bipolar Sox10-MCS5::eGFP+ cell showing a reduction in holding current variance in response to the blockers of glutamate receptors APV (40 μM) and NBQX (20 μM). Recordings were repeated in 5 cells from 2 independent inductions with similar results.
We next sought to determine whether hOLS-derived oligodendrocyte lineage cells mature electrophysiologically. As OPCs mature to multipolar oligodendrocytes, expression of voltage-gated sodium and potassium channels is progressively reduced31,32. We performed whole cell current clamp recordings from bipolar and multipolar Sox10-MCS5::eGFP+ cells in slices of hOLS from days 108 to 175 (Fig. 4c). We observed that the membrane response to current injection of bipolar cells, but not multipolar cells, was highly non-linear, indicating expression of voltage-dependent sodium and potassium channels in the former. In fact, the I-V curve of bipolar shaped cells was outwardly rectifying and therefore suggestive of slowly inactivating potassium currents (Fig. 4d, e; n= 10–13 cells, P< 0.001), and this difference was accompanied by complementary differences in input resistance and capacitance (Supplementary Fig. 4a, b). Moreover, during oligodendrocyte development, the release of glutamate by nearby neurons plays an instructive role in OPC differentiation33,34. We tested for sensitivity to released glutamate in voltage clamped Sox10-MCS5::eGFP+ cells and observed discrete inward-going events in approximately 70% of bipolar and multipolar cells (n= 13 and 10 cells, respectively; Fig. 4f). These events were blocked by application of 40 μM APV and 20 μM NBQX (n= 5 cells; Fig. 4f; Supplementary Fig. 4c), indicating that Sox10-MCS5::eGFP+ cells express ionotropic glutamate receptors that are spontaneously activated in hOLS.
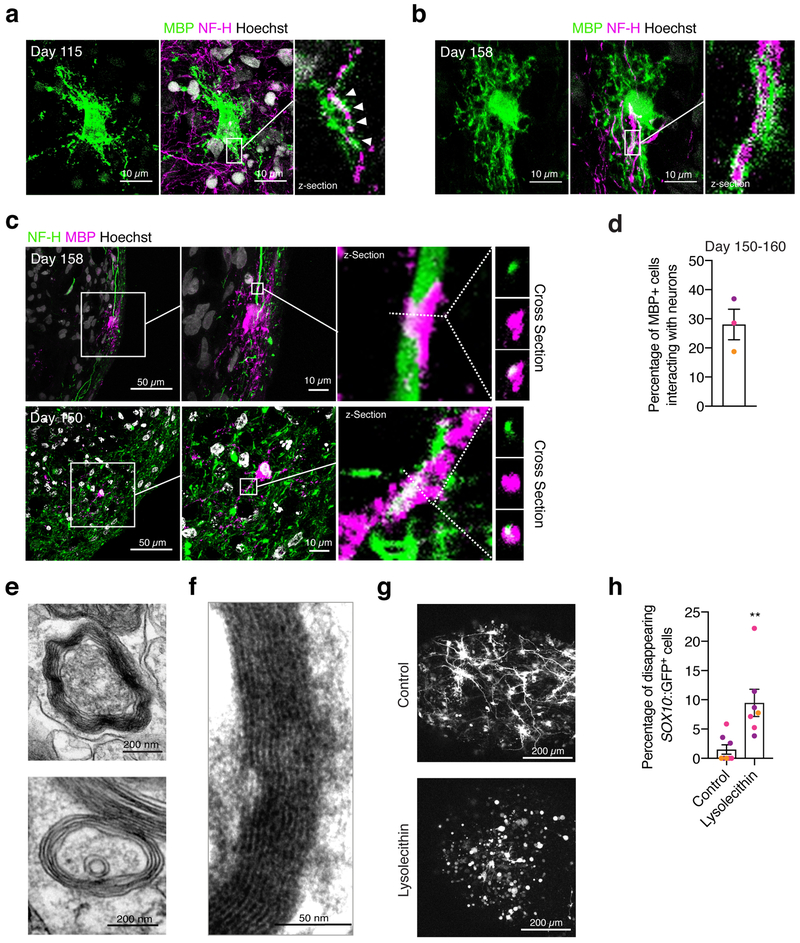
A hallmark of oligodendrocyte maturation is the ability to interact with and myelinate neuronal axons. We immunostained hOLS cryosections at day 115 for MBP and NF-H and observed the interaction of oligodendrocyte processes and nearby axons (Fig. 5a). Moreover, in hOLS cryosections obtained at day 150-158, we identified examples of MBP flanking neurofilament staining, indicative of myelination (Fig. 5b, c; Supplementary Fig. 4d). We found that at day 150–160, ~28% of MBP+ cells interacted with NF-H+ processes (Fig. 5d; n = 9 hOLS from 3 hiPS cell lines). To verify myelination, we performed transmission electron microscopy at day 100–170 in hOLS derived from 3 hiPS cell lines and found various stages of myelination, including lamellae of compact myelin surrounding axons (Fig. 5e, f; Supplementary Fig. 4e, f). Lastly, as a proof-of-principle that hOLS can be used to study oligodendrocyte loss or injury, we treated day 75–85 hOLS with a toxic phospholipid lysolecithin for 15–17 hours (Fig. 5g), which has been shown to cause demyelination in vivo and in explant cultures35–37. Monitoring by live imaging at 4–6 hours after treatment revealed changes in the morphology and loss of Sox10-MCS5::eGFP+ cells (Fig. 5g, h).
FIGURE 5. Oligodendrocyte-neuron interaction and myelination in hOLS.
a, b, Immunostaining of hOLS cryosections with MBP and NF-H (n=9 hOLS from 3 hiPS cell lines).
c, Example images of interactions between MBP+ cells and NF-H+ processes in cryosections at day 150–158 of in vitro culture imaged by confocal microscopy. The first and seconds panels of each row are max projections, the third panel of each row is an individual z-section, and the right most panels are cross sections.
d, Quantification of MBP+ cells that interact with NF-H+ neurons out of total MBP+ cells.
e, f, Transmission electron microscopy images of hOLS at day 103 of differentiation showing myelin in line 2242–1. Examples from hOLS derived from other hiPS cell lines are shown in Supplementary Fig. 4. Electron microscopy was performed on hOLS from 3 independent inductions with similar results.
g, Example images of Sox10-MCS5::eGFP+ cells in untreated and lysolecithin-treated hOLS after 13 hours of live imaging.
h, Percentage of Sox10-MCS5::eGFP+ cells that disappear during the 13-hour live imaging experiment following lysolecithin treatment (n=8 untreated hOLS from 3 hiPS cell lines, n=7 lysolecithin-treated hOLS from 3 hiPS cell lines, two-tailed Mann-Whitney test, **P=0.001).
Data are mean ± s.e.m.
Scale bars, 200 μm (g) 50 μm (c left panel) 10 μm (a, b, c middle panel) 200 nm (e), 50 nm (f).
DISCUSSION
We demonstrate the generation of human oligodendrocyte lineage cells in a 3D cellular platform that includes multiple stages of oligodendrocyte development, migration, and myelination. Our system produces neurons and astrocytes that spatially and temporally overlap with oligodendrocytes, allowing interaction between cell types. This model has several advantages over existing methods to produce oligodendrocytes in vitro, including the existence of multiple mature cell types (Supplementary Table 2). Additionally, the 3D nature of this platform more closely resembles the cytoarchitecture and cellular milieu that oligodendrocytes would encounter in vivo. There is increasing evidence of the importance of interactions between oligodendrocytes and astrocytes and between oligodendrocytes and neurons during development both through secreted factors and physical interactions5,8–11, and the recapitulation of these interactions may be essential to building more realistic in vitro models of brain function and dysfunction.
To validate the stages of oligodendrocyte development in hOLS, we performed deep single cell RNA-seq and identified a progression of oligodendrocyte lineage cells from dividing progenitors to mature oligodendrocytes. We additionally compared this transcriptional data to the signature of primary human oligodendrocytes isolated from human cerebral cortex and found that they clustered closely. Future studies are necessary to confirm transcriptional similarity in additional hiPS cell lines and with larger cell numbers.
The transcriptional maturation of oligodendrocyte lineage cells in hOLS was accompanied by functional changes as assessed by electrophysiology and electron microscopy. The maturation and similarity of oligodendrocytes in hOLS to primary adult samples suggests that this model can be applied to answer basic questions regarding human oligodendrocyte development, as demonstrated by the expression of white matter disease-related genes in single cells derived from hOLS. In order to study later stages of oligodendrocyte maturation and myelination, future methods need to be developed to better quantify the extent of compact myelination in 3D culture. Quantifying wrapping by traditional electron microscopy techniques is difficult due to inconsistency in the directionality of axons in hOLS. However, these current limitations may be resolved by incorporating scaffolds to guide projecting axons in hOLS or by performing high-throughput electron microscopy to image larger areas of hOLS and obtain more accurate quantifications of myelination.
A remaining obstacle in the use of 3D cultures to study disease processes is the inherent inter- and intra-organoid variability that complicates the comparison between control and disease-related cultures38. Strategies to limit the variability between organoids include directing differentiation to specific brain regions, limiting the use of embedding in extracellular matrices that can induce spurious fates, using cell specific reporters for imaging cell types of interest, and using single cell sequencing19,39. In hOLS, the variability in OPC yield is higher at earlier stages and decreases over time. Interestingly, we also found that transcriptional variability is lower at later stages of hCS differentiation40. This suggests that, for future applications, allowing hOLS to mature to around day 100 would result in higher consistency.
This hOLS method can be used to live image multiple stages of oligodendrocyte development over long periods of time in vitro to dissect the physical mechanisms of OPC migration and myelination. Here, as a proof of principle, we demonstrate that treatment with lysolecithin induces oligodendrocyte cell loss and that this process can be monitored by live imaging. Future studies can apply this cellular model to study leukodystrophies and other disorders affecting myelination in the central nervous system. The hOLS have the potential to be fused with other brain region-specific spheroids to derive brain assembloids to study the migration and differentiation of oligodendrocyte-lineage cells in different brain regions. hOLS may also potentially be combined with autologous patient-derived immune cells to study neuro-immune dysfunction, such as in multiple sclerosis. The personalized nature of the hiPS cell-derived platform will further allow the study of these cellular processes in the context of disease, such as vanishing white matter disorder12, or environmental alterations, such as the effect of hypoxic injury on the premature brain41. The scalability of hOLS also makes this system amenable to genetic and small molecule screens to discover modulators of human myelination and identify novel therapeutics.
METHODS
Culture of hiPS cells
The hiPS cell lines used in this study were validated using standardized methods as previously described42,43. Cultures were tested for and maintained mycoplasma free. A total of 7 control hiPS cell lines derived from fibroblasts collected from 7 subjects were used for experiments (Supplementary Table 1). The hiPS cell line H20961 was derived by the Gilad laboratory44. Approval was obtained from the Stanford IRB panel and informed consent was obtained from all subjects.
Generation of hCS and hOLS from hiPS cells
hiPS cells were cultured on vitronectin-coated plates (5 μg/mL, Thermo Fisher Scientific, A14700) in Essential™ 8 media (Thermo Fisher Scientific, A1517001). Cells were passaged every 4 days with 0.5 mM EDTA (Life Technologies, pH 8.0).
For the generation of 3D neural spheroids, hiPS cells were incubated with accutase (Innovate Cell Technologies, AT104) at 37ºC for 7 minutes and dissociated into single cells. To obtain uniformly-sized spheroids, AggreWell-800™ (Stemcell Technologies, 34815) containing 300 microwells were used. Approximately 3 × 106 single cells were added per AggreWell-800™ well in Essential™ 8 medium supplemented with the ROCK inhibitor Y-27632 (10 μM, EMD Chemicals, S1049), centrifuged to capture the cells in the microwells and incubated at 37ºC with 5% CO2. After 18–24 hours, spheroids consisting of approximately 10,000 cells were harvested from each microwell by pipetting medium in the well up and down with a cut pipet tip and transferred into ultra-low attachment plastic dishes (Thermo Fisher Scientific, 3262) in Essential™ 6 medium (Thermo Fisher Scientific, A1516401) supplemented with two SMAD pathway inhibitors– dorsomorphin (DM, 2.5 μM, Sigma-Aldrich, P5499-CONF) and SB-431542 (10 μM, R&D systems, 1614). For the first five days, Essential™ 6 medium was changed every day and supplemented with DM and SB-431542.
To generate hOLS, on day 6 in suspension the spheroids were transferred to differentiation and maintenance medium (DMM) containing DMEM/F12 (Thermo Fisher Scientific, 11330–057), B-27 Supplement without vitamin A (Thermo Fisher Scientific, 12587010), N2 Supplement (Thermo Fisher Scientific, 17502048), minimum essential media (MEM) Non-Essential Amino Acids (1:100, Thermo Fisher Scientific, 11140076), GlutaMax (1:100, Thermo Fisher Scientific, 35050079), human insulin (25 μ g ml−1, Sigma-Aldrich, I9278-5ML), β-mercaptoethanol (0.1 mM; Sigma-Aldrich M3148), and penicillinstreptomycin (1:100, Thermo Fisher Scientific, 15140163). The DMM medium was supplemented with 20 ng/ml EGF (R&D Systems, 236-EG-01M) and 20 ng/ml basic fibroblast growth factor (bFGF) (Peprotech, 100–26) for 19 days (until day 24) with daily medium change in the first 10 days, and every other day for the subsequent 9 days. The Wnt pathway inhibitor IWP-2 (5 μM, Selleckchem, S7085) was added from day 4 until day 24, and the small molecule SHH pathway smoothened agonist (SAG, 1 μM, Millipore Sigma, 566660) was added from day 12 to day 24. From day 25 to day 36, hOLS were cultured in DMM supplemented with T3 (60 ng/mL, Sigma Aldrich, T2877), biotin (100 ng/mL, Sigma Aldrich, B4639), NT-3 (20 ng/mL, Peprotech, 450–03), BDNF (20 ng/mL, Peprotech, 450–02), cAMP (1 μM, Sigma Aldrich, D0627), HGF (5 ng/mL, Peprotech, 315–23), IGF-1 (10 ng/mL, VWR, 100–11), and PDGF-AA (10 ng/mL, R&D Systems, 221-AA). From day 37 onwards, hOLs were cultured in complete DMM (DMM supplemented with T3, biotin, cAMP, and ascorbic acid (AA, 20 μg/mL, Wako Pure Chemical, 323–44822). From day 17 to day 43 media changes were performed every other day. From day 44 onwards media changes were preformed every 4–5 days.
The generation of hCS from hiPS cells was performed similarly to a method we previously described20. To generate hCS, on day 6 in suspension the microwell-generated spheroids were transferred to neural medium (NM) containing Neurobasal A (Thermo Fisher Scientific, 10888022), B-27 Supplement without vitamin A (Life Technologies, 12587), GlutaMax (Life Technologies, 1:100), and penicillin-Streptomycin (1:100, Thermo Fisher Scientific, 15140163). NM was supplemented with 20 ng/ml EGF (R&D Systems) and 20 ng/ml basic fibroblast growth factor (bFGF) (R&D Systems) for 19 days (until day 24) with daily medium change in the first 10 days, and every other day for the subsequent 9 days. To promote differentiation of the neural progenitors into neurons, NM was supplemented with 20 ng/ml BDNF (Peprotech, 450–02) and 20 ng/ml NT-3 (Peprotech, 450–03) with medium changes every other day. From day 44, only NM without growth factors was used for medium changes every 4 days.
Cryopreservation
hOLS were fixed in 4% paraformaldehyde (PFA) overnight. They were then washed in PBS and transferred to 30% sucrose for 3–7 days. Subsequently, they were rinsed in optimal cutting temperature (OCT) compound and 30% sucrose (1:1) and embedded in OCT and 30% sucrose (1:1) and transferred into embedding medium (Tissue-Tek OCT Compound 4583, Sakura Finetek) and stored at −80 °C. For immunofluorescence, 12– to 14–μm-thick sections were cut using a Leica cryostat.
Immunofluorescence
Cryosections were washed with PBS to remove excess OCT and blocked in 10% normal donkey serum (NDS, Abcam, AB7475), 0.3% Triton X-100 (Sigma Aldrich, X100) diluted in PBS for 1 hour at room temperature. The sections were then incubated overnight at 4°C with primary antibodies diluted in PBS containing 10% NDS and 0.3% Triton X-100. PBS was used to wash off the primary antibodies and the cryosections were incubated with secondary antibodies in PBS with 10% NDS and 0.3% Triton X-100 for 1 hour. Dissociated cultures on glass coverslips were fixed for in 4% PFA in PBS for 15 minutes at 4 °C and then rinsed for 5 min with PBS. The coverslips were then treated with 0.1% Triton-X100 in PBS for 15 min at room temperature except for when staining for PDGFRα. The coverslips were blocked for 1 hour at room temperature with 5% NDS in PBS and then incubated overnight at 4°C with primary antibodies diluted in PBS containing 5% NDS. PBS was used to wash off the primary antibodies and the cryosections were incubated with secondary antibodies in PBS with 5% NDS for 1 hour. The following primary antibodies were used for immunofluorescence: anti-OLIG245 (rabbit, 1:500, Millipore, AB9610), anti-NKX2–246 (mouse, 1:200, DSHB, 74.5A5), anti-PDGFRα47 (rabbit, 1:500, Santa Cruz, sc-338), anti-O448 (mouse, 1:500, R&D systems, MAB1326), anti-O149 (mouse, 1:200, R&D systems, MAB1327), anti-MBP50 (rat, 1:300, Millipore, MAB386), anti-GFAP51 (rabbit, 1:1000, Agilent DAKO, Z0334), anti-Neurofilament-H 200K52 (mouse, 1:500, Abcam, AB7795), anti-GABA53 (rabbit, 1:1000, Sigma, A2052), anti-MAP254 (guinea pig, 1:2500 on coverslips, 1:10000 on cryosections, Synaptic Systems, 188004). Alexa Fluor dyes (Life Technologies) were used at 1:1000 dilution on cryosections and 1:2000 on coverslips for amplification of the signal. Nuclei were visualized with Hoechst 33258 (Life Technologies, H3549). Cryosections were mounted for microscopy on glass slides using Aquamount (Polysciences Inc, 18606) and imaged on a Zeiss M1 Axioscope, Leica TCS SP8 confocal microscope or Keyence BZ-X710. Images were processed in ImageJ (Fiji Version 2.0.0). For the analysis in Fig. 5d, the proportion of MBP+ cells overlapping for more than 1 μm with NF-H in 0.5 μm z-sections were counted.
Dissociation and immunopanning of hOLS
For the enzymatic dissociation of hOLS for culture in monolayer and immunocytochemistry, hOLS were incubated with accutase for 30 minutes at 37 °C, washed with 10% fetal bovine serum (Thermo Fisher Scientific, 16000–044) in Hank’s Balanced Salt Solution (Thermo Fisher Scientific, 14180046) and gently triturated using a P200 pipette. Cells were centrifuged and resuspended in complete DMM. The cells were plated on glass coverslips (15 mm, Warner Instruments, 640713) coated with poly-L-ornithine (50 μg/mL, Sigma-Aldrich, P3655) and laminin (5 μg/mL, Sigma-Aldrich, L2020) at a density of around 1 spheroid per two coverslips.
To dissociate and isolate O4+ cells from hOLS for single-cell profiling, we adapted a previously published protocol used for primary human fetal brain tissue25. Ten cm plastic Petri plates (Thomas Scientific, 351058) were coated with 20 μL of anti-mouse IgG (Thermo Fisher, A-21042) in 7 mL of 50 mM Tris-HCl pH9.5 (Fisher Scientific, T1095) at 4°C overnight. The following day, each plate was rinsed 3 times with PBS and then coated with 1.78 μL of anti-O4 hybridoma (obtained from the B.A. Barres laboratory at Stanford University25) in 2.55 mL of 0.02% BSA diluted in PBS (Caisson Labs, PBL02) at room temperature for at least 3 hours and rinsed 4–5 times with PBS before use. Up to 6 spheroids per immunopanning plate were dissociated with accutase as described above and resuspended in 10 mL of 0.02% BSA. The resulting suspension was added to a pre-coated plastic Petri culture dish for at least 40 minutes with gentle occasional swirling. The supernatant was then removed from the O4-treated plates and the plates were rinsed with PBS 6–8 times to removed unattached cells. O4+ cells were detached from the plates using 0.25% trypsin (Thermo Fisher Scientific, 25200056) for 5 minutes at 37 °C. Cells were washed in DMEM (Thermo Fisher Scientific, 10313–039) containing fetal bovine serum and centrifuged for 5 minutes at 1200 rpm, and resuspended in 0.02% BSA and supplemented with Y-27632 (10 μM, EMD Chemicals, S1049). The suspension was filtered through a 40 μm strainer (Fisher Scientific, 352340) and single cells were sorted into lysis buffer 4 μl each in 96-well plates (Bio-Rad, HSP9631). The lysis buffer contains 4 U RNase inhibitor (40 U/μl, Clontech, 2313B), 0.05% Triton, 2.5 mM dNTP, 2.5 μM Oligo-dT30VN (IDT, RNase-free HPLC purification)55, 4 enzyme units (U) RNase inhibitor…2.5 mM deoxynucleotide triphosphates (dNTP)… external RNA control consortium (ERCC) RNA Spike-In Mix (1:2.4 X 107 diluted, Thermal Fisher Scientific, 4456740). Plates were briefly vortexed and spun down before being stored in –80˚C freezer until downstream processing.
Single cell RNAseq library preparation
Sorted cells (n= 332) in 96-well plates were thawed on ice and immediately used for library preparation following the Smartseq2 protocol55 with modifications. Briefly, plates were incubated at 72˚C for 3 minutes, during which RNA molecules with polyA tails were annealed with Oligo-dT30VN, and then they were immediately chilled on ice. Reverse transcription mixture was added 6 μl per well so that the final solution contained 95 enzyme units (U) SMARTScribe™ Reverse Transcriptase (100 U/μl, Clontech, 639538), 10 enzyme units (U) RNase inhibitor (40 U/μl), 1XFirst-Strand buffer, 5 mM DTT, 1 M Betaine, 6 mM MgCl2, 1 μM template switching oligos (TSO) (Exiqon, RNase-free HPLC purification)55. RT was performed at 42˚C for 90 minutes, followed by 70˚C, 5 minutes. To amplify the whole transcriptome, 15 μl of DNA amplification mixture was added with the final solution containing 1X KAPA HIFI Hotstart Master Mix (Kapa Biosciences, KK2602), 0.1 μM ISPCR Oligo 55, and 0.56 enzyme units (U) lambda exonuclease (5 U/μl, New England BioLabs, M0262S). cDNA was amplified using (1) 37˚C 30 minutes; (2) 95˚C 3 minutes; (3) 21 cycles of 98˚C 20 seconds, 67˚C 15 seconds, 72˚C 4 minutes; (4) 72˚C 5 minutes. Amplified cDNA was then purified using PCRClean DX beads (~0.7 volume, Aline Biosciences, C-1003–50), and reconstituted in 20 μl EB buffer. The cDNA quality and quantity were assessed using a Fragment Analyzer (AATI, High Sensitivity NGS Fragment Analysis Kit:1 bp - 6000 bp), and samples with concentrations below 0.08 ng/μl or abnormal peak patterns were filtered out. A total of 295 cDNA samples (retaining 88.9% of initially sorted) were diluted down to 0.15 ng/μl (only if above it) and pooled into a 384-well plate using a Mosquito X1 (TTP Labtech). Library preparation was performed using Nextera XT DNA Sample Prep Kit (Illumina, FC-131–1096) and Nextera XT 384 dual index primers, with the help of a Mosquito HTS machine for liquid transfer. For the tagmentation step, 0.4 μl cDNA (30–60 pg), 0.4 μl Tagment Enzyme and 0.8 μl Tagment DNA Buffer were mixed and incubated at 55˚C for 10 minutes, followed by neutralization (0.4 μl Neutralize Tagment Buffer). Indexes (0.5 μM each) were added along with Nextera PCR Master Mix to a final volume of 4 μl each well. PCR was performed following: (1) 72˚C 3 minutes; (2) 95˚C 30 seconds; (3) 10 cycles of 95˚C 10 seconds, 55˚C 30 seconds, 72˚C 1 minutes; (4) 72˚C 5 minutes. All samples were pooled (0.5 μl each) together into a 1.5 ml Eppendorf tube, and the mixed libraries were purified twice with PCRClean DX beads (~0.7 volume each time). The final sample was reconstituted in 50 μl EB buffer, and the concentration and peaks were measured by Qubit and Bioanalyzer.
Single-cell RNA Sequencing and QC
In total, 332 single cells using 101 bp paired end reads were sequenced on a HiSeq 4000 (Illumina). Raw reads were preprocessed and aligned to the human genome (hg38) using the pipeline described in Darmanis et al 201556, which includes Spliced Transcripts Alignment to a Reference (STAR) alignment and feature counts. As a quality metric, we first performed hierarchical clustering on all cells using a list of housekeeping genes and removed any cells with uniformly low expression across all genes. We separated the resulting dendrogram into two clusters containing cells that passed or failed this quality control (QC). All downstream analyses were performed using only the 295 cells that passed QC. After quality control and mapping, there was an average of 509,376 mapped reads per cell. These 295 cells were added to data from single cells collected in previous studies (adult cells from Darmanis et al. 201556, hCS from Sloan et al. 201757) for a total of n= 1473 cells. Cells from each of these studies were collected using identical protocols and sequencing methods. To control for unwanted variation in batch effects, all cells were normalized using58 with the following normalization genes: CYC1, EIF4A2, SDHA, ACTB, UBC, TOP1, RPL13A, GAPDH59.
Dimensionality reduction, clustering, and Monocle analysis
All data analysis was performed using the R software (https://www.r-project.org/). Dimensionality reduction was performed in three steps. First, we calculated the overdispersion of each gene as described in Fan et al. 201660. We then selected the top 2000 over-dispersed genes and constructed a cell-to-cell distance matrix (1 minus absolute correlation) of all cells (using either O4 immunopanned alone, or O4 immunopanned cells along with primary human single cells from Darmanis et al. 201556 and Sloan et al. 201757. The distance matrix was reduced to two dimensions using t-SNE23. Clustering of groups of similar cells was performed on the two-dimensional t-SNE space using k-means. The lineage tree for the single-cells was constructed using the Monocle algorithm 2.0 as described in Trapnell et al. 201424. The lineage trees included all O4 immunopanned single cells as well as primary adult human OPCs and oligodendrocytes from Darmanis et al. 201556. t-SNE plots were colored by hiPS cell line using metadata provided in GEO accession GSE115011. This metadata was also used to determine the proportion of dividing, immature (OPCs and NFOs), and mature myelinating oligodendrocytes (as defined by the k-means clustering of the t-SNE space described above) attributed to each hiPS cell line. Correlations between hiPS cell lines were calculated using Pearson correlation values between the log normalized gene expression data for each group.
Viral labeling and assembly of neural spheroids
The viral infection of the 3D neural spheroids was performed as previously described20,61. In brief, hOLS were transferred to a 1.5 ml microcentrifuge Eppendorf tube containing 300 μl complete DMM with virus and incubated overnight. The next day, hOLS were transferred into fresh complete DMM in ultralow attachment plates. Lentivirus (lenti-Sox10-MCS5::eGFP+; construct reported and received from F.J. Sim30) was generated by transfecting HEK293T cells with Lipofectamine 2000 (Thermo Fisher Scientific, 11668019) and concentrating the supernatant with the Lenti-X concentrator (Takara Bio, 631232) 72 hours later.
Live cell imaging and analysis of Sox10-MCS5::eGFP + cell migration
The migration of Sox10-MCS5::eGFP+ cells was imaged for 8–12 hours under environmentally controlled conditions (37 °C, 5% CO2) in intact hOLS using a confocal microscope with a motorized stage (Leica SP8). The hOLS were transferred to a well of a 96-well glass-bottom plates (Thomas Scientific, 4580) in 200 μl of complete DMM and incubated in an environmentally controlled chamber for 15–30 minutes before imaging. During a given recording session, up to 6 hOLS were imaged at a depth of 60–70 μm and at a rate of 13–20 minutes per frame.
Real-time quantitative PCR (qPCR)
mRNA was isolated using the RNeasy Mini kit (Qiagen, 74106) and RNase-Free DNAse I kit (Thermo Fisher Scientific, 18068–015), and template cDNA was prepared by reverse transcription using the SuperScript III First-Strand Synthesis SuperMix for qRT–PCR (Thermo Fisher Scientific, 11752250). qPCR was performed using SYBR Green (Thermo Fisher Scientific, 4312704) on a ViiA7 machine (Applied Biosystems, Life Technologies). Primers used are listed in Supplementary Table 3.
Transmission electron microscopy
hOLS were fixed in 2% glutaraldehyde and 4% PFA in 0.1M Sodium Cacodylate buffer, pH 7.4 for 1 hour at room temperature. After fixation samples are then moved to 4°C for immediate processing. The buffer was removed and replaced with 1% OsO4 in double distilled H2O and the hOLS were gently shaken for 1 hour at 4 °C. Samples were washed 3 times for 5 minutes each with cold double distilled H2O. After the third rinse the hOLS were kept in 1% Uranyl acetate in double distilled H2O, stain for 2 hours to overnight. The hOLS were then dehydrated by rinsing for 10 minutes each in 50% ethanol at 4°C, 70% ethanol at 4°C, 95% ethanol at room temperature, and 100% ethanol at room temperature. A second 10 minutes rinse in 100% ethanol was performed followed by a 15 minute rinse in acetonitrile at room temperature. The hOLS were transferred to 1:1 acetonitrile/epon for 1 hour and then to 1:2 acetonitrile/epon overnight. The following day the hOLS were placed in 100% epon for 2–3 hours and then placed in molds filled with 100% epon and allowed to settle for 4 hours to overnight. The molds were then polymerized in a 65°C oven for 24 hours. Samples were sectioned and imaged used a JEOL JEM1400 120 kV transmission electron microscope.
Electrophysiology
Sections of hOLS at day 96–175 of differentiation were obtained using an approach we previously described20. Briefly, hOLS were incubated in bicarbonate-buffered artificial cerebrospinal fluid (aCSF) at 23°C and equilibrated with a mixture of 95% O2 and 5% CO2. The aCSF solution contained: 126 mM NaCl, 26 mM NaHCO3, 10 mM glucose, 2.5 mM KCl, 1.25 mM NaH2PO4, 1 mM MgSO4 and 2 mM CaCl2. Slicing was performed using a Leica VT1200 vibratome. Immediately after sectioning, slices were moved to a circulation chamber containing oxygenated aCSF at room temperature.
For patch-clamp recording, cells were identified by the presence of the Sox10-MCS5::eGFP fluorescent reporter using an upright microscope (Scientifica). Recording electrodes of borosilicate glass had a resistance of 4–6 MΩ when filled with internal solution. The internal solution contained: 145 mM K-gluconate, 0.1 mM CaCl2, 2.5 mM MgCl2, 10 mM HEPES, 0.2 mM EGTA, 4 mM Na-phosphocreatine. Glutamatergic currents were blocked by application of NBQX (20 μM, Tocris) and APV (100 μM, Tocris), which were added to the bathing solution. Electrical simulation was delivered using a bipolar tungsten electrode (FHS) placed 100–200 μm away from a recorded cell. Stimulations were delivered to slices for 0.1 milliseconds at 300 μV and separated by at least 10 seconds. Voltage gated sodium channels were blocked by application of tetrodotoxin (1 μM, Tocris). All recordings were performed at room temperature (25o). Measurements were corrected for a liquid junction potential of 12 mV.
Data was collected using a 1550A digitizer (Molecular Devices), a 700B patch-clamp amplifier (Molecular Devices) and acquired with the pClamp 10.6 software (Molecular Devices). Data were low-pass filtered at 10 kHz and digitized at 20 kHz. Averaging, digital subtraction of null traces, and current peak detection were made using Clampfit (Molecular Devices). Data were fitted using Origin (OriginLab).
Lysolecithin treatment and live imaging
Lysolecithin was dissolved at a concentration of 0.25 mg/mL in complete DMM and added to day 70–75 hOLS in 1.5 mL eppendorf tubes for 15–17 hours at 37°C. Following treatment, hOLS were rinsed twice with complete DMM. Untreated and treated hOLS were transferred to a well of a 96-well glass-bottom plates (Thomas Scientific, 4580) in 200 μl of complete DMM 4–6 hours after rinsing, and incubated in an environmentally controlled chamber for 15–30 minutes before imaging. During a given recording session, hOLS were imaged at a depth of 60–70 μm and at a rate of 10–15 minutes per frame for 12 hours. Quantification of Sox10-MCS5::eGFP+ disappearing cells was performed blind.
Statistics and reproducibility
Data are presented as mean ± s.e.m. unless otherwise indicated. Distribution of the raw data was tested for normality of distribution; statistical analyses were performed using the Student’s t-test (two-sided), Mann–Whitney U-test (two-sided), or Kruskall-Wallis tests. Sample sizes were estimated empirically. One hOLS sample was removed in the qPCR analysis of VGLUT1 in Supplementary Fig. 1j following Grubb’s test for outliers (P<0.05). No statistical methods were used to determine sample size, but sample sizes for each type of experiment in this study are consistent with previously published work20,21,40,57,62,63. For lysolecithin treatment experiments, samples were randomly distributed between the two condition and data collection and analysis were performed blinded, and unblinded for all other experiments.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
Gene expression data has been deposited in the Gene Expression Omnibus (GEO) (see “Accession codes” section). The data that support the findings of this study are available on request from the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
This paper is dedicated to the memory of our wonderful colleague and mentor Ben A. Barres. We thank B.A. Barres, B. Zuchero and members of the Pasca laboratory for scientific input, J. Perrino (Stanford Cell Sciences Imaging Facility) for support with electron microscopy, as well as F. Sim (University of Buffalo) for providing the Sox10-MCS5::eGFP plasmid. This work was supported by the US National Institute of Health (NIH) BRAINS Award (MH107800), the MQ Fellow Award, the NYSCF Robertson Stem Cell Investigator Award, the Stanford Wu Tsai Neurosciences Institute’s Brain Rejuvenation Project and the Human Brain Organogenesis Project, the Kwan Research Fund and the California Institute of Regenerative Medicine (CIRM), the Child Health Research Institute Pilot Award, the NARSAD Independent Investigator Award from the Brain and Behavior Research Foundation (to S.P.P.); the National Science Foundation (NSF) Graduate Research Fellowship and the Bio-X Stanford Interdisciplinary Graduate Fellowship (to R.M.M.); Stanford Medicine’s Dean’s Fellowship (to Y.M.); NIMH T32GM007365, F30MH106261 and Bio-X Predoctoral Fellowship (to or supporting S.A.S.).
Footnotes
ACCESSION CODES
Gene expression data is available in the Gene Expression Omnibus (GEO) under accession numbers GSE115011.
COMPETING INTERESTS STATEMENT
Stanford University has filed a provisional patent application that covers the generation of myelinating oligospheroids for studying human development and disease (US patent application number 15/953,197).
REFERENCES
- 1.Nave KA Myelination and the trophic support of long axons. Nat Rev Neurosci 11, 275–283, doi: 10.1038/nrn2797 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Menichella DM et al. Genetic and physiological evidence that oligodendrocyte gap junctions contribute to spatial buffering of potassium released during neuronal activity. J Neurosci 26, 10984–10991, doi: 10.1523/JNEUROSCI.0304-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funfschilling U et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521, doi: 10.1038/nature11007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai X et al. The trophic role of oligodendrocytes in the basal forebrain. J Neurosci 23, 5846–5853 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orthmann-Murphy JL, Freidin M, Fischer E, Scherer SS & Abrams CK Two distinct heterotypic channels mediate gap junction coupling between astrocyte and oligodendrocyte connexins. J Neurosci 27, 13949–13957, doi: 10.1523/JNEUROSCI.3395-07.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore CS, Abdullah SL, Brown A, Arulpragasam A & Crocker SJ How factors secreted from astrocytes impact myelin repair. J Neurosci Res 89, 13–21, doi: 10.1002/jnr.22482 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Simons M & Trajkovic K Neuron-glia communication in the control of oligodendrocyte function and myelin biogenesis. J Cell Sci 119, 4381–4389, doi: 10.1242/jcs.03242 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Barres BA, Schmid R, Sendnter M & Raff MC Multiple extracellular signals are required for long-term oligodendrocyte survival. Development 118, 283–295 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Lin SC & Bergles DE Synaptic signaling between GABAergic interneurons and oligodendrocyte precursor cells in the hippocampus. Nat Neurosci 7, 24–32, doi: 10.1038/nn1162 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Bergles DE, Roberts JD, Somogyi P & Jahr CE Glutamatergic synapses on oligodendrocyte precursor cells in the hippocampus. Nature 405, 187–191, doi: 10.1038/35012083 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Hardy R & Reynolds R Neuron-oligodendroglial interactions during central nervous system development. J Neurosci Res 36, 121–126, doi: 10.1002/jnr.490360202 (1993). [DOI] [PubMed] [Google Scholar]
- 12.van der Knaap MS, Pronk JC & Scheper GC Vanishing white matter disease. The Lancet. Neurology 5, 413–423, doi: 10.1016/S1474-4422(06)70440-9 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Wolswijk G Oligodendrocyte survival, loss and birth in lesions of chronic-stage multiple sclerosis. Brain 123 (Pt 1), 105–115 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Douvaras P & Fossati V Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat Protoc 10, 1143–1154, doi: 10.1038/nprot.2015.075 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Numasawa-Kuroiwa Y et al. Involvement of ER stress in dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes. Stem Cell Reports 2, 648–661, doi: 10.1016/j.stemcr.2014.03.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stacpoole SR et al. High yields of oligodendrocyte lineage cells from human embryonic stem cells at physiological oxygen tensions for evaluation of translational biology. Stem Cell Reports 1, 437–450, doi: 10.1016/j.stemcr.2013.09.006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell 12, 252–264, doi: 10.1016/j.stem.2012.12.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogberg HT et al. Toward a 3D model of human brain development for studying gene/environment interactions. Stem Cell Res Ther 4 Suppl 1, S4, doi: 10.1186/scrt365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasca SP The rise of three-dimensional human brain cultures. Nature 553, 437–445, doi: 10.1038/nature25032 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Pasca AM et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature methods 12, 671–678, doi: 10.1038/nmeth.3415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birey F et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59, doi: 10.1038/nature22330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sloan SA, Andersen J, Pasca AM, Birey F & Pasca SP Generation and assembly of human brain region-specific three-dimensional cultures. Nat Protoc, doi: 10.1038/s41596-018-0032-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Maaten L & Hinton G Visualizing Data using t-SNE. J Mach Learn Res 9, 2579–2605 (2008). [Google Scholar]
- 24.Trapnell C et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386, doi: 10.1038/nbt.2859 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53, doi: 10.1016/j.neuron.2015.11.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crow YJ et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet 38, 910–916, doi: 10.1038/ng1842 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Gomez-Ospina N in GeneReviews((R)) (eds Adam MP et al. ) (1993). [Google Scholar]
- 28.Austin J et al. Studies in globoid (Krabbe) leukodystrophy (GLD). V. Controlled enzymic studies in ten human cases. Arch Neurol 23, 502–512 (1970). [DOI] [PubMed] [Google Scholar]
- 29.Bergles DE & Richardson WD Oligodendrocyte Development and Plasticity. Cold Spring Harb Perspect Biol 8, a020453, doi: 10.1101/cshperspect.a020453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pol SU et al. Sox10-MCS5 enhancer dynamically tracks human oligodendrocyte progenitor fate. Exp Neurol 247, 694–702, doi: 10.1016/j.expneurol.2013.03.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sontheimer H, Trotter J, Schachner M & Kettenmann H Channel expression correlates with differentiation stage during the development of oligodendrocytes from their precursor cells in culture. Neuron 2, 1135–1145 (1989). [DOI] [PubMed] [Google Scholar]
- 32.Livesey MR et al. Maturation and electrophysiological properties of human pluripotent stem cell-derived oligodendrocytes. Stem Cells 34, 1040–1053, doi: 10.1002/stem.2273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karadottir R & Attwell D Neurotransmitter receptors in the life and death of oligodendrocytes. Neuroscience 145, 1426–1438, doi: 10.1016/j.neuroscience.2006.08.070 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karadottir R, Cavelier P, Bergersen LH & Attwell D NMDA receptors are expressed in oligodendrocytes and activated in ischaemia. Nature 438, 1162–1166, doi: 10.1038/nature04302 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birgbauer E, Rao TS & Webb M Lysolecithin induces demyelination in vitro in a cerebellar slice culture system. J Neurosci Res 78, 157–166, doi: 10.1002/jnr.20248 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Blakemore WF, Eames RA, Smith KJ & McDonald WI Remyelination in the spinal cord of the cat following intraspinal injections of lysolecithin. J Neurol Sci 33, 31–43 (1977). [DOI] [PubMed] [Google Scholar]
- 37.Hall SM The effect of injections of lysophosphatidyl choline into white matter of the adult mouse spinal cord. J Cell Sci 10, 535–546 (1972). [DOI] [PubMed] [Google Scholar]
- 38.Amin ND & Pasca SP Building Models of Brain Disorders with Three-Dimensional Organoids. Neuron 100, 389–405, doi: 10.1016/j.neuron.2018.10.007 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Di Lullo E & Kriegstein AR The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci 18, 573–584, doi: 10.1038/nrn.2017.107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon Se-Jin, et al. "Reliability of human cortical organoid generation." Nature methods 16.1 (2019): 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volpe JJ The encephalopathy of prematurity--brain injury and impaired brain development inextricably intertwined. Semin Pediatr Neurol 16, 167–178, doi: 10.1016/j.spen.2009.09.005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 42.Pasca SP et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med 17, 1657–1662, doi: 10.1038/nm.2576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yazawa M et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 471, 230–234, doi:nature09855 [pii] 10.1038/nature09855 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallego Romero I et al. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. eLife 4, e07103, doi: 10.7554/eLife.07103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchetto MC et al. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell 3, 649–657, doi: 10.1016/j.stem.2008.10.001 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Xue Y et al. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat Neurosci 19, 807–815, doi: 10.1038/nn.4297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium HD i. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci 20, 648–660, doi: 10.1038/nn.4532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renner M et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J 36, 1316–1329, doi: 10.15252/embj.201694700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang SM et al. Efficient induction of oligodendrocytes from human embryonic stem cells. Stem Cells 25, 419–424, doi: 10.1634/stemcells.2005-0482 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Deborde S et al. Schwann cells induce cancer cell dispersion and invasion. J Clin Invest 126, 1538–1554, doi: 10.1172/JCI82658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao D et al. Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat Commun 9, 2865, doi: 10.1038/s41467-018-05209-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clarke KE et al. A robust and reproducible human pluripotent stem cell derived model of neurite outgrowth in a three-dimensional culture system and its application to study neurite inhibition. Neurochem Int 106, 74–84, doi: 10.1016/j.neuint.2016.12.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greber B et al. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J 30, 4874–4884, doi: 10.1038/emboj.2011.407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amin H et al. Electrical Responses and Spontaneous Activity of Human iPS-Derived Neuronal Networks Characterized for 3-month Culture with 4096-Electrode Arrays. Front Neurosci 10, 121, doi: 10.3389/fnins.2016.00121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181, doi: 10.1038/nprot.2014.006 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Darmanis S et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA 112, 7285–7290, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sloan SA et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779–790.e6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Risso D, Ngai J, Speed TP & Dudoit S Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32, 896–902, doi: 10.1038/nbt.2931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Penna I et al. Selection of candidate housekeeping genes for normalization in human postmortem brain samples. Int J Mol Sci 12, 5461–5470, doi: 10.3390/ijms12095461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fan J et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nature methods 13, 241–244, doi: 10.1038/nmeth.3734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deverman BE et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol 34, 204–209, doi: 10.1038/nbt.3440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas CA et al. Modeling of TREX1-Dependent Autoimmune Disease using Human Stem Cells Highlights L1 Accumulation as a Source of Neuroinflammation. Cell Stem Cell, doi: 10.1016/j.stem.2017.07.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mariani J et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390, doi: 10.1016/j.cell.2015.06.034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data has been deposited in the Gene Expression Omnibus (GEO) (see “Accession codes” section). The data that support the findings of this study are available on request from the corresponding author.