Abstract
TFEB is overexpressed in TFEB-rearranged renal cell carcinomas as well as in renal tumors with amplifications of TFEB at 6p21.1. As recent literature suggests that renal tumors with 6p21.1 amplification behave more aggressively than those with rearrangements of TFEB, we compared relative TFEB gene expression in these tumors.
This study included 37 TFEB-altered tumors: 15 6p21.1-amplified and 22 TFEB-rearranged (including 5 cases from The Cancer Genome Atlas dataset). TFEB status was verified using a combination of FISH (n=27) or comprehensive molecular profiling (n=13) and digital droplet PCR was used to quantify TFEB mRNA expression in 6p21.1-amplified (n=9) and TFEB-rearranged renal tumors (n=19). These results were correlated with TFEB immunohistochemistry.
TFEB-altered tumors had higher TFEB expression when normalized to B2M (mean: 168.9%, n=28), compared to non-TFEB-altered controls (mean: 7%, n=18, p=0.005). Interestingly, TFEB expression in tumors with rearrangements (mean: 224.7%, n=19) was higher compared to 6p21.1-amplified tumors (mean: 51.2%, n=9; p=0.06). Of note, classic biphasic morphology was only seen in TFEB-rearranged tumors and when present correlated with 6.8-fold higher TFEB expression (p=0.00004).
Our results suggest that 6p21.1 amplified renal tumors show increased TFEB gene expression but not as much as t(6;11) renal tumors. These findings correlate with the less consistent/diffuse expression of downstream markers of TFEB activation (cathepsin K, melan A, HMB45) seen in the amplified neoplasms. This suggests that the aggressive biologic behavior of 6p21.1 amplified renal tumors might be secondary to other genes at the 6p21.1 locus that are co-amplified, such as VEGFA and CCND3, or other genetic alterations.
Keywords: TFEB, Renal Cell Carcinoma, 6p21.1 Amplification, Rearrangement
Introduction
Transcription Factor EB (TFEB), a member of the microphthalmia-associated transcription factor family, when overexpressed, is thought to act as a key oncogenic driver in renal cell carcinomas (1, 2). The mechanism of TFEB overexpression that was initially described in TFEB-driven renal tumors involved structural rearrangements such as t(6;11), where the complete TFEB coding sequence was retained in most cases (1, 3–5). Subsequently, genomic amplifications at the chromosome 6p21.1 locus were described where TFEB overexpression secondary to copy number increases emerged as an alternate pathogenic mechanism in TFEB-driven renal tumors (2, 6–12).
Recent studies have highlighted that 6p21.1 (TFEB) amplified tumors may be biologically distinct from TFEB rearranged tumors as 6p21.1-amplified tumors show amplification of other oncogenes at the same locus, such as VEGFA and CCND3 (8, 9, 11, 12). In addition, initial reports suggest that the incidence of regional and distant metastasis in 6p21.1 (TFEB) amplified tumors may be higher than in TFEB rearranged tumors, which are thought to have a more indolent clinical course (2, 8, 9, 11–13).
The assessment of TFEB gene expression in these tumors has been pursued in a limited number of studies given that, until recently, the primary mechanism of TFEB overexpression, as a driver event, was thought to involve only gene rearrangements (5). Therefore, to better understand the pathogenesis of these tumors, TFEB gene expression status was assessed in both TFEB amplified and rearranged tumors. These results were then correlated with various clinicopathologic features including morphology, background genomic alterations and biologic behavior in a relatively large cohort of cases.
Materials and Methods
Patient Specimens
Six renal tumors with 6p21.1 (TFEB) amplifications, including 3 cases with TFEB rearrangements in the setting of a concurrent amplification at this locus, were identified from previously reported The Cancer Genome Atlas datasets (2, 8, 14, 15). This included an institutional case with COL21A1-TFEB rearrangement which was further characterized in this study (Figure 1).
Figure 1: Methods.
Graphical representation of different methodologies used to interrogate renal tumors with 6p21.1 (TFEB) amplifications and rearrangements, including molecular profiling (whole exome sequencing, targeted panels), fluorescent in situ hybridization (FISH), RNA sequencing (RNASeq, anchored multiplex technology), digital PCR (dPCR) and immunohistochemistry (IHC). Fusion partners for TFEB rearranged genes have been indicated. TCGA: The Cancer Genome Atlas.
This study was approved by the Memorial Sloan Kettering Cancer Center institutional review board and included cases that were profiled using a comprehensive molecular profiling strategy (Memorial Sloan Kettering Cancer Center Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT)), as part of an institutional clinical cancer genomics initiative (16, 17). Separate IRB approvals were obtained at contributing institutions. Furthermore, tumors identified using fluorescence in situ hybridization studies were contributed from the consultation files of one of the authors (PA). In addition to 6 previously reported cases from The Cancer Genome Atlas, 12 renal tumors with 6p21.1 (TFEB) amplifications and 19 t(6;11) tumors were analyzed in this study (Figure 1). Specifically 9 (of 12) new cases of 6p21.1 (TFEB) amplification and 18 (of 19) cases with t(6;11) have not been previously reported in the literature (2, 18).
Immunohistochemistry and Fluorescence in Situ Hybridization
Immunohistochemistry for TFEB was performed using an automated staining platform (Bond-III, Leica Biosystems, Buffalo Grove, IL). A mouse monoclonal TFEB antibody was used as a primary reagent (clone: C-6, Santa Cruz Biotechnology, Dallas, TX) at a concentration of 0.2μg/ml. A polymeric secondary kit (Refine, Leica) was used for the detection of the primary antibody. Fluorescence in situ hybridization was performed as previously described using probes that cover and flank the TFEB gene on 6p21.1 (2, 18, 19). Nuclei with incomplete signals were omitted from the score and the threshold for rearrangements and amplifications included >20% split signals and >10:1 ratio of TFEB signal to reference, respectively.
Next Generation Sequencing-Based Copy Number Assessment: 6p21.1 (TFEB) Amplifications
Details of the MSK-IMPACT assay have been previously reported (16, 17). The MSK-IMPACT assay involves hybridization capture-based library preparation followed by deep sequencing of select non-coding regions and 6,614 protein-coding exons of 468 genes. Genome-wide copy number assessment is facilitated by homogenous distribution of single nucleotide polymorphism tiling probes across the genome. Based on previously reported criteria, gains were defined as a fold change ≥1.5 and <2.0, while amplifications were defined as a fold change ≥ 2.0 (20–22).
Digital Droplet PCR for Assessment of TFEB Gene Expression Status
RNA extraction was performed as previously described (23). Specifically, RNA was extracted from a minimum of three, 5-micron thick, sections of formalin fixed paraffin embedded tissue following review of the corresponding H&E stained slide by a single pathologist (SG) and manual macrodissection of areas of interest. Specific steps included the addition of 10 μl of mineral oil to each slide prior to macrodissection and the further addition of 800 μl of mineral oil for deparaffinization and RNA extraction using the RNeasy FFPE Kit and protocol (Qiagen). RNA quantification was performed using the Qubit Broad Range RNA Assay Kit (Life Tech.) and 2ng of template was used in subsequent reactions. The reaction mixtures for downstream processing included the One-Step RT-ddPCR Advanced Kit (Bio-Rad, CA) and PrimePCR- ddPCR- Expression Probe Assay for (human) TFEB (Bio-Rad, CA; Chromosome Location: 6:41655765–41658411; RefSeq: NC_000006.11; Amplicon Length: 64). The QX200- Droplet Reader and QuantaSoft- Software (Bio-Rad, CA) were used to identify droplets containing amplification products using the FAM channel. TFEB gene expression was normalized to the expression of a corresponding housekeeping gene (B2M). All samples were tested in duplicate and samples were considered failures if a low number of total droplets (<10,000) was identified. Negative (no template) controls included water (n=4), while low positive controls included previously characterized cell free RNA samples (n=4). Non TFEB-altered controls (n=18) included non-neoplastic renal parenchyma (n=13) and renal tumors with no TFEB alterations detected on comprehensive molecular profiling using MSK-IMPACT (oncocytoma, n=2; clear cell renal cell carcinoma, n=3). A cutoff of 20% (TFEB: B2M) was selected retrospectively to distinguish tumors with TFEB alterations compared to those without TFEB alterations as this level of expression had a specificity of 100% in distinguishing these tumor types.
In a single case, t(6;11) was confirmed using anchored multiplex technology for fusion detection (Archer- FusionPlex-), as previously described (23).
Literature Review and Data Extraction from The Cancer Genome Atlas Datasets
The publicly available cBioPortal.32e34 platform was used to analyze data from The Cancer Genome Atlas and other public datasets (24). Six renal tumors with 6p21.1 (TFEB) amplifications were identified from previously reported The Cancer Genome Atlas datasets (2, 8, 14, 15).
Statistical Analysis
Continuous variables were evaluated with frequency counts and percentages. Tests used for the assessment of statistical significance were two-sided and p<0.05 was considered statistically significant.
Results
Next Generation Sequencing Results
All 5 renal tumors with 6p21.1 amplifications identified using MSK-IMPACT showed amplification of CCND3 and/or VEGFA at this locus. The results of molecular profiling for 7 cases (5 TFEB amplification and 2 TFEB rearrangement) have been listed in Table1.
Table1.
Molecular Profiling of TFEB Rearranged and 6p21.1 (TFEB) Amplified Tumors
| Tumor Mutation Burden (mt/MB) | Somatic Mutations | Copy Number Changes (Excluding TFEB) | Structural Variants | |
|---|---|---|---|---|
| TFEB Rearranged Tumors | ||||
| Case 2* | 0.9 mt/MB | FAT1 p.I2239K | - | *In Frame Fusion: MALAT1 (Exon1) and TFEB (Exon3) |
| Case 18 | 0.0 mt/MB | - | - | (TFEB rearrangement was detected using FISH) |
| 6p21.1 (TFEB) Amplified Tumors | ||||
| Case 25 | 1.0 mt/MB | NOTCH4 p.S79N | Focal Amplifications: PIM1, CCND3 and VEGFA (6p21.1); SMAD3 and MAP2K1 (15q22.31) Broad Losses: 3p/6p/8p/15p/16q Broad Gains: 1q/8q/12p/17/20 |
- |
| Case 26 | 8.9 mt/MB |
TP53 p.R273C; ERRFI1 p.E101V; LATS2 p.K138N; LATS2 p.Q148H; MAP2K1 p.N78S; MLL2 p.S1484F; PBRM1 p.F872fs; VHL p.S72fs SETD2 splicing variant (c.7432–1G>T); |
Focal Gain: CCND3 (6p21.1) Broad Losses: 2/3p/8p/12p/13q/16q/19/21 Broad Gains: 5q/7/17q |
LATS2 rearrangement: c.1006_1900–1285inv |
| Case 32 | 1.8 mt/MB | RNF43 p.G24V; STAG2 p.G129R | Focal Amplifications: CCND3 and VEGFA (6p21.1); RECQL4, PRDM14 and AGO2 (8q24.3) Broad Losses: 2q/8p/20q Broad Gains: 1q/8q/10p/12q/16/17/19/20/21 |
- |
| Case 33 | 5.9 mt/MB |
AKT3 p.W98fs; AXL p.K477M; DNMT3A p.G642*; MEN1 p.W126*; MGA p.L1003fs; NTRK3 p.L591V |
Focal Amplifications: CCND3 and VEGFA (6p21.1) Broad Losses: 11q/16q/22q Broad Gains: 12p/16q |
- |
| Case 34 | 3.0 mt/MB |
ASXL1 p.F252I; EP300 p.H844fs; YES1 p.D454fs |
Focal Amplifications: PIM1, CCND3 and VEGFA (6p21.1) Focal Deletions: E2F3 (6p22.3); CDKN2B, CDKN2Ap16INK4A and CDKN2Ap14ARF (9p21.3) Broad Losses: 2q/6q/17p/18q Broad Gains: 1q/2p/3q/4q/5/7/8/11p/12p/14q/16p/17q/19q/20q/22q |
- |
mt/MB: mutations/megabase; FISH: fluorescence in situ hybridization. The median tumor mutation burden assessed by MSK-IMPACT for all patients with renal cell carcinoma (n=718) is 3.9 mt/Mb.
Prior Neuroblastoma showed a PALB2 p.S537L alteration and no germline alterations were detected. TFEB rearrangement was detected using FISH and anchored multiplex PCR.
Clinical Features: TFEB rearranged and 6p21.1 (TFEB) Amplified Renal Tumors
Clinical features of patients diagnosed with renal tumors harboring TFEB alterations have been listed in Table 2 and summarized in Table 3. No significant gender predilection was identified. Of note, most of the tumors occurred in adults with only 5 (of 37, 14%) being diagnosed in individuals under the age of 25. Tumors with 6p21.1 (TFEB) amplifications, when compared to those with TFEB rearrangements, tended to be larger (mean size: 9.2cm vs 6.3cm; p=0.04). In addition, amongst a total of 15 cases where clinical follow-up was available, 6p21.1 (TFEB) amplified tumors had a higher incidence of documented regional or distant metastasis (88% vs 29%; p=0.04).
Table2.
Clinicopathologic Features: Case Details
| Case No. | Age | Gender | Size (cm) | pT | N | M | Documented Metastasis (At Diagnosis or Follow Up) | Anti-VEGF Therapy | Previously Reported |
|---|---|---|---|---|---|---|---|---|---|
| TFEB Rearranged Tumors | |||||||||
| 1 | 51 | Female | 14.2 | pTx | Nx | Mx | NA | NA | − |
| 2 | 12 | Male | 5.4 | pT1b | N0 | M0 | − | NA | − |
| 3 | 78 | Female | 3.5 | pT1a | N0 | M0 | − | NA | − |
| 4 | 49 | Female | 2 | pTx | Nx | Mx | NA | NA | − |
| 5 | 38 | Female | 5.3 | pTx | Nx | Mx | NA | NA | − |
| 6 | 59 | Female | 1.9 | pTx | Nx | Mx | NA | NA | − |
| 7 | 72 | Female | 4 | pT3a | Nx | Mx | NA | NA | − |
| 8 | 60 | Female | 4.5 | pT1b | Nx | Mx | NA | NA | − |
| 9 | 68 | Male | 2.8 | pT1a | Nx | Mx | NA | NA | Smith et al, 2014 |
| 10 | 55 | Male | 9.5 | pT3b | Nx | M1 | + | NA | − |
| 11 | 23 | Female | 3.5 | pT1 | Nx | Mx | NA | NA | − |
| 12 | 35 | Female | NA | pTx | Nx | Mx | NA | NA | − |
| 13 | 61 | Female | 5 | pTx | Nx | Mx | NA | NA | − |
| 14 | 23 | Female | NA | pTx | Nx | Mx | NA | NA | − |
| 15 | 51 | Male | 14.6 | pTx | Nx | Mx | NA | NA | − |
| 16 | 24 | Male | 6.4 | pT1b | Nx | Mx | NA | NA | − |
| 17 | 43 | Female | 10.3 | pTx | Nx | Mx | NA | NA | − |
| 18 | 41 | Female | 5.8 | pT1b | Nx | Mx | − | NA | − |
| TFEB Rearranged Tumors With 6p21.1 (TFEB) Amplifications | |||||||||
| 19 (t(6;11) & Amplification) | 82 | Female | 4.5 | pT1b | Nx | Mx | NA | NA | − |
| 20 (TCGA-A3-3313-01; TFEB-KHDRBS2) | 59 | Male | 4.5 | pT1b | Nx | Mx | − | NA | TCGA* |
| 21 (TCGA-B9-A69E-01; TFEB-CADM2) | 71 | Male | 8 | pT3a | Nx | Mx | − | NA | TCGA* |
| 22 (TCGA-BQ-7048; COL21A1-TFEB) | 64 | Male | 11 | pT3a | N0 | M0 | + | + | TCGA* |
| 6p21.1 (TFEB) Amplified Tumors | |||||||||
| 23 | 64 | Male | 2 | pT1a | Nx | Mx | NA | NA | − |
| 24 | 61 | Female | NA | pT3a | Nx | Mx | NA | NA | − |
| 25 | 23 | Female | 7 | pT1b | Nx | M0 | + | NA | Argani et al, 2016 |
| 26 | 53 | Male | 8.5 | pT3a | Nx | M0 | + | − | − |
| 27 | 68 | Male | 13 | pTx | Nx | Mx | NA | NA | − |
| 28 | 65 | Male | NA | pTx | Nx | Mx | NA | NA | Argani et al, 2016 |
| 29 | 66 | Female | 14 | pT3a | Nx | Mx | NA | NA | − |
| 30 | 78 | Female | 12 | pT3b | Nx | Mx | NA | NA | Argani et al, 2016 |
| 31 | 73 | Female | 14.4 | pTx | Nx | Mx | NA | NA | − |
| 32 | 73 | Male | 5 | pT1b | Nx | M0 | − | NA | − |
| 33 | 61 | Male | 13 | pT2b | Nx | M1 | + | + | − |
| 34 | 58 | Male | 9.1 | pT3b | N1 | M1 | + | + | − |
| 35 (TCGA-UZ-A9PQ-01) | 59 | Male | 9.2 | pT3a | N1 | Mx | + | NA | TCGA* |
| 36 (TCGA-GL-7966–01) | 28 | Female | 6.5 | pT3a | N1 | Mx | + | NA | TCGA* |
| 37 (TCGA-Q2-A5QZ-01) | 61 | Female | 5.3 | pT3a | N0 | Mx | + | NA | TCGA* |
Table3.
Clinicopathologic Features: Summarized Data
| TFEB Rearranged Tumors | TFEB Rearranged Tumors With 6p21.1 (TFEB) Amplifications | 6p21.1 Amplified Tumors | All TFEB Altered tumors | |
|---|---|---|---|---|
| Total Number of Cases | 22* | 4** | 15 | 37 |
| Age < 25 years | 4/22 (Mean: 51years) | 0/4 (Mean: 58years) | 1/15 (Mean: 59years) | 5/37 (Mean: 54years) |
| Gender (Male: Female) | 8:14 | 3:1 | 8:7 | 16:21 |
| Size (mean, cm) | 6.3 (n=20) | 6.4 (n=4) | 9.2 (n=13); p=0.04 | 7.4 (n=33) |
| ≥pT3 | 4/13 (31%) | 2/4 | 8/11 (73%); p=0.1 | 12/24 |
| N1 | 0/3 | 0/2 | 1/3 | 3/7 |
| M1 | 2/4 | 1/1 | 2/5 | 4/9 |
| Documented Distant/Regional Metastasis at Diagnosis or Follow Up | 2/7 (29%) | 1/3 | 7/8 (88%); p=0.04 | 9/15 |
| Anti-VEGF Therapy | Documented in 1 Case | Documented in 1 Case | Documented in 2 Cases | Documented in 3 Cases |
VEGF: Vascular Endothelial Growth Factor
Rearrangement partners include MALAT1 (n=19), COL21A1 (n=1), CADM2 (n=1) and KHDRBS2 (n=1).
Rearrangement partners include MALAT1 (n=1), COL21A1 (n=1), CADM2 (n=1) and KHDRBS2 (n=1).
Statistical analysis performed using Fishers exact test.
VEGFA amplifications have been reported to be present in renal tumors with 6p21.1 amplifications and therapy with anti-VEGF agents was documented for 2 patients with a 6p21.1 amplification and for 1 patient with a TFEB amplification in the background of a TFEB rearrangement, in our study (9, 11, 12). The first patient with a 6p21.1 amplification was a 61-year-old male who was diagnosed with a 13cm pT2b renal cell carcinoma and subsequently developed radiographic evidence of lung metastasis at 18 months of follow up. He was initially managed with bevacizumab and everolimus, and subsequently treated with other therapeutic agents (sunitinib, sorafenib, temsirolimus and nivolumab). Disease progression involved radiographic evidence of metastatic disease involving the lungs, mediastinal and hilar lymph nodes, liver and soft tissue. He eventually died of disease related complications at 55 months of follow up post-nephrectomy. The second patient with a 6p21.1 amplification was a 58-year-old male with a pT3b tumor with widely metastatic disease including extensive retroperitoneal lymphadenopathy and multiple hepatic metastasis. He was initially treated with pazopanib, followed by cabozantinib (VEGF inhibitor) and had stable disease for approximately 8 months. He eventually developed progressive disease and died of disease related complications at 1 year of follow up, post-initiation of therapy with pazopanib. The third patient with a TFEB amplification, in the background of a COL21A1-TFEB rearrangement, was a 64-year-old male with a pT3a tumor. This patient developed mediastinal lymphadenopathy at approximately 9 months of follow up. Initial management was with sunitinib followed by temsirolimus (tyrosine kinase inhibitor). Further disease progression involved osseous metastases and subsequently pulmonary metastasis. Subsequent management included immunotherapeutic agents such as nivolumab (immune checkpoint inhibitor). This patient is currently alive with stable disease at 187 days of follow up.
As these three cases represent limited reports of VEGF-directed therapy in patients with renal tumors that harbor TFEB alterations, no definitive conclusions could be drawn regarding therapeutic efficacy.
Histopathologic Features: TFEB rearranged and 6p21.1 (TFEB) Amplified Renal Tumors
Histologic features of patients diagnosed with renal tumors harboring TFEB alterations have been listed in Table 4 and summarized in Table 5. Representative images of these tumors have been depicted in Figures 2 and 3. These tumors were evaluated for features such as classic biphasic morphology which is characterized by tumor cells with clear to reticulated cytoplasm arranged in nests, alveoli or acini. Within these structures were smaller cells surrounding basement membrane-like material. Most of these cases showed immunohistochemical evidence of either cathepsin K, melan A or HMB45 expression (25 of 26, 96%) and tubulopapillary architectural patterns were seen in a third of these cases (12 of 36, 33.3%).
Table4.
Methods & Histopathology: Case Details
| Case No. | Digital PCR (TFEB/ B2M Percentage) |
IHC: TFEB | IHC: CathepsinK/ MelanA/ HMB45 |
Morphology | Biphasic Features |
Cytoplasmic Features |
WHO/ISUP Grade |
Sarcomatoid/Rhabdoid Features/Coagulative Necrosis |
Pigment/ Calcifications |
|---|---|---|---|---|---|---|---|---|---|
| TFEB Rearranged Tumors | |||||||||
| 1 | 2.4 | NA | CathepsinK+, MelanA+, HMB45+ | Tubulopapillary | − | Eosinophilic | 3 | Necrosis+ | − |
| 2 | 148.9 | + | CathepsinK+ | Acinar | + | Clear | 3 | − | − |
| 3 | 297.8 | + | CathepsinK+ | Solid | + | Clear | 3 | − | − |
| 4 | 313.2 | NA | CathepsinK+, MelanA+, HMB45− | Nested | − | Clear | 2 | − | − |
| 5 | 124.4 | NA | CathepsinK+, MelanA+, HMB45+ | Acinar | + | Clear | 2 | − | − |
| 6 | 196 | NA | CathepsinK+, MelanA+ | Nested | + | Clear | 2 | − | Calcification+ |
| 7 | 2.9 | + | CathepsinK+, MelanA+, HMB45+ | Tubulopapillary | − | Eosinophilic | 4 | Sarcomatoid+ | Pigment+ |
| 8 | 46.4 | + | CathepsinK+ | Tubulopapillary | − | Eosinophilic | 2 | − | − |
| 9 | 38.6 | + | CathepsinK+, MelanA+ | Solid | − | Clear | 2 | − | − |
| 10 | 34.8 | + | CathepsinK+ | Nested | − | Clear | 3 | Necrosis+ | − |
| 11 | 632.1 | NA | CathepsinK+, MelanA+, HMB45+ | Nested | + | Clear | 4 | Rhabdoid+ | − |
| 12 | 76.2 | NA | CathepsinK+, HMB45+ | Other | − | Clear | 1 | − | − |
| 13 | 26.2 | NA | CathepsinK+ | Tubulopapillary | − | Clear | 2 | Necrosis+ | − |
| 14 | 217.3 | NA | CathepsinK+, MelanA+ | Acinar | + | Clear | 2 | − | − |
| 15 | 40.9 | NA | CathepsinK+, MelanA+, HMB45− | Solid | − | Clear | 2 | − | Calcification+ |
| 16 | 804.2 | NA | CathepsinK+, MelanA+ | Solid | + | Clear | 1 | − | − |
| 17 | 890 | NA | CathepsinK+, MelanA+, HMB45+ | Nested | + | Clear | 1 | − | Pigment+, Calcification+ |
| 18 | NA | + | CathepsinK+, MelanA+ | Solid | − | Clear | 2 | − | − |
| TFEB Rearranged Tumors With 6p21.1 (TFEB) Amplifications | |||||||||
| 19 (t(6;11) & Amplification) | 72.7 | NA | CathepsinK+, MelanA+, HMB45− | Nested | − | Eosinophilic | 3 | Necrosis+ | − |
| 20 (TCGA-A3-3313-01; TFEB-KHDRBS2) | NA | NA | NA | Nested | − | Clear | 3 | NA | NA |
| 21 (TCGA-B9-A69E-01; TFEB-CADM2) | NA | NA | NA | Tubulopapillary | − | Eosinophilic | 2 | NA | − |
| 22 (TCGA-BQ-7048; COL21A1-TFEB) | 304.3 | + | NA | Tubulopapillary | + | Eosinophilic | 3 | − | Calcification+ |
| 6p21.1 (TFEB) Amplified Tumors | |||||||||
| 23 | 3.7 | NA | CathepsinK+, MelanA+, HMB45− | Tubulopapillary | − | Eosinophilic | 3 | − | − |
| 24 | 82.2 | NA | CathepsinK+, MelanA+, HMB45− | Tubulopapillary | − | Eosinophilic | 4 | Necrosis+, Rhabdoid+ | Pigment+ |
| 25 | 169.8 | NA | NA | Nested | − | Eosinophilic | 4 | − | − |
| 26 | 29.6 | − | NA | Nested | − | Clear | 4 | Necrosis+, Sarcomatoid+ | − |
| 27 | 10.5 | NA | CathepsinK+, MelanA+, HMB45+ | Tubulopapillary | − | Eosinophilic | 3 | − | − |
| 28 | 85.4 | NA | CathepsinK+, MelanA+, HMB45− | Nested | − | Eosinophilic | 3 | − | − |
| 29 | 7.9 | NA | CathepsinK−, MelanA−, HMB45− | Nested | − | Clear | 3 | Necrosis+ | − |
| 30 | 66.5 | + (weak) | CathepsinK+, MelanA+, HMB45− | Tubulopapillary | − | Eosinophilic | 3 | Necrosis+ | − |
| 31 | 5.3 | NA | MelanA+, HMB45− | Solid | − | Eosinophilic | 3 | Necrosis+ | − |
| 32 | NA | NA | NA | Tubulopapillary | − | Eosinophilic | 3 | − | − |
| 33 | NA | NA | NA | Papillary | − | Eosinophilic | 3 | − | − |
| 34 | NA | NA | NA | NA | NA | NA | NA | − | NA |
| 35 (TCGA-UZ-A9PQ-01) | NA | NA | NA | Other | − | Eosinophilic | 3 | NA | NA |
| 36 (TCGA-GL-7966–01) | NA | NA | NA | Tubulopapillary | − | Eosinophilic | 3 | NA | NA |
| 37 (TCGA-Q2-A5QZ-01) | NA | NA | NA | Papillary | − | Eosinophilic | 3 | Necrosis+ | NA |
Table5.
Methods & Histopathology: Summarized Data
| TFEB Rearranged Tumors | TFEB Rearranged Tumors With 6p21.1 (TFEB) Amplifications | 6p21.1 (TFEB) Amplified Tumors | All TFEB Altered tumors | |
|---|---|---|---|---|
| Total Number of Cases | 22* | 4** | 15 | 37 |
| Digital PCR (TFEB/B2M > 20%) | 17/19 (90%) | 2/2 | 5/9 (56%); p=0.06 | 22/28 (78.6%) |
| Immunohistochemistry (Diffuse Positivity: TFEB) | 8/8 (100%) | 1/1 | 1/2 (50%) (weak staining) | 9/10 (90%) |
| Immunohistochemistry (CathepsinK/ MelanA/ HMB45) | 19/19 (100%) | 1/1 | 6/7 (85.7%) | 25/26 (96%) |
| Morphology: Papillary | 0/22 (0%) | 0/4 | 2/14 (14.2%); p=0.14 | 2/36 (5.5%) |
| Morphology: Tubulopapillary | 6/22 (27.3%) | 2/4 | 6/14 (42.8%); p=0.47 | 12/36 (33.3%) |
| Morphology: Solid | 5/22 (22.7%) | 0/4 | 1/14 (7.1%); p=0.37 | 6/36 (16.7%) |
| Morphology: Nested | 7/22 (31.8%) | 2/4 | 4/14 (28.6%); p=1.0 | 11/36 (30.5%) |
| Morphology: Acinar | 3/22 (13.6%) | 0/4 | 0/14 (0%); p=0.27 | 3/36 (8.3%) |
| Morphology: Biphasic Features | 9/22 (41%) | 1/4 | 0/14 (0%); p=0.006 | 9/36 (25%) |
| Cytoplasm: Predominantly Eosinophilic | 6/22 (27.3%) | 1/3 | 12/14 (85.7%); p=0.0016 | 18/36 (50%) |
| Cytoplasm: Predominantly Clear | 16/22 (72.7%) | 1/4 | 2/14 (14.3%); p=0.0016 | 18/36 (50%) |
| ≥WHO/ISUP Grade3 | 9/22 (41%) | 1/3 | 14/14 (100%); p=0.0003 | 23/36 (63.8%) |
| Rhabdoid Features | Documented in 1 Case | - | Documented in 1 Case | Documented in 2 Cases |
| Sarcomatoid Features | Documented in 1 Case | - | Documented in 1 Case | Documented in 2 Cases |
| Coagulative Necrosis | Documented in 4 Cases | Documented in 1 Case | Documented in 6 Cases | Documented in 10 Cases |
| Pigment | Documented in 2 Cases | - | Documented in 1 Case | Documented in 3 Cases |
| Calcification | Documented in 4 Cases | Documented in 1 Case | - | Documented in 4 Cases |
WHO: World Health Organization; ISUP: International Society of Urologic Pathology
Rearrangement partners include MALAT1 (n=19), COL21A1 (n=1), CADM2 (n=1) and KHDRBS2 (n=1).
Rearrangement partners include MALAT1 (n=1), COL21A1 (n=1), CADM2 (n=1) and KHDRBS2 (n=1).
Statistical analysis performed using Fishers exact test.
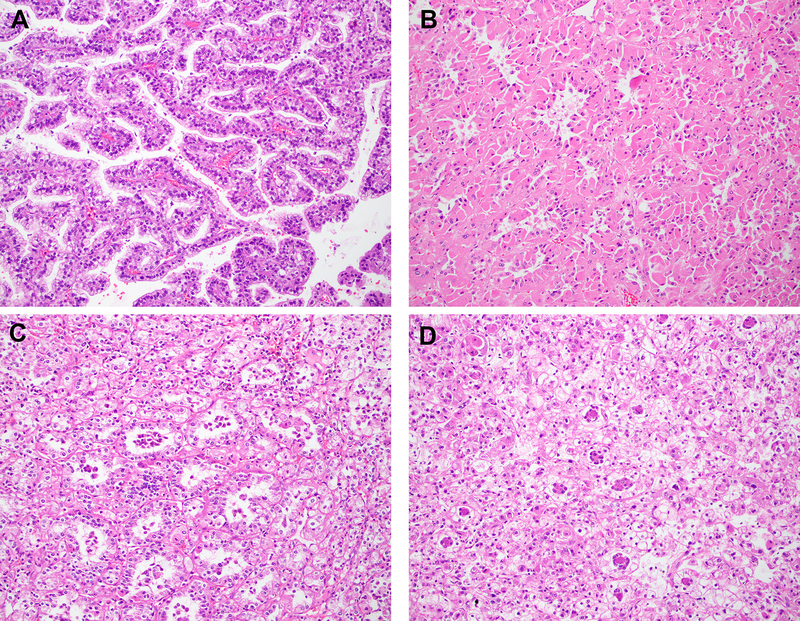
Figure 2: Histopathology.
Representative H&E stained images of TFEB-rearranged renal tumors and corresponding digital PCR quantification for TFEB are depicted ((a) papillary, TFEB/B2M=26%; (b) tubulopapillary with prominent cytoplasmic eosinophilia, TFEB/B2M=46%; (c, d) classic biphasic; × 200 magnification, c: TFEB/B2M=217% and d: TFEB/B2M=890%).
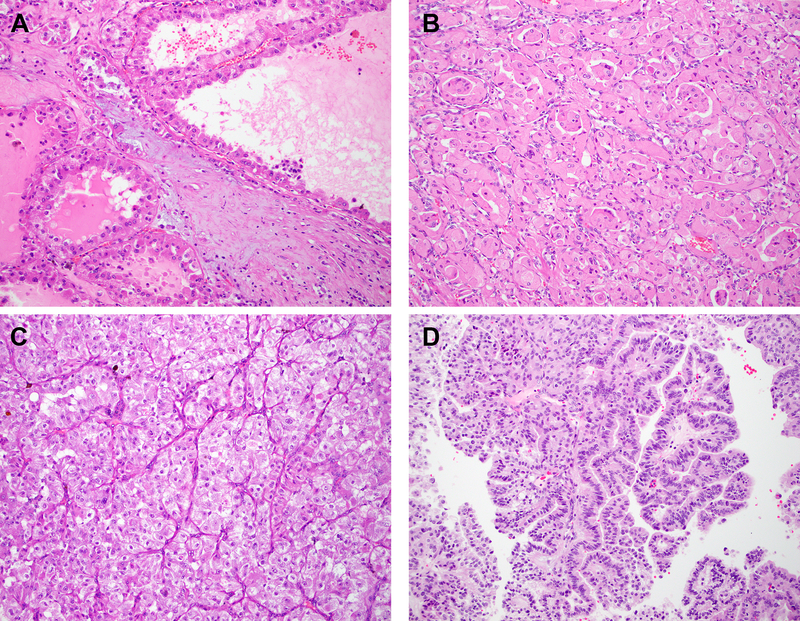
Figure 3: Histopathology.
Representative H&E stained images of 6p21.1 (TFEB)-amplified renal tumors and corresponding digital PCR quantification for TFEB are depicted ((a) cystic, TFEB/B2M=10.5%; (b) tubulopapillary, TFEB/B2M=66.5%; (c) alveolar/acinar, TFEB/B2M=85.4%; (d) papillary, × 200 magnification, digital PCR not performed).
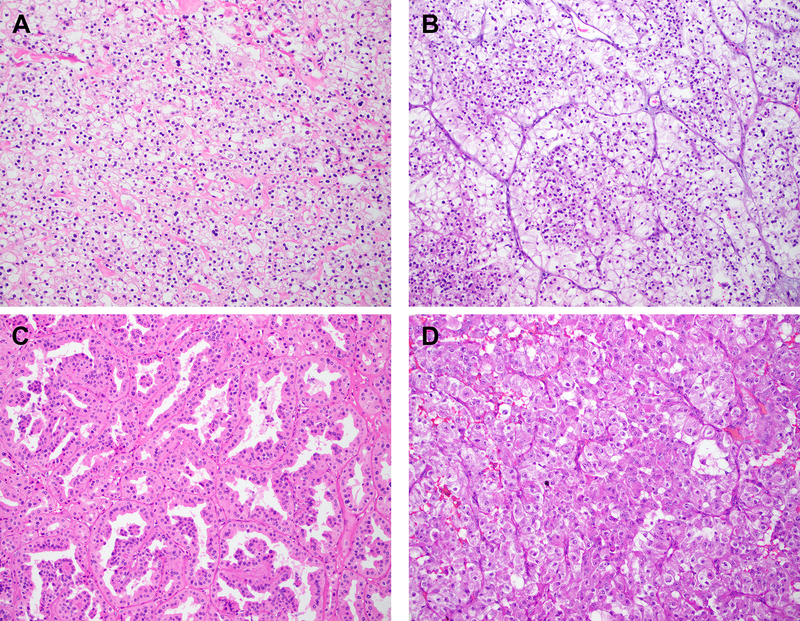
Cytologic features that helped in the separation of 6p21.1 (TFEB) amplified and TFEB-rearranged tumors included the presence of prominent cytoplasmic eosinophilia in the former (85.7% vs 27.3%; p=0.0016). Interestingly, while cytoplasmic eosinophilia was often seen in 6p21.1 amplified tumors, clear cytoplasm was frequently seen in TFEB-rearranged tumors (72.7% vs 14.3%; p=0.0016) (Figure 4). Classic biphasic morphology was not seen in any 6p21.1 (TFEB) amplified renal tumor (0/14, 0%) and was seen in almost half of all cases with TFEB rearrangements (9 of 22, 41%; p=0.006). Therefore, when biphasic features are present, they are helpful in morphologically separating these two entities. Other features that did not reveal any statistically significant differences included papillary, tubulopapillary, solid, nested and acinar architectural patterns. Tumors with tubulopapillary, solid and alveolar/acinar architectural patterns have been illustrated in Figure 4. The presence of pigment and calcifications were documented in a limited number of cases. Similarly, features associated with adverse outcomes in renal tumors such as coagulative tumor necrosis, rhabdoid and sarcomatoid transformation were seen in occasional cases with both 6p21.1 amplifications and TFEB rearrangements. Interestingly, all 6p21.1 amplified tumors were at least WHO/ISUP grade3 or higher (14 of 14 cases, 100%) when compared to TFEB-rearranged tumors (9 of 22, 41%; p=0.0003).
Figure 4: Histopathology.
Representative H&E stained images of TFEB-rearranged renal tumors with clear cytoplasm ((a) solid architecture; (b) alveolar/acinar architecture; × 200 magnification) and 6p21.1 (TFEB) amplified renal tumors with cytoplasmic eosinophilia ((c) tubulopapillary architecture; (d) solid to alveolar/acinar architecture; × 200 magnification) have been depicted.
TFEB Gene Expression: TFEB rearranged and 6p21.1 (TFEB) Amplified Renal Tumors
The results of TFEB gene expression profiling using digital PCR was correlated with immunohistochemistry for TFEB protein. Non-neoplastic renal parenchyma revealed a low-level of TFEB gene expression and this correlated with the nuclear expression of TFEB in a subset of renal tubules in all cases that were tested (Figure 5a–d). Preliminary results suggest that TFEB protein expression is restricted to distal tubules and future studies are needed to confirm this observation. Interestingly, the background level of TFEB gene expression in renal tumors that lacked TFEB alterations trended lower compared to non-neoplastic renal parenchyma (mean: 3.9% vs 8.2%; Figure 5e). TFEB gene expression in renal tumors with (n=28) and without (n=5) known TFEB alterations was assessed relative to TFEB gene expression in non-neoplastic renal parenchyma. In contrast to tumors that lacked TFEB amplifications/rearrangements, most tumors with TFEB alterations showed higher gene expression compared to non-neoplastic renal parenchyma (Figure 5d, f; p=0.0005).
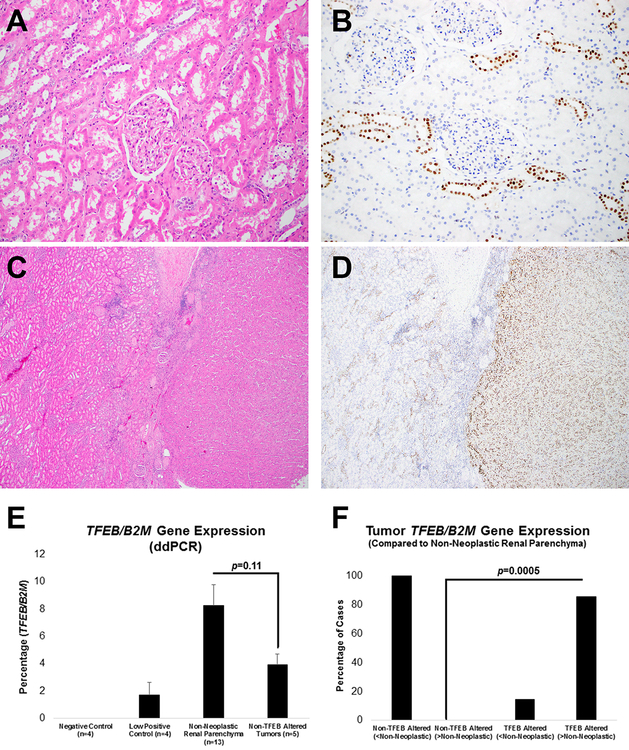
Figure 5: Histopathology, Immunohistochemistry and Digital Droplet PCR (Non-Neoplastic Renal Tissue).
Representative H&E stained images of non-neoplastic renal parenchyma adjacent to tumors with TFEB alterations is depicted ((a) × 200 magnification, 7.5% TFEB gene expression by digital PCR; (c) × 40 magnification, 3.7% TFEB gene expression by digital PCR). Corresponding immunostaining for TFEB shows scattered distal tubules with nuclear expression ((b) × 200 magnification; (d) × 40 magnification, with approximately >12-fold TFEB gene expression by digital PCR in the adjacent TFEB-rearranged renal tumor). Non-neoplastic renal parenchyma (n=13), when compared to non TFEB-altered renal tumors (n=5), showed a trend towards higher TFEB gene expression ((e) digital PCR). TFEB gene expression (normalized to B2M) was measured using digital PCR in both renal tumor types (with or without TFEB alterations) and in non-neoplastic renal parenchyma. Tumors that lacked TFEB amplifications/rearrangements showed lower expression compared to non-neoplastic renal parenchyma (f). In contrast, most tumors with TFEB alterations showed higher expression compared to non-neoplastic renal parenchyma (f).
Unlike immunohistochemistry, digital PCR to assess TFEB gene expression did not account for tumor heterogeneity. Two cases have been highlighted where the macrodissected tumor included either a sarcomatoid component (Figure 6a–d) or a second population of intra-luminal smaller viable cells (Figure 6e–f) that completely lacked nuclear TFEB protein expression by immunohistochemistry. Of note, the case with intra-luminal smaller cells did not show classic biphasic morphology characterized by smaller cells surrounding basement membrane-like material (Figure 6e–f). These elements likely contributed to a lower estimate for overall TFEB gene expression for both tumors.
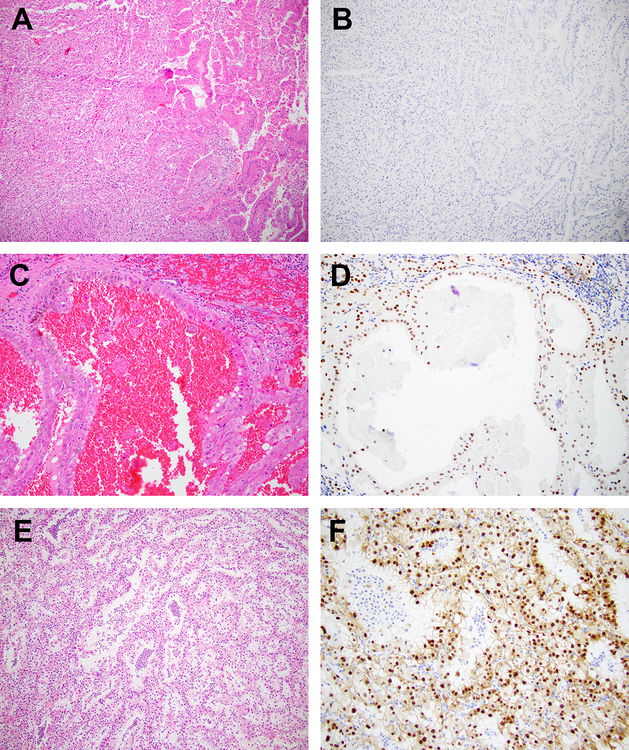
Figure 6: Histopathology and Immunohistochemistry (Tumor Heterogeneity).
Representative H&E stained images of a TFEB-rearranged renal tumor is depicted ((a) × 200 magnification, showing transition to sarcomatoid areas; (c) × 200 magnification, no sarcomatoid transformation). Corresponding immunostaining for TFEB shows absent expression in areas of sarcomatoid transformation (b) compared to nuclear expression in areas without sarcomatoid transformation (d). TFEB gene expression by digital PCR in both areas combined was 2.9%. Representative H&E stained image of a second TFEB-rearranged renal tumor ((e) × 40 magnification, 149% TFEB gene expression by digital PCR) shows a second population of intra-luminal, smaller viable cells that lack TFEB expression by immunohistochemistry (f).
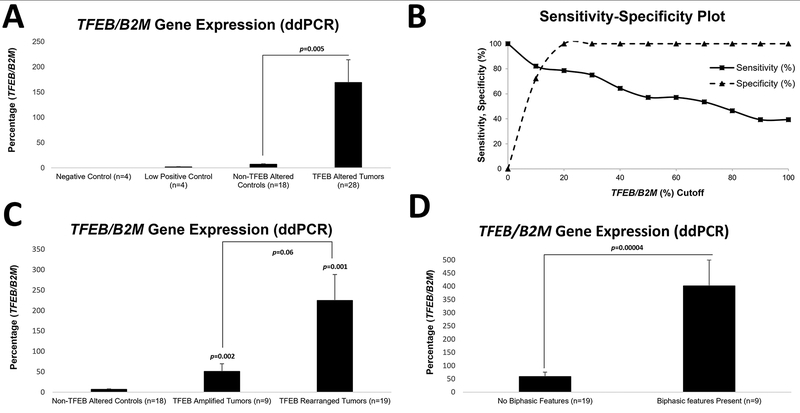
As expected, renal tumors with known TFEB alterations had significantly higher TFEB gene expression when compared to appropriate controls (Figure 7a). A cutoff of 20% TFEB gene expression had a sensitivity of 78.6% (22 of 28 cases) for detecting these cases and a specificity of 100% for discriminating these from non TFEB-altered tumors (n=18) (Figure 7b). Of note, cases with TFEB amplification tended to have lower expression of this gene compared to cases with rearrangements (mean: 51.2% vs 224.7%, p=0.06; Figure 7c and Table 5).
Figure 7: Digital Droplet PCR to Quantify TFEB Gene Expression.
Mean values of TFEB gene expression, comparing renal tumors with 6p21.1 (TFEB) amplification/TFEB rearrangement to non-TFEB altered controls is shown in (a). The relative sensitivity and specificity, for every 10% incremental increase in TFEB gene expression (normalized to B2M, digital PCR), for the detection of renal tumors with TFEB alterations is depicted (b). Comparisons of TFEB gene expression between renal tumors with 6p21.1 (TFEB) amplifications and TFEB rearrangements (c) and TFEB-altered renal tumors with or without biphasic morphologic features (d) is depicted.
Amongst tumors with known TFEB alterations, correlation of morphology with TFEB gene expression revealed that those with classic biphasic histologic features had significantly higher TFEB gene expression (mean: 401.7% vs 58.7%, p=0.00004; Figure 7d). A case with a known COL21A1-TFEB rearrangement occurring in the background of a 6p21.1 (TFEB) amplification (TCGA-BQ-7048) is highlighted in support of this observation (Figure 8a–f) (15). Two morphologically distinct areas from the same tumor were separately macrodissected and profiled for TFEB gene expression status. Areas with clear cell change showed lower gene and protein expression (Figure 8a–b) compared to areas that had features reminiscent of an oncocytoma (Figure 8c–d) adjacent to tumor with biphasic morphologic features (Figure 8e–f).
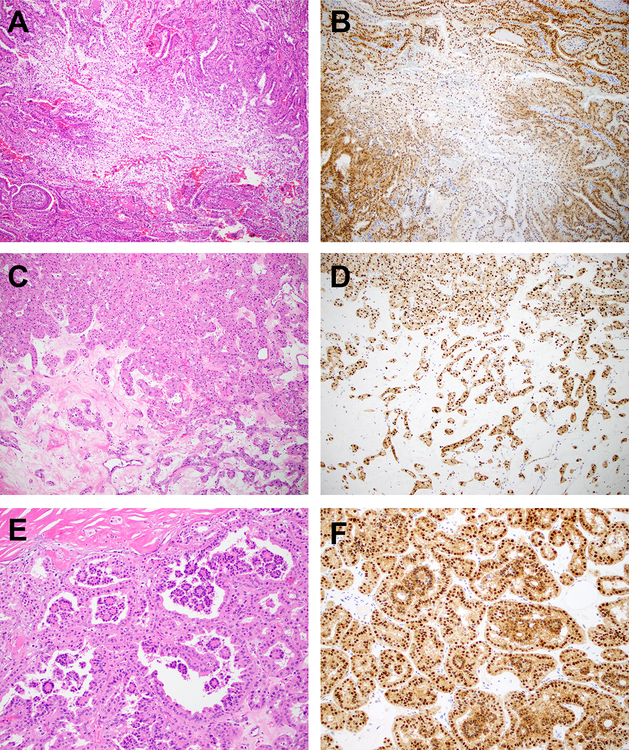
Figure 8: Histopathology and Immunohistochemistry (TFEB Rearranged (COL21A1-TFEB) and 6p21.1 (TFEB) Amplified Tumor).
Representative H&E and TFEB immunohistochemistry is shown for two morphologically distinct areas of a renal tumor with a COL21A1-TFEB rearrangement and associated 6p21.1 (TFEB) amplification which were separately macrodissected and profiled for TFEB gene expression status. An area of clear cell change is depicted ((a) × 100 magnification, (b) low/absent TFEB nuclear expression by immunohistochemistry, 72% TFEB gene expression by digital PCR). Other areas of the same tumor show features reminiscent of an oncocytoma ((c) × 100 magnification) and classic biphasic morphology that is characteristic of t(6;11) renal tumors ((e) × 100 magnification). These areas showed strong TFEB nuclear expression by immunohistochemistry (d, f) and higher (304%) TFEB gene expression by digital PCR.
Discussion
TFEB-overexpressing renal tumors were initially described in pediatric patient populations and are being increasingly identified in adult patients. This observation is supported by the results of our study (1). Amongst TFEB-overexpressing tumors, those harboring 6p21.1 (TFEB) amplifications were only recently described and emerging data suggests that these tumors have a more aggressive clinical course compared to TFEB-rearranged tumors (11).
In this regard, the English language literature was reviewed to determine the relative incidence of renal tumors with 6p21.1 (TFEB) amplifications and TFEB rearrangements, as well as documented cases of regional and distant metastatic disease for both tumor types (Table 6). This information was combined with results of the current study. For tumors harboring TFEB rearrangements we identified at least 13 cases with regional or distant metastasis (13, 18, 19, 25–31). These reports of aggressive disease were identified from approximately 106 cases (12%) reported between 1996 to 2019 and many of the reported cases had limited long-term follow up (1, 3–7, 10, 13–15, 18, 19, 25–50). In contrast, within a much shorter span of time (2014–2019) at least 25 6p21.1 (TFEB) amplified renal tumors with regional or distant metastasis have been reported (2, 6, 8–12, 14, 15, 31, 51). These reports were identified amongst 57 cases (44%) and a limited long-term follow up was documented for many of these cases as well (2, 6–12, 14, 15, 31, 47, 48, 51–53). The results of our study support this trend as well. Furthermore, at least 8 of 16 (50%) reported cases of renal tumors with lower level gains (defined as <10:1 ratio of TFEB signal to reference) at the 6p21.1 locus exhibited aggressive behavior (9, 11). Therefore, as recognition of this entity grows, future studies may be needed to refine diagnostic definitions, taking into consideration both tumor biology and outcomes.
Table6.
Reports of Regional and Distant Metastasis in Tumors with Molecular Confirmation of 6p21.1 (TFEB) Amplification or TFEB Rearrangement
| TFEB Rearranged Tumors | 6p21.1 Amplified Tumors (With or Without TFEB Rearrangements) | ||
|---|---|---|---|
| Reference | Total Cases | Reference | Total Cases |
| Pecciarini et al, 2007 | 1 | Peckova et al, 2014 | 1 |
| Camparo et al, 2008 | 1 | Argani et al, 2016 | 4 |
| Ishihara et al, 2011 | 1 | TCGA Malouf et al, 2014 TCGA Research Network, 2016 Argani et al, 2016 Williamson et al, 2017 Gupta et al, 2017 Mendel et al, 2018 Calio et al, 2018 |
4 |
| Argani et al, 2012 | 2 | Gupta et al, 2017* | 5 |
| Inamura et al, 2012 | 1 | Qu et al, 2017 | 1 |
| Smith et al, 2014 | 1 | Skala et al, 2017 | 2 |
| Hora et al, 2014 | 1 | Mendel et al, 2018 | 3 |
| Lilleby et al, 2015 | 1 | Calio et al, 2018* | 1 |
| Calio et al, 2017 | 2 | Kojima et al, 2019 | 1 |
| Kojima et al, 2019 | 1 | Current study | 3 |
| Current Study | 1 | ||
| Total Cases (1996 to 2019) | 13 (≈12.3%) | Total Cases (2014 to 2019) | 25 (≈43.9%) |
| Approximate Total Number of Reported Cases | 106 | Approximate Total Number of Reported Cases | 57 |
8 of 16 (≈50%) cases with low level gains exhibited regional or distant metastasis. TCGA: The Cancer Genome Atlas.
Broad molecular characterization of these tumors in prior studies had reported chromosome 3p loss/VHL alterations in a total of 7 such cases and two cases in our study showed 3p loss/VHL alteration (The Cancer Genome Atlas/Williamson et al, n=5; Mendel et al, n=2; MSK-IMPACT/current study, n=2; Table1) (7, 8, 12, 15). Overall, no recurrent alteration other than amplified oncogenes at 6p21.1 were identified in 5 such cases. It could therefore be hypothesized that the primary driver alterations in these tumors include TFEB and oncogenes present at the 6p21.1 locus (8, 12, 14, 15). VEGFA and CCND3 at the 6p21.1 locus are two candidate genes which could promote aggressive biologic behavior in these tumors and a few studies have documented alterations of the former in 6p21.1 amplified tumors (7, 9, 11, 12). However, the level of TFEB overexpression in these tumors was an unanswered question. Herein we have correlated TFEB gene expression with various clinicopathologic features to better understand the biology of these tumors.
Our current series of 37 cases (including 5 cases from The Cancer Genome Atlas dataset) was used to evaluate for histopathologic features that might be helpful in diagnosing these tumors. Our study suggests that renal tumors with alterations of the TFEB gene show significant morphologic heterogeneity. These results suggest that some morphologic features may be helpful in screening renal tumors for further characterization with ancillary immunohistochemical and molecular techniques, to establish a more precise diagnosis. Specifically, our results suggest that 6p21.1 (TFEB) amplified tumors exhibit tubulopapillary architecture and prominent cytoplasmic eosinophilia in close to half of these cases. Features helpful in the separation of TFEB amplified and rearranged tumors include cytoplasmic eosinophilia and high WHO/ISUP grade (≥ grade 3) in the former. Interestingly, classic biphasic morphology characterized by smaller cells surrounding basement membrane-like material was only seen in TFEB-rearranged tumors (41% of cases) and was not identified in any case with isolated 6p21.1 amplifications. This trend is consistent with what has been reported in the literature for (non-TCGA) 6p21.1 amplified cases (Argani et al: 2/8 cases; Williamson et al: 0/3 cases; Gupta et al: 3/11 cases; Skala et al: 0/6 cases; Mendel et al: 0/3 cases; Calio et al: 0/3 cases) (2, 8–12). Furthermore, tumors that exhibited classic biphasic features tended to have on average a 6.8-fold higher level of TFEB gene expression.
It is, however, important to note that some cases have a TFEB-rearrangement in the background of a 6p21.1 amplification and it may be challenging to identify rearrangement events in such cases using break-apart fluorescent in situ hybridization probes (7, 14, 15). Herein, we highlight one such case with a COL21A1-TFEB rearrangement that exhibited classic biphasic features (15). Finally, immunohistochemistry for either cathepsin K/ melan A or HMB45 was an effective screen and was able to identify >95% of cases for confirmatory testing.
At a cutoff of 20% TFEB: B2M expression, the sensitivity of the digital PCR assay in discriminating TFEB mRNA over-expressing tumors from non-TFEB overexpressing tumors was 78.6%, while maintaining a specificity of 100%. The lower sensitivity, at lower levels of TFEB gene expression, can in part be attributed to the inclusion of elements of non-neoplastic renal parenchyma in macrodissected tissue for downstream analysis. Other confounding variables, determined by correlations with corresponding immunohistochemistry, include tumor heterogeneity and TFEB gene expression in lymphoid infiltrates.
Important observations included a trend of lower TFEB gene expression in 6p21.1 (TFEB) amplified tumors (n=9, mean expression: 51.2%) compared to TFEB-rearranged tumors (n=19, mean expression: 224.7%, p=0.06). This was correlated with over 40% of the 6p21.1 amplified tumors showing TFEB gene expression under the 20% cutoff and with at least 2 cases lacking expression of screening markers (cathepsin K, melan A, HMB45, n=1; TFEB, n=1) by immunohistochemistry. These results suggest that the aggressive behavior of 6p21.1 (TFEB) amplified tumors may not be linked to higher levels of TFEB gene expression in these tumors. Our study therefore adds to the scientific literature by helping to establish a rationale for identifying other pathogenic alterations that may drive aggressive behavior in 6p21.1 (TFEB) amplified tumors.
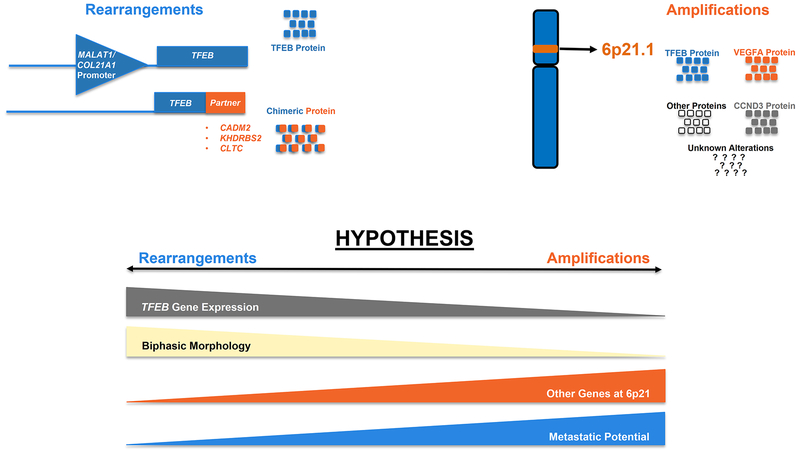
In summary, our model for differing pathogenic mechanisms in 6p21.1 (TFEB) amplified compared to TFEB-rearranged tumors is shown in Figure 9. Our data suggests that TFEB-amplified tumors have lower (TFEB) gene expression compared to TFEB-rearranged tumors. This, however, does not explain the more aggressive biologic behavior of these tumors, which might be better explained by other alterations including the amplification of additional oncogenes at the 6p21.1 locus. Finally, classic biphasic morphology appears to be primarily seen in TFEB-rearranged tumors and is correlated with significantly higher levels of TFEB gene expression.
Figure 9: Schematic Representation: Correlation of TFEB Gene Expression Status with Tumor Biology.
A graphical representation of the hypothesized spectrum of TFEB gene expression, biphasic morphologic features, 6p21.1 gene expression correlated with metastatic potential in renal tumors with TFEB rearrangements compared to those with 6p21.1 (TFEB) amplifications is depicted.
Acknowledgement
The authors would like to thank Jessica M. Menzel, Jennifer Posada and Christine Moon for administrative assistance.
Disclosures: The authors of this article have no relevant financial relationships with commercial interests to disclose. This study was supported in part through NIH/NCI Cancer Center Support grant P30CA008748.
References
- 1.Argani P, Hawkins A, Griffin CA, et al. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol. 2001;158:2089–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argani P, Reuter VE, Zhang L, et al. TFEB-amplified renal cell carcinomas: an aggressive molecular subset demonstrating variable melanocytic marker expression and morphologic heterogeneity. Am J Surg Pathol. 2016;40:1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dijkhuizen T, Berg EV, Störkel S, et al. Two cases of renal cell carcinoma, clear cell type, revealing a t(6;11)(p21;q13). Cancer Genetics; 1996:141. [Google Scholar]
- 4.van Asseldonk M, Schepens M, de Bruijn D, et al. Construction of a 350-kb sequence-ready 11q13 cosmid contig encompassing the markers D11S4933 and D11S546: mapping of 11 genes and 3 tumor-associated translocation breakpoints. Genomics. 2000;66:35–42. [DOI] [PubMed] [Google Scholar]
- 5.Kuiper RP, Schepens M, Thijssen J, et al. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet. 2003;12:1661–1669. [DOI] [PubMed] [Google Scholar]
- 6.Peckova K, Vanecek T, Martinek P, et al. Aggressive and nonaggressive translocation t(6;11) renal cell carcinoma: comparative study of 6 cases and review of the literature. Ann Diagn Pathol. 2014;18:351–357. [DOI] [PubMed] [Google Scholar]
- 7.Durinck S, Stawiski EW, Pavia-Jimenez A, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015;47:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williamson SR, Grignon DJ, Cheng L, et al. Renal cell carcinoma with chromosome 6p amplification including the TFEB gene: a novel mechanism of tumor pathogenesis? Am J Surg Pathol. 2017;41:287–298. [DOI] [PubMed] [Google Scholar]
- 9.Gupta S, Johnson SH, Vasmatzis G, et al. TFEB-VEGFA (6p21.1) co-amplified renal cell carcinoma: a distinct entity with potential implications for clinical management. Mod Pathol. 2017;30:998–1012. [DOI] [PubMed] [Google Scholar]
- 10.Skala SL, Xiao H, Udager AM, et al. Detection of 6 TFEB-amplified renal cell carcinomas and 25 renal cell carcinomas with MITF translocations: systematic morphologic analysis of 85 cases evaluated by clinical TFE3 and TFEB FISH assays. Mod Pathol. 2018;31:179–197. [DOI] [PubMed] [Google Scholar]
- 11.Calio A, Brunelli M, Segala D, et al. VEGFA amplification/increased gene copy number and VEGFA mRNA expression in renal cell carcinoma with TFEB gene alterations. Mod Pathol. 2018. [DOI] [PubMed] [Google Scholar]
- 12.Mendel L, Ambrosetti D, Bodokh Y, et al. Comprehensive study of three novel cases of TFEB-amplified renal cell carcinoma and review of the literature: Evidence for a specific entity with poor outcome. Genes Chromosomes Cancer. 2018;57:99–113. [DOI] [PubMed] [Google Scholar]
- 13.Calio A, Brunelli M, Segala D, et al. t(6;11) renal cell carcinoma: a study of seven cases including two with aggressive behavior, and utility of CD68 (PG-M1) in the differential diagnosis with pure epithelioid PEComa/epithelioid angiomyolipoma. Mod Pathol. 2018;31:474–487. [DOI] [PubMed] [Google Scholar]
- 14.Malouf GG, Su X, Yao H, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res. 2014;20:4129–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research N, Linehan WM, Spellman PT, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med. 2016;374:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith NE, Illei PB, Allaf M, et al. t(6;11) renal cell carcinoma (RCC): expanded immunohistochemical profile emphasizing novel RCC markers and report of 10 new genetically confirmed cases. Am J Surg Pathol. 2014;38:604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argani P, Yonescu R, Morsberger L, et al. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am J Surg Pathol. 2012;36:1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross DS, Zehir A, Cheng DT, et al. Next-generation assessment of Human Epidermal Growth Factor Receptor 2 (ERBB2) amplification status: clinical validation in the context of a hybrid capture-based, comprehensive solid tumor genomic profiling assay. J Mol Diagn. 2017;19:244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta S, Vanderbilt CM, Cotzia P, et al. JAK2, PD-L1, and PD-L2 (9p24.1) amplification in metastatic mucosal and cutaneous melanomas with durable response to immunotherapy. Hum Pathol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta S, Vanderbilt CM, Cotzia P, et al. Next-generation sequencing-based assessment of JAK2, PD-L1, and PD-L2 copy number alterations at 9p24.1 in breast cancer: potential implications for clinical management. J Mol Diagn. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu G, Benayed R, Ho C, et al. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pecciarini L, Cangi MG, Lo Cunsolo C, et al. Characterization of t(6;11)(p21;q12) in a renal-cell carcinoma of an adult patient. Genes Chromosomes Cancer. 2007;46:419–426. [DOI] [PubMed] [Google Scholar]
- 26.Camparo P, Vasiliu V, Molinie V, et al. Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol. 2008;32:656–670. [DOI] [PubMed] [Google Scholar]
- 27.Ishihara A, Yamashita Y, Takamori H, et al. Renal carcinoma with (6;11)(p21;q12) translocation: report of an adult case. Pathol Int. 2011;61:539–545. [DOI] [PubMed] [Google Scholar]
- 28.Inamura K, Fujiwara M, Togashi Y, et al. Diverse fusion patterns and heterogeneous clinicopathologic features of renal cell carcinoma with t(6;11) translocation. Am J Surg Pathol. 2012;36:35–42. [DOI] [PubMed] [Google Scholar]
- 29.Hora M, Urge T, Travnicek I, et al. MiT translocation renal cell carcinomas: two subgroups of tumours with translocations involving 6p21 [t (6; 11)] and Xp11.2 [t (X;1 or X or 17)]. Springerplus. 2014;3:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilleby W, Vlatkovic L, Meza-Zepeda LA, et al. Translocational renal cell carcinoma (t(6;11)(p21;q12) with transcription factor EB (TFEB) amplification and an integrated precision approach: a case report. J Med Case Rep. 2015;9:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima F, Kuroda N, Matsuzaki I, et al. Aggressive TFEB-rearranged renal cell carcinoma mimicking chromophobe and clear cell renal cell carcinoma. Pathol Int. 2019. [DOI] [PubMed] [Google Scholar]
- 32.Davis IJ, Hsi BL, Arroyo JD, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A. 2003;100:6051–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argani P, Lae M, Hutchinson B, et al. Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am J Surg Pathol. 2005;29:230–240. [DOI] [PubMed] [Google Scholar]
- 34.Argani P, Lae M, Ballard ET, et al. Translocation carcinomas of the kidney after chemotherapy in childhood. J Clin Oncol. 2006;24:1529–1534. [DOI] [PubMed] [Google Scholar]
- 35.Hora M, Hes O, Urge T, et al. A distinctive translocation carcinoma of the kidney [“rosette-like forming,” t(6;11), HMB45-positive renal tumor]. Int Urol Nephrol. 2009;41:553–557. [DOI] [PubMed] [Google Scholar]
- 36.Zhan HQ, Wang CF, Zhu XZ, et al. Renal cell carcinoma with t(6;11) translocation: a patient case with a novel Alpha-TFEB fusion point. J Clin Oncol. 2010;28:e709–713. [DOI] [PubMed] [Google Scholar]
- 37.Zhong M, De Angelo P, Osborne L, et al. Translocation renal cell carcinomas in adults: a single-institution experience. Am J Surg Pathol. 2012;36:654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao Q, Liu B, Cheng L, et al. Renal cell carcinomas with t(6;11)(p21;q12): A clinicopathologic study emphasizing unusual morphology, novel alpha-TFEB gene fusion point, immunobiomarkers, and ultrastructural features, as well as detection of the gene fusion by fluorescence in situ hybridization. Am J Surg Pathol. 2012;36:1327–1338. [DOI] [PubMed] [Google Scholar]
- 39.Rao Q, Zhang XM, Tu P, et al. Renal cell carcinomas with t(6;11)(p21;q12) presenting with tubulocystic renal cell carcinoma-like features. Int J Clin Exp Pathol. 2013;6:1452–1457. [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuura K, Inoue T, Kai T, et al. Molecular analysis of a case of renal cell carcinoma with t(6;11)(p21;q12) reveals a link to a lysosome-like structure. Histopathology. 2014;64:306–309. [DOI] [PubMed] [Google Scholar]
- 41.Chaste D, Vian E, Verhoest G, et al. Translocation renal cell carcinoma t(6;11)(p21;q12) and sickle cell anemia: first report and review of the literature. Korean J Urol. 2014;55:145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falzarano SM, McKenney JK, Montironi R, et al. Renal cell carcinoma occurring in patients with prior neuroblastoma: a heterogenous group of neoplasms. Am J Surg Pathol. 2016;40:989–997. [DOI] [PubMed] [Google Scholar]
- 43.Saleeb RM, Srigley JR, Sweet J, et al. Melanotic MiT family translocation neoplasms: expanding the clinical and molecular spectrum of this unique entity of tumors. Pathol Res Pract. 2017;213:1412–1418. [DOI] [PubMed] [Google Scholar]
- 44.Williamson SR, Eble JN, Palanisamy N. Sclerosing TFEB-rearrangement renal cell carcinoma: a recurring histologic pattern. Hum Pathol. 2017;62:175–179. [DOI] [PubMed] [Google Scholar]
- 45.Suarez-Vilela D, Izquierdo-Garcia F, Mendez-Alvarez JR, et al. Renal translocation carcinoma with expression of TFEB: presentation of a case with distinctive histological and immunohistochemical features. Int J Surg Pathol. 2011;19:506–509. [DOI] [PubMed] [Google Scholar]
- 46.Cajaiba MM, Dyer LM, Geller JI, et al. The classification of pediatric and young adult renal cell carcinomas registered on the children’s oncology group (COG) protocol AREN03B2 after focused genetic testing. Cancer. 2018;124:3381–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyvekens N, Rechsteiner M, Fritz C, et al. Histological and molecular characterization of TFEB-rearranged renal cell carcinomas (In Press). Virchows Arch. 2019. [DOI] [PubMed] [Google Scholar]
- 48.Martin EE, Mehra R, Jackson-Cook C, et al. Renal cell carcinoma with TFEB translocation versus unclassified renal cell carcinoma with TFEB amplification. 2017;22:305–312. [Google Scholar]
- 49.Kuroda N, Yorita K, Sasaki N, et al. Clinicopathological study of 5 cases of renal cell carcinoma with t(6;11)(p21;q12). Pol J Pathol. 2017;68:66–72. [DOI] [PubMed] [Google Scholar]
- 50.Petersson F, Vanecek T, Michal M, et al. A distinctive translocation carcinoma of the kidney; “rosette forming,” t(6;11), HMB45-positive renal tumor: a histomorphologic, immunohistochemical, ultrastructural, and molecular genetic study of 4 cases. Hum Pathol. 2012;43:726–736. [DOI] [PubMed] [Google Scholar]
- 51.Qu X, Tretiakova MS, Chen Y, et al. TFEB amplification renal cell carcinoma detected by chromosome genomic array testing: a case report for diagnosis of a novel entity. Journal of Clinical and Molecular Pathology. 2017;1:7 Available from: iMedPub, Hyderabad, India. Accessed March12, 2019. [Google Scholar]
- 52.Andeen NK, Qu X, Antic T, et al. Clinical utility of chromosome genomic array testing for unclassified and advanced-stage renal cell carcinomas. Arch Pathol Lab Med. 2018. [DOI] [PubMed] [Google Scholar]
- 53.Mayoral Guisado C, Gómez Durán Á, Agustín Benítez López D, et al. [TFEB-amplified renal cell carcinoma. A case report and review of the literature]. Rev Esp Patol. 2018;51:248–252. [DOI] [PubMed] [Google Scholar]