Introduction
Directed evolution is an effective approach to engineering enzymes and proteins for industrial, medical, and biotech applications [1,2] and was recently recognized with a Nobel Prize in Chemistry for its extraordinary practical impact. What is perhaps less well-known is the role directed evolution has played in elucidating and testing evolutionary mechanisms and theories of gene adaptation [3–6]. Understanding how a gene evolves is traditionally done retrospectively, by examining natural sequences and structures and working backwards to reconstruct descent and key evolutionary intermediates [7–11]. However, inferred histories are incomplete, can never be fully validated, and usually represent an N=1 experiment, as the conditions of natural evolution do not systematically repeat. By accelerating the process of evolution in the laboratory, directed evolution offers a way to study the evolution of genes in the forward direction, enabling researchers to observe adaptation in controlled environments, often in many replicates, and armed with the ability to sample and characterize the entire “fossil record” of each experiment. Such studies have yielded critical insights into the mechanisms by which genes, particularly proteins, evolve [12–15], the importance of stability in protein evolvability [16–21], the catalytic promiscuity of enzymes in the evolution of new activities [22–25], the complex fitness landscapes of proteins [26–35], and the role of neutral drift and fluctuating environments in crossing fitness valleys [36–39], to name only a few – and has solidified directed evolution as a powerful tool for understanding adaptation [5,6,40–42].
Despite these significant successes, there are limitations to using classical directed evolution techniques to study evolutionary mechanisms. Commonly, directed evolution mimics natural evolution by subjecting one or more genes of interest (GOIs) to multiple rounds of ex vivo diversification (e.g. error-prone PCR), transformation into cells, and selection [2]. Each round of this process represents a step in an adaptive trajectory but requires significant manual intervention that restricts the extent and scale of experiments. This keeps three tantalizing categories of experiments largely outside our reach. First are experiments requiring the traversal of long mutational pathways such as ambitious adaptations or studies aimed at probing gene evolution under varying conditions over extended periods of time. Indeed, most directed evolution experiments reach outcomes less than 5–10 non-synonymous mutations away from the parent sequence [16,38,43], with some exceptions that study the effects of extensive mutagenesis in one or few rounds [44–47]. Second are experiments requiring high statistical power through replication, such as studying drug resistance pathways, comparing the effects of different conditions on adaptation, detecting rare outcomes and rare adaptive trajectories, or mapping rugged fitness landscapes. Currently, most directed evolution experiments are limited to only a few replicates [39,43,48]. Third are experiments that wish to capture or test complex population dynamics, since the technical idiosyncrasies of transformation and ex vivo diversification can cause population bottlenecks and perturb dynamics in artificial ways that influence evolutionary trajectories.
To address these limitations, synthetic biologists are working to establish a new paradigm in directed evolution through the construction of so-called continuous evolution systems. Continuous evolution achieves diversification of GOIs in vivo such that manual rounds of ex vivo diversification, transformation, and in vivo GOI expression and selection are not needed [49–52]. Instead, rapid diversification of GOIs occurs concurrently with their expression and functional selection, converting labor-intensive stepwise directed evolution processes into ones requiring only the serial passaging of cells under selection conditions. This allows for evolution experiments that require long mutational pathways, large-scale replication, and the ability to capture complex population dynamics, categories particularly useful for testing evolutionary mechanisms and theory (Figure 1). In this opinion, we will briefly discuss the current state of continuous evolution systems and present their early successes and potential in reinvigorating the use of directed evolution to study basic questions in gene and protein adaptation.
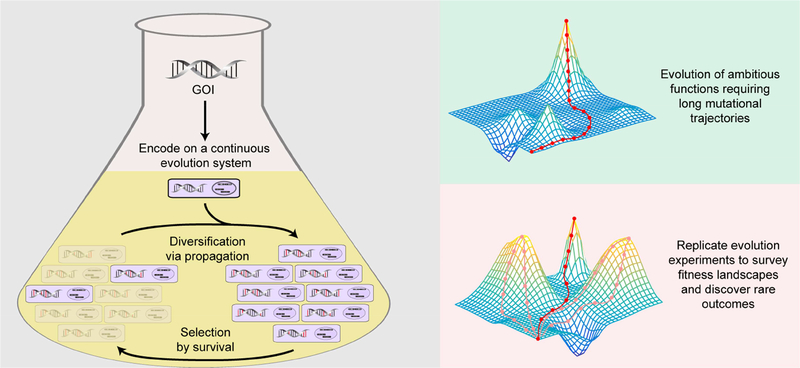
Figure 1.
Continuous in vivo evolution systems enable the rapid continuous diversification of genes of interest in multiple replicate cultures. Through coupling continuous diversification with selection, simply passaging cultures can drive protein evolution on laboratory timescales. This allows proteins to achieve ambitious functions that may require high numbers of mutation (>10–20). Further, the ability to run replicate evolution experiments allows for the detailed mapping of fitness landscapes, discovery of rare outcomes, exploration of multiple environmental conditions and population structures, and statistical power in testing evolutionary reproducibility and basic evolutionary theories.
State of continuous evolution systems
We will center our discussion in this section around three key properties that continuous evolution systems should have: targeting, durability, and scalability. There are others, discussed in depth elsewhere [52,53], but targeting, durability, and scalability are uniquely important if the goal is to study mechanisms of how a GOI evolves, as these three properties enable the rapid exploration of long mutational trajectories with statistical power.
First, targeting. Certainly, if one wishes to explore how a GOI evolves, one will not want other loci contributing to the evolved function. But beside this practical reason for targeting is a deeper one. Very high rates of diversification are needed to see adaptation at the gene level on laboratory timescales, but such high rates harm or destroy host genomes, since there is a general inverse relationship between the rate at which an information polymer can be mutated under selection for function and its size [54–57]. For example, a GOI of size 1 kb can likely withstand a continuous mutation rate on order ~10−3 substitutions per base (s.p.b.) while a host genome of size 107 bp (e.g. for Saccharomyces cerevisiae) will likely accumulate a lethal mutation every generation at mutation rates around ~10−6 s.p.b. and experience clear fitness defects at mutation rates above ~10−8 s.p.b. [53,55]. Indeed, mutation rates of microbes and mammalian cells are evolutionarily optimized to be in the 10−9-10−10 s.p.b. range to prevent deterioration of fitness through high mutational loads over time [57–59]. Therefore, to evolve a GOI rapidly in vivo, mutations must be targeted to the GOI with extreme specificity. Most continuous evolution systems achieve incomplete targeting of GOIs relative to host genomes and other DNA [60–62], but two systems have either managed to achieve complete targeting or avoid the problems of genomic mutation physically [49,53]. OrthoRep, developed in our lab, consists of an orthogonal DNA polymerase (DNAP)-plasmid pair in S. cerevisiae that can mutate target GOIs at ~10−5 s.p.b. without any increase in host genomic rates (~10−10 s.p.b.) [53]; and phage assisted continuous evolution (PACE) elevates phage genome mutation rates along with host mutation rates, but ingeniously disregards host mutation effects by removing host Escherichia coli cells fast enough to prevent host propagation but slow enough to ensure phage propagation [49,63].
Second, durability. Ideally, a continuous evolution system will mutate target GOIs indefinitely so that long mutational paths (e.g. >10 non-synonymous mutations) can be traversed over extended periods of strong selection or more complex sequences of selection that mimic natural evolution [64,65]. So far, both OrthoRep and PACE have proven to be quite durable – we have used OrthoRep in several evolution experiments to evolve GOIs for >300 generations and still observe rapid adaptation, accumulating 10–20 mutations (manuscripts in preparation); and PACE has been used in experiments that adapt over hundreds of phage generations, accumulating 10–20 mutations [64,65]. Durability in other continuous evolution systems [60–62,66–73] remain untested, but one can predict durability based on the architecture of the system. For example, in OrthoRep, the only way a GOI gets replicated is through an error-prone DNAP, encoded on a host plasmid or genome that doesn’t experience elevated mutagenesis. This, combined with the fact that OrthoRep achieves complete mutational targeting to avoid selection against elevated mutation rates through mutational loads on the genome, favors durability. Likewise, in PACE, durability is favored, because the only way a phage genome encoding a GOI is replicated is through error-prone means.
Third, scalability. Especially important for experimental evolution, a continuous evolution system should ideally be scalable in nature. Evolving a GOI with a large number of replicates is crucial for observing low frequency events [53,74,75], inferring beneficial mutations [76,77], and determining the extent to which evolutionary trajectories are reproducible [31,78]. Continuous evolution systems that are fully in vivo, such as OrthoRep, offer scalablity, because evolution experiments can be carried out simply through serial passaging, amenable to extensive replication or parallelization [60,61,71,72]. Although PACE usually requires chemostat or turbidostat setups that limit scale, recent experiments demonstrate that PACE may be conducted via bulk passaging without such setups and should be amenable to extensive replication [79].
In short, a number of continuous evolution systems, including OrthoRep and PACE, are at a stage of development where they should be able to routinely drive GOI evolution at the speeds, durations, and scale required to study mechanisms of gene evolution through forward evolution experiments.
Early applications of continuous evolution to studying evolutionary pathways and mechanisms
PACE has been the most successful continuous evolution system for proteins to date. In PACE, a GOI is encoded on a phage’s genome and through coupling an improvement in the GOI’s function to phage survival and infectivity, GOIs with beneficial mutations rapidly propagate in a pool of E. coli. By having a continual influx of E. coli at a rate that is between the doubling time of phage and E. coli, GOIs can rapidly accumulate mutations while mutated host cells are removed. Although most PACE experiments have focused on protein engineering applications, some have aimed to understand the details of evolutionary mechanisms. In 2013, Leconte and colleagues examined the effect of selection stringency and rate of mutagenesis, key parameters in evolutionary theory, on the evolution of T7 RNA polymerase (RNAP) towards recognition of the T3 promoter [80]. While the effect of mutation rate on adaptive pathways has been studied in other contexts [81–87], PACE enabled Leconte et al. to isolate the effects of mutation rate on a single gene in freely evolving replicate cultures [80]. They demonstrated that high mutation rates resulted in more reproducible fixation, possibly by increasing the frequency in which superior mutations are accessed, as consistent with predictions made in silico [84]. Further, Leconte et al. showed that the strength of selection resulted in substantial differences in adaptive trajectories with stronger selection favoring lower diversity and higher reproducibility as expected. Significantly, only with the benefit of replicate experiments were Leconte et al. able to show that while mutational patterns indeed appeared across replicates, both the specific adaptive mutations undergoing fixation, and even successful adaptation itself, can be stochastic in nature.
In a separate study, Dickinson et al. used PACE to explore contingency in evolution [88]. Previous research has shown that while the stepwise evolution of a single gene or a small set of genes could be practically deterministic [26,31,89], convergent evolution from dissimilar proteins and histories can lead to vastly different sequences, structures, and activities [32,90,91]. In their 2013 study, Dickinson et al. asked just how much dissimilarity in the history of a protein’s evolution was needed to result in significant changes in evolutionary outcomes. With PACE, they were able to conduct replicate evolution experiments where T7 RNAP was first diverged to recognize either the T3 or SP6 promoter, and subsequently pressured to recognize the same final promoter, a hybrid T3/SP6 promoter. Dickenson et al. impressively showed that the divergent evolutionary steps were sufficient to drastically alter the mutational trajectory as well as the maximum catalytic efficiency of the final evolved enzymes. Specifically, populations of T7 RNAP that were first evolved for T3 promoter recognition evolved lower activity for the final promoter compared to populations first evolved for the SP6 promoter, with differences persisting even after extensive continued selection (~40 generations at high mutational load) for recognition of the final promoter. This study elegantly shows the importance of contingency in evolution and how historical effects are not easily forgotten through strong selection alone. Further, through characterization of mutations present in different experiments, Dickenson et al. identified a key epistatic interaction between two mutations that act to isolate two outcomes from each other. With these results, PACE gives us a sense of the types of questions in basic evolutionary biology that continuous evolution can address, questions that would be difficult to study with traditional directed evolution techniques. Although these examples focus only on T7 RNAP, PACE is generalizable to the evolution of any function that can be linked to the expression or activity of an essential phage coat protein [63]. Indeed, in molecular-engineering-motivated PACE experiments, many other proteins have been evolved [2,49,64,65,79], and additional basic evolution experiments on proteins besides T7 RNAP are surely underway.
Another continuous evolution technology, OrthoRep, has recently enabled a detailed mapping of adaptive trajectories on a fitness landscape, including low probability events, and demonstrated the effects of epistasis and clonal interference on the reproducibility of adaptation [92]. OrthoRep uses an orthogonal error-prone DNA polymerase-plasmid pair in S. cerevisiae to achieve targeted mutagenesis of GOIs [53,92]. By encoding Plasmodium falciparum dihydrofolate reductase (PfDHFR) on OrthoRep, Ravikumar et al. rapidly evolved resistant pyrimethamine-resistant PfDHFR variants simply by passaging 0.5 mL yeast cultures in media containing increasing concentrations of pyrimethamine. Owing to the scalability of OrthoRep, this experiment was easily repeated 90 times to abundantly sample adaptive trajectories. From this, Ravikumar et al. uncovered a more complex fitness landscape than previously realized, including new mutants as resistant as those widely studied. One mutant occurred frequently due to a highly adaptive first-step mutation (S108N) that exhibited a conflict with a highly-adaptive later mutation (D54N), making most sequences containing both S108N and D54N non-functional. Yet C59R and/or Y57H were able to resolve this conflict between S108N and D54N. This led to convergence of adaptive trajectories across replicates. However, in a few replicates, rare mutations steered populations towards other equally-fit outcomes, including ones lacking S108N, and suboptimal local fitness peaks. Since these alternative variants are expected to respond differently to secondary drugs, population structures and strategies that favor rare mutational pathways may be important for drug schedule design, which we are currently exploring. In short, by exploiting rapid and scalable continuous evolution, one can explore adaptation on rugged fitness landscapes to tease out both the stochastic and deterministic nature of evolution.
Future potential
Continuous evolution systems hold great promise in studying the mechanisms and pathways of gene adaptation. The early studies described above give a glimpse into how continuous evolution can be used to carry out controlled forward evolution experiments that discover and map interesting regions of fitness landscapes, test the reproducibility of adaptation, and compare how different parameters of evolution and selection schedules result in different mutational trajectories and outcomes. As more researchers use continuous evolution to carry out forward evolution experiments with previously inaccessible speed, depth, and scale, significant insights should be made. These should not only include exquisite details of how specific genes adapt through the interplay among mutations, but also general insights into the most fundamental questions in molecular evolution – the reproducibility of adaptation [93,94], how fitness valleys are crossed [95,96], the importance of fluctuating environments or population structure in adaptation [97], the prevalence and role of epistasis in protein evolution [98–100], the existence of tradeoffs among different gene functions [19,41], the determinants of evolvability [40,101], the high prevalence of certain folds or structures in enzymes [102–104], the evolutionary basis of protein-protein interactions [105,106], and the role of both intracellular and environmental conditions in dictating how a gene adapts [107,108]. With the number of powerful systems available and ongoing development in each, such as the inclusion of gene-specific sexual recombination into OrthoRep (unpublished data), continuous evolution should become a staple technology for probing the fundamentals of adaptation.
Acknowledgements
We thank the National Institutes of Health (1DP2GM119163-01) the Sloan Research Fellowship for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turner NJ: Directed evolution drives the next generation of biocatalysts. Nat Chem Biol 2009, 5:567–573. [DOI] [PubMed] [Google Scholar]
- 2.Packer MS, Liu DR: Methods for the directed evolution of proteins. Nat Rev Genet 2015, 16:379–394. [DOI] [PubMed] [Google Scholar]
- 3.Peisajovich SG, Tawfik DS: Protein engineers turned evolutionists. Nat Methods 2007, 4:991–994. [DOI] [PubMed] [Google Scholar]
- 4.Bloom JD, Arnold FH: In the light of directed evolution: Pathways of adaptive protein evolution. Proc Natl Acad Sci 2009, 106:9995–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romero PA, Arnold FH: Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol 2009, 10:866–876.** Review covering some of the key findings on protein adaptation using directed evolution.
- 6.Arnold FH, Wintrode PL, Miyazaki K, Gershenson A: How enzymes adapt: Lessons from directed evolution. Trends Biochem Sci 2001, 26:100–106. [DOI] [PubMed] [Google Scholar]
- 7.Merkl R, Sterner R: Ancestral protein reconstruction: Techniques and applications. Biol Chem 2016, 397:1–21. [DOI] [PubMed] [Google Scholar]
- 8.Koshi JM, Goldstein RA: Probabilistic reconstruction of ancestral protein sequences. J Mol Evol 1996, 42:313–320. [DOI] [PubMed] [Google Scholar]
- 9.Zuckerkandl E, Pauling L: Molecules as Documents of Evolutionary History. J Theor Biol 1965, 8:357–366. [DOI] [PubMed] [Google Scholar]
- 10.Salemi M, Vandamme A-M (Eds): The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny. Cambridge University Press, Cambridge, UK; 2003. [Google Scholar]
- 11.Nei M, Kumar S: Molecular Evolution and Phylogenetics. Oxford University Press; 2000. [Google Scholar]
- 12.Tokuriki N, Jackson CJ, Afriat-Jurnou L, Wyganowski KT, Tang R, Tawfik DS: Diminishing returns and tradeoffs constrain the laboratory optimization of an enzyme. Nat Commun 2012, 3:1257–1259. [DOI] [PubMed] [Google Scholar]
- 13.Yadid I, Kirshenbaum N, Sharon M, Dym O, Tawfik DS: Metamorphic proteins mediate evolutionary transitions of structure. Proc Natl Acad Sci 2010, 107:7287–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peisajovich SG, Rockah L, Tawfik DS: Evolution of new protein topologies through multistep gene rearrangements. Nat Genet 2006, 38:168–174. [DOI] [PubMed] [Google Scholar]
- 15.Khersonsky O, Malitsky S, Rogachev I, Tawfik DS: Role of chemistry versus substrate binding in recruiting promiscuous enzyme functions. Biochemistry 2011, 50:2683–2690. [DOI] [PubMed] [Google Scholar]
- 16.Bloom JD, Labthavikul ST, Otey CR, Arnold FH: Protein stability promotes evolvability. Proc Natl Acad Sci 2006, 103:5869–5874.** An important study that identified the destabilizing effects of beneficial mutations in directed evolution studies and how a selection for stability can predispose an enzyme for further functional gains.
- 17.Bloom JD, Raval A, Wilke CO: Thermodynamics of neutral protein evolution. Genetics 2007, 175:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokuriki N, Tawfik DS: Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 2009, 19:596–604. [DOI] [PubMed] [Google Scholar]
- 19.Tokuriki N, Stricher F, Serrano L, Tawfik DS: How protein stability and new functions trade off. PLoS Comput Biol 2008, 4:35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Minasov G, Shoichet BK: Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol 2002, 320:85–95. [DOI] [PubMed] [Google Scholar]
- 21.Beadle BM, Shoichet BK: Structural bases of stability-function tradeoffs in enzymes. J Mol Biol 2002, 321:285–296. [DOI] [PubMed] [Google Scholar]
- 22.Bloom JD, Romero PA, Lu Z, Arnold FH: Neutral genetic drift can alter promiscuous protein functions, potentially aiding functional evolution. Biol Direct 2007, 2:7–10.* This study showed that enzymes can easily achieve a wide range of promiscuous functions.
- 23.Kaltenbach M, Emond S, Hollfelder F, Tokuriki N: Functional Trade-Offs in Promiscuous Enzymes Cannot Be Explained by Intrinsic Mutational Robustness of the Native Activity. PLoS Genet 2016, 12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khersonsky O, Tawfik DS: Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective. Annu Rev Biochem 2010, 79:471–505. [DOI] [PubMed] [Google Scholar]
- 25.Aharoni A, Gaidukov L, Khersonsky O, Gould SM, Roodveldt C, Tawfik DS: The “evolvability” of promiscuous protein functions. Nat Genet 2005, 37:73–76.* This study demonstrated that when an enzyme develops promiscuous functions, it does not necessarly result in a deterioration of the original function.
- 26.Weinreich DM, Delaney NF, Depristo M a, Hartl DL: Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science 2006, 312:111–114. [DOI] [PubMed] [Google Scholar]
- 27.Tan L, Serene S, Chao HX, Gore J: Hidden randomness between fitness landscapes limits reverse evolution. Phys Rev Lett 2011, 106:1–4. [DOI] [PubMed] [Google Scholar]
- 28.Hietpas RT, Jensen JD, Bolon DNA: Experimental illumination of a fitness landscape. Proc Natl Acad Sci 2011, 108:7896–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS: Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 2006, 444:929–932. [DOI] [PubMed] [Google Scholar]
- 30.Haddox HK, Dingens AS, Hilton SK, Overbaugh J, Bloom JD: Mapping mutational effects along the evolutionary landscape of HIV envelope. Elife 2018, 7:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Visser JAGM, Krug J: Empirical fitness landscapes and the predictability of evolution. Nat Rev Genet 2014, 15:480–490. [DOI] [PubMed] [Google Scholar]
- 32.Salverda MLM, Dellus E, Gorter FA, Debets AJM, van der Oost J, Hoekstra RF, Tawfik DS, de Visser JAGM: Initial mutations direct alternative pathways of protein evolution. PLoS Genet 2011, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellus-Gur E, Elias M, Caselli E, Prati F, Salverda MLM, De Visser JAGM, Fraser JS, Tawfik DS: Negative epistasis and evolvability in TEM-1 β-lactamase - The thin line between an enzyme’s conformational freedom and disorder. J Mol Biol 2015, 427:2396–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bank C, Matuszewski S, Hietpas RT, Jensen JD: On the (un)predictability of a large intragenic fitness landscape. Proc Natl Acad Sci 2016, 113:14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaltenbach M, Jackson CJ, Campbell EC, Hollfelder F, Tokuriki N: Reverse evolution leads to genotypic incompatibility despite functional and active site convergence. Elife 2015, 4:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amitai G, Gupta RD, Tawfik DS: Latent evolutionary potentials under the neutral mutational drift of an enzyme. HFSP J 2007, 1:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith WS, Hale JR, Neylon C: Applying neutral drift to the directed molecular evolution of a -glucuronidase into a -galactosidase: Two different evolutionary pathways lead to the same variant. BMC Res Notes 2011, 4:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bershtein S, Goldin K, Tawfik DS: Intense Neutral Drifts Yield Robust and Evolvable Consensus Proteins. J Mol Biol 2008, 379:1029–1044.* This study demonstrates that while maintaining function, an enzyme develops mutations that allow it tolerate destabilizing mutations when evolving a novel function.
- 39.Steinberg B, Ostermeier M: Environmental changes bridge evolutionary valleys. Sci Adv 2016, 2:1–9.* This study uses a clever band-pass selection to identify selection conditions for a beta-lactamase, TEM-1, that allow crossing of fitness valleys.
- 40.Soskine M, Tawfik DS: Mutational effects and the evolution of new protein functions. Nat Rev Genet 2010, 11:572–582. [DOI] [PubMed] [Google Scholar]
- 41.Tawfik DS: Accuracy-rate tradeoffs: How do enzymes meet demands of selectivity and catalytic efficiency? Curr Opin Chem Biol 2014, 21:73–80. [DOI] [PubMed] [Google Scholar]
- 42.Tokuriki N, Tawfik DS: Protein Dynamism and Evolvability. Science 2012, 324:203–207. [DOI] [PubMed] [Google Scholar]
- 43.Goldsmith M, Tawfik DS: Enzyme engineering: reaching the maximal catalytic efficiency peak. Curr Opin Struct Biol 2017, 47:140–150.* This review summarizes some of the key lessons learned via directed evolution and provides guidelines for evolving functional proteins, especially when a fitness plateau is suspected.
- 44.Drummond DA, Iverson BL, Georgiou G, Arnold FH: Why high-error-rate random mutagenesis libraries are enriched in functional and improved proteins. J Mol Biol 2005, 350:806–816. [DOI] [PubMed] [Google Scholar]
- 45.Zaccolo M, Gherardi E: The effect of high-frequency random mutagenesis on in vitro protein evolution: A study on TEM-1 β-lactamase. J Mol Biol 1999, 285:775–783. [DOI] [PubMed] [Google Scholar]
- 46.Kunichika K, Hashimoto Y, Imoto T: Robustness of hen lysozyme monitored by random mutations. Protein Eng 2002, 15:805–809. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Ruff AJ, Arlt M, Schwaneberg U: Casting epPCR (cepPCR): A simple random mutagenesis method to generate high quality mutant libraries. Biotechnol Bioeng 2017, 114:1921–1927. [DOI] [PubMed] [Google Scholar]
- 48.Tokuriki N, Tawfik DS: Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 2009, 459:668–673. [DOI] [PubMed] [Google Scholar]
- 49.Badran AH, Liu DR: In vivo continuous directed evolution. Curr Opin Chem Biol 2015, 24:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.d’Oelsnitz S, Ellington A: Continuous directed evolution for strain and protein engineering. Curr Opin Biotechnol 2018, 53:158–163. [DOI] [PubMed] [Google Scholar]
- 51.Zheng X, Xing XH, Zhang C: Targeted mutagenesis: A sniper-like diversity generator in microbial engineering. Synth Syst Biotechnol 2017, 2:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wellner A, Ravikumar A, Liu CC: Continuous evolution of proteins in vivo. 2018:in revision.* This review discusses points of distinction among and potential of current continuous in vivo evolution systems.
- 53.Ravikumar A, Arzumanyan GA, Obadi MKA, Javanpour AA, Liu CC: Scalable continuous evolution of genes at mutation rates above genomic error thresholds. Cell 2018, 175:1–12.** In this study, the authors describe a continuous in vivo evolution system, which they use to demonstrate evolution of a target protein in 90 replicates, revealing interesting mutational pathways to adaptation and rare outcomes.
- 54.Bull JJ, Sanjuan R, Wilke CO: Theory of Lethal Mutagenesis for Viruses. J Virol 2007, 81:2930–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herr AJ, Ogawa M, Lawrence NA, Williams LN, Eggington JM, Singh M, Smith RA, Preston BD: Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet 2011, 7:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C: Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 2001, 412:331–333. [DOI] [PubMed] [Google Scholar]
- 57.Nowak M, Schuster P: Error Thresholds of Replication in Finite Populations Mutation Frequencies and the Onset of Muller’s Ratchet. J Theor Biol 1989, 137:375–395. [DOI] [PubMed] [Google Scholar]
- 58.Felsenstein J: The evolutionary advantage of recombination. Genetics 1976, 83:845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller HJ: The Relation of Recombination to Mutational Advance. Mutat Res 1964, 1:2–9. [DOI] [PubMed] [Google Scholar]
- 60.Halperin SO, Tou CJ, Wong EB, Modavi C, Schaffer DV., Dueber JE: CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 2018, 560:248–252. [DOI] [PubMed] [Google Scholar]
- 61.Moore CL, Papa LJ, Shoulders MD: A Processive Protein Chimera Introduces Mutations across Defined DNA Regions in Vivo. J Am Chem Soc 2018, 140:11560–11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Camps M, Naukkarinen J, Johnson BP, Loeb LA: Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci U S A 2003, 100:9727–9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esvelt KM, Carlson JC, Liu DR: A system for the continuous directed evolution of biomolecules. Nature 2011, 472:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Packer MS, Rees HA, Liu DR: Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat Commun 2017, 8:956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Badran AH, Guzov VM, Huai Q, Kemp MM, Vishwanath P, Kain W, Nance AM, Evdokimov A, Moshiri F, Turner KH, et al. : Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature 2016, 533:58–63.** This study demonstrates the ability of PACE to easily achieve 14+ mutations in a single gene.
- 66.Smith SN, Wang Y, Baylon JL, Singh NK, Baker BM, Tajkhorshid E, Kranz DM: Changing the peptide specificity of a human T-cell receptor by directed evolution. Nat Commun 2014, 5:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fabret C, Poncet S, Danielsen S, Borchert TV., Ehrlich SD, Janniere L: Efficient gene targeted random mutagenesis in genetically stable Escherichia coli strains. Nucleic Acids Res 2000, 28:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang CL, Harper RA, Wabl M: Genome-wide somatic hypermutation. Proc Natl Acad Sci 2004, 101:7352–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romanini DW, Peralta-Yahya P, Mondol V, Cornish VW: A heritable recombination system for synthetic darwinian evolution in yeast. ACS Synth Biol 2012, 1:602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finney-Manchester SP, Maheshri N: Harnessing mutagenic homologous recombination for targeted mutagenesis in vivo by TaGTEAM. Nucleic Acids Res 2013, 41:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Crook N, Abatemarco J, Sun J, Wagner JM, Schmitz A, Alper HS: In vivo continuous evolution of genes and pathways in yeast. Nat Commun 2016, 7:13051–13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hess GT, Frésard L, Han K, Lee CH, Li A, Cimprich KA, Montgomery SB, Bassik MC: Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 2016, 13:1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X: Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 2016, 13:1029–1035. [DOI] [PubMed] [Google Scholar]
- 74.Burke MK, Dunham JP, Shahrestani P, Thornton KR, Rose MR, Long AD: Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 2010, 467:587–590. [DOI] [PubMed] [Google Scholar]
- 75.Imhof M, Schlotterer C: Fitness effects of advantageous mutations in evolving Escherichia coli populations. Proc Natl Acad Sci 2001, 98:1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, Desai MM: Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 2013, 500:571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF: Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 2009, 461:1243–1247. [DOI] [PubMed] [Google Scholar]
- 78.Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ: Different trajectories of parallel evolution during viral adaptation. Science 1999, 285:422–424. [DOI] [PubMed] [Google Scholar]
- 79.Bryson DI, Fan C, Guo LT, Miller C, Söll D, Liu DR: Continuous directed evolution of aminoacyl-tRNA synthetases. Nat Chem Biol 2017, 13:1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leconte AM, Dickinson BC, Yang DD, Chen I a, Allen B, Liu DR: A Population-Based Experimental Model for Protein Evolution: Effects of Mutation Rate and Selection Stringency on Evolutionary Outcomes. Biochemistry 2013, 52:1490–1499.** Phage assisted continuous evolution allowed the authors to demonstrate, across replicate evolution experiments, the deterministic and stochastic nature of evolution of the T7 RNAP for recognition of the T3 promoter.
- 81.Elena SF, Wilke CO, Ofria C, Lenski RE: Effects of population size and mutation rate on the evolution of mutational robustness. Evolution 2007, 61:666–674. [DOI] [PubMed] [Google Scholar]
- 82.Rouzine IM, Rodrigo A, Coffin JM: Transition between Stochastic Evolution and Deterministic Evolution in the Presence of Selection: General Theory and Application to Virology. Microbiol Mol Biol Rev 2001, 65:151–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vahdati AR, Sprouffske K, Wagner A: Effect of Population Size and Mutation Rate on the Evolution of RNA Sequences on an Adaptive Landscape Determined by RNA Folding. Int J Biol Sci 2017, 13:1138–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szendro IG, Franke J, de Visser JAGM, Krug J: Predictability of evolution depends nonmonotonically on population size. Proc Natl Acad Sci 2013, 110:571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang X, Mu B, Huang Z, Zhang M, Wang X, Tao S: Impacts of mutation effects and population size on mutation rate in asexual populations: A simulation study. BMC Evol Biol 2010, 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Daugherty PS, Chen G, Iverson BL, Georgiou G: Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. Proc Natl Acad Sci 2000, 97:2029–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Dijk T, Hwang S, Krug J, De Visser AGM, Zwart MP: Mutant library size and the repeatability of selection for antibiotic resistance. Phys Biol 2017, 14:1–29. [DOI] [PubMed] [Google Scholar]
- 88.Dickinson BC, Leconte AM, Allen B, Esvelt KM, Liu DR: Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc Natl Acad Sci 2013, 110:9007–9012.** This study demonstrated the ability of continuous evolution to simultaneously and effortlessly probe multiple paths and conditions of evolution.
- 89.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE: Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci 2006, 103:9107–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuriyan J, Krishna TSR, Wong L, Guenther B, Pahler A, Williams CH, Model P: Convergent evolution of similar function in two structurally divergent enzymes. Nature 1991, 352:172–174. [DOI] [PubMed] [Google Scholar]
- 91.Graumann P, Marahiel MA: A case of convergent evolution of nucleic acid binding modules. BioEssays 1996, 18:309–315. [DOI] [PubMed] [Google Scholar]
- 92.Ravikumar A, Arrieta A, Liu CC: An orthogonal DNA replication system in yeast. Nat Chem Biol 2014, 10:175–177. [DOI] [PubMed] [Google Scholar]
- 93.Achaz G, Rodriguez-Verdugo A, Gaut BS, Tenaillon O: The Reproducibility of Adaptation in the Light of Experimental Evolution with Whole Genome Sequencing In Ecological Genomics. Advances in Experimental Medicine and Biology, vol 781 Edited by Landry CR, Aubin-Horth N. Springer, Dordrecht; 2014:211–231. [DOI] [PubMed] [Google Scholar]
- 94.Lässig M, Mustonen V, Walczak AM: Predicting evolution. Nat Ecol Evol 2017, 1:1–9. [DOI] [PubMed] [Google Scholar]
- 95.Svensson E, Calsbeek R (Eds): The Adaptive Landscape in Evolutionary Biology. Oxford University Press; 2012. [Google Scholar]
- 96.Obolski U, Ram Y, Hadany L: Key issues review: Evolution on rugged adaptive landscapes. Reports Prog Phys 2018, 81:1–31. [DOI] [PubMed] [Google Scholar]
- 97.Wright S: The Roles of Mutation, Inbreeding, Crossbreeding and Selection in Evolution. Proc 6th Int Congr Genet 1932, 1:356–366. [Google Scholar]
- 98.Starr TN, Thornton JW: Epistasis in protein evolution. Protein Sci 2016, 25:1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ: Empirical fitness landscapes reveal accessible evolutionary paths. Nature 2007, 445:383–6. [DOI] [PubMed] [Google Scholar]
- 100.Storz JF: Compensatory mutations and epistasis for protein function. Curr Opin Struct Biol 2018, 50:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pal C, Papp B, Lercher MJ: An integrated view on protein evolution. Nat Rev Genet 2006, 7:337–348. [DOI] [PubMed] [Google Scholar]
- 102.Tóth-Petróczy Á, Tawfik DS: The robustness and innovability of protein folds. Curr Opin Struct Biol 2014, 26:131–138. [DOI] [PubMed] [Google Scholar]
- 103.Glasner ME, Gerlt JA, Babbitt PC: Evolution of enzyme superfamilies. Curr Opin Chem Biol 2006, 10:492–497. [DOI] [PubMed] [Google Scholar]
- 104.Storz JF: Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet 2016, 17:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Juan D, Pazos F, Valencia A: Emerging methods in protein co-evolution. Nat Rev Genet 2013, 14:249–261. [DOI] [PubMed] [Google Scholar]
- 106.Szurmant H, Weigt M: Inter-residue, inter-protein and inter-family coevolution: bridging the scales. Curr Opin Struct Biol 2018, 50:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melbinger A, Vergassola M: The Impact of Environmental Fluctuations on Evolutionary Fitness Functions. Sci Rep 2015, 5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Poelwijk FJ, De Vos MGJ, Tans SJ: Tradeoffs and optimality in the evolution of gene regulation. Cell 2011, 146:462–470. [DOI] [PubMed] [Google Scholar]