Abstract
Paclitaxel is a common chemotherapy drug associated with the development of chronic paclitaxel-induced peripheral neuropathy (PIPN). PIPN is associated with neuroinflammatory mechanisms in pre-clinical studies. Here, we evaluated for differential gene expression (DGE) in peripheral blood between breast cancer survivors with and without PIPN and for neuroinflammatory (NI) related signaling pathways and whole-transcriptome profiles from other experiments. Pathway impact analysis identified 8 perturbed NI related pathways. Expression profile analysis found 15 experiments having similar whole-transcriptome profiles of DGE related to neuroinflammation and PIPN. These findings suggest that perturbations in pathways associated with neuroinflammation are found in cancer survivors with PIPN.
Keywords: Taxanes, Neuroinflammation, Cytokines, Chemokines, Neuropathy, Breast cancer, Survivor, Paclitaxel, Differential gene expression, Pathway analysis
Graphical abstract
Highlights
-
•
Paclitaxel-induced peripheral neuropathy (PIPN) is associated with Paclitaxel treatment
-
•
Differential gene expression was associated with PIPN in breast cancer survivors.
-
•
Perturbations of neuroinflammatory-related pathways were identified between survivors.
-
•
Transcriptome profile was similar to other pre-clinical and clinical studies.
1. Introduction
Paclitaxel is an extremely effective chemotherapeutic agent for the treatment of breast, ovarian, and lung cancer (Kudlowitz and Muggia, 2013). However, paclitaxel-induced peripheral neuropathy (PIPN), a major dose-limiting toxicity, occurs in 59% to 87% of patients who receive this drug (Jones et al., 2005; Sarosy et al., 1992). Paclitaxel is an anti-tubulin drug that causes microtubule stabilization. In the peripheral nervous system, administration of paclitaxel results in distal axonal degeneration, secondary demyelination, and nerve fiber loss.(Gornstein and Schwarz, 2014; Sahenk et al., 1994).
A growing body of evidence has implicated neuroinflammation in the development of PIPN (Makker et al., 2017; Wang et al., 2012). While most chemotherapy (CTX) drugs do not cross the blood-brain-barrier, they readily penetrate the blood-nerve-barrier and bind to and accumulate in dorsal root ganglia (DRG) and peripheral axons.(Gornstein and Schwarz, 2014; Park et al., 2013) In addition to its direct neurotoxic effects, CTX can induce neuroinflammation through activation of immune and immune-like glial cells. In fact, a growing body of preclinical evidence suggests that immune cells (e.g., macrophages, lymphocytes) and glial cells (e.g., Schwann cells) in the peripheral nervous system (PNS) and astrocytes and microglia in the central nervous system (CNS) play an important role in the induction and maintenance of neuropathic pain.(Krames, 2014; Zhang et al., 2017) This activation of the immune system results in the release of inflammatory mediators, within the DRG and dorsal horn, (Krames, 2014) that enhances neuronal excitability and results in pain hypersensitivity in peripheral neurons (Makker et al., 2017).
Not all patients develop neurotoxicity, which suggests that molecular factors may play a role in the development of PIPN. In fact, in our recent report, using RNA-seq data from breast cancer patients who did (n = 25) and did not (n = 25) develop PIPN,(Kober et al., 2018b) we identified nine perturbed pathways that were associated with various aspects of mitochondrial dysfunction including oxidative stress, iron homeostasis, mitochondrial fission, apoptosis, and autophagy. In this paper, we extend these findings and describe differentially perturbed pathways, as well as whole transcriptome profiles of differential gene expression (DGE), associated with neuroinflammation, in the same sample of breast cancer survivors with (n = 25) and without (N = 25) chronic PIPN.
2. Materials and methods
2.1. Study design
In this study, we used two different approaches to evaluate whole transcriptome data for patterns of DGE in biological pathways associated with neuroinflammation and PIPN (Fig. 1 ). The first approach utilized pathway impact analysis (PIA), where well-defined signaling pathways are evaluated for perturbations using the significance and magnitude of gene-gene interactions from DGE tests. For the second approach (i.e., Expression Profile Analysis (EPA)), we compared our whole transcriptome pattern of DGE between breast cancer survivors with and without CIPN to a database of publicly available and well annotated gene expression experiments to identify other systems and studies with similar patterns. For this analysis, we used demographic, clinical, and gene expression data from our previous study (Kober et al., 2018b).
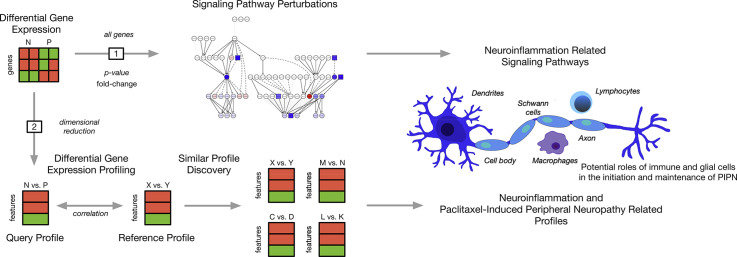
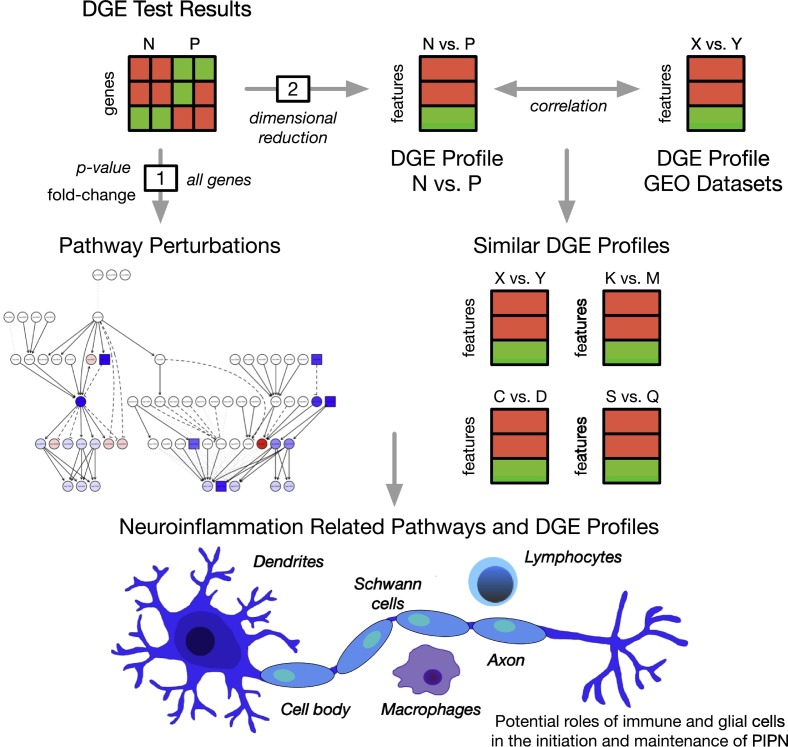
Fig. 1.
An overview of the analytic approach used to evaluate for neuroinflammation related pathways and gene expression experiments associated with paclitaxel-induced peripheral neuropathy (PIPN). Differential gene expression (DGE) in peripheral blood was found between breast cancer survivors with (P) and without (N) paclitaxel-induced peripheral neuropathy and evaluated for (1) perturbed neuroinflammation related signaling pathways using pathway impact analysis and (2) experiments from the gene expression omnibus (GEO) with similar whole-transcriptome profiles of differential gene expression related to neuroinflammation and PIPN using expression profile analysis. Taken together, our results suggest that perturbations in pro- and anti-inflammatory pathways associated with neuroinflammation are found in cancer survivors with PIPN.
2.2. Acquisition and processing of gene expression data
The methods for the gene expression analysis are described in detail elsewhere.(Kober et al., 2018b) Gene expression of total RNA isolated from peripheral blood was assayed using RNA-seq. Gene expression was summarized as counts per gene and used as input for the PIA and EPA analyses.
2.3. Pathway impact analysis
Differential gene expression was quantified using general linear models (Kober et al., 2018b). These DGE analyses were adjusted for demographic (i.e., age, employment status) and clinical (i.e., Alcohol Use Disorders Identification Test (AUDIT)(Bohn et al., 1995) score, body mass index (BMI), Karnofsky Performance Score (KPS)(Karnofsky, 1977; Karnofsky et al., 1948; Schnadig et al., 2008) score) characteristics that differed between the PIPN groups, as well as for technical variability (e.g., potential batch effects). The DGE results were summarized as the log fold change and p-value for each gene. Then, PIA was used to evaluate for perturbations in well-defined signaling pathways. PIA includes potentially important biological factors (e.g., gene-gene interactions, flow signals in a pathway, pathway topologies), the magnitude (i.e., log fold-change), and p-values from the differential expression (DE) analysis (reviewed in (Mitrea et al., 2013)). The PIA included the results of the DE analysis for all genes (i.e., cutoff free) to determine probability of pathway perturbations (pPERT) using Pathway Express.(Draghici et al., 2007).
A total of 208 signaling pathways were defined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.(Aoki-Kinoshita and Kanehisa, 2007) Sequence loci data were annotated with Entrez gene IDs. The gene symbols were annotated using the HUGO Gene Nomenclature Committee resource database.(Gray et al., 2013) We assessed for significance of the PIA using a strict false discovery rate (FDR) of <1 under the Benjamini-Hochberg (BH) procedure.(Benjamini and Hochberg, 1995; Hochberg and Benjamini, 1990) Finally, we evaluated these results specifically for pathways related to neuroinflammation.
2.4. Expression profile analysis
To evaluate the whole transcriptome pattern of DGE in relationship to neuroinflammation and PIPN, we compared these findings with independent and well annotated publicly available data sets of gene expression. We utilized ProfileChaser(Engreitz et al., 2010) to develop a transcriptome wide ‘profile’ of the DGE pattern between the PIPN and no PIPN groups and compared this profile to all of the NCBI Gene Expression Omnibus (GEO) curated GEO DataSets (GDS) available. These GDS include gene expression data from a wide variety of tissues, species, and study designs. Each study is annotated with a number of characteristics termed as ‘factors’ (e.g., sample ID, age, cell line, treatment group).
ProfileChaser generates reference profiles from all of the pairwise comparisons of all of the factors defined in a GDS. Then, it compares the query dataset to identify similar reference profiles. In this way, ProfileChaser identifies experiments with similar biological conditions to the query experiment. By performing this evaluation of all available GDS and their annotated factors, we limit any bias from a priori filtering. Gene level summaries of RNA abundance were annotated with ENTREZ ID and estimated as the log transformed reads per mean thousand (i.e., log2(RPMK+1)). We assessed for significance of the EPA using a strict FDR of 5% under the BH procedure.(Benjamini and Hochberg, 1995; Hochberg and Benjamini, 1990) Using our previous method,(Kober et al., 2016) we evaluated the significant findings for interpretability based on the experimental design of the reference profile.
3. Results
3.1. Differences in demographic, clinical, pain, sensation, and balance characteristics
As previously reported (Kober et al., 2018b), survivors with PIPN were significantly older (p = .006) and were more likely to be unemployed (p = .022) In terms of clinical characteristics, survivors with PIPN had: a lower AUDIT score (p = .012), a higher body mass index (BMI; p = .017), and a lower KPS score (p < .001). Of note, no between group differences were found in the total cumulative dose of paclitaxel received or in the percentage of patients who had a dose reduction or delay due to PIPN (see Supplementary Table 1, Supplementary Table 2).
Supplementary Table 3 summarizes the self-reported pain characteristics of the survivors with PIPN. Worst pain severity was reported as 6.3 (±2.1) out of ten and the duration of PIPN was 3.8 (±3.9) years.
Supplementary Table 3 summarizes the self-reported pain characteristics of the survivors with PIPN. Worst pain severity was reported as 6.3 (±2.1) out of ten and the duration of PIPN was 3.8 (±3.9) years.
Survivors with PIPN had a higher number of lower extremity sites with loss of light touch, cold, and pain sensations (all, p < .05). Vibration thresholds were significantly higher in the PIPN group (p = .009, Supplementary Table 4).
Survivors with PIPN were more likely to report trouble with balance (p < .001) as well as higher severity and distress (both p < .05) scores associated with balance problems. In addition, these survivors reported worse Timed Get Up and Go (p = .001) and worse Fullerton Advanced Balance (p = .004) scores (Supplementary Table 4).
3.2. PIA of the whole transcriptome
Successful annotation with ENTREZ IDs was performed for 11,174 unique genes. Fold changes and p-values from the DE analysis for these genes were included in the pathway perturbation analysis of the 208 KEGG signaling pathways. PIA identified 53 KEGG signaling pathways that were significantly perturbed between the PIPN groups after correction for multiple hypothesis testing at a conservative FDR of 1% (adjusted perturbation p-value <.01). Of these, 8 KEGG signaling pathways were related to neuroinflammation (Table 1 ).
Table 1.
Significantly perturbed neuroinflammatory related KEGG pathways between breast cancer survivors with and without paclitaxel-induced peripheral neuropathy.
| Pathway ID | Pathway name | Total perturbation | Adjusted pPerta |
|---|---|---|---|
| hsa04060 | Cytokine-cytokine receptor interaction | 6.37 | 0.004 |
| hsa04064 | NF-kappa B signaling pathway | 4.50 | 0.005 |
| hsa04727 | GABAergic synapse | 4.63 | 0.007 |
| hsa04920 | Adipocytokine signaling pathway | 9.13 | 0.008 |
| hsa04657 | IL-17 signaling pathway | 3.15 | 0.010 |
| hsa04621 | NOD-like receptor signaling pathway | 11.32 | 0.004 |
| hsa04152 | AMPK signaling pathway | 15.14 | 0.004 |
| hsa04350 | TGF-beta signaling pathway | 10.30 | 0.010 |
Abbreviations: AMPK = adenosine monophosphate-activated protein kinase, GABA = gamma amino butyric acid, IL = interleukin, KEGG = Kyoto Encyclopedia of Genes and Genomes, NF = nuclear factor, NOD = nucleotide-binding and oligomerization domain, TGF = transforming growth factor.
pPert: Perturbation p-value adjusted using the Benjamini-Hochberg method.
3.3. EPA of the whole transcriptome
Whole transcriptome profiles were evaluated for similarity in DGE between our sample of survivors with and without PIPN and the 280 profiles across 84 unique GDS that were identified using EPA (Supplementary File 1). After an evaluation of the study designs of these 280 profiles, 27 profiles from 15 unique GDS and 13 published studies were identified to be associated with neuroinflammation and PIPN. These findings were organized into six categories based on their similar biological characteristics (Table 2 ).
Table 2.
Whole-transcriptome gene expression GEO datasets with similar profiles to breast cancer survivors with CIPN vs. without CIPN.
| GEO ID | Referencea | Organism | Primary comparison | Primary factor | Score | q-value | Tissue |
|---|---|---|---|---|---|---|---|
| Pre-clinical models of neuropathic pain | |||||||
| GDS2159 | M.m. | Sham SCI vs. naïve | Protocol | 0.428 | 0.025 | SC | |
| GDS2159 | M.m. | 28d vs 0 h post SCI | Time | 0.383 | 0.037 | SC | |
| GDS2159 | M.m. | 7d vs 0 h post SCI | Time | 0.379 | 0.037 | SC | |
| GDS2159 | M.m. | Moderate SCI vs. naïve | Protocol | 0.374 | 0.037 | SC | |
| GDS2439 | R.n. | 28d vs 50d after L5 SNL | Time | 0.352 | 0.046 | DRG | |
| GDS259 | 12,666,113 | R.n. | 3d vs 2d after SCI at T9 | Time | 0.352 | 0.056 | SC |
| GDS339 | 12,666,113 | R.n. | 3d vs 2d after SCI at T9 | Time | 0.348 | 0.047 | SC |
| Response to infection | |||||||
| GDS1028 | 15,655,079 | H.s. | SARS vs control | Disease state | 0.399 | 0.031 | PBMC |
| GDS1499 | 15,897,992 | H.s. | Control vs. HIV-1, drug regimen not indicated | Protocol | 0.401 | 0.031 | PBMC |
| GDS1499 | 15,897,992 | H.s. | HIV-1 seronegative vs HIV-1 seropositive | Disease state | 0.400 | 0.033 | PBMC |
| GDS1499 | 15,897,992 | H.s. | Control vs. HIV-1, drug naïve | Protocol | 0.353 | 0.045 | PBMC |
| GDS1971 | 30,638,864 | H.s. | Complicated malaria vs healthy | Disease state | 0.569 | 0.007 | PWB |
| GDS1971 | 30,638,864 | H.s. | Uncomplicated malaria vs healthy | Disease state | 0.453 | 0.020 | PWB |
| GDS2362 | 16,988,231 | H.s. | Presymptomatic vs experimentally acquired malaria | Uninfected | 0.469 | 0.017 | PBMC |
| Neurological condition | |||||||
| GDS1311 | 16,043,692 | H. s. | HD symptomatic vs normal | Disease state | 0.463 | 0.018 | PWB |
| GDS1311 | 16,043,692 | H. s. | HD presymptomatic vs. normal | Disease state | 0.464 | 0.025 | PWB |
| Exercise-induced effects | |||||||
| GDS2310 | 16,990,507 | H.s. | After exhaustive vs. before moderate exercise | Time | 0.680 | 0.003 | WBC |
| GDS2310 | 16,990,507 | H.s. | After exhaustive vs. before exhaustive exercise | Time | 0.654 | 0.003 | WBC |
| GDS2310 | 16,990,507 | H.s. | After exhaustive vs. after moderate exercise | Time | 0.557 | 0.008 | WBC |
| GDS2310 | 16,990,507 | H.s. | After moderate vs. before moderate exercise | Time | 0.517 | 0.011 | WBC |
| GDS2417 | H.s. | Post-exercise vs. pre-exercise | Protocol | 0.383 | 0.036 | WBC | |
| GDS962 | 15,194,674 | H.s. | 60 min after vs before exercise | Time | 0.501 | 0.012 | PBMC |
| Inflammatory bowel disease | |||||||
| GDS1615 | 16,436,634 | H.s. | Crohn's disease vs normal | Disease state | 0.487 | 0.015 | PBMC |
| GDS1615 | 16,436,634 | H.s. | Ulcerative colitis vs. normal | Disease state | 0.419 | 0.027 | PBMC |
| Hematopoiesis | |||||||
| GDS2321 | 18,268,278 | H.s. | G-CSF vs pegylated G-CSF | Agent | 0.412 | 0.028 | CD34(+) cells |
| GDS2959 | H.s. | G-CSF vs untreated | Agent | 0.536 | 0.010 | PWB | |
| GDS781 | H.s. | G-CSF treated vs untreated | Agent | 0.429 | 0.025 | PBMC | |
DRG = dorsal root ganglia, G-CSF = granulocyte-colony stimulating factor, GDS = Geo dataset, GEO = gene expression omnibus, H.s. = Homo sapiens, HD = Huntington's disease, HIV = Human Immunodeficiency Virus, M.m. = Mus musculus, PBMC = peripheral blood mononuclear cells, PWB = peripheral whole blood, R.n. = Rattus norvegicus, SARS = severe acute respiratory syndrome, SC = spinal cord, SCI = spinal cord injury, SNL = spinal nerve ligation, WBC = white blood cells.
PubMed ID (if known).
4. Discussion
4.1. Perturbed neuroinflammation-related pathways associated with PIPN
This study is the first to provide molecular evidence that a number of neuroinflammatory mechanisms identified in preclinical models of neuropathic pain and/or PIPN(Flatters et al., 2017; Ma et al., 2018) are perturbed in cancer survivors with PIPN. As noted in a review on the role of cytokines in chemotherapy-induced peripheral neuropathy (CIPN),(Wang et al., 2012) pro-inflammatory cytokines and chemokines play a critical role in the development and maintenance of painful peripheral neuropathy. Consistent with our findings of a differential perturbation in the cytokine-cytokine receptor interaction pathway, several lines of pre-clinical evidence suggest that the administration of paclitaxel is associated with the activation of immune and immune-like cells in both the PNS and CNS. For example, following the administration of intravenous paclitaxel in rats, the number of activated macrophages in the DRG, peripheral nerves, and Schwann cells increased and was associated with allodynia and hyperalgesia.(Peters et al., 2007) Macrophage infiltration results in the production and release of a number of cytokines (e.g., tumor necrosis factor alpha (TNF-α), interleukin 6 (IL6)) and chemokines. In addition, Schwann cells release TNF-α, IL6, prostaglandin E2 (PGE2), and adenosine triphosphate (ATP).(Ozturk et al., 2005) All of these molecules contribute to neuroinflammation.(Sommer and Kress, 2004; Tofaris et al., 2002).
Additional evidence from preclinical models of peripheral neuropathy supports our finding of perturbations in the nuclear factor (NF)-κβ signaling pathway. For example, building on the observation that NF-κβ immunoreactive neurons are increased on the ipsilateral side of a partial sciatic nerve injury,(Ma and Bisby, 1998) the effects of NF-κβ in PIPN were evaluated in a rodent model.(Kamei et al., 2017) In this study, paclitaxel increased the protein levels of spinal phosphorylated NF-κβ and an NF-κβ inhibitor attenuated mechanical hyperalgesia. The authors concluded that the activation of NF-κβ mediates paclitaxel-induced hyperalgesia. In another preclinical study,(Gui et al., 2018) the intraperitoneal administration of paclitaxel induced the activation of NF-κβ; the upregulation of TNF-α, IL1β, and IL6; and astrocyte activation in the spinal cord.
Another perturbed pathway in our study was the IL17 signaling pathway. The IL17 family is a subset of cytokines that play crucial roles in both acute and chronic inflammatory responses.(Isailovic et al., 2015) IL17 is produced by CD4+ T cells and appears to play a role in inflammatory(Lubberts, 2008) and neuropathic(Day et al., 2014; Kim and Moalem-Taylor, 2011) pain. In the most recent preclinical study,(Sun et al., 2017) expression of IL17 in the spinal cord was evaluated following spinal nerve ligation. During the maintenance phase of neuropathic pain, mRNA expression levels of IL17, IL1β, and IL6 were significantly increased in the dorsal horn of the spinal cord of animals with the nerve injury. The authors concluded that IL17 may contribute to neuropathic pain by promoting the proliferation of astrocytes and the secretion of pro-inflammatory cytokines. While no studies were found on the role of IL17 in CIPN, astrocytes appear to play a role in the development of this chronic pain condition.(Robinson and Dougherty, 2015; Robinson et al., 2014).
In terms of perturbations in the adipocytokine signaling pathway, leptin is an important regulator of energy intake and metabolic rate. In addition, leptin influences the normal functioning of the immune system through the stimulation of cytokine production and chemotaxis. In addition, leptin expression can be regulated by cytokines.(Carniglia et al., 2017) While no studies were found on associations between leptin and CIPN, in a rat model of chronic constriction injury, administration of a leptin antagonist prevented the development of injury-induced mechanical allodynia and thermal hyperalgesia.(Lim et al., 2009) In a mouse model of partial sciatic nerve ligation,(Maeda et al., 2009) leptin production was induced in the adipocytes that were present in the epineurium of the injured nerve and leptin was necessary for tactile allodynia associated with the ligation. In addition, leptin enhanced the production of cyclo‑oxygenase (COX)-2, inducible nitric oxide synthase (iNOS), and matrix metalloprotease-9 from macrophages that suggests that adipokines activate macrophages that contribute to the development of neuropathic pain. However, in another preclinical study, leptin exhibited protective effects in a model of sciatic nerve injury.(Fernandez-Martos et al., 2012).
The various members of the nucleotide-binding and oligomerization domain (NOD)-like receptor signaling pathway play pivotal roles in the recognition of intracellular ligands. Of particular interest is the growing body of evidence that suggests that one member of this pathway (i.e., NOD-like receptor protein 3 (NLRP3)) is involved in various types of neuropathic pain.(Khan et al., 2018; Li et al., 2018; Pan et al., 2018; Pu et al., 2018; Tonkin et al., 2018; Xu et al., 2019) Of note, in a recent preclinical study of PIPN in rats,(Jia et al., 2017) paclitaxel increased the expression of and activated fragments of caspase-1 and IL1β which suggests activation of the NLRP3 inflammasome. The expression of NLRP3 was located in CD68 macrophages that infiltrated the L4–5 DRG and sciatic nerve. The authors concluded that their findings suggest that the administration of paclitaxel induced mitochondrial damage and reactive oxygen species production that resulted in the activation of the NLRP3 inflammasome in peripheral nerves and contributes to PIPN (Jia et al., 2017).
While no preclinical studies were identified that implicate the transforming growth factor (TGF)-β signaling pathway in the development of PIPN, this pathway includes a number of structurally related cytokines that have a wide spectrum of cellular functions. Within the nervous system, TGF-β1 controls the proliferation of neurons and regulates neuronal survival.(Krieglstein et al., 1998) In astroglia, TGF-β1 exerts anti-inflammatory and immunosuppressive effects.(Bottner et al., 2000) In preclinical studies, TGF-β1 prevented the development of neuropathic pain associated with traumatic nerve injury and reversed previously established neuropathic pain.(Chen et al., 2016; Chen et al., 2013; Echeverry et al., 2009).
Gamma amino butyric acid (GABA) is the most abundant inhibitory neurotransmitter within the CNS. In various models of neuropathic pain, decreases in GABA receptor-mediated inhibitory synaptic transmission in dorsal horn neurons occurs following nerve injury.(Gwak and Hulsebosch, 2011; Yin et al., 2018) In addition, in a mouse model of PIPN,(Braz et al., 2015) the intraspinal transplantation of cortical precursors of GABAergic interneurons from the embryonic medial ganglionic eminence reversed both the mechanical allodynia and heat hyperalgesia associated with the administration of paclitaxel.
As noted in a recent review,(Asiedu et al., 2016) the adenosine monophosphate-activated protein kinase (AMPK) signaling pathway is a novel target for the alleviation of neuropathic pain. AMPK activators inhibit signaling pathways that are known to promote changes peripheral nociceptors that result in chronic pain. In the only study that evaluated the role of AMPK in CIPN,(Mao-Ying et al., 2014) the co-administration of platinum with metformin, a drug that activates AMPK, prevented the increased latency associated with detection of an adhesive patch, as well as the decrease in the density of intra-dermal nerve fibers following the administration of CTX in a mouse model.
4.2. Whole-transcriptome profiles of DGE
Using EPA, we identified similar profiles in experimental systems of inflammatory responses, exercise-induced effects, and pre-clinical models of pain (see Table 2). In terms of inflammatory responses, similar profiles of DGE were found between our survivors with and without PIPN and patients with SARS,(Reghunathan et al., 2005) HIV-1,(Ockenhouse et al., 2005) and malaria(Boldt et al., 2019; Ockenhouse et al., 2006) who were compared to healthy or pre-symptomatic individuals. As noted above, neuroinflammation is thought to play a major role in the development and maintenance of PIPN (Makker, Duffy, 2017, Wang et al., 2012). The similarities between our chronic PIPN profile and these pathogen-mediated inflammatory profiles suggests that systemic inflammatory processes may play a role in the development and maintenance of PIPN.
In terms of the findings related to exercise-mediated effects, limited evidence exists that exercise benefits some patients with various types of neuropathic pain,(Dobson et al., 2014) including CIPN.(Kleckner et al., 2018) In our study, compared to survivors without PIPN (87.5%), only 60.0% of the survivors with PIPN reported that they exercised on a regular basis (p = .051). In the EPA, the DGE profiles in the blood from healthy volunteers that was taken before and after increasing levels of exercise (Buttner et al., 2007; Connolly et al., 2004) were similar to the profile in our survivors with and without PIPN. Additional research is warranted on this association because in our two previous analyses of CIPN(Miaskowski et al., 2017) and PIPN(Kober et al., 2018a) in larger samples of survivors who were heterogenous in terms of their cancer diagnoses and/or CTX regimens, no associations were found between the use of regular exercise and the occurrence of neuropathic pain.
Of note, seven profiles from two pre-clinical models of neuropathic pain (i.e., spinal cord injury (SCI)(Shiao and Lee-Kubli, 2018) and L5 spinal nerve ligation (SNL)(Chung et al., 2004; Kim and Chung, 1992)) share a similar whole transcriptome profile of DGE with our cohort. It is important to note that while our survivors' DGE profile was derived using blood samples, the preclinical findings came from a different species (i.e., rodents) and different tissues (i.e., spinal cord, DRG). Gene expression differences across tissues are mainly driven by a small number of genes,(Mele et al., 2015) suggesting that gene expression changes in one tissue may provide insights into gene expression patterns in another tissue. Changes in gene expression in whole blood have been used as a marker for neuronal injury.(Tang et al., 2003) In addition, gene expression patterns in blood, hippocampus, and pre-frontal cortex show broad levels of co-expression.(Witt et al., 2013) Given the recent interest in evaluating for concordance and discordance in the expression of genes that represent known biomarkers and therapies across tissues(Kosti et al., 2016), the controversy about whether changes in GE in peripheral blood reflects changes within the PNS and CNS (Colleoni and Sacerdote, 2010; Seok et al., 2013; Jaggi et al., 2011), and our findings of similarities between human whole blood and pre-clinical models of neuropathic pain in neuronal tissue (i.e., rat DRG (GDS2439) and mouse SC (GDS2159)), future research should evaluate for genes that are similar and unique to these tissue types in relation to PIPN. Another point worth noting is that in some of the preclinical studies that were identified using EPA, differences in GE were evaluated days to months after the nerve injury. This evaluation time frame is consistent with a more chronic pain phenotype and perhaps is more relevant to our cohort of survivors who had PIPN for 3.8 (±3.9) years. The EPA analysis did not evaluate for any specific preclinical or human studies of CIPN or PIPN.
4.3. Conclusions
Taken together, these findings suggest that in addition to perturbed pathways associated with mitochondrial dysfunction (Kober et al., 2018b), perturbations in pro- and anti-inflammatory pathways associated with neuroinflammation are found in cancer survivors with PIPN. Furthermore, whole transcriptome profiles of DGE patterns in preclinical and clinical studies provide additional support for our findings.
4.4. Limitations
Several limitations need to be considered. While our sample size was relatively small, we had an extremely well characterized sample of breast cancer survivors with and without PIPN. Of note, no differences were found in the total cumulative dose of paclitaxel that these survivors received. Although not significantly different between groups in this study, exercise regularity may contribute to differences in differential gene expression between breast cancer survivors with and without PIPN. Given that an evaluation of differences in gene expression from DRG neurons cannot be done in living individuals, like others,(Langjahr et al., 2018; Uceyler et al., 2007) we evaluated for differences in RNA expression using peripheral blood. Some limitations of using whole blood to evaluate for patterns of expression in other tissues include: blood shows the largest number of uniquely expressed genes (Mele et al., 2015); blood has the smallest number of expressed transcripts across tissues;(Consortium, 2015) and baseline blood does not share similar whole-transcriptome pattern with DRG, as well as other CNS or PNS tissue.(Ray et al., 2018).
4.5. Future research
This study is the first to provide molecular evidence that neuroinflammatory mechanisms identified in preclinical models of various types of neuropathic pain including CIPN (Makker, Duffy, 2017; Wang et al., 2012), are found in peripheral blood leukocytes of cancer survivors with persistent PIPN. Despite the limitations of the analyses of RNA from blood, we did observe profiles of DGE and perturbations in these processes, which suggest persistent damage and/or changes in the PNS. Given that these survivors had PIPN for approximately four years, our findings suggest that chronic neuroinflammation is involved in the maintenance of this neuropathic pain condition. Future studies need to evaluate for differences in epigenetic changes (i.e., methylation, microRNA) between survivors with and without PIPN, which may reflect changes in regulation patterns.
The following are the supplementary data related to this article.
– Differences in Demographic Characteristics Between Breast Cancer Survivors With and Without Paclitaxel-Induced Neuropathy.
– Differences in Clinical Characteristics Between Breast Cancer Survivors With and Without Paclitaxel-Induced Peripheral Neuropathy.
– Pain Characteristics of the Breast Cancer Survivors With Paclitaxel-Induced Peripheral Neuropathy.
– Differences in Sensation and Balance Measures Between Breast Cancer Survivors With and Without Paclitaxel-Induced Peripheral Neuropathy.
– Results of the ProfileChaser analysis.
Declaration of Competing Interest
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgements
This study was funded by the National Cancer Institute (NCI, CA151692) and the American Cancer Society (ACS, IRG-97-150-13). Dr. Miaskowski is supported by grants from the ACS and NCI (CA168960). This project was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 TR000004. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Recruitment was facilitated by Dr. Susan Love Research Foundation's Army of Women® Program. Anatol Sucher managed the storage and processing of the biospecimens. Kheo P. Kober contributed to the artwork used in the manuscript.
References
- Aoki-Kinoshita K.F., Kanehisa M. Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. (Clifton, NJ) 2007;396:71–91. doi: 10.1007/978-1-59745-515-2_6. [DOI] [PubMed] [Google Scholar]
- Asiedu M.N., Dussor G., Price T.J. Targeting AMPK for the alleviation of pathological pain. Exs. 2016;107:257–285. doi: 10.1007/978-3-319-43589-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Bohn M.J., Babor T.F., Kranzler H.R. The alcohol use disorders identification test (AUDIT): validation of a screening instrument for use in medical settings. J. Stud. Alcohol. 1995;56:423–432. doi: 10.15288/jsa.1995.56.423. [DOI] [PubMed] [Google Scholar]
- Boldt A.B.W., van Tong H., Grobusch M.P., Kalmbach Y., Dzeing Ella A., Kombila M. The blood transcriptome of childhood malaria. EBioMedicine. 2019;40:614–625. doi: 10.1016/j.ebiom.2018.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottner M., Krieglstein K., Unsicker K. The transforming growth factor-betas: structure, signaling, and roles in nervous system development and functions. J. Neurochem. 2000;75:2227–2240. doi: 10.1046/j.1471-4159.2000.0752227.x. [DOI] [PubMed] [Google Scholar]
- Braz J.M., Wang X., Guan Z., Rubenstein J.L., Basbaum A.I. Transplant-mediated enhancement of spinal cord GABAergic inhibition reverses paclitaxel-induced mechanical and heat hypersensitivity. Pain. 2015;156:1084–1091. doi: 10.1097/j.pain.0000000000000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner P., Mosig S., Lechtermann A., Funke H., Mooren F.C. Exercise affects the gene expression profiles of human white blood cells. J. Appl. Physiol. 2007;102:26–36. doi: 10.1152/japplphysiol.00066.2006. (1985) [DOI] [PubMed] [Google Scholar]
- Carniglia L., Ramirez D., Durand D., Saba J., Turati J., Caruso C. Neuropeptides and microglial activation in inflammation, pain, and neurodegenerative diseases. Mediat. Inflamm. 2017;2017:5048616. doi: 10.1155/2017/5048616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N.F., Huang S.Y., Chen W.F., Chen C.H., Lu C.H., Chen C.L. TGF-beta1 attenuates spinal neuroinflammation and the excitatory amino acid system in rats with neuropathic pain. J. Pain. 2013;14:1671–1685. doi: 10.1016/j.jpain.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Chen N.F., Chen W.F., Sung C.S., Lu C.H., Chen C.L., Hung H.C. Contributions of p38 and ERK to the antinociceptive effects of TGF-beta1 in chronic constriction injury-induced neuropathic rats. J. Headache Pain. 2016;17:72. doi: 10.1186/s10194-016-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.M., Kim H.K., Chung K. Segmental spinal nerve ligation model of neuropathic pain. Methods Mol Med. 2004;99:35–45. doi: 10.1385/1-59259-770-X:035. [DOI] [PubMed] [Google Scholar]
- Colleoni M., Sacerdote P. Murine models of human neuropathic pain. Biochim. Biophys. Acta. 2010;1802:924–933. doi: 10.1016/j.bbadis.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Connolly P.H., Caiozzo V.J., Zaldivar F., Nemet D., Larson J., Hung S.P. Effects of exercise on gene expression in human peripheral blood mononuclear cells. J. Appl. Physiol. 2004;97:1461–1469. doi: 10.1152/japplphysiol.00316.2004. (1985) [DOI] [PubMed] [Google Scholar]
- Consortium G.T. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day Y.J., Liou J.T., Lee C.M., Lin Y.C., Mao C.C., Chou A.H. Lack of interleukin-17 leads to a modulated micro-environment and amelioration of mechanical hypersensitivity after peripheral nerve injury in mice. Pain. 2014;155:1293–1302. doi: 10.1016/j.pain.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Dobson J.L., McMillan J., Li L. Benefits of exercise intervention in reducing neuropathic pain. Front. Cell. Neurosci. 2014;8:102. doi: 10.3389/fncel.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draghici S., Khatri P., Tarca A.L., Amin K., Done A., Voichita C. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverry S., Shi X.Q., Haw A., Liu H., Zhang Z.W., Zhang J. Transforming growth factor-beta1 impairs neuropathic pain through pleiotropic effects. Mol. Pain. 2009;5:16. doi: 10.1186/1744-8069-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J.M., Morgan A.A., Dudley J.T., Chen R., Thathoo R., Altman R.B. Content-based microarray search using differential expression profiles. BMC Bioinforma. 2010;11:603. doi: 10.1186/1471-2105-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martos C.M., Gonzalez P., Rodriguez F.J. Acute leptin treatment enhances functional recovery after spinal cord injury. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatters S.J.L., Dougherty P.M., Colvin L.A. Clinical and preclinical perspectives on chemotherapy-induced peripheral neuropathy (CIPN): a narrative review. Br. J. Anaesth. 2017;119:737–749. doi: 10.1093/bja/aex229. [DOI] [PubMed] [Google Scholar]
- Gornstein E., Schwarz T.L. The paradox of paclitaxel neurotoxicity: mechanisms and unanswered questions. Neuropharmacology. 2014;76(Pt A):175–183. doi: 10.1016/j.neuropharm.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Gray K.A., Daugherty L.C., Gordon S.M., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–D552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y., Zhang J., Chen L., Duan S., Tang J., Xu W. Icariin, a flavonoid with anti-cancer effects, alleviated paclitaxel-induced neuropathic pain in a SIRT1-dependent manner. Mol. Pain. 2018;14 doi: 10.1177/1744806918768970. (1744806918768970) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwak Y.S., Hulsebosch C.E. GABA and central neuropathic pain following spinal cord injury. Neuropharmacology. 2011;60:799–808. doi: 10.1016/j.neuropharm.2010.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Isailovic N., Daigo K., Mantovani A., Selmi C. Interleukin-17 and innate immunity in infections and chronic inflammation. J. Autoimmun. 2015;60:1–11. doi: 10.1016/j.jaut.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Jaggi A.S., Jain V., Singh N. Animal models of neuropathic pain. Fundam. Clin. Pharmacol. 2011;25:1–28. doi: 10.1111/j.1472-8206.2009.00801.x. [DOI] [PubMed] [Google Scholar]
- Jia M., Wu C., Gao F., Xiang H., Sun N., Peng P. Activation of NLRP3 inflammasome in peripheral nerve contributes to paclitaxel-induced neuropathic pain. Mol. Pain. 2017;13 doi: 10.1177/1744806917719804. (1744806917719804) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.E., Erban J., Overmoyer B., Budd G.T., Hutchins L., Lower E. Randomized phase III study of docetaxel compared with paclitaxel in metastatic breast cancer. J. Clin. Oncol. 2005;23:5542–5551. doi: 10.1200/JCO.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Kamei J., Hayashi S., Sakai A., Nakanishi Y., Kai M., Ikegami M. Rikkunshito prevents paclitaxel-induced peripheral neuropathy through the suppression of the nuclear factor kappa B (NFkappaB) phosphorylation in spinal cord of mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnofsky D. Plenum Press; New York: 1977. Performance Scale. [Google Scholar]
- Karnofsky D., Abelmann W.H., Craver L.V., Burchenal J.H. The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer. 1948;1:634–656. [Google Scholar]
- Khan N., Kuo A., Brockman D.A., Cooper M.A., Smith M.T. Pharmacological inhibition of the NLRP3 inflammasome as a potential target for multiple sclerosis induced central neuropathic pain. Inflammopharmacology. 2018;26:77–86. doi: 10.1007/s10787-017-0401-9. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Chung J.M. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain. 1992;50:355–363. doi: 10.1016/0304-3959(92)90041-9. [DOI] [PubMed] [Google Scholar]
- Kim C.F., Moalem-Taylor G. Interleukin-17 contributes to neuroinflammation and neuropathic pain following peripheral nerve injury in mice. J. Pain. 2011;12:370–383. doi: 10.1016/j.jpain.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Kleckner I.R., Kamen C., Gewandter J.S., Mohile N.A., Heckler C.E., Culakova E. Effects of exercise during chemotherapy on chemotherapy-induced peripheral neuropathy: a multicenter, randomized controlled trial. Support Care Cancer. 2018;26:1019–1028. doi: 10.1007/s00520-017-4013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober K.M., Dunn L., Mastick J., Cooper B., Langford D., Melisko M. Gene expression profiling of evening fatigue in women undergoing chemotherapy for breast cancer. Biol. Res. Nurs. 2016;18:370–385. doi: 10.1177/1099800416629209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober K.M., Mazor M., Abrams G., Olshen A., Conley Y.P., Hammer M. Phenotypic characterization of paclitaxel-induced peripheral neuropathy in cancer survivors. J. Pain Symptom Manag. 2018 Dec;56(6) doi: 10.1016/j.jpainsymman.2018.08.017. 908-919.e3, Epub 2018 Aug 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober K.M., Olshen A., Conley Y.P., Schumacher M., Topp K., Smoot B. Expression of mitochondrial dysfunction-related genes and pathways in paclitaxel-induced peripheral neuropathy in breast cancer survivors. Mol. Pain. 2018;14 doi: 10.1177/1744806918816462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosti I., Jain N., Aran D., Butte A.J., Sirota M. Cross-tissue analysis of gene and protein expression in normal and cancer tissues. Sci. Rep. 2016;6:24799. doi: 10.1038/srep24799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krames E.S. The role of the dorsal root ganglion in the development of neuropathic pain. Pain Med. 2014;15:1669–1685. doi: 10.1111/pme.12413. [DOI] [PubMed] [Google Scholar]
- Krieglstein K., Henheik P., Farkas L., Jaszai J., Galter D., Krohn K. Glial cell line-derived neurotrophic factor requires transforming growth factor-beta for exerting its full neurotrophic potential on peripheral and CNS neurons. J. Neurosci. 1998;18:9822–9834. doi: 10.1523/JNEUROSCI.18-23-09822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlowitz D., Muggia F. Defining risks of taxane neuropathy: insights from randomized clinical trials. Clin. Cancer Res. 2013;19:4570–4577. doi: 10.1158/1078-0432.CCR-13-0572. [DOI] [PubMed] [Google Scholar]
- Langjahr M., Schubert A.L., Sommer C., Uceyler N. Increased pro-inflammatory cytokine gene expression in peripheral blood mononuclear cells of patients with polyneuropathies. J. Neurol. 2018;265:618–627. doi: 10.1007/s00415-018-8748-4. [DOI] [PubMed] [Google Scholar]
- Li S.J., Zhang Y.F., Ma S.H., Yi Y., Yu H.Y., Pei L. The role of NLRP3 inflammasome in stroke and central poststroke pain. Medicine (Baltimore) 2018;97:e11861. doi: 10.1097/MD.0000000000011861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G., Wang S., Zhang Y., Tian Y., Mao J. Spinal leptin contributes to the pathogenesis of neuropathic pain in rodents. J. Clin. Invest. 2009;119:295–304. doi: 10.1172/JCI36785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubberts E. IL-17/Th17 targeting: on the road to prevent chronic destructive arthritis? Cytokine. 2008;41:84–91. doi: 10.1016/j.cyto.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Ma W., Bisby M.A. Increased activation of nuclear factor kappa B in rat lumbar dorsal root ganglion neurons following partial sciatic nerve injuries. Brain Res. 1998;797:243–254. doi: 10.1016/s0006-8993(98)00380-1. [DOI] [PubMed] [Google Scholar]
- Ma J., Kavelaars A., Dougherty P.M., Heijnen C.J. Beyond symptomatic relief for chemotherapy-induced peripheral neuropathy: targeting the source. Cancer. 2018 Jun 1;124(11):2289–2298. doi: 10.1002/cncr.31248. Epub 2018 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Kiguchi N., Kobayashi Y., Ikuta T., Ozaki M., Kishioka S. Leptin derived from adipocytes in injured peripheral nerves facilitates development of neuropathic pain via macrophage stimulation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13076–13081. doi: 10.1073/pnas.0903524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makker P.G., Duffy S.S., Lees J.G., Perera C.J., Tonkin R.S., Butovsky O. Characterisation of immune and neuroinflammatory changes associated with chemotherapy-induced peripheral neuropathy. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Ying Q.L., Kavelaars A., Krukowski K., Huo X.J., Zhou W., Price T.J. The anti-diabetic drug metformin protects against chemotherapy-induced peripheral neuropathy in a mouse model. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miaskowski C., Mastick J., Paul S.M., Topp K., Smoot B., Abrams G. Chemotherapy-induced neuropathy in cancer survivors. J. Pain Symptom Manag. 2017;54:204–218. doi: 10.1016/j.jpainsymman.2016.12.342. (e2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea C., Taghavi Z., Bokanizad B., Hanoudi S., Tagett R., Donato M. Methods and approaches in the topology-based analysis of biological pathways. Front. Physiol. 2013;4:278. doi: 10.3389/fphys.2013.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockenhouse C.F., Bernstein W.B., Wang Z., Vahey M.T. Functional genomic relationships in HIV-1 disease revealed by gene-expression profiling of primary human peripheral blood mononuclear cells. J. Infect. Dis. 2005;191:2064–2074. doi: 10.1086/430321. [DOI] [PubMed] [Google Scholar]
- Ockenhouse C.F., Hu W.C., Kester K.E., Cummings J.F., Stewart A., Heppner D.G. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect. Immun. 2006;74:5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk G., Erdogan E., Anlar O., Kosem M., Taspinar M. Effect of leukemia inhibitory factor in experimental cisplatin neuropathy in mice. Cytokine. 2005;29:31–41. doi: 10.1016/j.cyto.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Pan Z., Shan Q., Gu P., Wang X.M., Tai L.W., Sun M. miRNA-23a/CXCR4 regulates neuropathic pain via directly targeting TXNIP/NLRP3 inflammasome axis. J. Neuroinflammation. 2018;15:29. doi: 10.1186/s12974-018-1073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.B., Goldstein D., Krishnan A.V., Lin C.S., Friedlander M.L., Cassidy J. Chemotherapy-induced peripheral neurotoxicity: a critical analysis. CA Cancer J. Clin. 2013;63:419–437. doi: 10.3322/caac.21204. [DOI] [PubMed] [Google Scholar]
- Peters C.M., Jimenez-Andrade J.M., Jonas B.M., Sevcik M.A., Koewler N.J., Ghilardi J.R. Intravenous paclitaxel administration in the rat induces a peripheral sensory neuropathy characterized by macrophage infiltration and injury to sensory neurons and their supporting cells. Exp. Neurol. 2007;203:42–54. doi: 10.1016/j.expneurol.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Pu S., Li S., Xu Y., Wu J., Lv Y., Du D. Role of receptor-interacting protein 1/receptor-interacting protein 3 in inflammation and necrosis following chronic constriction injury of the sciatic nerve. Neuroreport. 2018;29:1373–1378. doi: 10.1097/WNR.0000000000001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P., Torck A., Quigley L., Wangzhou A., Neiman M., Rao C. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain. 2018;159:1325–1345. doi: 10.1097/j.pain.0000000000001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reghunathan R., Jayapal M., Hsu L.Y., Chng H.H., Tai D., Leung B.P. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 2005;6:2. doi: 10.1186/1471-2172-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.R., Dougherty P.M. Spinal astrocyte gap junction and glutamate transporter expression contributes to a rat model of bortezomib-induced peripheral neuropathy. Neuroscience. 2015;285:1–10. doi: 10.1016/j.neuroscience.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C.R., Zhang H., Dougherty P.M. Astrocytes, but not microglia, are activated in oxaliplatin and bortezomib-induced peripheral neuropathy in the rat. Neuroscience. 2014;274:308–317. doi: 10.1016/j.neuroscience.2014.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahenk Z., Barohn R., New P., Mendell J.R. Taxol neuropathy. Electrodiagnostic and sural nerve biopsy findings. Arch. Neurol. 1994;51:726–729. doi: 10.1001/archneur.1994.00540190110024. [DOI] [PubMed] [Google Scholar]
- Sarosy G., Kohn E., Stone D.A., Rothenberg M., Jacob J., Adamo D.O. Phase I study of taxol and granulocyte colony-stimulating factor in patients with refractory ovarian cancer. J. Clin. Oncol. 1992;10:1165–1170. doi: 10.1200/JCO.1992.10.7.1165. [DOI] [PubMed] [Google Scholar]
- Schnadig I.D., Fromme E.K., Loprinzi C.L., Sloan J.A., Mori M., Li H. Patient-physician disagreement regarding performance status is associated with worse survivorship in patients with advanced cancer. Cancer. 2008;113:2205–2214. doi: 10.1002/cncr.23856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J., Warren H.S., Cuenca A.G., Mindrinos M.N., Baker H.V., Xu W. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao R., Lee-Kubli C.A. Neuropathic pain after spinal cord injury: challenges and research perspectives. Neurotherapeutics. 2018;15:635–653. doi: 10.1007/s13311-018-0633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C., Kress M. Recent findings on how proinflammatory cytokines cause pain: peripheral mechanisms in inflammatory and neuropathic hyperalgesia. Neurosci. Lett. 2004;361:184–187. doi: 10.1016/j.neulet.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Sun C., Zhang J., Chen L., Liu T., Xu G., Li C. IL-17 contributed to the neuropathic pain following peripheral nerve injury by promoting astrocyte proliferation and secretion of proinflammatory cytokines. Mol. Med. Rep. 2017;15:89–96. doi: 10.3892/mmr.2016.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Nee A.C., Lu A., Ran R., Sharp F.R. Blood genomic expression profile for neuronal injury. J. Cereb. Blood Flow Metab. 2003;23:310–319. doi: 10.1097/01.WCB.0000048518.34839.DE. [DOI] [PubMed] [Google Scholar]
- Tofaris G.K., Patterson P.H., Jessen K.R., Mirsky R. Denervated Schwann cells attract macrophages by secretion of leukemia inhibitory factor (LIF) and monocyte chemoattractant protein-1 in a process regulated by interleukin-6 and LIF. J. Neurosci. 2002;22:6696–6703. doi: 10.1523/JNEUROSCI.22-15-06696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin R.S., Bowles C., Perera C.J., Keating B.A., Makker P.G.S., Duffy S.S. Attenuation of mechanical pain hypersensitivity by treatment with Peptide5, a connexin-43 mimetic peptide, involves inhibition of NLRP3 inflammasome in nerve-injured mice. Exp. Neurol. 2018;300:1–12. doi: 10.1016/j.expneurol.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Uceyler N., Rogausch J.P., Toyka K.V., Sommer C. Differential expression of cytokines in painful and painless neuropathies. Neurology. 2007;69:42–49. doi: 10.1212/01.wnl.0000265062.92340.a5. [DOI] [PubMed] [Google Scholar]
- Wang X.M., Lehky T.J., Brell J.M., Dorsey S.G. Discovering cytokines as targets for chemotherapy-induced painful peripheral neuropathy. Cytokine. 2012;59:3–9. doi: 10.1016/j.cyto.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt S.H., Sommer W.H., Hansson A.C., Sticht C., Rietschel M., Witt C.C. Comparison of gene expression profiles in the blood, hippocampus and prefrontal cortex of rats. In Silico Pharmacol. 2013;1:15. doi: 10.1186/2193-9616-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wang Q., Jiang W., Yu S., Zhang S. MiR-34c ameliorates neuropathic pain by targeting NLRP3 in a mouse model of chronic constriction injury. Neuroscience. 2019;399:125–134. doi: 10.1016/j.neuroscience.2018.12.030. [DOI] [PubMed] [Google Scholar]
- Yin Y., Yi M.H., Kim D.W. Impaired autophagy of GABAergic interneurons in neuropathic pain. Pain Res. Manag. 2018;2018:9185368. doi: 10.1155/2018/9185368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.J., Jiang B.C., Gao Y.J. Chemokines in neuron-glial cell interaction and pathogenesis of neuropathic pain. Cell. Mol. Life Sci. 2017;74:3275–3291. doi: 10.1007/s00018-017-2513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
– Differences in Demographic Characteristics Between Breast Cancer Survivors With and Without Paclitaxel-Induced Neuropathy.
– Differences in Clinical Characteristics Between Breast Cancer Survivors With and Without Paclitaxel-Induced Peripheral Neuropathy.
– Pain Characteristics of the Breast Cancer Survivors With Paclitaxel-Induced Peripheral Neuropathy.
– Differences in Sensation and Balance Measures Between Breast Cancer Survivors With and Without Paclitaxel-Induced Peripheral Neuropathy.
– Results of the ProfileChaser analysis.