Abstract
Background:
Previously, we identified four depression subtypes defined by distinct functional connectivity alterations in depression-related brain networks, which in turn predicted clinical symptoms and treatment response. Optogenetic fMRI offers a promising approach for testing how dysfunction in specific circuits gives rise to subtype-specific, depression-related behaviors. However, this approach assumes that there are robust, reproducible correlations between functional connectivity and depressive symptoms—an assumption that was not extensively tested in previous work.
Methods:
First, we comprehensively re-evaluate the stability of canonical correlations between functional connectivity and symptoms (N=220 subjects), using optimized approaches for large-scale statistical hypothesis testing, and we validate methods for improving estimation of latent variables driving brain-behavior correlations. Having confirmed this necessary condition, we review recent advances in optogenetic fMRI and illustrate one approach to formulating hypotheses regarding latent subtype-specific circuit mechanisms and testing them in animal models.
Results:
Correlations between connectivity features and clinical symptoms are robustly significant, and CCA solutions tested repeatedly on held-out data generalize. However, they are sensitive to data quality, preprocessing, and clinical heterogeneity, which can reduce effect sizes. Generalization can be markedly improved by adding L2-regularization, which decreases estimator variance, increases canonical correlations in left-out data, and stabilizes feature selection. These improvements are useful for identifying candidate circuits for optogenetic interrogation in animal models.
Conclusions:
Multi-view, latent-variable approaches like CCA offer a conceptually useful framework for discovering stable patient subtypes by synthesizing multiple clinical and functional measures. Optogenetic fMRI holds promise for testing hypotheses regarding latent, subtype-specific mechanisms driving depressive symptoms and behaviors.
Keywords: depression, machine learning, depression subtypes, optogenetic fMRI, neuroimaging, biomarkers
Depression is a heterogeneous neuropsychiatric syndrome that is thought to be caused by multiple distinct and interacting neurobiological mechanisms that may play unique roles in various patient subgroups (1–6). Pioneering work identified melancholic, atypical, seasonal, and other clinical subtypes of depression, defined by symptoms or clinical characteristics that tend to co-occur (7–11), but it has been challenging to identify neurobiological correlates that could be used as biomarkers. An alternative strategy for parsing heterogeneity would involve subgrouping patients based on objective biological, cognitive, or behavioral substrates, and then testing whether they predict clinical symptoms and outcomes—an approach with proven utility in psychosis, autism, and other disorders (12–17), and more recently, in depression (18–22).
Our prior work identified four neurophysiological subtypes of depression defined by distinct functional connectivity alterations in limbic and frontostriatal brain networks, which in turn predicted distinct clinical symptom profiles (18). We used canonical correlation analysis (CCA) to identify linear combinations of resting state functional connectivity (RSFC) features that predicted linear combinations of clinical symptoms, both of which could be used for either defining patient subtypes or for rating individual patients along continuous dimensions that capture unique aspects of brain dysfunction, consistent with multiple previous studies identifying correlations between RSFC features, symptoms, and diagnostic status (23–32). However, subtype-specific connectivity patterns were complex, and as in other studies (18–21), it remains unclear how connectivity alterations in specific circuits mediate particular symptoms and behaviors. Addressing this issue will require new approaches aimed at “bridging the causality gap” (33) by experimentally manipulating specific circuits and testing for effects on behavior.
The primary goal of this work is to illustrate one such approach to formulating hypotheses regarding subtype-specific circuit mechanisms driving depressive behaviors in patients and then testing homologues in animal models using optogenetic fMRI. Importantly, this approach assumes that RSFC alterations capture an important latent component of depression pathophysiology that reliably predicts symptoms and behavior. However, a recent preprint raised questions about this central assumption by showing that CCA involving high-dimensional neuroimaging data tends to overfit and suggesting that RSFC-behavior correlations may not be reliable (34). Thus, a necessary prior goal is to test the assumption that we can reliably reproduce latent variables underlying RSFC-behavior correlations.
We begin by comprehensively re-evaluating whether RSFC alterations are stably related to depressive symptoms using optimized approaches for large-scale statistical testing. We find that correlations between RSFC features and clinical symptoms are robustly significant, and further, that latent variable (CCA) solutions tested repeatedly on held-out data generalize, but tend to overfit with increasing numbers of features. To overcome this obstacle, we show that generalization can be markedly improved by adding L2-regularization. Having confirmed these key assumptions, we review recent advances in optogenetic fMRI and illustrate how it could be used to causally interrogate latent subtype-specific circuit mechanisms driving particular depression-related behaviors, integrating results from our recent subtyping work with published optogenetic fMRI studies. We also discuss pros and cons of this method, relative to lesion analyses and non-invasive brain stimulation methods that can be applied directly in humans.
Methods and Materials
Subjects.
The analyses reported in Figs. 1–3 were designed to re-evaluate our approach in (18) using state-of-the-art statistical methods to test whether depression-related RSFC alterations are significant and stable predictors of clinical symptoms. Therefore, these analyses were conducted in the same “subtype-discovery sample” used in (18), which comprised N=220 subjects meeting DSM-IV criteria for a diagnosis of (unipolar) major depressive disorder and currently experiencing an active, non-psychotic major depressive episode at the time of the fMRI scan (see Table 1 for details). In addition, in order to better understand whether differences between this sample (summarized in Supp. Table 1) and the sample used in Ref. (34) may have influenced their power to detect statistically significant RSFC-clinical symptom correlations, we conducted supplementary analyses in a separate sample of N=184 subjects (acquired during on-going studies at Cornell and Toronto) that more closely resembles their dataset. See Supplementary Methods for further details on subjects and MRI data acquisition.
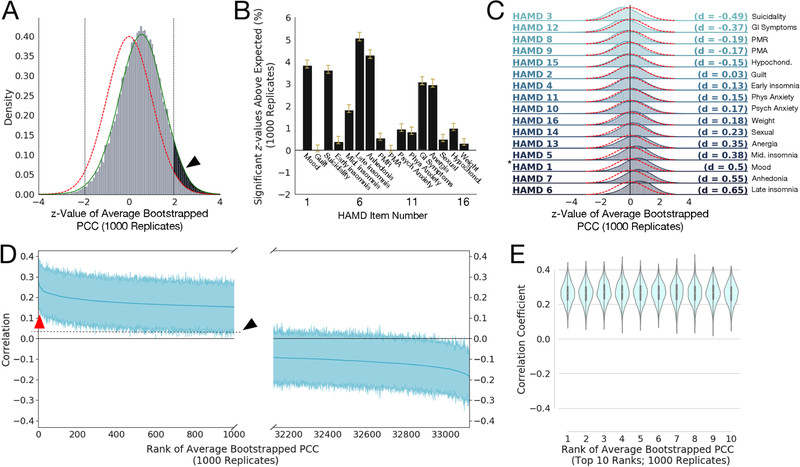
Figure 1: Robust correlations exist between Resting State Functional Connectivity Features (RSFCs) and clinical measures (HAMD).
A) Histogram of z-values for the 33,123 average Pearson Correlation Coefficients (PCCs) between RSFC features and HAMD item 1 (“depressed mood”) scores, averaged over 1000 bootstrap replicates, and compared to a standard Gaussian distribution (red line) and with a smoothed kernel density estimate plotted over the histogram (green line; see Methods). The black arrow and shaded region show the area of the z-values that exceeds the area expected for a standard normal distribution (outside the two-sided significance criterion of z > ± 1.96 for p < 0.05; shown as vertical dotted black lines). Note the empirical distribution of z-values has sample mean and standard deviation , respectively, with the lower-than-expected sample variance resulting from correlations among the statistics; we correct for the effects of such inter-statistic correlations using the procedure in (35)(see Methods). B) Bar plots of the mean percentage of z-values that exceeded that expected by chance (e.g., the percentage above 2.5%, shown as the shaded black area in A for HAMD 1) for 1000 bootstrap replicates. Yellow whiskers on the bars denote 95% confidence intervals (corrected for multiple comparisons and data correlation using Bonferroni-Holm and (35), respectively). We see that HAMD measures 1, 3, 5, 6, 7, 12, and 13 have mean significant percentages well in excess of 1% more than expected under the null hypothesis. Anhedonia also encompasses work activities; PMR = pscyhomotor retardation; PMA = psychomotor agitation; psych = psychological anxiety symptoms; phys = physiological anxiety symptoms; GI = gastrointestinal symptoms; anergia also encompassses somatic symptoms; hypochond = hypochondriasis; weight = weight loss or weight gain. C) Histograms of z-values like that shown in A for all 16 HAMD clinical measures considered, ordered by effect size (Cohen’s d, given at right of each plot; magnitudes between 0.2 and 0.5 are considered small to medium effect sizes, between 0.5 and 0.8 are considered medium to large effect sizes; calculated between the smoothed z-value distributions like the green line in Fig. 1A with the standard normal). Red dotted lines denote the standard normal distribution. Asterisk (*) marks the distribution for HAMD1 shown in A. Symptom abbreviations as in B. D) Bootstrapped PCCs for HAMD measure 1 for the 1000 most positive (left) and 1000 most negative (right) RSFC features (shaded regions shows 95% percentile-bootstrap confidence interval for the mean), ordered by mean correlation (thick blue line). Red arrow points to top 10 most positive-ranked RSFC features (shown in E); note both have confidence intervals excluding zero, indicating that while they cover an appreciable range, they are significantly different than zero across the 1000 bootstrap replicates and thus somewhat stable across bootstrap replicates. The black arrow and dotted line show the upward shift resulting from the positive shift of the distribution shown by the black arrow in panel A. E) Violin plot (with superimposed boxplots showing 1st and 3rd quartiles as black bar and the median as white point) of the top ten positive ranked RSFCs by average PCC to HAMD measure 1 (corresponding to red arrow in D), with mean 95% confidence intervals ± SD of [0.148 ± 0.0152, 0.376 ± 0.0119]. Note these look very similar, suggesting the rank order could easily change across replicates.
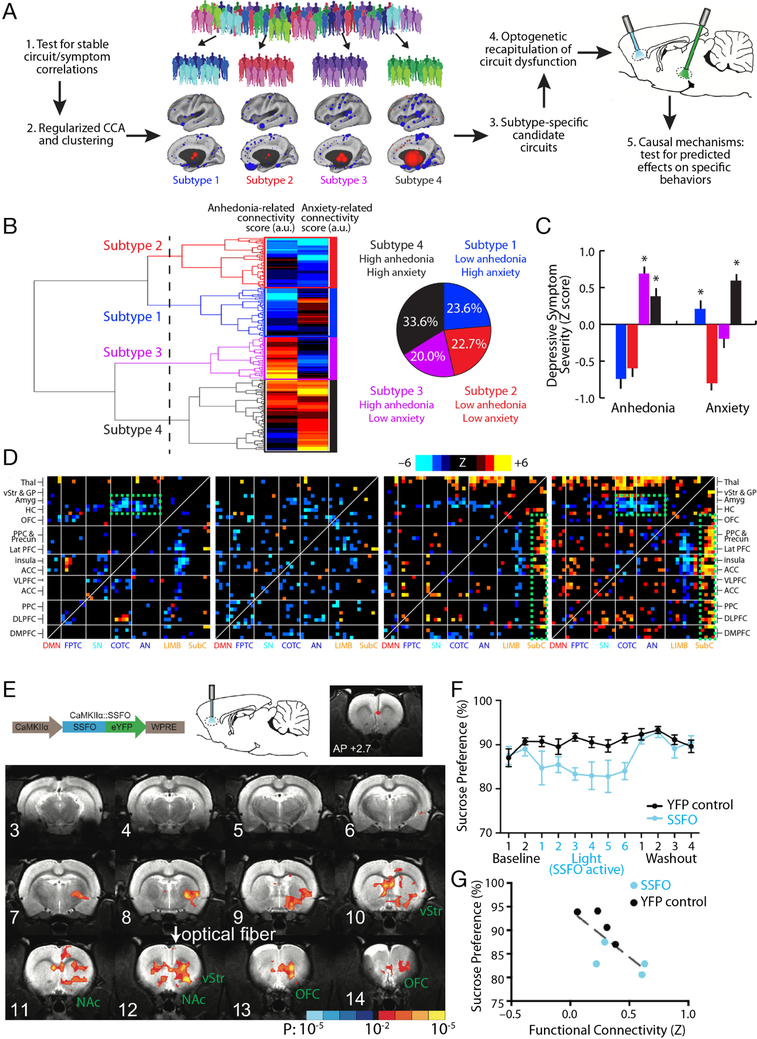
Figure 3. Optogenetic fMRI for interrogating subtype-specific circuit mechanisms in depression.
A) Schematic illustration of a model for formulating hypotheses regarding subtype-specific mechanisms driving depressive symptoms and behaviors, and testing them in animal models using optogenetic fMRI. By first testing for robust and stable RSFC-clinical symptom correlations as in Fig. 1 and then using CCA and hierarchical clustering, relatively homogeneous subgroups of a heterogeneous MDD sample can be identified. These subgroups can be used to identify subtype-specific candidate circuits (see main text), and ofMRI can be used to test hypotheses about dysfunction in specific circuits driving specific behaviors, while also validating whether the RSFC effects evoked by the optogenetic manipulation resemble those observed in human subjects. B) In Ref. (18), hierarchical clustering on two canonical variates representing anhedonia- and anxiety-related RSFC revealed at least four clusters of patients in these two dimensions. The height of each linkage in the dendrogram represents the distance between the clusters joined by that link. The dashed line denotes 20 times the mean distance between pairs of subjects within a cluster. C) The four subtypes predicted significant group differences in anhedonia and anxiety (P < 0.005, Kruskal Wallis ANOVA) as indexed by item-level responses on the HAMD (item 7 and 11, respectively). Symptom severities are Z-scored with respect to the mean and standard deviation of all patients in the sample. Error bars = S.E.M. D) Heatmaps depicting subtype-specific patterns of altered functional connectivity for the top 50 neuroanatomical ROIs with the most subtype-specific RSFC features by Kruskal Wallis ANOVA. The color scale represents Wilcoxon rank sum test scores for the difference between patients in each subtype and matched healthy controls. The green boxes denote RSFC features discussed in the main text. For additional details on panels B-D, see Ref. (18). E) In Ref. (55), a viral vector (AAV/CaMKIIa/SSFO) driving SSFO expression in projection neurons was injected into mPFC, and an optical fiber implanted over the mPFC target was used to activate (blue light) and inactivate (amber light) the opsin during alternating rsfMRI scanning periods (300 s per scan). SSFO activation induced a pattern of increased functional connectivity between an mPFC seed (denoted by the red dot) and a network of structures depicted here, where colors denote the Z statistic (and associated P value) for RSFC changes in the opsin-on vs. opsin-off conditions (N = 4 rats, 14 runs). NAc = nucleus accumbens; OFC = orbitofrontal cortex; vStr = ventral striatum. F) Subjects (N = 8 SSFO rats, blue; N = 10 control rats, black) were assessed on the sucrose preference test during a 2-day baseline period, followed by 6 days with SSFO activated, followed by a 4-day “washout” period with SSFO off. SSFO activation reduced sucrose preference behavior (F(11,176) = 2.56, P = 0.0051, two-way repeated measures ANOVA), compared to subjects expressing a YFP control construct. G) Individual differences in RSFC between the mPFC seed and the ventral striatum correlated with sucrose preference behavior (R2 = 0.56, P = 0.03). Panels B-D and E-G were adapted from Refs. (18) and (55), respectively. See corresponding references for additional details.
Table 1. Subject demographics, medication status and psychiatric comorbidities.
The analyses in Figs. 1–3 were implemented in the same dataset used in Ref. (18). Subjects recruited at the Toronto and Cornell sites were matched for age (p = 0.41), sex (p = 0.87), and depression severity (HAMD17 total score, p = 0.11). *Psychiatric medications listed as “Other” included benzodiazepines, non-benzodiazepine sedative-hypnotics, stimulants, and thyroid hormone. **Psychiatric comorbidities listed as “Other” included obsessive compulsive disorder, attention-deficit/hyperactivity disorder, Asperger Syndrome, and Tourette’s Syndrome.
| Toronto Sample | Cornell Sample | |
|---|---|---|
| Number of Subjects | 124 | 96 |
| Age (mean) | 40.4 years | 42.1 years |
| Sex | 57.3% female | 58.3% female |
| HAMD17 Total Score (mean) | 20.4 | 19.3 |
| Psychiatric Medications | ||
| Antidepressant | 59.7% | 57.3% |
| Mood Stabilizer | 16.9% | 17.7% |
| Antipsychotic | 17.7% | 15.6% |
| Other* | 45.2% | 42.7% |
| Psychiatric Comorbidities | ||
| Generalized Anxiety Disorder | 4.8% | 5.2% |
| Post-traumatic Stress Disorder | 6.5% | 4.2% |
| Social Anxiety Disorder | 4.8% | 4.2% |
| Panic Disorder | 2.4% | 3.1% |
| Other** | 4.0% | 3.1% |
fMRI Data Preprocessing and RSFC Quantification.
Preprocessing was identical to the procedure defined in our previous report (18), and is described in the Supplementary Methods.
Data Analysis.
The stability and significance of correlations between RSFC features and HAMD clinical symptoms was assessed by calculating the 33,123 Pearson correlation coefficients (PCCs) between each RSFC feature and each of 16 of the HAMD item-level measures on 1000 bootstrap replicates in order to estimate the variance of these correlations (item 17 was excluded, having zero variance in many replicates). We then followed the procedure of of Efron and colleagues (35), using correlation-corrected z-values and bootstrapping to calculate the percentage of correlations that exceeded chance level. See Supplementary Methods for further details.
Canonical correlation analysis (36, 37) was performed between clinical measures and a selected subset of screened RSFC features (those with highest Spearman correlation) as previously described (18). Due to this feature screening step, we use validation on held-out data in subsequent analyses to avoid overly optimistic correlation estimates due to training-set overfitting. To better stabilize CCA coefficients, L2-regularized CCA (38) was also applied. This approach uses two regularization parameters, λX and λY to regularize the estimated covariance matrices for the RSFC and clinical features, respectively. To find the best combination of these two variables, a grid search over possible values of the parameters and number of features was conducted, with 1000 RCCA fits found for each parameter combination. For each set of parameters, model fitting was done on training data and then assessed via the magnitude of the 1st canonical correlation coefficient on held-out validation “test” data, using the same procedure as above for standard CCA. See Supplementary Materials for further details.
Results
Testing for Robust Correlations between RSFCs and Clinical Symptoms.
We began with a modern approach to a classical problem: establishing the existence and strength of correlations between brain and behavior using mass univariate statistics. Examining number, strength, and effect size of these correlations gives us a strong basis from which to begin more complicated multivariate analyses (like CCA), and convinces us of the utility of doing so. Furthermore, understanding the structure of univariate correlations between RSFC and clinical symptoms gives us insight into what kind of challenges might present themselves in the multivariate setting. First, we correlated each RSFC feature with each HAMD clinical symptom, and estimated the number of z-values for the resulting RSFC-symptom Pearson correlation coefficients (PCCs) that exceeded the threshold of significance expected by chance (Fig. 1), after correcting for correlations between RSFCs across subjects and for large-scale correlations and multiple comparisons (see Supplementary Methods). We were also interested in establishing the variance of the number of significant correlations: is it stable, or do small changes in the data collection conditions translate to large changes in the number of correlations that are found to be significant (indicating unstable correlation estimates)?
To estimate the variance of the number of correlations above the significance threshold, we used the bootstrap (39), resampling the RSFC and clinical data for each subject to generate 1000 bootstrap replicate data sets, and then ran the z-value procedure from (35) on each. A representative result for HAMD item 1 (HAMD1: “depressed mood”)) is shown in Fig. 1A, with the shaded region showing the number of significant RSFC feature–HAMD1 correlations above the number expected by chance. We generated confidence intervals for the significant z-value estimates with the “percentile bootstrap” and corrected for the 16 multiple comparisons across the HAMD clinical features using the Holm-Bonferroni procedure, yielding the results shown in Fig. 1B, with effect sizes in Fig. 1C (40). Seven HAMD measures had median significant percentages (number of correlations more than expected by chance) well in excess of 1% (representing hundreds of significant correlations), and overall, 14 out of the 16 z-value distributions showed reliable, significant shifts in correlations.
We also examined the range of RSFC–HAMD correlations. Fig. 1D shows the 1000 most positive (left) and 1000 most negative (right) correlations, ordered by the average PCC across bootstrap replicates (solid blue line), with 95% confidence intervals (percentile bootstrap). All of the 1000 most positive PCCs and a substantial fraction of the 1000 most negative PCCs had confidence intervals excluding zero, but there was also a significant range over which different bootstrap replicates might yield different orderings of the coefficients. This is illustrated in Fig. 1E, showing violin plots detailing the distribution of PCCs for the 10 most positive PCCs: the distributions were significantly different from zero, but looked relatively exchangeable, such that their ranking would change over bootstrap replicates. Thus, a large number of very similar variables could result in highly variable feature selection, with implications for CCA discussed below.
RSFC-clinical symptom correlations are sensitive to clinical sampling and preprocessing decisions.
A recent preprint (34) reported the results of an analysis similar to our earlier work (18) and concluded that RSFC-clinical symptom correlations were not significant, which would seem to contradict the findings reported in Fig. 1. However, there were several important differences between these two studies (see Supplementary Table 1 for details), especially in their clinical sample characteristics and preprocessing pipelines. Of note, the sample in Ref. (34) included N=187 subjects scanned on four different scanners (versus N=220 subjects scanned on just two scanners in our previous work, yielding a larger number of subjects per scanner and potentially more stable corrections for scanner-related differences). Among other differences, Ref. (34) did not directly control for scanner-related differences, and their sample was also more clinically heterogeneous (including MDD, generalized anxiety disorder, social phobia, or panic disorder with no specified requirements for active depressive symptoms vs. currently active, treatment-resistant MDD in our work). By testing for RSFC-clinical symptom correlations in this more heterogeneous sample, the approach in Ref. (34) assumes that the mechanisms driving these correlations are the same across these disorders, but this may not be true. For example, it is possible that different mechanisms may drive anxiety symptoms in MDD compared with panic disorder, in which case an analysis of subjects with mixed diagnoses could yield smaller effect sizes and unstable results in held-out data.
To test whether these clinical sample and preprocessing differences could influence their power to detect robust RSFC-clinical symptom correlations, we repeated the analysis reported in Fig. 1 in a second more clinically heterogeneous sample of N=184 subjects with MDD or an anxiety disorder, scanned on one of four scanners, and preprocessed exactly as in Ref. (34). (see Supplementary Methods) The results in Supplementary Fig. 1 show that small but statistically significant RSFC-clinical symptom correlations are still detectable for 10 of 16 symptoms (vs. 14 of 16 in Fig. 1B), but these associations are modest, with uniformly small effect sizes (d=0.21–0.29 for 5 symptoms, d<0.2 for all others). These results are consistent with the interpretation that distinct mechanisms give rise to RSFC-clinical symptom correlations across these heterogeneous disorders and that preprocessing decisions may be important.
Stable canonical correlations between RSFC features and clinical symptoms.
CCA (36, 37) is a classical multi-view statistical approach that we (18) and others (21) have used to find latent linear combinations of RSFC measures and clinical features (canonical variates [CVs]) that are maximally correlated with each other. In principle, CCA is a potentially useful approach for discovering subtypes of depression (or a dimensional rating system) anchored in brain network dysfunction and for identifying potential latent targets for optogenetic and other causal investigations: it provides a generalizable, low-dimensional representation of the relationship between neuroimaging and clinical features in the form of a simplified summary of the interesting structure between them. However, traditional CCA has some potential weaknesses, particularly on large-scale, correlated data. In particular, CCA coefficients become unstable in the presence of multicollinearity (i.e. significant correlations between variables, as we might suspect between RSFC features and HAMD symptoms) (38). Further, CCA can only operate on as many variables as there are observations, so that feature selection is necessary prior to applying CCA in order to reduce the 33,123 RSFC measures to a number less than or equal to the number of subjects in the study (38). Despite this, CCA yielded promising results in recent studies (21) and in the data presented in our previous work (18). However, the stability of CCA solutions was not integral to the other analyses in our previous study (18) and thus was not directly assessed.
To test this, we resampled the data 1000 times (without replacement) into training (90% of subjects) and validation (“test”) sets (the remaining 10%), and assessed CCA stability by comparing the resulting canonical correlations in the first CV subspace, across increasing numbers of RSFC features (Supp. Methods). Fig. 2A shows that standard CCA overfits: the training correlations gradually approached 0.9, while the test correlations increased initially but then decreased towards 0.1. The variance of the distributions for test canonical correlations was large, but the best fit had a median canonical correlation of 0.557 (IQR=0.456–0.642), suggesting that the approach is promising.
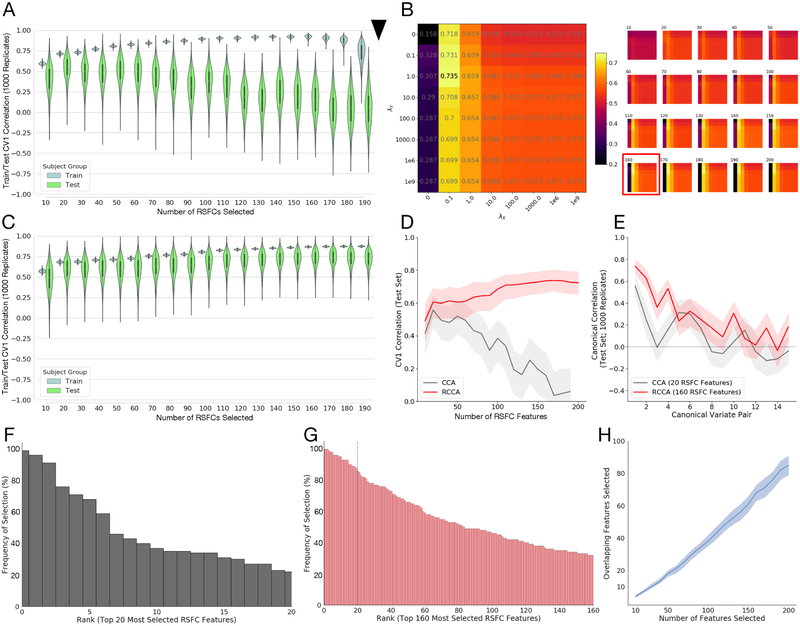
Figure 2: Stable train/test canonical correlations between RSFC features and clinical measures are improved by regularization.
A) Violin plots (with superimposed boxplots) of correlations between the first canonical variates of a standard Canonical Correlation Analysis (CCA) on training data (90% of subjects) and test data (10% of subjects) for a range of features (10 to 190 by increments of 10) selected using the correlation method proposed in (18), with this procedure bootstrapped 1000 times for each number of features to yield the plotted distributions. Feature selection and CCA fitting was done on training data, separately for each bootstrap replicate, and then estimated CCA coefficients applied to the selected features in the held-out validation set to obtain test correlations. Test correlations for CCA peak at 20 features selected. Black arrow: standard CCA cannot be fit to more correlations than there are observations (in this case 90% of n=220, or 198 subjects). B) Median test rates fit over a grid of regularization parameters λX, λY for each number of features selected. (Left) The grid corresponding to the best test correlations corresponding to using 160 RSFC features. The color of each square in the grid corresponds to the median test correlation (also printed in grey in the center of each square; colorbar at right gives hue values). (Right) Similar grids for other numbers of RSFC features (number of features selected shown above grid, test correlations shown in color only, not text). The best fit (160 features; shown on the left) is boxed in red box in the full set of fits on the right. Fitting more than 198 coefficients is possible. C) Violin plots (with superimposed boxplots) of correlations between the first canonical variates of the Regularized Canonical Correlation Analysis (RCCA) with the best regularization parameters (λX = 0.1, λY = 1, NF = 160) on training data (90% of subjects) and test data (10% of subjects) for the various numbers of features selected using the correlation method proposed in (18) (resampled 1000 times), as in A. Fitting more than 198 coefficients is possible. D) Test rates for the first canonical variate (CV1) as a function of the number of features selected for CCA (grey) and RCCA (red); shaded region shows 1st through 3rd quartile for the replicate fits. E) Test correlations between canonical variates 1–15 for the best fit from A (CCA fit in grey; 20 features), and the best fit from C (RCCA fit in red; 160 features); shaded region shows 1st through 3rd quartile for the replicate fits. F) Ordered (by top rank) histogram of the top 20 features chosen by the feature selection approach (from (18)) showing the percentage of times they were chosen across the 1000 subsampled replicate data sets. Just 3 features are selected more than 80% of the time. G) Ordered (by top rank) histogram of the top 160 features chosen by the feature selection approach showing the percentage of times they were chosen across 1000 subsampled replicates. 25 features appearing more than 80% of the time, dotted line denotes top 20 features; compare with F. H) Number of overlapping features in all pairwise combinations of 100 randomly chosen replicates as a function of number of features selected (dark blue line shows median and shaded region 1st through 3rd quartile across replicates). The median number of overlapping features selected increases approximately linearly with the total number of features selected.
We hypothesized that these results might be stabilized via L2-regularization applied to the CCA coefficients associated with both the RSFC and clinical features, as both were multicollinear. L2-regularization (the “ridge” penalty (41)) induces a small downward bias in coefficient magnitude in exchange for a potentially large reduction in coefficient variance (42). In regularized CCA (RCCA), we shrink both the sample covariance matrix for the RSFC features and for the clinical measures toward the identity matrix by replacing them with and , respectively (38). This requires specifying the value of the two regularization parameters λX and λY for each RCCA fit. To assess the effects of these parameters on fit quality, we fit each of our RCCA models over a grid of λX and λY, with each parameter taking values in set {0, 0.1, 1, 10, 100, 1000, 1e6, 1e9}.
Fig. 2B depicts the median canonical correlation results on the held-out test data (over 1000 replicates) and shows that a small amount of regularization of the RSFC feature coefficients greatly improved the test canonical correlations. To a lesser extent, regularization of the HAMD coefficients also benefits fit, with a peak median test canonical correlation at λX = 0.1 and λY = 1.0 of 0.735 (IQR=0.665–0.797). Compared to the CCA fit in Fig. 2A, the test canonical correlations for the best RCCA (fit at λX = 0.1, λY = 1.0) had lower variance, remained above zero, and improved with increasing number of features (Fig. 2C–D). Furthermore, if we examine the stability of test correlations between additional canonical variates (Fig. 2E), we see that RCCA uniformly outperforms CCA (at its best performance at 20 RSFC features) for the first 4 sets of canonical variates. Thus regularization of both RSFC and HAMD feature coefficients stabilizes and improves low-dimensional co-embedding of neuroimaging and clinical measures.
As noted above, Fig. 1D showed that a large number of very similar variables could result in highly variable feature selection across bootstrap replicates. Fig. 2F and G respectively show the ranked distributions of which RSFC features were chosen by the screening procedure over the 1000 subsamples when selecting the top 20 features (the optimum for traditional CCA in Fig. 2A) vs. the top 160 features (the optimum for RCCA in Fig. 2C). Having just 20 RSFC features (Fig. 2F) means just 3 features are selected more than 80% of the time, whereas having 160 features results in 25 features appearing more than 80% of the time. In Fig. 2H, we ran pairwise comparisons looking at how many features appeared in both of two replicates (randomly choosing 100 of the subsample replicates), and found that the number of consistently selected features increased linearly with the total number of features selected. Thus, stabilizing CCA with regularization allows the model to leverage more features than standard CCA, yielding a broader set of more reliable features that result in higher out-of-sample test correlations.
Discussion
Together, these results support the hypothesis that RSFC alterations capture an important component of the pathophysiology of depression and are robust and reliable predictors of specific symptoms in actively depressed MDD patients. In particular, as shown in our previous work (18), CCA in this sample revealed two canonical variates, respectively predicting individual differences in 1) anhedonia and psychomotor slowing (HAMD items 7–8) and 2) anxiety and insomnia (HAMD items 4–5,11). Individual patients, in turn, could be clustered into subgroups defined by relatively homogeneous patterns of altered functional connectivity in these two dimensions, which predicted distinct clinical symptom and treatment response profiles (18)(Fig. 3). Other groups have reported similarly promising results for parsing diagnostic heterogeneity based on task-related and rsfMRI, clinical symptoms, and neuropsychological profiles in affective disorders (19–22), as well as in psychosis and ADHD (12, 43–45). For example, Price et al. identified two sexually dimorphic subgroups of patients with depression that differed with respect to RSFC in the default mode network and predicted individual differences in comorbid anxiety and history of recurrence (19). More recently, Xia et al. used sparse CCA in a sample of 663 youths with mixed diagnoses to identify four dimensions of altered functional connectivity predicting mood symptoms, psychosis, fear, and externalizing behavior (21). Importantly, they went on to replicate these findings in an independent sample of 336 subjects, providing further support for the assumption that stable latent-variable relationships between RSFC and clinical symptoms could be used to develop more biologically homogeneous diagnostic labels.
Of course, in all of these studies, it remains unclear whether RSFC alterations reflect changes in specific circuits driving depression-related behaviors, or are merely correlated with them. Optogenetic tools offer one approach to addressing this question. Over the last ten years, optogenetic studies have begun to define causal relationships between circuit function and behavior (46–51), with important implications for both neurological (52–54) and psychiatric diseases (49, 55–61). Importantly, these methods can also be integrated with functional MRI and other noninvasive neuroimaging techniques that are widely used in humans, offering new opportunities for testing hypotheses and predictions derived from human neuroimaging studies (55, 62). Below, we review these developments, illustrate one model for testing such hypotheses, and discuss important caveats and limitations relative to other approaches.
Optogenetic fMRI for testing subtype-specific circuit mechanisms in depression.
First introduced in 2010 (62), this approach combines high-field fMRI with photoactivatable opsins to manipulate the activity of genetically defined cellular subtypes and test for local and global effects on neuronal activity and brain network function. The initial report by Lee et al. (62) underscored two of the most important and commonly implemented applications of optogenetic fMRI (ofMRI). First, it showed how ofMRI could be used to glean mechanistic insights into the neurophysiological basis of the fMRI BOLD signal—a critical issue for interpreting the results of clinical neuroimaging studies. This report (62) showed that optogenetic stimulation of neocortical or thalamic excitatory neurons was sufficient to drive local BOLD signal responses, informing an ongoing debate about the nature of the neuronal signals and cellular subtypes that underlie the BOLD signal. Subsequent ofMRI studies showed that the BOLD signal is more strongly correlated with local spiking activity than with the local field potential (63) and is driven by the effects of neuronal activity on cerebral venules (64). Recent studies have also shown how inhibitory interneurons and astrocytes contribute to the BOLD signal, independently of activity in excitatory pyramidal neurons and through distinct mechanisms (65, 66). Second, Lee et al. (62) went on to show how ofMRI could be used for whole-brain functional circuit mapping, by optogenetically manipulating the activity of excitatory pyramidal cells in a specific brain area and testing for downstream BOLD signal effects. More recent studies extended this approach to map the functional networks activated by specific circuits (e.g. dorsal vs. ventral hippocampus)(67–72) and by specific cellular subtypes (e.g. dopaminergic vs. glutamatergic cells in the VTA; serotonergic responses to fluoxetine and acute stress)(73–76), often with surprising results that could not be predicted based solely on mapping the axonal projection fields of a given brain region (71, 76). Other studies are defining new methods for integrating ofMRI with two-photon microscopy and head-fixed behavior (77, 78).
Of particular relevance for translational neuroscience studies, ofMRI methods can also be used to recapitulate disease-related pathophysiological processes and evaluate their impact on brain networks and behavior. To this end, we illustrate one approach for formulating hypotheses regarding subtype-specific mechanisms driving depression-related behaviors, and testing them in animal models using ofMRI (Fig. 3A), drawing on two recently published works. In this model, rsfMRI is used to identify candidate circuits that predict specific symptoms and behaviors in patients. ofMRI, in turn, can be used to recapitulate and validate these connectivity changes in functionally related circuits in rodents, and test for causal effects on associated behaviors. One approach to identifying promising candidate circuits involves searching for connectivity alterations and clinical symptoms that tend to co-occur. For example, in our previous work (18), hierarchical clustering on the two canonical variates described above revealed at least four clusters or subtypes (Fig. 3B), predicting group differences in multiple symptoms, especially anhedonia and anxiety (Fig. 3C). Group differences in anhedonia and anxiety, in turn, were associated with functional differences in depression-related brain networks (Fig. 3D).
These subtype-specific patterns were complex; however, qualitatively, two observations stood out. First, Subtypes 1 and 4 were associated with increased anxiety and connectivity deficits in fronto-amygdala circuits (Fig. 3D: green boxes), which have been implicated in the regulation of fear memories and the cognitive reappraisal of negative emotional states (79–82). Second, Subtypes 3 and 4 were associated with increased anhedonia and hyperconnectivity between the medial prefrontal cortex, ventral striatum, and other frontostriatal circuits that have been implicated in reward processing, effort valuation, and motivation (6, 27, 83–89).
Optogenetic tools provide one means of testing whether altering functional connectivity in these circuits is sufficient for driving specific depression-related behaviors. Stable step function opsins (SSFOs) are particularly useful in this context, in that they were designed to achieve stable, partial depolarization on a timescale of minutes (49), suitable for use in resting state fMRI analyses of low-frequency signal fluctuations, but still immediately reversible, enabling within-subject statistical comparisons. Furthermore, by partially depolarizing neurons and rendering them responsive to their physiological inputs, they can in principle be used to reversibly modulate functional connectivity in specific circuits and cell types.
A recent ofMRI study by Ferenczi et al. (55) provides evidence consistent with the hypothesis that increased functional connectivity in a specific frontostriatal network, qualitatively similar to the pattern observed in Subtypes 3 and 4, is sufficient to drive anhedonic behavior in rats. In this study, SSFO was expressed in CaMKIIa+ projection neurons in the medial prefrontal cortex (mPFC), and rsfMRI was used to quantify functional connectivity changes elicited by SSFO activation in the mPFC (Fig. 3E). SSFO activation increased functional connectivity between the mPFC target and a network of structures including the ventral striatum, nucleus accumbens, orbitofrontal cortex, anterior cingulate cortex, and thalamus (Fig. 3E), qualitatively similar to many of the areas exhibiting increased connectivity in Subtypes 3 and 4. SSFO modulation of mPFC projection cells was also sufficient to drive anhedonia-like behavior in the sucrose preference test (Fig. 3F–G).
Importantly, this approach also provides a means of testing how circuits interact to produce anhedonic behavior. Ferenczi et al. (55) went on to show that mPFC and the ventral tegmental area (VTA) compete to influence processing in striatum. VTA stimulation drove a striatal BOLD response that predicted reward-seeking behavior, while SSFO modulation of mPFC excitability suppressed the striatal response to VTA stimulation and disrupted reward processing. Of course, these findings do not necessarily indicate that the same mechanism is involved in driving anhedonic behavior in Subtypes 3 and 4. Rather, they show that this particular pattern of frontostriatal hyperconnectivity, elicited by increasing the excitability of mPFC projection neurons, is sufficient to disrupt reward-seeking behavior. Future studies could test whether these subtypes are associated with hyperexcitability in mPFC; with deficits in striatal reward reactivity; and with abnormal interactions between VTA, mPFC, and striatum. Likewise, new viral tools for targeting opsin expression to topologically defined projection neuron subtypes with increased ease and efficiency (90–92) will enable more targeted investigations that modulate connectivity between specific nodes in this frontostriatal network.
The example in Fig. 3 illustrates one approach to formulating hypotheses about candidate circuits for optogenetic study, based on qualitatively similar connectivity alterations that co-occur with specific symptoms across subtypes. However, candidate circuits could also be identified in a data-driven way, especially with larger sample sizes. Indeed, multi-view, latent-variable methods like RCCA are well suited to this purpose, as reliable latent variables underlying brain-behavior correlations and discovered by RCCA suggest targets for optogenetic interrogation in rodent experiments, which could test whether symptom dimensions can indeed be dissociated by modulating the candidate neural targets. Including sparsity constraints as in (21) may further refine candidate targets for optogenetic interrogation using RCCA.
Caveats and Limitations.
It is also worth noting some important caveats associated with this approach. First, Fig. 2A underscores how CCA has a tendency to overfit when combined with a feature selection step. Therefore, when screening is used to pre-select features for further analysis, careful training and test validation are necessary to generate models that perform well in held-out data and to avoid overfitting. Second, the feature selection approach used here is adequate for identifying stable and robust associations between RSFC features and clinical symptoms, but other approaches (e.g. nonlinear multi-view and/or sparse methods) could yield superior results.
Third, these approaches may be highly sensitive to clinical sample characteristics (e.g. distinct circuit mechanisms may be at play in active depression, depression in remission, and various anxiety disorders), as well as to medication status, data quality, head motion, and other sources of global signal artifacts. Therefore, it is important to optimize and validate preprocessing methods and other data quality controls, based on the goals of a given study. Medication status is an especially important issue: our sample was treatment resistant, and most subjects were taking at least one psychiatric medication at the time of their scans (Supp. Table 1). The subtypes did not differ by medication status, indicating that the subtyping results were not likely driven by medication usage per se (18). However, several studies indicate that antidepressants and other psychotropic medications have significant and varied effects on RSFC measures (93–98). Therefore, future studies will be needed to systematically characterize medication effects on resting state networks and to evaluate the extent to which our results would generalize to unmedicated patients, non-treatment resistant patients, and first episode patients.
Fourth, categorical subtyping is just one approach to parsing diagnostic heterogeneity, and the 4-cluster solution in Fig. 3B is not the only solution. Rather, as discussed in (18), this 4-cluster solution was stable and clinically useful (predicting clinical symptoms and treatment response), but also most likely constrained by features of the subtype discovery dataset, especially sample size and the available clinical data. Item-level HAMD responses provide a relatively coarse, ordinal rating of a limited set of depressive symptoms, and future studies will surely benefit from incorporating more precise rating scales designed to measure specific constructs, as well as objective behavioral measures. Likewise, although a model anchored in categorical subtypes provides a familiar and clinically useful heuristic for clinicians to parse diagnostic heterogeneity, other methods might be superior. One alternative approach that warrants further examination would substitute a multi-dimensional rating system for categorical subtype diagnoses.
Finally, although we focus here on ofMRI, this approach has some limitations, and others should also be considered. First, it is unclear whether RSFC measures are interpretable in the same way in rodents and primates. A growing body of work highlights qualitative cross-species similarities (99), including a reliable RSFC signal that correlates with low-frequency (delta) power (100, 101); robust resting state functional networks (67, 102–105); and a neuroanatomically similar default mode network in both rats and mice (103, 106). However, cross-species differences are also evident. For example, the rodent default mode network lacks a neuroanatomical correlate of the primate posterior cingulate areas 23 and 31 (99, 106). Likewise, other rsfMRI studies comparing the topology of the mouse, macaque, and human brain have identified reliably conserved properties (e.g. “rich club” connectivity) but also important differences (e.g. the probability that highly connected “hubs” are connected to other “hubs”) (106). Second, some brain circuits in primates may not have clear homologs in rodents. For example, the prefrontal cortex exhibits a host of cytoarchitectonic, topological, and molecular differences in rodents vs. primates (107), and multimodal association cortex occupies a much larger proportion of the human brain (108). Third, rodent models of human behavior are inherently limited to behaviors that are well conserved across species (109), and even superficially similar behaviors and cognitive processes may be implemented by different mechanisms across species (110, 111). Consequently, studies drawing parallels between brain circuits and behavior in rodents vs. humans must be interpreted with care, and some human brain circuits and behaviors are simply not well modeled in the mouse. In these cases, other approaches such as concurrent TMS/fMRI (112–114) and new methods for analyzing interactions between brain lesions and their relationship to behavior (115) may be superior for testing causality in the human brain directly (33).
Conclusions.
These caveats notwithstanding, the results in Figs. 1–3 and the accompanying review highlight the potential for integrating clinical neuroimaging analyses with ofMRI approaches to formulate and test hypotheses regarding latent, subtype-specific mechanisms underlying depression-related behavior. RCCA can be used to discover robust and stable latent associations between functional connectivity and behavior, linking specific circuits with specific clinical symptom combinations that may be differentially involved in individual MDD patients. ofMRI, in turn, provides a powerful tool for testing hypotheses derived from clinical neuroimaging data; for implicating specific patterns of network dysfunction as causal mechanisms, not just functional correlates; and for isolating the contributions of specific network nodes and circuits and studying their interactions.
Supplementary Material
Acknowledgments.
The authors gratefully acknowledge Dr. Karl Deisseroth and Dr. Emily Ferenczi for granting permission to adapt selected figure panels from Ref. (55), for presentation in Fig. 3. They also thank Amanda Buch for assistance with illustrating Fig. 3. This work was supported by grants from the National Institute of Mental Health, the One Mind Institute, the Klingenstein-Simons Foundation Fund, the Rita Allen Foundation, the Whitehall Foundation, the Dana Foundation, the Brain and Behavior Research Foundation (NARSAD), and the Hartwell Foundation. L.G. was supported by the Simons Foundation Society of Fellows. M.J.D. has received research grants from Neuronetics and Tal Medical, Inc. All other authors report no biomedical financial interests or other potential conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. (2010): Research Domain Criteria (RDoC): Toward a New Classification Framework for Research on Mental Disorders. Am J Psychiatry. 167: 748–751. [DOI] [PubMed] [Google Scholar]
- 2.Insel TR, Cuthbert BN (2015): Brain disorders? Precisely. Science 348: 499–500. [DOI] [PubMed] [Google Scholar]
- 3.Oathes DJ, Patenaude B, Schatzberg AF, Etkin A (2015): Neurobiological Signatures of Anxiety and Depression in Resting-State Functional Magnetic Resonance Imaging. Biol Psychiatry. 77: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodkind M, Eickhoff SB, Oathes DJ, Jiang Y, Chang A, Jones-Hagata LB, et al. (2015): Identification of a Common Neurobiological Substrate for Mental Illness. JAMA Psychiatry. 2015/02/05. doi: 10.1001/jamapsychiatry.2014.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson RJ, Pizzagalli D, Nitschke JB, Putnam K (2002): Depression: Perspectives from affective neuroscience. Annu Rev Psychol. 53: 545–574. [DOI] [PubMed] [Google Scholar]
- 6.Pizzagalli DA (2014): Depression, Stress, and Anhedonia: Toward a Synthesis and Integrated Model In: Cannon TD, Widiger T, editors. Annual Review of Clinical Psychology, Vol 10, Annual Review of Clinical Psychology. (Vol. 10), pp 393–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong ML, Kling MA, Munson PJ, Listwak S, Licinio J, Prolo P, et al. (2000): Pronounced and sustained central hypernoradrenergic function in major depression with melancholic features: relation to hypercortisolism and corticotropin-releasing hormone. Proc Natl Acad Sci U S A. 97: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold PW, Chrousos GP (2002): Organization of the stress system and its dysregulation in melancholic and atypical depression: high vs low CRH/NE states. Mol Psychiatry. 7: 254–275. [DOI] [PubMed] [Google Scholar]
- 9.Carroll BJ, Feinberg M, Greden JF, Tarika J, Albala AA, Haskett RF, et al. (1981): A SPECIFIC LABORATORY TEST FOR THE DIAGNOSIS OF MELANCHOLIA-STANDARDIZATION, VALIDATION, AND CLINICAL UTILITY. Arch Gen Psychiatry. 38: 15–22. [DOI] [PubMed] [Google Scholar]
- 10.Lewy AJ, Sack RL, Miller LS, Hoban TM (1987): Antidepressant and Circadian Phase-Shifting Effects of Light. Science. 235: 352–354. [DOI] [PubMed] [Google Scholar]
- 11.Schatzberg AF, Posener JA, DeBattista C, Kalehzan BM, Rothschild AJ, Shear PK (2000): Neuropsychological deficits in psychotic versus nonpsychotic major depression and no mental illness. Am J Psychiatry. 157: 1095–1100. [DOI] [PubMed] [Google Scholar]
- 12.Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD, et al. (2016): Identification of Distinct Psychosis Biotypes Using Brain-Based Biomarkers. Am J Psychiatry. 173: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill SK, Reilly JL, Keefe RSE, Gold JM, Bishop JR, Gershon ES, et al. (2013): Neuropsychological impairments in schizophrenia and psychotic bipolar disorder: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study. Am J Psychiatry. 170: 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999): Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 23: 185–188. [DOI] [PubMed] [Google Scholar]
- 15.Baron-Cohen S, Wheelwright S (2004): The empathy quotient: an investigation of adults with Asperger syndrome or high functioning autism, and normal sex differences. J Autism Dev Disord. 34: 163–175. [DOI] [PubMed] [Google Scholar]
- 16.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. (2007): Strong association of de novo copy number mutations with autism. Science. 316: 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. (2010): Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 466: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. (2017): Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 23: 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price RB, Gates K, Kraynak TE, Thase ME, Siegle GJ (2017): Data-Driven Subgroups in Depression Derived from Directed Functional Connectivity Paths at Rest. Neuropsychopharmacology. 42: 2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price RB, Lane S, Gates K, Kraynak TE, Horner MS, Thase ME, Siegle GJ (2017): Parsing Heterogeneity in the Brain Connectivity of Depressed and Healthy Adults During Positive Mood. Biol Psychiatry. 81: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia CH, Ma Z, Ciric R, Gu S, Betzel RF, Kaczkurkin AN, et al. (2018): Linked dimensions of psychopathology and connectivity in functional brain networks. Nat Commun. 9: 3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grisanzio KA, Goldstein-Piekarski AN, Wang MY, Rashed Ahmed AP, Samara Z, Williams LM (2018): Transdiagnostic Symptom Clusters and Associations With Brain, Behavior, and Daily Function in Mood, Anxiety, and Trauma Disorders. JAMA Psychiatry. 75: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, et al. (2007): Resting-state functional connectivity in major depression: Abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry. 62: 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pezawas L, Meyer-Lindenberg A, Drabant EM, Verchinski BA, Munoz KE, Kolachana BS, et al. (2005): 5-HTTLPR polymorphism impacts human cingulate-amygdala interactions: a genetic susceptibility mechanism for depression. Nat Neurosci. 8: 828–834. [DOI] [PubMed] [Google Scholar]
- 25.Sheline YI, Price JL, Yan Z, Mintun MA (2010): Resting-state functional MRI in depression unmasks increased connectivity between networks via the dorsal nexus. Proc Natl Acad Sci U S A. 107: 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheline YI, Barch DM, Price JL, Rundle MM, Vaishnavi SN, Snyder AZ, et al. (2009): The default mode network and self-referential processes in depression. Proc Natl Acad Sci U S A. 106: 1942–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutson B, Bhanji JP, Cooney RE, Atlas Lauren Y., Gotlib IH (2008): Neural responses to monetary incentives in major depression. Biol Psychiatry. 63: 686–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liston C, Chen AC, Zebley BD, Drysdale AT, Gordon R, Leuchter B, et al. (2014): Default Mode Network Mechanisms of Transcranial Magnetic Stimulation in Depression. Biol Psychiatry. 76: 517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga-Barke EJS (2009): Default-mode brain dysfunction in mental disorders: a systematic review. Neurosci Biobehav Rev. 33: 279–296. [DOI] [PubMed] [Google Scholar]
- 30.Anand A, Li Y, Wang Y, Wu J, Gao S, Bukhari L, et al. (2005): Activity and connectivity of brain mood regulating circuit in depression: a functional magnetic resonance study. Biol Psychiatry. 57: 1079–1088. [DOI] [PubMed] [Google Scholar]
- 31.Kaiser RH, Andrews-Hanna JR, Wager TD, Pizzagalli DA (2015): Large-Scale Network Dysfunction in Major Depressive Disorder: A Meta-analysis of Resting-State Functional Connectivity. JAMA Psychiatry. 72: 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McTeague LM, Huemer J, Carreon DM, Jiang Y, Eickhoff SB, Etkin A (2017): Identification of Common Neural Circuit Disruptions in Cognitive Control Across Psychiatric Disorders. Am J Psychiatry. 174: 676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etkin A (2018): Addressing the Causality Gap in Human Psychiatric Neuroscience. JAMA Psychiatry. 75: 3–4. [DOI] [PubMed] [Google Scholar]
- 34.Dinga R, Schmaal L, Penninx B, van Tol MJ, Veltman D, van Velzen L, et al. (2018, September 14): Evaluating the evidence for biotypes of depression: attempted replication of Drysdale et al. 2017 bioRxiv. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efron B (2010): Correlated z-values and the accuracy of large-scale statistical estimates. J Am Stat Assoc. 105: 1042–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotelling H (1936): Relations Between Two Sets of Variates. Biometrika. 28: 321–377. [Google Scholar]
- 37.Hardoon DR, Szedmak S, Shawe-Taylor J (2004): Canonical correlation analysis: an overview with application to learning methods. Neural Comput. 16: 2639–2664. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez I, Dejean S (n.d.): CCA: An R Package to Extend Canonical Correlation Analysis. . Retrieved from https://core.ac.uk/download/pdf/6303071.pdf.
- 39.Efron B, Tibshirani RJ (1994): An Introduction to the Bootstrap. CRC Press. [Google Scholar]
- 40.Cohen J (1988): Statistical power analysis for the behavioral sciences 2nd edn. .
- 41.Hoerl AE, Kennard RW (1970): Ridge Regression: Biased Estimation for Nonorthogonal Problems. Technometrics. 12: 55–67. [Google Scholar]
- 42.Hastie T, Tibshirani R, Friedman J (2009): The Elements of Statistical Learning: Data Mining, Inference, and Prediction, Second Edition, 2nd ed Springer Series in Statistics. Springer-Verlag; New York. [Google Scholar]
- 43.Karalunas SL, Fair D, Musser ED, Aykes K, Iyer SP, Nigg JT (2014): Subtyping attention-deficit/hyperactivity disorder using temperament dimensions: toward biologically based nosologic criteria. JAMA Psychiatry. 71: 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Cao H, Chén OY, Chung Y, Forsyth JK, McEwen SC, Gee DG, et al. (2018): Cerebellothalamo-cortical hyperconnectivity as a state-independent functional neural signature for psychosis prediction and characterization. Nat Commun. 9: 3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z, Xu Y, Xu T, Hoy CW, Handwerker DA, Chen G, et al. (2014): Brain network informed subject community detection in early-onset schizophrenia. Sci Rep. 4: 5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K (2005): Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 8: 1263–1268. [DOI] [PubMed] [Google Scholar]
- 47.Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, et al. (2009): Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 459: 663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F, Wang L-P, Brauner M, Liewald JF, Kay K, Watzke N, et al. (2007): Multimodal fast optical interrogation of neural circuitry. Nature. 446: 633–639. [DOI] [PubMed] [Google Scholar]
- 49.Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, et al. (2011): Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 477: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, et al. (2010): High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 463: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grosenick L, Marshel JH, Deisseroth K (2015): Closed-loop and activity-guided optogenetic control. Neuron. 86: 106–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L (2007): Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 450: 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K (2009): Optical deconstruction of parkinsonian neural circuitry. Science. 324: 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kravitz AV, Freeze BS, Parker PRL, Kay K, Thwin MT, Deisseroth K, Kreitzer AC (2010): Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 466: 622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferenczi EA, Zalocusky KA, Liston C, Grosenick L, Warden MR, Amatya D, et al. (2016): Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 351: aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Covington HE, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, et al. (2010): Antidepressant Effect of Optogenetic Stimulation of the Medial Prefrontal Cortex. Journal of Neuroscience. 30: 16082–16090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaudhury D, Walsh JJ, Friedman AK, Juarez B, Ku SM, Koo JW, et al. (2013): Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature. 493: 532–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tye KM, Prakash R, Kim SY, Fenno LE, Grosenick L, Zarabi H, et al. (2011): Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 471: 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM, et al. (2012): Input-specific control of reward and aversion in the ventral tegmental area. Nature. 491: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stuber GD, Sparta DR, Stamatakis AM, van Leeuwen WA, Hardjoprajitno JE, Cho S, et al. (2011): Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature. 475: 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ciocchi S, Herry C, Grenier F, Wolff SBE, Letzkus JJ, Vlachos I, et al. (2010): Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 468: 277–282. [DOI] [PubMed] [Google Scholar]
- 62.Lee JH, Durand R, Gradinaru V, Zhang F, Goshen I, Kim D-S, et al. (2010): Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 465: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kahn I, Knoblich U, Desai M, Bernstein J, Graybiel AM, Boyden ES, et al. (2013): Optogenetic drive of neocortical pyramidal neurons generates fMRI signals that are correlated with spiking activity. Brain Res. 1511: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu X, He Y, Wang M, Merkle H, Dodd SJ, Silva AC, Koretsky AP (2016): Sensory and optogenetically driven single-vessel fMRI. Nat Methods. 13: 337–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vazquez AL, Fukuda M, Kim S-G (2018): Inhibitory Neuron Activity Contributions to Hemodynamic Responses and Metabolic Load Examined Using an Inhibitory Optogenetic Mouse Model. Cereb Cortex. 28: 4105–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takata N, Sugiura Y, Yoshida K, Koizumi M, Hiroshi N, Honda K, et al. (2018): Optogenetic astrocyte activation evokes BOLD fMRI response with oxygen consumption without neuronal activity modulation. Glia. 66: 2013–2023. [DOI] [PubMed] [Google Scholar]
- 67.Liang Z, Watson GDR, Alloway KD, Lee G, Neuberger T, Zhang N (2015): Mapping the functional network of medial prefrontal cortex by combining optogenetics and fMRI in awake rats. Neuroimage. 117: 114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan RW, Leong ATL, Ho LC, Gao PP, Wong EC, Dong CM, et al. (2017): Low-frequency hippocampal-cortical activity drives brain-wide resting-state functional MRI connectivity. Proc Natl Acad Sci U S A. 114: E6972–E6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leong ATL, Chan RW, Gao PP, Chan Y-S, Tsia KK, Yung W-H, Wu EX (2016): Long-range projections coordinate distributed brain-wide neural activity with a specific spatiotemporal profile. Proc Natl Acad Sci U S A. 113: E8306–E8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weitz AJ, Fang Z, Lee HJ, Fisher RS, Smith WC, Choy M, et al. (2015): Optogenetic fMRI reveals distinct, frequency-dependent networks recruited by dorsal and intermediate hippocampus stimulations. Neuroimage. 107: 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takata N, Yoshida K, Komaki Y, Xu M, Sakai Y, Hikishima K, et al. (2015): Optogenetic Activation of CA1 Pyramidal Neurons at the Dorsal and Ventral Hippocampus Evokes Distinct Brain-Wide Responses Revealed by Mouse fMRI. PLOS One. 10: e0121417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benekareddy M, Stachniak TJ, Bruns A, Knoflach F, von Kienlin M, Künnecke B, Ghosh A (2018): Identification of a Corticohabenular Circuit Regulating Socially Directed Behavior. Biol Psychiatry. 83: 607–617. [DOI] [PubMed] [Google Scholar]
- 73.Bernal-Casas D, Lee HJ, Weitz AJ, Lee JH (2017): Studying Brain Circuit Function with Dynamic Causal Modeling for Optogenetic fMRI. Neuron. 93: 522–532.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee HJ, Weitz AJ, Bernal-Casas D, Duffy BA, Choy M, Kravitz AV, et al. (2016): Activation of Direct and Indirect Pathway Medium Spiny Neurons Drives Distinct Brain-wide Responses. Neuron. 91: 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brocka M, Helbing C, Vincenz D, Scherf T, Montag D, Goldschmidt J, et al. (2018): Contributions of dopaminergic and non-dopaminergic neurons to VTA-stimulation induced neurovascular responses in brain reward circuits. Neuroimage. 177: 88–97. [DOI] [PubMed] [Google Scholar]
- 76.Grandjean J, Corcoba A, Kahn MC, Upton AL, Deneris ES, Seifritz E, et al. (2019): A brain-wide functional map of the serotonergic responses to acute stress and fluoxetine. Nat Commun. 10: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han Z, Chen W, Chen X, Zhang K, Tong C, Zhang X, et al. (2019): Awake and behaving mouse fMRI during Go/No-Go task. Neuroimage. 188: 733–742. [DOI] [PubMed] [Google Scholar]
- 78.Desjardins M, Kılıç K, Thunemann M, Mateo C, Holland D, Ferri CGL, et al. (2018): Awake Mouse Imaging: From Two-Photon Microscopy to Blood Oxygen-Level Dependent Functional Magnetic Resonance Imaging. Biol Psychiatry Cogn Neurosci Neuroimaging. . doi: 10.1016/j.bpsc.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wager TD, Davidson ML, Hughes BL, Lindquist MA, Ochsner KN (2008): Prefrontal-subcortical pathways mediating successful emotion regulation. Neuron. 59: 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Milad MR, Quirk GJ (2002): Neurons in medial prefrontal cortex signal memory for fear extinction. Nature. 420: 70–74. [DOI] [PubMed] [Google Scholar]
- 81.Phelps EA, Delgado MR, Nearing KI, LeDoux JE (2004): Extinction learning in humans: role of the amygdala and vmPFC. Neuron. 43: 897–905. [DOI] [PubMed] [Google Scholar]
- 82.Ochsner KN, Gross JJ (2005): The cognitive control of emotion. Trends Cogn Sci. 9: 242–249. [DOI] [PubMed] [Google Scholar]
- 83.Pizzagalli DA, Holmes AJ, Dillon DG, Goetz EL, Birk JL, Bogdan R, et al. (2009): Reduced Caudate and Nucleus Accumbens Response to Rewards in Unmedicated Individuals With Major Depressive Disorder. Am J Psychiatry. 166: 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kvitsiani D, Ranade S, Hangya B, Taniguchi H, Huang JZ, Kepecs A (2013): Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature. 498: 363–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hillman KL, Bilkey DK (2012): Neural encoding of competitive effort in the anterior cingulate cortex. Nat Neurosci. 15: 1290–1297. [DOI] [PubMed] [Google Scholar]
- 86.Croxson PL, Walton ME, O’Reilly JX, Behrens TEJ, Rushworth MFS (2009): Effort-based cost-benefit valuation and the human brain. J Neurosci. 29: 4531–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Treadway MT, Bossaller NA, Shelton RC, Zald DH (2012): Effort-based decision-making in major depressive disorder: a translational model of motivational anhedonia. J Abnorm Psychol. 121: 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schultz W, Dayan P, Montague PR (1997): A neural substrate of prediction and reward. Science. 275: 1593–1599. [DOI] [PubMed] [Google Scholar]
- 89.Cardinal RN, Parkinson JA, Hall J, Everitt BJ (2002): Emotion and motivation: the role of the amygdala, ventral striatum, and prefrontal cortex. Neurosci Biobehav Rev. 26: 321–352. [DOI] [PubMed] [Google Scholar]
- 90.Tervo DGR, Hwang B-Y, Viswanathan S, Gaj T, Lavzin M, Ritola KD, et al. (2016): A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 92: 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo L, Callaway EM, Svoboda K (2018): Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron. 98: 256–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, et al. (2015): Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell. 162: 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anand A, Li Y, Wang Y, Gardner K, Lowe MJ (2007): Reciprocal effects of antidepressant treatment on activity and connectivity of the mood regulating circuit: An fMRI study. J Neuropsychiatry Clin Neurosci. 19: 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anand A, Li Y, Wang Y, Wu JW, Gao SJ, Bukhari L, et al. (2005): Antidepressant effect on connectivity of the mood-regulating circuit: An fMRI study. Neuropsychopharmacology. 30: 1334–1344. [DOI] [PubMed] [Google Scholar]
- 95.Scheidegger M, Walter M, Lehmann M, Metzger C, Grimm S, Boeker H, et al. (2012): Ketamine decreases resting state functional network connectivity in healthy subjects: implications for antidepressant drug action. PLoS One. 7: e44799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McCabe C, Mishor Z (2011): Antidepressant medications reduce subcortical-cortical resting-state functional connectivity in healthy volunteers. Neuroimage. 57: 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Posner J, Hellerstein DJ, Gat I, Mechling A, Klahr K, Wang Z, et al. (2013): Antidepressants normalize the default mode network in patients with dysthymia. JAMA Psychiatry. 70: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L, Xia M, Li K, Zeng Y, Su Y, Dai W, et al. (2015): The effects of antidepressant treatment on resting-state functional brain networks in patients with major depressive disorder. Hum Brain Mapp. 36: 768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gozzi A, Schwarz AJ (2016): Large-scale functional connectivity networks in the rodent brain. Neuroimage. 127: 496–509. [DOI] [PubMed] [Google Scholar]
- 100.Lu H, Zuo Y, Gu H, Waltz JA, Zhan W, Scholl CA, et al. (2007): Synchronized delta oscillations correlate with the resting-state functional MRI signal. Proc Natl Acad Sci U S A. 104: 18265–18269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zuo X-N, Di Martino A, Kelly C, Shehzad ZE, Gee DG, Klein DF, et al. (2010): The oscillating brain: complex and reliable. Neuroimage. 49: 1432–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hutchison RM, Mirsattari SM, Jones CK, Gati JS, Leung LS (2010): Functional networks in the anesthetized rat brain revealed by independent component analysis of resting-state FMRI. J Neurophysiol. 103: 3398–3406. [DOI] [PubMed] [Google Scholar]
- 103.Lu H, Zou Q, Gu H, Raichle ME, Stein EA, Yang Y (2012): Rat brains also have a default mode network. Proc Natl Acad Sci U S A. 109: 3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liang Z, King J, Zhang N (2012): Anticorrelated resting-state functional connectivity in awake rat brain. Neuroimage. 59: 1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grandjean J, Schroeter A, Batata I, Rudin M (2014): Optimization of anesthesia protocol for resting-state fMRI in mice based on differential effects of anesthetics on functional connectivity patterns. Neuroimage. 102 Pt 2: 838–847. [DOI] [PubMed] [Google Scholar]
- 106.Stafford JM, Jarrett BR, Miranda-Dominguez O, Mills BD, Cain N, Mihalas S, et al. (2014): Large-scale topology and the default mode network in the mouse connectome. Proc Natl Acad Sci U S A. 111: 18745–18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ongur D, Price JL (2000): The organization of networks within the orbital and medial prefrontal cortex of rats, monkeys and humans. Cereb Cortex. 10: 206–219. [DOI] [PubMed] [Google Scholar]
- 108.Buckner RL, Krienen FM (2013): The evolution of distributed association networks in the human brain. Trends Cogn Sci. 17: 648–665. [DOI] [PubMed] [Google Scholar]
- 109.Nestler EJ, Hyman SE (2010): Animal models of neuropsychiatric disorders. Nat Neurosci. 13: 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Redish AD (2004): Addiction as a computational process gone awry. Science. 306: 1944–1947. [DOI] [PubMed] [Google Scholar]
- 111.Sweis BM, Abram SV, Schmidt BJ, Seeland KD, MacDonald AW, Thomas MJ, Redish AD (2018): Sensitivity to “sunk costs” in mice, rats, and humans. Science. 361: 178–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruff CC, Blankenburg F, Bjoertomt O, Bestmann S, Freeman E, Haynes J-D, et al. (2006): Concurrent TMS-fMRI and psychophysics reveal frontal influences on human retinotopic visual cortex. Curr Biol. 16: 1479–1488. [DOI] [PubMed] [Google Scholar]
- 113.Hanlon CA, Dowdle LT, Moss H, Canterberry M, George MS (2016): Mobilization of Medial and Lateral Frontal-Striatal Circuits in Cocaine Users and Controls: An Interleaved TMS/BOLD Functional Connectivity Study. Neuropsychopharmacology. 41: 3032–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen AC, Oathes DJ, Chang C, Bradley T, Zhou Z-W, Williams LM, et al. (2013): Causal interactions between fronto-parietal central executive and default-mode networks in humans. Proc Natl Acad Sci U S A. 110: 19944–19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fox MD (2018): Mapping Symptoms to Brain Networks with the Human Connectome. N Engl J Med. 379: 2237–2245. [DOI] [PubMed] [Google Scholar]
- 116.Carp J (2013): Optimizing the order of operations for movement scrubbing: Comment on Power et al. Neuroimage. 76: 436–438. [DOI] [PubMed] [Google Scholar]
- 117.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE (2013): Steps toward optimizing motion artifact removal in functional connectivity MRI; a reply to Carp. Neuroimage. 76: 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. (2011): Functional Network Organization of the Human Brain. Neuron. 72: 665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Holm S (1979): A Simple Sequentially Rejective Multiple Test Procedure. Scand Stat Theory Appl. 6: 65–70. [Google Scholar]
- 120.Grosenick L, Klingenberg B, Katovich K, Knutson B, Taylor JE (2013): Interpretable whole-brain prediction analysis with GraphNet. Neuroimage. 72: 304–321. [DOI] [PubMed] [Google Scholar]
- 121.Hunter JD (2007): Matplotlib: A 2D Graphics Environment. Comput Sci Eng. 9: 90–95. [Google Scholar]
- 122.Perez F, Granger BE (2007): IPython: A System for Interactive Scientific Computing. Computing in Science Engineering. 9: 21–29. [Google Scholar]
- 123.Oliphant TE (2007): SciPy: Open source scientific tools for Python. Computing in Science and Engineering. 9: 10–20. [Google Scholar]
- 124.Oliphant TE (2006): A guide to NumPy. (Vol. 1), Trelgol Publishing USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.