Abstract
Recently, MR simulators gain popularity because of unnecessary radiation exposure of CT simulators being used in radiation therapy planning. We propose a method for pseudo CT estimation from MR images based on a patch-based random forest. Patient-specific anatomical features are extracted from the aligned training images and adopted as signatures for each voxel. The most robust and informative features are identified using feature selection to train the random forest. The well-trained random forest is used to predict the pseudo CT of a new patient. This prediction technique was tested with human brain images and the prediction accuracy was assessed using the original CT images. Peak signal-to-noise ratio (PSNR) and feature similarity (FSIM) indexes were used to quantify the differences between the pseudo and original CT images. The experimental results showed the proposed method could accurately generate pseudo CT images from MR images. In summary, we have developed a new pseudo CT prediction method based on patch-based random forest, demonstrated its clinical feasibility, and validated its prediction accuracy. This pseudo CT prediction technique could be a useful tool for MRI-based radiation treatment planning and attenuation correction in a PET/MRI scanner.
Keywords: Pseudo CT, MRI, random forest, patch
1. INTRODUCTION
Computed tomography (CT), with its high availability and geometric accuracy, has proven an invaluable tool for cancer diagnosis and treatment. Since a CT voxel’s intensity directly relates to its attenuation of X-rays, CT images provide a means of estimating tissue electron density, which, in turn, is necessary to simulate radiation transport. For this reason, CT is a cornerstone of both radiotherapy planning and attenuation correction of Positron Emission Tomography (PET) images. Recently, there has been increasing interest in replacing CT with magnetic resonance (MR) in MRI-only radiotherapy treatment planning as well as in PET/MR attenuation correction. The first application is in certain types of radiotherapy planning, where CT’s limited soft tissue contrast stunts accurate tumor delineation whereas MR provides valuable guidance [1]; such cases include stereotactic radiosurgery of brain disorders and radiotherapy treatment of prostate cancer [2, 3]. The second application is in combined PET/MR imaging currently transitioning to reach its full clinical potential, where attenuation correction is necessary but CT acquisition would require an additional scan [4]. Therefore, there is an increasing clinical need to reliably estimate electron densities (called pseudo CT images) in these applications based on the MR images. The benefits of pseudo CT images include reducing medical cost, sparing the patient from X-ray exposure, eliminating registration errors between MR and CT images and simplifying clinical workflow. These issues greatly motivate the development of methods for estimate of CT image from the corresponding MR image.
Several methods have been investigated to estimate electron density information using MRI [5–19], which can be characterized into four categories: segmentation-based, atlas-based, pseudo-CT based and sequence-based methods. Each has its advantages and limitations. (1) In segmentation-based methods [6, 14, 16, 17, 19, 20], MR images are manually or automatically segmented into different tissue types (bone, soft tissue and air) with predefined electron density or attenuation coefficient. The main drawback of these approaches is the inter-observer inconsistency of the manual contours or the segmentation errors of the auto-contours [21]. (2) In atlas-based methods [15, 22], a deformable image registration is used to map a sample patient or atlas with known tissue label or electron density (typically derived from CT) to an individual’s MR image. The electron density map for this individual can then be obtained. These methods could suffer from inherent registration errors due to inter-patient anatomical differences. (3) In pseudo-CT based methods [5, 8, 10, 12, 18], tissue properties are directly characterized based on the MR image intensity and anatomy. However, due to the lack of a one-to-one relation between electron density and MR image intensity, this approach could lead to ambiguous results. In particular, the differentiation of bone from air has been challenging because of their similar and short T2 characteristics (both appear dark on MRI) [23]. (4) In MRI sequence-based methods [9, 13, 24], MR sequences such as the ultra-short echo time (UTE) sequence are used to visualize bony anatomy. However, the currently available image quality of UTE imaging is still far from satisfactory. Moreover, the non-standard MR sequence also adds considerable scan time, which may introduce more patient motion and discomfort. In this study we proposed to develop a learning-based method to derive electron density from standard clinical MRI. In this approach we integrated patch-based anatomical signature into machine learning framework to predict pseudo CT using MRI.
2. METHODS
Suppose we have a set of pairs of MR and CT training images. For each pair, the CT image is used as the regression target of the MR image. We further assume that the training data have been preprocessed by removing noise and uninformative regions, and have been aligned. The proposed prediction method consists of two major stages: the training stage and the prediction stage. During the training stage, patch-based anatomical features are extracted from the registered training images with patient-specific information, and the most robust and informative CT-MR features are identified by feature selection to train a random forest. During the prediction stage, we extract the selected features from the new (target) MR image and feed them into the well-trained forests for CT image prediction. The four major steps are briefly described below.
2.1. Image Alignment for Dataset
Before doing prediction, some preprocessing is performed for all CT and MR images in dataset, which includes noise reduction, inhomogeneity correction, and inter-subject intensity normalization. Such preprocessing steps are to improve the accuracy of the following prediction. The intra-subject registration is used to align each pair of CT and MR images of the same subject. We perform rigid and deformable registration with B-Splines [25], using mutual information as a similarity measure. The inter-subject registration is used to align each CT-MR pair images among the different subjects. During the inter-subject alignment processing of training set, we first select one MR-CT pair image as the template, and align other MR images to the template MR image. We use the corresponding transformation obtained from training MR image alignment to align the corresponding CT images of each subject to the template CT images. Since the CT images of each training image are available, we again align each training CT image to the template CT images to optimize the alignment of the training set, and then apply the corresponding transformation obtained from training CT image alignment to align the corresponding MR images to the template MR images. For each newly acquired MR image, all aligned training images in training set are rigidly registered to this new image. The rigid registration is used to obtain the spatial deformation field between the new MR image and training MR images. The same transformations are applied to the CT in training set.
2.2. Patch-based Feature Extraction
Patch-based representation has been widely used as voxel anatomical signature in computer vision and medical image analysis [5, 26–28]. The principle of the conventional patch-based representation is to first define a small image patch centered at each voxel and then use the voxel intensities of image patch as the anatomical signature of each voxel. However, due to the noise and anatomical complexity of MR images, patch-based representation using voxel intensities alone may not be able to effectively distinguish the voxels of different tissue in MRI. We propose to use a patch-based anatomical features are used as signature for each voxel to characterize the image appearance. Two types of images features – the Gabor wavelet feature and the local binary pattern (LBP) feature – are extracted from a small image patch centered at each voxel of each aligned training image. Gabor can provide complementary anatomical information to each other, and LBP can capture texture information from the input image. The 16 Gabor feature is used in this study. The LBP feature is extracted in three resolution levels and it has a dimension 30. Therefore, for each voxel, it is represented by a 46 dimensional feature signature.
2.3. Patch Preselection Based on Features
Patch preselection is applied here among all candidate patches, in order to reduce the computational burden and improve prediction accuracy by excluding irrelevant patches [26]. Based on the above features, we can obtain patch-based representation of each voxel. It should be noted that the patch-based anatomical signature may contain noisy and redundant features which will affect the performance in excluding the irrelevant patches. Therefore, feature selection should be performed to identify the most informative and salient features in the anatomical signature of each voxel [28]. The aim of feature selection is to select a small subset of most informative feature as anatomical signature, which can be well accomplished by enforcing the sparsity constraint during the regression process. Therefore, the feature selection problem can be formulated as a logistic sparse least-absolute- shrinkage-and-selection operator (LASSO) problem [29]. It is defined as,
| (1) |
where denotes the original feature signature of voxel . is the sparse coefficient vector, is the l1 norm, b is the intercept scalar, and λ is the regularization parameter. The first term is obtained by inputting the values of drawn samples and their original feature signatures to the function, and then takes the logarithm for maximum likelihood estimation. The second term is the l1 norm which aims to enforce the sparsity constraint for LASSO. Through minimizing the sparse LASSO energy function the features with superior discriminant power are selected. Based on the selected features, we can directly measure their discriminant power to exclude the irrelevant patches, based on the Fisher’s score.
2.4. Random Forest Training and Pseudo CT Prediction
The random forest classifier is a learning-based method for classification and regression problems, and has been widely adopted in medical imaging applications [30]. A random forest comprises of multiple decision trees. At each internal node of a tree, a feature is chosen to split the incoming training samples to maximize the information gain. Specifically, let be an input feature vector, and be its corresponding target value for regression. For a given internal node j and a set of samples , the information gain achieved by choosing the kth feature to split the samples in the regression problem is computed by:
| (2) |
| (3) |
| (4) |
where L and R denote the left and right child nodes, , , uk is the kth feature of feature vector u, is the splitting threshold chosen to maximize the information gain for the kth feature uk, and |•| is the cardinality of the set. H (S) denotes the variance of all target values in our regression problem. During the training stage, the splitting process is performed recursively until the information gain is not significant, or the number of training samples falling into one node is less than a pre-defined threshold. In the testing stage, to predict the CT image, the new given MR image follows the same sequence as learned during the training stage, to generate the final prediction.
3. EXPERIMENTS AND RESULTS
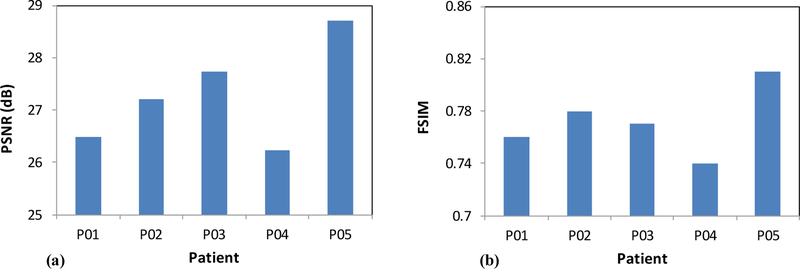
In order to test the proposed method, we applied our method to5 patients’ MR and CT data. All patients’ MR data were acquired using a Siemens MR scanner. Each 3D T1W MR data sets consisted of 256×256×147 voxels to cover the whole head and the voxel size was 1.0×1.0×1.0 mm3. All patients’ CT data were acquired using a Siemens CT scanner. Each 3D CT data sets consisted of 512×512×160 voxels to cover the whole head and the voxel size was 0.8×0.8×1.0 mm3. We performed leave-one-out cross-validation method to evaluate the proposed CT prediction algorithm. In other words, we used the 4 CT and MRI images as the training set and applied the proposed method to predict the remaining subject. Our pseudo CT were compared with the original CT images. In order to get a quantitative evaluation, we used peak signal-to-noise ratio (PSNR) [31] and feature similarity (FSIM) index [32]. Figure 1 shows an example of MR-based pseudo CT predicted by the proposed method. This pseudo CT is close to original CT images. Figure 2 shows the PSNR and FSIM between the pseudo and original CT for each patient. Overall the mean PSNR and FSIM were 27.27±1.00 and 0.77±0.03, which demonstrated the prediction accuracy of the proposed learning-based method.
Figure 1.
MR-based pseudo CT prediction results. Images from left to right are MRI, CT, pseudo CT and the difference between pseudo and original CT images.
Figure 2.
PSNR (a) and FSIM (b) between the pseudo and original CT for each patient.
4. CONCLUSION
In this paper, we propose a novel learning-based approach to predict a pseudo CT image from a MR image based on a random forest regression with a patch-based anatomical signature to effectively capture the relationship between the CT and MR images. We have demonstrated that the proposed method is capable of reliably predicting the CT image from the MR image. This pseudo CT prediction technique could be a useful tool for MRI-based radiation treatment planning and attenuation correction in PET/MRI scanner.
ACKNOWLEDGEMENTS
This research is supported in part by the Department of Defense (DoD) Prostate Cancer Research Program (PCRP) Award W81XWH-13–1-0269 and Dunwoody Golf Club Prostate Cancer Research Award, a philanthropic award provided by the Winship Cancer Institute of Emory University.
REFERENCES
- [1].Sjolund J, Forsberg D, Andersson M et al. , “Generating patient specific pseudo-CT of the head from MR using atlas-based regression,” Physics in Medicine and Biology, 60(2), 825–839 (2015). [DOI] [PubMed] [Google Scholar]
- [2].Stanescu T, Jans HS, Pervez N et al. , “A study on the magnetic resonance imaging (MRI)-based radiation treatment planning of intracranial lesions,” Physics in Medicine and Biology, 53(13), 3579–3593 (2008). [DOI] [PubMed] [Google Scholar]
- [3].Uha J, Merchant TE, Li YM et al. , “MRI-based treatment planning with pseudo CT generated through atlas registration,” Medical Physics, 41(5), (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bezrukov I, Mantlik F, Schmidt H et al. , “MR-Based PET Attenuation Correction for PET/MR Imaging,” Seminars in Nuclear Medicine, 43(1), 45–59 (2013). [DOI] [PubMed] [Google Scholar]
- [5].Andreasen D, Van Leemput K, Hansen RH et al. , “Patch-based generation of a pseudo CT from conventional MRI sequences for MRI-only radiotherapy of the brain,” Medical Physics, 42(4), 1596–1605 (2015). [DOI] [PubMed] [Google Scholar]
- [6].Berker Y, Franke J, Salomon A et al. , “MRI-Based Attenuation Correction for Hybrid PET/MRI Systems: A 4-Class Tissue Segmentation Technique Using a Combined Ultrashort-Echo-Time/Dixon MRI Sequence,” Journal of Nuclear Medicine, 53(5), 796–804 (2012). [DOI] [PubMed] [Google Scholar]
- [7].Beyer T, Weigert M, Quick HH et al. , “MR-based attenuation correction for torso-PET/MR imaging: pitfalls in mapping MR to CT data,” European Journal of Nuclear Medicine and Molecular Imaging, 35(6), 1142–1146 (2008). [DOI] [PubMed] [Google Scholar]
- [8].Burgos N, Cardoso MJ, Thielemans K et al. , “Attenuation Correction Synthesis for Hybrid PET-MR Scanners: Application to Brain Studies,” Ieee Transactions on Medical Imaging, 33(12), 2332–2341 (2014). [DOI] [PubMed] [Google Scholar]
- [9].Catana C, van der Kouwe A, Benner T et al. , “Toward Implementing an MRI-Based PET Attenuation-Correction Method for Neurologic Studies on the MR-PET Brain Prototype,” Journal of Nuclear Medicine, 51(9), 1431–1438 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gudur MSR, Hara W, Le QT et al. , “A unifying probabilistic Bayesian approach to derive electron density from MRI for radiation therapy treatment planning,” Physics in Medicine and Biology, 59(21), 6595–6606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hofmann M, Steinke F, Scheel V et al. , “MRI-Based Attenuation Correction for PET/MRI: A Novel Approach Combining Pattern Recognition and Atlas Registration,” Journal of Nuclear Medicine, 49(11), 1875–1883 (2008). [DOI] [PubMed] [Google Scholar]
- [12].Huynh T, Gao YZ, Kang JY et al. , “Estimating CT Image From MRI Data Using Structured Random Forest and Auto-Context Model,” Ieee Transactions on Medical Imaging, 35(1), 174–183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Keereman V, Fierens Y, Broux T et al. , “MRI-Based Attenuation Correction for PET/MRI Using Ultrashort Echo Time Sequences,” Journal of Nuclear Medicine, 51(5), 812–818 (2010). [DOI] [PubMed] [Google Scholar]
- [14].Martinez-Moller A, Souvatzoglou M, Delso G et al. , “Tissue Classification as a Potential Approach for Attenuation Correction in Whole-Body PET/MRI: Evaluation with PET/CT Data,” Journal of Nuclear Medicine, 50(4), 520–526 (2009). [DOI] [PubMed] [Google Scholar]
- [15].Schreibmann E, Nye JA, Schuster DM et al. , “MR-based attenuation correction for hybrid PET-MR brain imaging systems using deformable image registration,” Medical Physics, 37(5), 2101–2109 (2010). [DOI] [PubMed] [Google Scholar]
- [16].Schulz V, Torres-Espallardo I, Renisch S et al. , “Automatic, three-segment, MR-based attenuation correction for whole-body PET/MR data,” European Journal of Nuclear Medicine and Molecular Imaging, 38(1), 138–152 (2011). [DOI] [PubMed] [Google Scholar]
- [17].Steinberg J, Jia G, Sammet S et al. , “Three-region MRI-based whole-body attenuation correction for automated PET reconstruction,” Nuclear Medicine and Biology, 37(2), 227–235 (2010). [DOI] [PubMed] [Google Scholar]
- [18].Torrado-Carvajal A, Herraiz JL, Alcain E et al. , “Fast Patch-Based Pseudo-CT Synthesis from T1-Weighted MR Images for PET/MR Attenuation Correction in Brain Studies,” Journal of Nuclear Medicine, 57(1), 136–143 (2016). [DOI] [PubMed] [Google Scholar]
- [19].Wagenknecht G, Kops ER, Tellmann L et al. , “Knowledge-based Segmentation of Attenuation-relevant Regions of the Head in T1-weighted MR Images for Attenuation Correction in MR/PET Systems,” 2009 Ieee Nuclear Science Symposium Conference Record, Vols 1–5, 3338–3343 (2009). [Google Scholar]
- [20].Fei BW, Yang XF, Nye JA et al. , “MR/PET quantification tools: Registration, segmentation, classification, and MR-based attenuation correction,” Medical Physics, 39(10), 6443–6454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang XF, and Fei BW, “Multiscale segmentation of the skull in MR images for MRI-based attenuation correction of combined MR/PET,” Journal of the American Medical Informatics Association, 20(6), 1037–1045 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kops ER, and Herzog H, “Alternative methods for attenuation correction for PET images in MR-PET scanners,” 2007 Ieee Nuclear Science Symposium Conference Record, Vols 1–11, 4327–4330 (2007). [Google Scholar]
- [23].Yang XF, and Fei BW, “A multiscale and multiblock fuzzy C-means classification method for brain MR images,” Medical Physics, 38(6), 2879–2891 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hsu SH, Cao Y, Lawrence TS et al. , “Quantitative characterizations of ultrashort echo (UTE) images for supporting air-bone separation in the head,” Physics in Medicine and Biology, 60(7), 2869–2880 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang XF, Wu N, Cheng GH et al. , “Automated Segmentation of the Parotid Gland Based on Atlas Registration and Machine Learning: A Longitudinal MRI Study in Head-and-Neck Radiation Therapy,” International Journal of Radiation Oncology Biology Physics, 90(5), 1225–1233 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang XF, Jani AB, Rossi PJ et al. , “Patch-Based Label Fusion for Automatic Multi-Atlas-Based Prostate Segmentation in MR Images,” Medical Imaging 2016: Image-Guided Procedures, Robotic Interventions, and Modeling, 9786, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang XF, Jani AB, Rossi PJ et al. , “A MRI-CT Prostate Registration Using Sparse Representation Technique,” Medical Imaging 2016: Image-Guided Procedures, Robotic Interventions, and Modeling, 9786, (2016). [Google Scholar]
- [28].Yang XF, Rossi PJ, Jani AB et al. , “3D Transrectal Ultrasound (TRUS) Prostate Segmentation Based on Optimal Feature Learning Framework,” Medical Imaging 2016: Image Processing, 9784, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Aseervatham S, Antoniadis A, Gaussier E et al. , “A sparse version of the ridge logistic regression for large-scale text categorization,” Pattern Recognition Letters, 32(2), 101–106 (2011). [Google Scholar]
- [30].Criminisi A, and Shotton J, [Decision Forests for Computer Vision and Medical Image Analysis] Springer Publishing Company, Incorporated, (2013). [Google Scholar]
- [31].Yang X, and Fei B, “A wavelet multiscale denoising algorithm for magnetic resonance (MR) images,” Meas Sci Technol, 22(2), 25803 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang L, Zhang L, Mou XQ et al. , “FSIM: A Feature Similarity Index for Image Quality Assessment,” Ieee Transactions on Image Processing, 20(8), 2378–2386 (2011). [DOI] [PubMed] [Google Scholar]