Abstract
Deficient progranulin levels cause dose-dependent neurological syndromes: haploinsufficiency leads to frontotemporal lobar degeneration (FTLD) and nullizygosity produces adult-onset neuronal ceroid lipofuscinosis. Mechanisms controlling progranulin levels are largely unknown. To better understand progranulin regulation, we performed a genome-wide RNAi screen using an ELISA-based platform to discover genes that regulate progranulin levels in neurons. We identified 830 genes that raise or lower progranulin levels by at least 1.5-fold in Neuro2a cells. When inhibited by siRNA or some by submicromolar concentrations of small-molecule inhibitors, 33 genes of the druggable genome increased progranulin levels in mouse primary cortical neurons; several of these also raised progranulin levels in FTLD model mouse neurons. “Hit” genes regulated progranulin by transcriptional or posttranscriptional mechanisms. Pathway analysis revealed enrichment of hit genes from the autophagy–lysosome pathway (ALP), suggesting a key role for this pathway in regulating progranulin levels. Progranulin itself regulates lysosome function. We found progranulin deficiency in neurons increased autophagy and caused abnormally enlarged lysosomes and boosting progranulin levels restored autophagy and lysosome size to control levels. Our data link the ALP to neuronal progranulin: progranulin levels are regulated by autophagy and, in turn, progranulin regulates the ALP. Restoring progranulin levels by targeting genetic modifiers reversed FTLD functional deficits, opening up potential opportunities for future therapeutics development.
SIGNIFICANCE STATEMENT Progranulin regulates neuron and immune functions and is implicated in aging. Loss of one functional allele causes haploinsufficiency and leads to frontotemporal lobar degeneration (FTLD), the second leading cause of dementia. Progranulin gene polymorphisms are linked to Alzheimer's disease (AD) and complete loss of function causes neuronal ceroid lipofuscinosis. Despite the critical role of progranulin levels in neurodegenerative disease risk, almost nothing is known about their regulation. We performed an unbiased screen and identified specific pathways controlling progranulin levels in neurons. Modulation of these pathways restored levels in progranulin-deficient neurons and reversed FTLD phenotypes. We provide a new comprehensive understanding of the genetic regulation of progranulin levels and identify potential targets to treat FTLD and other neurodegenerative diseases, including AD.
Keywords: AD, autophagy, FTD, lysosome, progranulin
Introduction
Progranulin is a widely expressed secreted protein encoded by the GRN gene. In the brain, progranulin is expressed primarily by neurons and microglia involved in neuronal survival, neurite outgrowth, and synaptogenesis and is a key regulator of neuroinflammation (Ahmed et al., 2007; Chitramuthu et al., 2010; Cenik et al., 2012; Gass et al., 2012; Petkau et al., 2012; Petoukhov et al., 2013; Petkau and Leavitt, 2014). More than 60 mutations leading to haploinsufficiency of GRN have been identified and result in decreased progranulin levels. People with GRN haploinsufficiency develop the fatal neurodegenerative disease frontotemporal lobar degeneration (FTLD), the second most common cause of early-onset dementia after Alzheimer's disease (AD) (Rabinovici and Miller, 2010; Rademakers et al., 2012; Riedl et al., 2014). Rare individuals nullizygous for GRN develop neuronal ceroid lipofuscinosis, which has an onset much earlier than FTLD (Smith et al., 2012; Götzl et al., 2014). Recently, polymorphisms that decrease GRN expression were linked to increased risk for AD and boosting levels of progranulin in mouse models of AD suppresses neuroinflammatory-, pathological-, and cognitive-based disease phenotypes (Pereson et al., 2009; Hsiung et al., 2011; Kämäläinen et al., 2013; Perry et al., 2013; Minami et al., 2014; Chen et al., 2015; Hosokawa et al., 2015; Xu et al., 2017). Therefore, differing levels of progranulin deficiency in the brain lead to different neurodegenerative syndromes, showing that loss of progranulin function contributes to disease development.
Although progranulin is critically important in the brain, we still do not understand how progranulin is regulated and what its cellular functions are. Previous studies implicated cellular conditions such as inflammation and hypoxia as regulators of GRN transcription; however, the transcription factors directly mediating GRN expression in these conditions are unknown (Piscopo et al., 2010; Suh et al., 2012; Luo et al., 2014). A recent screen identified histone deacetylases (HDACs) as regulators of GRN transcription in Neuro2a (N2a) cells and in human iPSC-derived cortical neurons generated from FTLD patients harboring GRN mutations. The specific HDAC(s) and the mechanisms by which the HDACs affect GRN expression are not known (Cenik et al., 2011; Almeida et al., 2016). Other studies revealed that progranulin levels might be modulated through posttranscriptional mechanisms. For example, inhibition of lysosome function with bafilomycin A1 increased progranulin protein levels in neuronal cells, possibly through a mechanism involving the differential translation of GRN mRNAs with short versus long 5′ UTRs or by derepression of miRNA-regulated translation (Capell et al., 2011, 2014). Progranulin is a secreted protein and mechanisms affecting its secretion, uptake, and degradation may regulate progranulin levels. Sortilin, the first cell surface receptor identified for neuronal progranulin, regulates the uptake of extracellular progranulin and delivers it to lysosomes (Hu et al., 2010). Reducing sortilin levels increases levels of extracellular progranulin in neurons. Interestingly, progranulin itself regulates sortilin degradation and thus may regulate its own levels in an autocrine fashion to maintain the pathogenic progression of prostate cancer (Tanimoto et al., 2017). Similarly, prosaposin (PSAP) binds and targets extracellular progranulin to lysosomes; suppression of PSAP increased progranulin levels in neurons (Zhou et al., 2015).
Progranulin itself plays a functional role in lysosomes (Ahmed et al., 2010; Tanaka et al., 2013a,b, 2014, 2017; Lui et al., 2016; Beel et al., 2017; Chang et al., 2017; Evers et al., 2017). Loss of progranulin results in enlarged lysosomes with the aberrant accumulation of lipofuscin, an autofluorescent amalgam of predominantly oxidized protein and lipid degradation residues that accumulates with age or lysosome dysfunction (Ahmed et al., 2010; Tanaka et al., 2013a,b, 2014, 2017; Lui et al., 2016; Evers et al., 2017; Ward et al., 2017). Tanaka et al. (2017) suggested that progranulin regulates lysosome acidification, but whether this occurs in neurons and by what mechanism is not known (Tanaka et al., 2017). Progranulin may directly bind and modulate lysosomal enzymes such as cathepsin D (Beel et al., 2017). Finally, progranulin deficiency leads to dysregulated autophagy in cells, but it is unclear whether this is due to loss of function at the lysosome level or at earlier points in the ALP (Liu et al., 2015; Chang et al., 2017). Although it appears that progranulin levels can be regulated at multiple points in cells, we still do not know the identities of many of the key molecules and pathways involved. Also, it will be important to determine the functional impact of how and where progranulin levels are raised in the cell to preserve or restore progranulin functions essential for ameliorating the disease process.
In this study, we wanted to develop a more complete picture of the molecules and biological pathways that regulate progranulin levels and to better understand the functional effects of modulating progranulin levels in neurons. We performed an unbiased whole-genome RNAi screen by an ELISA-based strategy to identify genes that act at multiple steps, increasing progranulin levels in N2a cells. We found several genes from the druggable genome that raised progranulin levels when knocked down, many with known connections to the ALP. Multiple small-molecule inhibitors against four of the genes phenocopied RNAi-induced knock-down of those genes, raising progranulin levels in a dose-dependent manner at submicromolar concentrations. Progranulin itself regulates the ALP: we found that Grn-deficient neurons had an abnormally increased level of autophagy and enlarged lysosomes. Remarkably, increasing progranulin levels by inhibiting hit genes restored autophagy and lysosome size to control levels in neurons. We identified novel genetic modifiers of progranulin levels and functions in neurons, which suggest an autocrine loop exists in which progranulin levels are regulated by autophagy and yet progranulin itself is a regulator of the ALP, potentially modulating its own levels through regulating its degradation in neurons.
Materials and Methods
Reagents and antibodies.
Cells were treated with 17-AAG, N-oxalylglycine (NOG), or 3-methyladenine (Sigma-Aldrich); AUY922 (Selleckchem); AS1842856 or JMJD histone demethylase inhibitor III (Calbiochem/EMD Millipore); XRP44X (Tocris Bioscience) as indicated in the figure legends. High-sensitivity streptavidin HRP was from Pierce. Tetramethylbenzidine (TMB)-E solution was from EMD Millipore. The progranulin rabbit polyclonal antibody was generated by 21st Century Biochemicals (Protocol 2819) against a C-terminal polypeptide, Ac-CLRKKIPRWDMFLRDPVPRPLL, and was provided to us as a gift from R. Farese and the Consortium for Frontotemporal Dementia. The progranulin sheep polyclonal and the anti-sheep biotinylated IgG were from R&D Sciences. The TDP-43 rabbit polyclonal is from Proteintech. Lysotracker red was from Thermo Fisher Scientific. pGW1-EOS2-LC3 was created by replacing Dendra2-LC3 with a 1228-bp Nhe1-Mfe1 fragment containing EOS2-LC3 in the pGW1-Dendra2-LC3 plasmid as described previously (Tsvetkov et al., 2013). pGW1-EGFP was created as described previously (Arrasate et al., 2004).
N2a cell culture and transfection.
N2a cells were grown in DMEM high-glucose (Mediatech), supplemented with 10% FBS (Life Technologies), 1% penicillin/streptomycin (Life Technologies), and 1% GlutaMAX (Life Technologies) at 5% CO2 and 37°C. Cells were passaged when they reached 80% confluence. For cell transfections, N2a cells (25,000 cells/well) were reverse transfected in 96-well plates using lipofectamine 2000 and plated in DMEM + 1% FBS posttransfection to induce differentiation.
Primary neuron cell culture and transfection.
Cortical neurons were dissected from embryonic day 18 mouse pups and cultured at 125,000 cells/well for 4 d in vitro, as described previously (Barmada et al., 2014). Neurons were cultured from Grn± (Martens et al., 2012), GrnR493X/+, or WT control littermates (Nguyen et al., 2018). Equal numbers of male and female mice were used in the neuron cultures. Euthanasia for these experiments is entirely consistent with the recommendations of the Guidelines on Euthanasia of the American Veterinary Medical Association. Transfection of primary neurons was accomplished using Lipofectamine 2000 (Invitrogen). All of the transfections in primary neurons involved 25 nm siRNA and 0.5 μl of Lipofectamine 2000 per well unless otherwise noted. Cells were incubated with Lipofectamine–siRNA complexes for 40 min at 37°C before rinsing and maintained in Neurobasal-B27 growth medium (Life Technologies) + 2% B27 supplement (Life Technologies) + 1% penicillin/streptomycin (Life Technologies) + 1% GlutaMAX (Life Technologies) (Brewer et al., 1993). The remainder of the transfection protocol was per the manufacturer's suggestions.
Progranulin ELISA development.
The progranulin ELISA used a rabbit anti-mouse progranulin antibody, a sheep anti-mouse progranulin (R&D Systems), and biotin-labeled anti-sheep IgG antibody, streptavidin-linked HRP, and TMB/peroxide substrate. Recombinant mouse progranulin (R&D Systems) was used to generate standard curves with typical curve heights between 1.0 to 3.0 optical density (OD) units using a four-parameter logistics fit of the data points and with little background (<0.1 OD units) when validated using serum from either WT or Grn knock-out mice. Plates were coated overnight with the rabbit anti-progranulin polyclonal antibody and blocked with 1% BSA for 1 h at 37°C. Plates were incubated with analyte (culture supernatants, mouse serum, or recombinant mouse progranulin) for 1 h at 37°C, followed by sequential detection using the sheep anti-progranulin polyclonal antibody, anti-sheep biotinylated IgG antibody, and streptavidin–HRP conjugate. Mouse serum samples were diluted 1:100 for analyte levels to fall within the range of the assay. We did not detect progranulin in serum from Grn knock-out mice. The ELISA is sensitive and accurately detects nanogram amounts of extracellular progranulin from differentiated N2a cell or primary mouse cortical neuron culture supernatants. We miniaturized the assay to a 96-well format and then evaluated its performance characteristics.
Library screening.
We used an arrayed Dharmacon mouse siGENOME SMARTpool siRNA library from GE Lifesciences that targets the whole mouse genome. The genome-wide siRNA library targets ∼16,500 different Mus musculus genes with pools of four siRNAs designed against different regions of the same gene. To perform the screen, siRNAs were introduced into N2a cells by reverse transfection and extracellular progranulin levels were measured in culture supernatants 48 h after transfection. On each plate, three controls were included.
RNA extraction and qRT-PCR.
RNA was isolated using the RNeasy micro kit (Qiagen) following the manufacturer's instructions. Next, qRT-PCR was performed using the Superscript III First Strand Synthesis Supermix and following the manufacturer's instructions (Invitrogen). Finally, cDNA was diluted fourfold. For qRT-PCR, 4.5 μl of cDNA was combined with 5 μl of SYBR Green Master Mix (Applied Biosystems) and 0.5 μl of primer mixture (20 μm each primer). Samples were run on an Applied Biosystems 7900HT. Data were analyzed using the delta CT method. Values were normalized to housekeeping gene cyclophilin. Primers used were as follows: mouse cyclophilin forward: TGGAAGAGCACCAAGACAACA, mouse Cyclophilin reverse: TGCCGGAGTCGACAATGAT, mouse Grn forward: TGGTTCACACACGATGCGTTTCAC, mouse Grn reverse: AAAGGCAAAGACACTGCCCTGTTG.
Bioinformatics analysis.
Ingenuity Pathway Analysis (IPA) software (Qiagen) was used to determine significantly overrepresented pathways populated by the screen hits using the whole-mouse genome as the reference gene set (16,565 genes). Potential molecular networks with known connections to the ALP for the 33 validated hits, as well as other hits from the primary screen, were mapped using an annotated reference library for autophagy and lysosome genes in IPA (2000 genes).
Autophagy flux assay.
Turnover of EOS2-LC3 was performed by optical pulse labeling (OPL). EOS2 is a photoswitchable protein that we used in place of the photoswitchable protein Dendra2, which we described previously (Barmada et al., 2014). EOS2 has improved photostability, brightness, maturation kinetics, and acid sensitivity compared with Dendra2. cDNA for the autophagy upregulator Beclin1 was cotransfected into neurons as a positive control for induced/enhanced autophagy flux.
Results
Identification of modulators of progranulin levels from an unbiased genetic screen
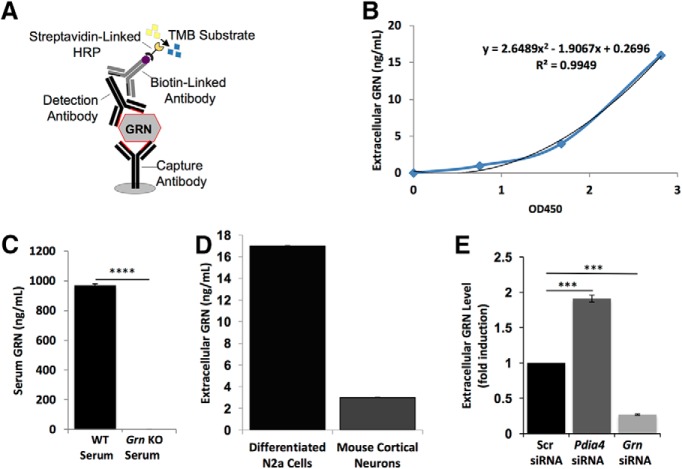
We sought to identify target genes that increased or decreased steady-state progranulin levels in neuronal cells by screening a mouse genome-wide siRNA library. We first designed and developed an ELISA to detect extracellular mouse progranulin protein in neuronal culture supernatants and to serve as the platform assay for the screen (Fig. 1). We reasoned that, by screening for steady-state extracellular progranulin levels, we could identify genes involved in most, if not all, aspects of progranulin production (e.g., transcription, translation, folding, trafficking, and secretion), uptake, and degradation. The 96-well format ELISA is sensitive and accurately detects nanogram amounts of extracellular progranulin from differentiated N2a cells or primary mouse cortical neuron culture supernatants (Fig. 1B,D). This ELISA is also specific: we detected no progranulin in serum from Grn knock-out mice (Fig. 1C). Pooled siGENOME siRNAs against Grn were used as a positive control for suppressors of progranulin levels (Z′ = 0.8), and pooled siGENOME siRNAs against Pdia4 (Z′ = 0.6) were used as a positive control for enhancers of progranulin levels; pooled siGENOME scrambled nontargeting siRNAs (Scr siRNA) were used as a negative control (Fig. 1E) (Almeida et al., 2011). Based on the excellent performance of the assay, we concluded that it was suitable for screening.
Figure 1.
Development of a sensitive mouse progranulin ELISA. A, Design of a sandwich ELISA to detect mouse progranulin (GRN). B, Representative standard curve for the GRN ELISA. C, Measurement of serum GRN level from a WT or a Grn knock-out (KO) mouse. D, Measurement of extracellular GRN levels in supernatants from undifferentiated N2a cells, differentiated N2a cells, or 15 d in vitro mouse primary cortical neuron cultures. E, Identification of controls for the siRNA screen. Shown are measurements of extracellular GRN levels in supernatants from cultured differentiated N2a cells transfected with either 50 nm Scr nontargeting siRNA, 50 nm Pdia4 siRNA, or 50 nm Grn siRNA. ***p < 0.001; ****p < 0.0001.
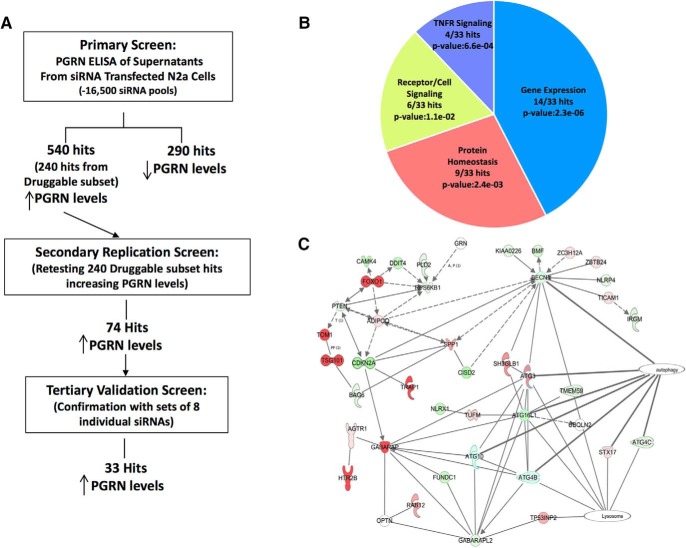
We performed the primary screen in N2a cells with the assay for progranulin levels using the arrayed Dharmacon siGENOME SMARTpool library, testing each pool of four siRNAs in a single pass (Fig. 2A). Primary screen hits were those genes that increased or decreased extracellular progranulin levels by at least ±1.5-fold relative to the mean level for that microtiter plate after siRNA knock-down. Approximately 830 siRNA pools modulated progranulin levels in the primary screen, corresponding to a hit rate of ∼5% (Fig. 2A).
Figure 2.
A genome-wide siRNA screen identifies modulators of progranulin levels in neuronal cells. A, Summary of the siRNA screen. B, Diagram of Gene Ontology analysis results. C, IPA reveals a hit network with connections to the ALP. Nodes (genes) and edges (gene relationships) are pictured. Edges are indicated as lines; a solid line indicates a direct interaction and a dashed line indicates an indirect interaction. The arrowhead indicates the direction of the interaction; that is, gene X acts on gene Y if the arrowhead points from gene X to gene Y. The saturation intensity of the node color indicates the degree of increased progranulin level (red) or decreased progranulin level (green) after siRNA knock-down of the hit gene compared with Scr siRNA-treated control cells. Genes in uncolored nodes were not identified in our screen and were integrated into the computationally generated networks on the basis of the evidence stored in the IPA knowledge memory indicating a relevance to this network. The node shapes denote enzymes, phosphatases, kinases, peptidases, G-protein coupled receptor, transmembrane receptor, cytokines, growth factor, ion channel, transporter, translation factor, nuclear receptor, transcription factor, and other.
Some hit genes may be false-positives due to off-target effects of the siRNAs. Therefore, we further generated a list of high-confidence hit genes based on the following rigorous validation strategy. We focused our validation efforts on hits from the “druggable genome” portion of the library because of their increased potential to be therapeutic targets. Specifically, 74 of the 240 hits from the druggable genome for which inhibition by siRNA led to increased progranulin levels (1.4% hit rate) were confirmed in a secondary screen by repeating the primary screen assay in triplicate (Fig. 2A). Next, to determine whether these 74 pools of siRNA that reproducibly induced increased progranulin levels did so by acting specifically on their intended gene target, we performed a twofold tertiary screen. First, the four individual siGENOME siRNAs making up the library pool of each hit were each tested separately in our primary screen assay. Next, any hits that passed testing with the original deconvolved siRNAs sets were tested with an additional set of four siRNAs. The new sets of four siRNAs were distinguished from the original four siRNAs of the library pool in that they were: (1) generated by a different proprietary synthesis method (OnTARGET) and (2) targeted new regions within each gene not recognized by the original four siRNAs. In this tertiary screen, a hit was validated as a bona fide genetic modifier of progranulin production if two of the four siRNAs of each set (four of the eight total) increased in progranulin levels. From the druggable genome subset, 33 of the 74 confirmed hits that increased progranulin when knocked down were validated in N2a cells, with seven or eight of the eight siRNAs leading to increased progranulin levels for many of the 33 genes. We conclude that these 33 genes represent bona fide (true) modifiers of progranulin levels (Table 1).
Table 1.
Druggable library genes that passed the tertiary screen in N2a cells
| Gene name | Accession no. | Fold change in PGRN levela | No. of siRNAs increasing [PGRN]b | |
|---|---|---|---|---|
| 1 | TSG101 | NM_021884 | 3.4 | 6/8 |
| 2 | ELK3 | NM_013508 | 2.6 | 6/8 |
| 3 | FGF17 | NM_008004 | 2.2 | 6/8 |
| 4 | BAG3 | NM_013863 | 2.2 | 5/8 |
| 5 | FOXD2 | NM_008593 | 2.1 | 6/8 |
| 6 | TRAP1 | NM_026508 | 2.1 | 8/8 |
| 7 | CLCA5 | NM_178697 | 1.9 | 8/8 |
| 8 | H2AFY3 | NM_207000 | 1.9 | 7/8 |
| 9 | ONECUT2 | NM_194268 | 1.9 | 8/8 |
| 10 | E2F5 | NM_007892 | 1.9 | 6/8 |
| 11 | FGF13 | NM_010200 | 1.9 | 7/8 |
| 12 | DOCK3 | NM_153413 | 1.8 | 6/8 |
| 13 | F5 | NM_007976 | 1.8 | 6/8 |
| 14 | JMJD6 | NM_033398 | 1.8 | 8/8 |
| 15 | HTR2B | NM_008311 | 1.8 | 5/8 |
| 16 | CREBBP | NM_00102543 | 1.8 | 5/8 |
| 17 | GRAP1 | NM_027817 | 1.8 | 6/8 |
| 18 | BCLAF1 | NM_00102539 | 1.8 | 3/8 |
| 19 | GPX1 | NM_008160 | 1.8 | 3/8 |
| 20 | GABARAP1 | NM_019749 | 1.7 | 5/8 |
| 21 | FBXW2 | NM_013890 | 1.7 | 8/8 |
| 22 | ATF7iP | NM_019426 | 1.7 | 7/8 |
| 23 | FOXO1 | NM_019739 | 1.7 | 8/8 |
| 24 | POU5F1 | NM_013633 | 1.7 | 3/8 |
| 25 | PRDM4 | NM_181650 | 1.7 | 5/8 |
| 26 | BAG4 | NM_026121 | 1.6 | 8/8 |
| 27 | BAG1 | NM_009736 | 1.6 | 6/8 |
| 28 | TOM1 | NM_011622 | 1.6 | 6/8 |
| 29 | SMAD1 | NM_008539 | 1.6 | 3/8 |
| 30 | CFB | NM_008198 | 1.6 | 5/8 |
| 31 | NFKBIA | NM_010907 | 1.5 | 4/8 |
| 32 | HSPB2 | NM_024441 | 1.5 | 5/8 |
| 33 | BAG5 | NM_027404 | 1.5 | 4/8 |
aFold change in PGRN levels relative to Neuro2a cells transfected with Scrambled control siRNAs.
bEach gene was validated with the original set of four individual siGENOME siRNAs (deconvolved screen pools) and a second set of four unique, individual OnTARGET siRNAs to give a total set of eight unique siRNAs tested against each gene.
Bioinformatics analysis revealed that some of the screen hits were overrepresented in pathways connected to immunomodulation via TNF/NF-κB signaling, hypoxia, cancer, and protein homeostasis (Fig. 2B, Table 2). Due to recent work from us and others showing that progranulin has functions as a lysosomal protein, we took a closer look at a group of the hits that had known roles or connections to the ALP and that formed a potential molecular network among themselves (Fig. 2C). We found that eight of the validated druggable genes (Gabarap1, Tom1, Tsg101, Foxo1, Sort1, Jmjd6, Elk3, and Trap1/HSP90L) have known roles in regulating autophagy and lysosomal sorting. GABARAP1 is a member of the LC3 family of proteins that mediate autophagosome formation and maturation and are also important in autophagosome-to-lysosome fusion (Jenzer et al., 2014; Albanesi et al., 2015; McEwan et al., 2015; Wang et al., 2015; Martens, 2016; Nguyen et al., 2016). TOM1 is an endosome protein that functions in autophagosome-to-lysosome fusion (Wang et al., 2010; Bond et al., 2011; Tumbarello et al., 2012, 2013). TSG101 is an ESCRT1 protein that is required for endosomal maturation, trafficking, and exosome secretion (Babst et al., 2000; Colombo et al., 2013). FOXO1 is a forkhead O family transcription factor that has been shown to regulate autophagy in response to stress (Zhao et al., 2010). Sortilin (SORT1) is a lysosomal sorting receptor that traffics proteases to the lysosome (Canuel et al., 2008). In addition, SORT1 mediates uptake of extracellular progranulin (Hu et al., 2010; Zheng et al., 2011). JMJD6 encodes a jumonji C-domain containing, bifunctional arginine demethylase and lysyl-hydroxylase (Chang et al., 2007; Webby et al., 2009; Boeckel et al., 2011; Mantri et al., 2011; Unoki et al., 2013; Heim et al., 2014; Poulard et al., 2014; Zhang et al., 2015; Walport et al., 2016). JMJD6 demethylates histones H3 and H4, potentially imparting epigenetic regulation of transcription. The arginine demethylase activity of Jmjd6 has been shown to target the stress granule nucleating protein G3BP1, where demethylation of G3BP1 promoted stress granule assembly in response to stress (Tsai et al., 2017). Jmjd6 also regulates RNA splicing by mediating 5-hydroxylation of U2AF2/U2AF65, affecting the pre-mRNA splicing activity of U2AF2/U2AF65 (Webby et al., 2009). Recently, Jmjd6 has been shown to promote autophagy in triple-negative breast cancer cells (Liu et al., 2019). Elk3 is an ETS transcription factor that functions as a transcriptional repressor and regulates gene expression during angiogenesis and hypoxia (Gross et al., 2008; Heo and Cho, 2014). Knock-down of ELK3 in a triple-negative breast cancer cell line led to a repression of autophagy via activation of the PI3K/Akt/mTOR pathway and, as a result, conferred sensitivity to the anticancer drug doxorubicin (Park et al., 2016). TRAP1/HSP90L is a mitochondrial chaperone protein and a member of the HSP90 family of heat shock proteins (Putcha et al., 2010; Takemoto et al., 2011; Altieri et al., 2012; Amoroso et al., 2012; Baldo et al., 2012; Matassa et al., 2012; Hong et al., 2013). TRAP1/HSP90L has been shown recently to act as a regulator of autophagy in lung cancer cells (Barbosa et al., 2018).
Table 2.
Significantly overrepresented pathways of top druggable subset hits
| Signaling pathway | p-value | |
|---|---|---|
| 1 | Protein ubiquitination pathway | 0.008 |
| 2 | Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 0.010 |
| 3 | NF-κB signaling | 0.012 |
| 4 | Molecular mechanisms of cancer | 0.014 |
| 5 | Actin cytoskeleton signaling | 0.014 |
| 6 | Glucocorticoid receptor signaling | 0.015 |
| 7 | Clathrin-mediated endocytosis signaling | 0.016 |
| 8 | PI3K/Akt signaling | 0.016 |
| 9 | B-cell receptor signaling | 0.017 |
| 10 | IL-6 signaling | 0.017 |
| 11 | Role of osteoblasts, osteoclasts, and chondrocytes in rheumatoid arthritis | 0.018 |
| 12 | Wnt/β-catenin signaling | 0.018 |
| 13 | Human embryonic stem cell pluripotency | 0.022 |
| 14 | Hypoxia signaling in the cardiovascular system | 0.031 |
| 15 | Mouse embryonic stem cell pluripotency | 0.032 |
| 16 | FGF signaling | 0.035 |
| 17 | Role of RIG1-like receptors in antiviral innate immunity | 0.044 |
| 18 | iNOS signaling | 0.046 |
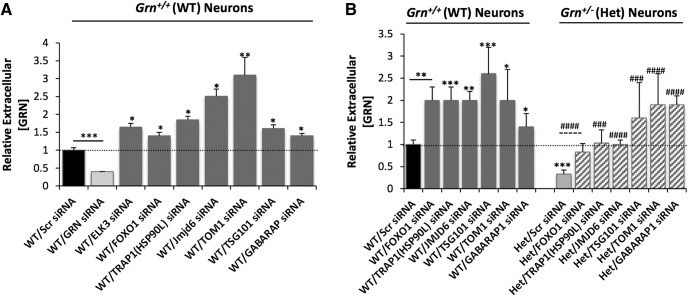
The original screen and validation steps were performed in N2a cells, an immortalized neuroblastoma cell line. To determine whether the genetic regulators of progranulin production identified in N2a cells are biologically relevant, we tested whether suppressing a subset of the hit genes increased progranulin levels in WT (Grn+/+) or progranulin haploinsufficient (Grn+/; Het) primary mouse cortical neurons, a physiologically relevant cell type that is affected in FTLD. We chose genes for which we identified commercially available small-molecule inhibitors (see below) or which had a role in the ALP, a recently identified pathway connected to progranulin. The ALP is a key contributing pathway for cellular proteostasis and dysfunction of this pathway is linked to neurodegenerative diseases, including FTLD. We measured extracellular progranulin in the culture supernatant by mouse primary cortical neurons with the ELISA and found that it was produced at concentrations ∼10-fold lower (∼1–10 ng/ml) than in N2a cells. Knock-down of Elk3, Jmjd6, Trap1/Hsp90L, Tom1, Tsg101, Foxo1, and Gabarap1 raised progranulin levels in WT mouse cortical neurons (Fig. 3A). Next, we tested a subset of the hits in cortical neurons that were cultured from FTD model mice harboring one functional allele of Grn (Grn±; Het) (Martens et al., 2012). The Grn± cortical neurons had ∼67% less extracellular progranulin than WT littermate controls, thus mimicking the (>50%) reduction in circulating progranulin levels seen in FTLD patients due to Grn haploinsufficiency (WT/Scr siRNA vs WT/Grn siRNA, Fig. 3A; WT/Scr siRNA vs Het/Scr siRNA, Fig. 3B). Strikingly, knock-down of Foxo1, Trap1/Hsp90L, Jmjd6, Tom1, Tsg101, or Gabarap1 raised progranulin levels in the Grn± haploinsufficient cortical neurons to the WT control level or greater (Fig. 3B).
Figure 3.
Genetic inhibition of select hits with siRNAs affects progranulin levels in primary neurons cultured from WT or FTLD model mice. A, Representative ELISA data showing siRNAs against Elk3, Foxo1, Trap1/Hsp90L, Jmjd6, Tom1, Tsg101, or Gabarap1 increase progranulin levels in WT primary mouse neurons. B, Representative ELISA data showing siRNAs against Foxo1, Trap1/Hsp90L, Jmjd6, Tom1, Tsg101, or Gabarap1 increase progranulin levels in Grn-deficient primary cortical neurons from a WT sibling (left side of graph) or FTLD-model mouse (right side of graph). Scr siRNA is used as a baseline control in all experiments. Three independent experiments performed (shown: mean ± SD). Statistical test: Student's t test, *p < 0.05; **p < 0.01; ***p < 0.001; ###p < 0.001; ####p < 0.0001.
Small-molecule inhibitors enhance progranulin levels in N2a cells
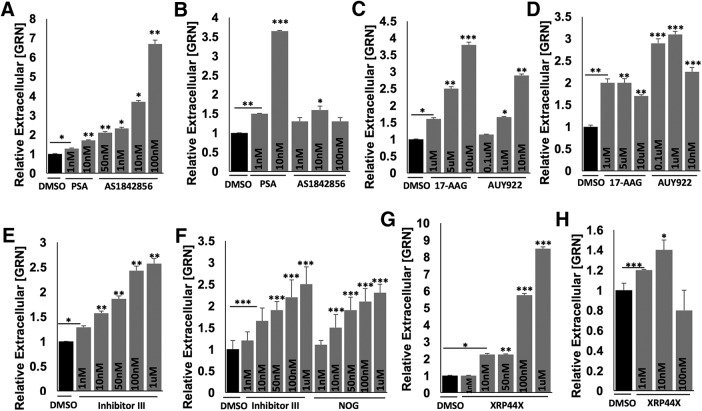
Compared with an siRNA, pharmacological inhibitors offer an orthogonal way to inhibit a target, providing another layer of validation to increase confidence in a hit gene. We identified and tested the six commercially available small-molecule inhibitors against four of the validated druggable hit genes (Foxo1, Trap1/Hsp90L, Jmjd6, and Elk3) for their ability to mimic the increase in progranulin levels observed with siRNA knock-down of each of these genes. We first tested whether acute inhibition of the targets increased progranulin levels after 2 h of treatment. Acute inhibition is less likely to be significantly confounded by adaptive or detrimental changes that cells might undergo over longer periods of treatment and that indirectly affect progranulin levels. Therefore, acute inhibition may better represent the direct effects of a compound on its intended target. We also tested for sustained effects by monitoring progranulin levels after 24 h of inhibition.
Psammaplysene A (PSA) and AS1842856, two small-molecule inhibitors of FOXO1, increased progranulin levels in N2a cells (Fig. 4A,B) (Mojsilovic-Petrovic et al., 2009; Nagashima et al., 2010). We found that after a 2 h acute inhibition in N2a cells, PSA significantly and potently increased progranulin levels in a dose-dependent manner (Fig. 4A). AS1842856, which blocks FOXO1 activity via a different mechanism, also significantly and potently increased progranulin levels in a dose-dependent manner after 2 h of inhibition (Fig. 4A). Both inhibitors showed a sustained increase in progranulin levels when tested after 24 h of inhibition (Fig. 4B). AS1842856 is of particular interest. It is orally available and used extensively to investigate the in vivo role of FOXO1 for therapeutic indications.
Figure 4.
Pharmacologic inhibition of FOXO1, TRAP1/HSP90L, JMJD6, or ELK3 affects progranulin levels in N2a cells. Representative ELISA data showing small-molecule inhibitors against FOXO1: (A) 2 h and (B) 24 h treatment with PSA or AS1842856; TRAP1/HSP90L: (C) 2 h and (D) 24 h treatment with 17AAG or AUY922; JMJD6: (E) 2 h treatment with Inhibitor III (Inh III) and (F) 24 h treatment with Inhibitor III (Inh III) or NOG; ELK3: (G) 2 h and (H) 24 h treatment with XRP44X increase progranulin levels in N2a cells in a dose-dependent manner after treatment with indicated concentrations. Three independent experiments performed (shown: mean ± SD). Statistical test: Student's t test, *p < 0.05; **p < 0.01; ***p < 0.001.
HSP90 inhibitors have been shown to block TRAP1 function (Putcha et al., 2010; Takemoto et al., 2011; Altieri et al., 2012; Amoroso et al., 2012; Baldo et al., 2012; Matassa et al., 2012; Hong et al., 2013). Two HSP90 inhibitors, 17AAG and AUY922, increased progranulin levels in N2a cells (Fig. 4C,D) (Hong et al., 2013). Acute 2 h inhibition with 17AAG or AUY922 showed dose-dependent increases in progranulin levels (Fig. 4C). AUY922 (0.1 μm) was particularly effective, producing an almost threefold sustained increase in progranulin in N2a cells after a 24 h inhibition (Fig. 4D). Mechanistically, inhibiting TRAP1/HSP90L may enhance the folding, maturation, or trafficking of progranulin, leading to its subsequent secretion.
We next tested two inhibitors of JMJD6, inhibitor III and NOG. Inhibitor III showed a dose-dependent increase in progranulin levels in N2a cells after a 2 h inhibition (Fig. 4E). Both inhibitor III and NOG increased progranulin levels after 24 h inhibition (Fig. 4F).
Finally, we tested an inhibitor of ELK3 signaling, XRP44X, in N2a cells (Wasylyk et al., 2008; Semenchenko et al., 2016). Two hours of inhibition with higher doses of XRP44X showed more potent increases in progranulin levels (Fig. 4G), whereas a lower dose of XRP44X showed a modest increase in progranulin levels after a 24 h inhibition (Fig. 4H). The fact that multiple drugs that inhibit the products of several of the hit genes also recapitulate the effects of siRNAs validates the success of our screen and lends value to the strategy of identifying loss-of-function suppressors that could be leveraged to discover truly druggable targets with the same pharmacodynamic effect (raising progranulin levels). We identified six small-molecule inhibitors that targeted four of the hit genes and worked in a dose-dependent manner at micromolar or submicromolar concentrations, suggesting that new drugs can be developed against some of the other hit genes that might raise progranulin levels and even be clinical candidates. Interestingly, there might be many avenues to restore progranulin to normal levels.
Validated druggable genes regulate progranulin levels through different mechanisms
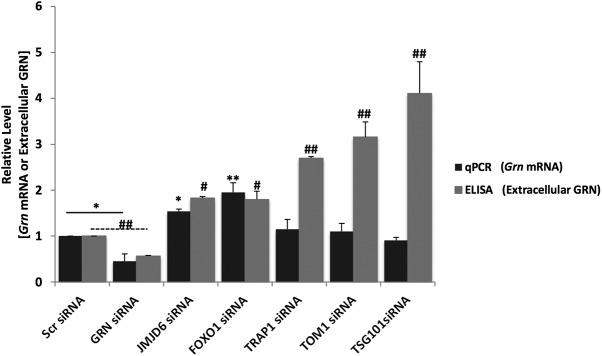
We next knocked down a subset of genes identified in our screen to determine whether they acted via transcriptional or posttranscriptional mechanisms to increase progranulin levels. N2a cells were transfected with siRNAs against Jmjd6, Foxo1, Trap1/Hsp90L, Tom1, or Tsg101. After a 48 h knock-down, RNA was harvested for qRT-PCR to measure the relative levels of Grn mRNA and ELISA to measure extracellular levels of progranulin protein in culture supernatants. siRNA knock-down of Jmjd6 or Foxo1 increased the relative abundance of Grn mRNA (Fig. 5); conversely, siRNA knock-down of Trap1/Hsp90L, Tom1, or Tsg101 did not, despite increasing extracellular progranulin levels (Fig. 5). Therefore, these validated genes parse into two distinct mechanistic categories: those that regulate progranulin primarily at the transcriptional level (Jmjd6; Foxo1) and those that regulate progranulin primarily at the protein level (Trap1/Hsp90L; Tom1; Tsg101), suggesting that progranulin levels are regulated by multiple mechanisms.
Figure 5.
Genetic inhibition of exemplary validated genes affects progranulin levels through transcriptional or posttranscriptional mechanisms in mouse cortical neurons. Mouse cortical neurons were transfected with Scr siRNA, Grn siRNA, Jmjd6 siRNA, Foxo1 siRNA, Trap1/Hsp90L siRNA, Tom1 siRNA, or Tsg101 siRNA, and cells were lysed for RNA extraction and qRT-PCR to measure Grn mRNA level, and conditioned media were used for ELISA to measure extracellular progranulin levels. Two independent experiments performed (shown: mean ± SD). Statistical test: Student's t test, *p < 0.05; **p < 0.01; #p < 0.05; ##p < 0.01.
Progranulin deficiency leads to aberrant autophagy and enlarged lysosomes in cortical neurons
Our bioinformatics analysis linked several hits to the ALP and recent work has shown that progranulin functions in lysosomes and affects autophagy (Chang et al., 2017). Adding extracellular progranulin repressed autophagy, whereas inhibiting Grn expression with siRNA caused an exaggerated increase in autophagy in hepatocytes (Liu et al., 2015). Alternatively, complete loss of progranulin in neurons was shown to repress the selective autophagy pathways xenophagy and aggrephagy, possibly through an impairment of ALP signaling (Chang et al., 2017). Progranulin is localized to lysosomes and may be important for lysosome acidification, may be targeted for degradation, or both. This raises the question, is progranulin simply a substrate for the ALP or does it have an important role regulating lysosome structure and function? As a substrate for and a regulator of the ALP, progranulin may act in an autocrine manner to regulate its own levels in neurons. To address this possibility, we interrogated the effect of inhibiting the ALP on progranulin levels and the effect of knocking down Grn on the ALP in neurons.
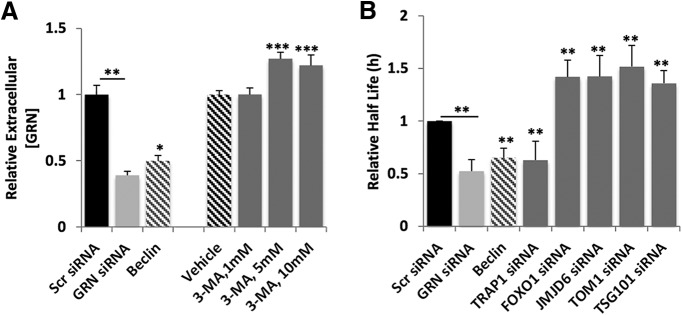
We directly tested whether the ALP regulates progranulin levels by modulating the pathway with a pharmacological inhibitor, 3-methyladenine (3-MA), and measuring extracellular levels of progranulin from neurons. 3-MA increased extracellular progranulin, whereas overexpressing a genetic inducer of autophagy, Beclin1, decreased progranulin secretion (Fig. 6A). Interestingly, Osaka et al. (2015) also found that 3-MA led to increased progranulin secretion in N2a cells. These data suggest that blocking the ALP stabilizes progranulin, possibly switching it into a secretory pathway and increasing its release from neurons.
Figure 6.
Reduced progranulin levels are linked to dysregulated autophagy in mouse cortical neurons. A, ELISA measuring extracellular progranulin levels show that known modulators of autophagy (Beclin cDNA; 5 and 10 mm 3-MA) affect progranulin levels in mouse cortical neurons 48 h after transfection or 24 h after treatment with either vehicle or 3-MA. B, Measurement of autophagy flux using the EOS2-LC3 photoswitchable probe cotransfected into mouse cortical neurons with Scr siRNA, Grn siRNA, Beclin cDNA, Trap1/Hsp90L siRNA, Foxo1 siRNA, Jmjd6 siRNA, Tom1 siRNA, or Tsg101 siRNA. Statistical test: Student's t test, *p < 0.05; **p < 0.01; ***p < 0.001.
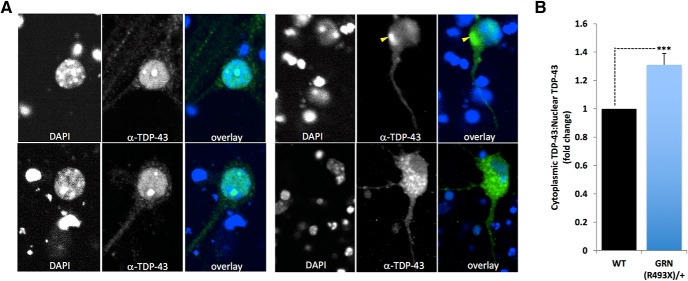
We next used a sensitive OPL assay to measure autophagic flux in primary mouse cortical neurons using robotic microscopy and longitudinal imaging of a photoswitchable fluorescent protein reporter, EOS2-LC3. We reported the use of Dendra2-LC3 to measure autophagy in neurons (Tsvetkov et al., 2010). Here, we used another photoswitchable probe, EOS2, a more sensitive and efficient probe than Dendra2 (Zhang et al., 2012). Both Dendra2 and EOS2 emit green fluorescence. Photoswitching of Dendra2 or EOS2 proteins results in an irreversible conversion of green to red fluorescence. We fused EOS2 to LC3, a protein that becomes conjugated to phosphatidylethanolamine and inserted into the membranes of autophagophores as they become autophagosomes. Longitudinal imaging of the red fluorescence intensity over time in photoswitched cells expressing EOS2-LC3 allows for half-life calculations to be determined as a surrogate measure of autophagic flux. WT mouse cortical neurons were cotransfected with a morphology marker, EGFP, the autophagy probe EOS2-LC3, and siRNA against Grn. Control neurons were transfected with EGFP, EOS2-LC3, and Scr nontargeting siRNA or Beclin 1 cDNA. Beclin 1 reduces the half-life of Dendra2-LC3 (Barmada et al., 2014). Knock-down of Grn by siRNA, which reduced progranulin levels by >50% (Fig. 6A), resulted in an upregulation of autophagic flux in mouse cortical neurons, as indicated by a reduction in EOS2-LC3 half-life, similar in level to that induced by Beclin 1 overexpression (Fig. 6B). Interestingly, this exaggerated autophagy response could be suppressed by coexpression of siRNAs against Foxo1, Jmjd6, Tom1, or Tsg101, the knock-down of which led to increased progranulin levels (Fig. 6B). An upregulation in autophagy is surprising given that progranulin deficiency leads to the abnormal cytoplasmic accumulation and aggregation of TDP-43, which would be consistent with an impairment in proteostasis. In progranulin-deficient cultured cortical neurons, we found that endogenous TDP-43 abnormally accumulates in cytoplasmic punctate structures, similar to the cytopathological features of the human disease (Fig. 7). This suggests that selective autophagy (aggrephagy) may be compromised by progranulin deficiency, whereas general autophagy is increased possibly as a compensatory reaction to accumulating toxic proteins and lipids.
Figure 7.
TDP-43 aggregates accumulate in the cytoplasm of PGRN-deficient cortical neurons cultured from a Grn493X/+ FTLD-model mouse. A, Confocal images of cultured cortical neurons (26 d in vitro) stained with DAPI (blue) and a TDP-43 antibody (green). Left top and bottom, Representative images of control littermate WT neurons. Right top and bottom, Representative images of Grn-deficient (Grn493X/+) neurons. Scale bar, 5 μm B, Quantitation of the ratio of cytoplasmicTDP-43:nuclear TDP-43. Statistical test: Student's t test, ***p < 0.001.
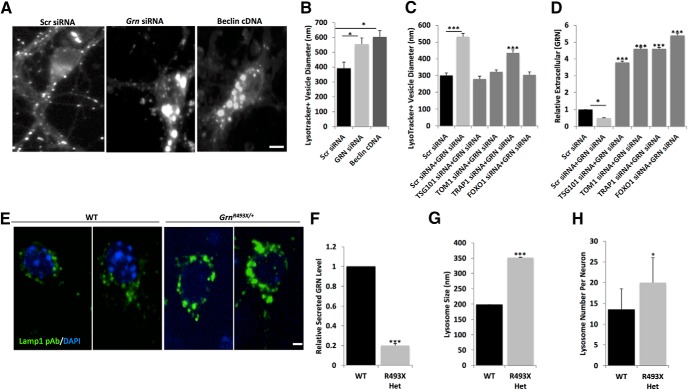
Exaggerated autophagy may also be linked to a progressive impairment of lysosome function (Elrick and Lieberman, 2013). We investigated whether lysosomes were compromised in progranulin-deficient neurons, which may contribute to the abnormal cytoplasmic aggregation of TDP-43. Using LysoTracker, a pH-sensitive dye, we observed that Grn knock-down in mouse cortical neurons resulted in enlarged lysosomes, similar to a genetic induction of autophagy by Beclin 1 overexpression (Fig. 8A,B). Coexpression of siRNAs targeting Tsg101, Tom1, or Foxo1 in progranulin-deficient neurons increased progranulin levels and suppressed lysosome enlargement (Fig. 8C,D). We also found that lysosomes were enlarged and increased in number in cultured progranulin-deficient GrnR493X/+ rodent cortical neurons (Fig. 8E–H).
Figure 8.
Reduced progranulin levels lead to enlarged lysosomes in mouse cortical neurons. A, Images of mouse WT primary cortical neurons cotransfected with pGW1EGFP and Scr siRNA, Grn siRNA, or Beclin cDNA and stained with Lysotracker red. Scale bar, 10 μm. B, Diameters of Lysotracker red-positive structures measured in ImageJ. C, Diameters of Lysotracker red-positive structures measured in ImageJ for mouse cortical neurons transfected with Scr siRNA alone, Scr siRNA + Grn siRNA, Tsg101 siRNA + Grn siRNA, Tom1 siRNA+ Grn siRNA, Trap1/Hsp90L siRNA+ Grn siRNA, or Foxo1 siRNA+ Grn siRNA. D, Extracellular progranulin levels measured by ELISA in mouse cortical neurons transfected with Scr siRNA alone, Scr siRNA + Grn siRNA, Tsg101 siRNA + Grn siRNA, Tom1 siRNA+ Grn siRNA, Trap1/Hsp90L siRNA+ Grn siRNA, or Foxo1 siRNA+ Grn siRNA. Statistical test: Student's t test, *p < 0.05; ***p < 0.001. E, Representative images of LAMP1+ (green) lysosomes in control littermate WT neurons (left two panels) or in Grn-deficient (Grn493X/+) neurons (right two panels) also stained with DAPI (blue). Scale bar, 5 μm. Grn-deficient (Grn493X/+) neurons have reduced progranulin levels (F), increased lysosome size (G), and increased lysosome numbers (H). Statistical test: Student's t test, ***p < 0.001.
Rescue of the exaggerated autophagy flux and enlarged lysosomes does not appear to be limited to a particular modality by which progranulin levels are increased because increasing progranulin expression through a transcriptional mechanism (Foxo1 or Jmjd6 siRNA) was equally effective as increasing progranulin through a posttranscriptional mechanism (Trap1, Tsg101, or Tom1 siRNA). We conclude that boosting progranulin levels by different mechanisms restores normal autophagy flux, corrects lysosome enlargement, and therefore may be an effective therapeutic approach for FTLD.
Discussion
We found >800 genetic modifiers of progranulin levels in neurons and elucidated new molecular mechanisms of progranulin regulation. When inhibited, these genes raise or lower progranulin at the transcriptional or posttranscriptional level in N2a cells and primary neurons. Several hits boosted abnormally low progranulin to normal levels in progranulin-deficient primary cortical neurons cultured from Grn+/493X FTLD model mice and reversed autophagy and lysosomal defects associated with Grn haploinsufficiency. Six small-molecule inhibitors were available for four hits (FOXO1, TRAP1, JMJD6, and ELK3) and all increased progranulin at submicromolar levels in a dose-dependent manner, suggesting that some of the hits from this screen might be suitable targets for small-molecule therapeutics to effectively normalize progranulin levels.
By performing a genome-wide screen and by designing it to detect modifiers working by diverse transcriptional and posttranscriptional mechanisms, we aimed to get an unbiased and relatively comprehensive view of the genetic regulation of progranulin production. Our bioinformatics analysis revealed that the hits mostly fell into four major categories, including gene expression regulation, cell signaling, and TNF receptor signaling.
Protein homeostasis also was a major category accounting for >25% of the hits, including genes involved in endocytic trafficking, autophagy, and lysosomal function. For example, FOXO1 and Jmjd6 can induce autophagy (Zhao et al., 2010; Liu et al., 2019). GABARAP1, TSG101, and Tom1 each play roles in cargo trafficking to lysosomes. GABARAP1 is an autophagosome protein and is important for their formation, whereas TSG101 and Tom1 are endosomal proteins. TSG101 regulates maturation of endosomes into multivesicular bodies. Tom1 participates in autophagosome maturation and lysosome fusion through binding to myosin VI on autophagosomes (Tumbarello et al., 2012; Ryan and Tumbarello, 2018). Myosin VI also binds to optineurin and together they mediate secretory vesicle trafficking and fusion to the plasma membrane (Bond et al., 2011; Ryan and Tumbarello, 2018). Optineurin has been localized to vesicles containing progranulin and ubiquilin2 and overexpression of ubiquilin2 can alter secreted progranulin levels in N2a cells (Osaka et al., 2015). Overexpression of a mutant form of ubiquilin2 was linked to the nuclear-to-cytoplasmic mislocalization and aggregation of endogenous TDP-43 in N2a cells (Picher-Martel et al., 2015) and mutations in optineurin and ubiquilin2 are known to cause the human neurodegenerative disease amyotrophic lateral sclerosis (Osaka et al., 2015). The fact that these hits emerged independently from an unbiased screen and that they are enriched in the ALP strongly implicates the ALP in progranulin regulation.
At the same time, our results also indicate that progranulin regulates the ALP, suggesting the existence of a feedback loop that may be relevant to pathogenic mechanisms of FTD. Reduction of progranulin to model GRN FTLD caused the formation of abnormally large lysosomes, accelerated clearance of an autophagy reporter gene, and the abnormal accumulation of TDP-43. This is consistent with recent studies showing that reduced progranulin levels were correlated with dysfunctional autophagy and the subsequent accumulation of toxic forms of TDP-43 in neurons (Chang et al., 2017; Beel et al., 2018). Boosting progranulin levels helped reduce insoluble TDP-43 levels in vitro (Chang et al., 2017; Beel et al., 2018) and corrected lysosomal deficits and reversed behavioral deficits in a mouse model of FTLD (Arrant et al., 2017). Interestingly, we found that normalization of progranulin by targeting genetic modifiers from our screen rescued the cytopathological changes, indicating that progranulin deficiency likely causes these deficits and that they are reversible.
It remains unclear how progranulin deficiency leads to the accumulation of TDP-43 while also accelerating the clearance of the autophagy flux reporter. One potential way to reconcile these findings is if progranulin deficiency impairs the ALP in ways that selectively affect some cargo, such as TDP-43. Interestingly, upregulation of general autophagy with concurrent inhibition of selective autophagy has been seen in cells with the lysosomal storage disorder, Niemann–Pick type C (NPC). This may arise from the excessive delivery of cargo such as cholesterol and other lipids destined for degradation, which overburdens or even damages lysosomes (Pacheco and Lieberman, 2007; Pacheco et al., 2007; Elrick et al., 2012; Elrick and Lieberman, 2013) (Evers et al., 2017; Ward et al., 2017). Perhaps progranulin-deficient neurons have a similar underlying defect in lysosome function that inhibits clearance of specific cargo while triggering a compensatory feedback loop that upregulates autophagy more generally.
Surprisingly, knock-down of some of our validated hits reduced autophagic flux in progranulin-deficient neurons and restored lysosomes to normal size. Knocking down genes regulating vesicular trafficking to lysosomes may help slow or reduce lipid delivery to lysosomes reducing excessive demands on the pathway by cargo overload. Interestingly, inhibition of autophagy in NPC1-deficient cells reduced cholesterol storage and restored normal lysosome enzyme proteolysis (Elrick and Lieberman, 2013). In addition, inhibition of autophagy also promoted increased progranulin secretion in neuronal cells (Capell et al., 2011; Osaka et al., 2015). These findings underscore the critical need to define the nature of deficits in the ALP, which appear commonly in different neurodegenerative diseases, to determine whether induction or inhibition would likely be helpful, and to design strategies for effective interventions.
Footnotes
This work was supported by the National Institutes of Health (Grants 1R21NS093236-01A1, 1P01AG054407, and 1RF1AG058476), the Consortium for Frontotemporal Dementia Research, the Taube/Koret Center for Neurodegenerative Disease Research, and the J. David Gladstone Institutes. The Gladstone Institutes received support from the National Center for Research Resources (Grant RR18928). We thank Robert V. Farese (Harvard Medical School), Andrew D. Nguyen (Harvard Medical School), and Ping Zhou (Gladstone Institute of Cardiovascular Disease) for kindly providing the GRN± and GRNR493X/+ mice and the progranulin rabbit polyclonal antibody; Laura Mitic (UCSF) and members of the Finkbeiner lab for discussion and comments on these experiments; Megan Laurance (UCSF) for assistance with Ingenuity Pathway Analysis; Hui Wang for animal husbandry; Gary Howard for editorial assistance; and Kelley Nelson for administrative assistance. PSA was a gift from R.G. Kalb (Mojsilovic-Petrovic et al., 2009).
The authors declare no competing financial interests. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Ahmed Z, Mackenzie IR, Hutton ML, Dickson DW (2007) Progranulin in frontotemporal lobar degeneration and neuroinflammation. J Neuroinflammation 4:7. 10.1186/1742-2094-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z, Sheng H, Xu YF, Lin WL, Innes AE, Gass J, Yu X, Wuertzer CA, Hou H, Chiba S, Yamanouchi K, Leissring M, Petrucelli L, Nishihara M, Hutton ML, McGowan E, Dickson DW, Lewis J (2010) Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol 177:311–324. 10.2353/ajpath.2010.090915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanesi J, Wang H, Sun HQ, Levine B, Yin H (2015) GABARAP-mediated targeting of PI4K2A/PI4KIIalpha to autophagosomes regulates PtdIns4P-dependent autophagosome-lysosome fusion. Autophagy 11:2127–2129. 10.1080/15548627.2015.1093718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida S, Zhou L, Gao FB (2011) Progranulin, a glycoprotein deficient in frontotemporal dementia, is a novel substrate of several protein disulfide isomerase family proteins. PLoS One 6:e26454. 10.1371/journal.pone.0026454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida S, Gao F, Coppola G, Gao FB (2016) Suberoylanilide hydroxamic acid increases progranulin production in iPSC-derived cortical neurons of frontotemporal dementia patients. Neurobiol Aging 42:35–40. 10.1016/j.neurobiolaging.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri DC, Stein GS, Lian JB, Languino LR (2012) TRAP-1, the mitochondrial Hsp90. Biochim Biophys Acta 1823:767–773. 10.1016/j.bbamcr.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso MR, Matassa DS, Laudiero G, Egorova AV, Polishchuk RS, Maddalena F, Piscazzi A, Paladino S, Sarnataro D, Garbi C, Landriscina M, Esposito F (2012) TRAP1 and the proteasome regulatory particle TBP7/Rpt3 interact in the endoplasmic reticulum and control cellular ubiquitination of specific mitochondrial proteins. Cell Death Differ 19:592–604. 10.1038/cdd.2011.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrant AE, Filiano AJ, Unger DE, Young AH, Roberson ED (2017) Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain 140:1447–1465. 10.1093/brain/awx060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431:805–810. 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Babst M, Odorizzi G, Estepa EJ, Emr SD (2000) Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248–258. 10.1034/j.1600-0854.2000.010307.x [DOI] [PubMed] [Google Scholar]
- Baldo B, Weiss A, Parker CN, Bibel M, Paganetti P, Kaupmann K (2012) A screen for enhancers of clearance identifies huntingtin as a heat shock protein 90 (Hsp90) client protein. J Biol Chem 287:1406–1414. 10.1074/jbc.M111.294801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa IA, Vega-Naredo I, Loureiro R, Branco AF, Garcia R, Scott PM, Oliveira PJ (2018) TRAP1 regulates autophagy in lung cancer cells. Eur J Clin Invest 48. [DOI] [PubMed] [Google Scholar]
- Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, Shaw C, Chandran S, Finkbeiner S (2014) Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol 10:677–685. 10.1038/nchembio.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel S, Moisse M, Damme M, De Muynck L, Robberecht W, Van Den Bosch L, Saftig P, Van Damme P (2017) Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo. Hum Mol Genet 26:2850–2863. 10.1093/hmg/ddx162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beel S, Herdewyn S, Fazal R, De Decker M, Moisse M, Robberecht W, Van Den Bosch L, Van Damme P (2018) Progranulin reduces insoluble TDP-43 levels, slows down axonal degeneration and prolongs survival in mutant TDP-43 mice. Mol Neurodegener 13:55. 10.1186/s13024-018-0288-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckel JN, Guarani V, Koyanagi M, Roexe T, Lengeling A, Schermuly RT, Gellert P, Braun T, Zeiher A, Dimmeler S (2011) Jumonji domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. Proc Natl Acad Sci U S A 108:3276–3281. 10.1073/pnas.1008098108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond LM, Peden AA, Kendrick-Jones J, Sellers JR, Buss F (2011) Myosin VI and its binding partner optineurin are involved in secretory vesicle fusion at the plasma membrane. Mol Biol Cell 22:54–65. 10.1091/mbc.e10-06-0553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ (1993) Optimized survival of hippocampal neurons in B27-supplemented neurobasal, a new serum-free medium combination. J Neurosci Res 35:567–576. 10.1002/jnr.490350513 [DOI] [PubMed] [Google Scholar]
- Canuel M, Korkidakis A, Konnyu K, Morales CR (2008) Sortilin mediates the lysosomal targeting of cathepsins D and H. Biochem Biophys Res Commun 373:292–297. 10.1016/j.bbrc.2008.06.021 [DOI] [PubMed] [Google Scholar]
- Capell A, Liebscher S, Fellerer K, Brouwers N, Willem M, Lammich S, Gijselinck I, Bittner T, Carlson AM, Sasse F, Kunze B, Steinmetz H, Jansen R, Dormann D, Sleegers K, Cruts M, Herms J, Van Broeckhoven C, Haass C (2011) Rescue of progranulin deficiency associated with frontotemporal lobar degeneration by alkalizing reagents and inhibition of vacuolar ATPase. J Neurosci 31:1885–1894. 10.1523/JNEUROSCI.5757-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell A, Fellerer K, Haass C (2014) Progranulin transcripts with short and long 5′ untranslated regions (UTRs) are differentially expressed via posttranscriptional and translational repression. J Biol Chem 289:25879–25889. 10.1074/jbc.M114.560128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik B, Sephton CF, Dewey CM, Xian X, Wei S, Yu K, Niu W, Coppola G, Coughlin SE, Lee SE, Dries DR, Almeida S, Geschwind DH, Gao FB, Miller BL, Farese RV Jr, Posner BA, Yu G, Herz J (2011) Suberoylanilide hydroxamic acid (vorinostat) up-regulates progranulin transcription: rational therapeutic approach to frontotemporal dementia. J Biol Chem 286:16101–16108. 10.1074/jbc.M110.193433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik B, Sephton CF, Kutluk Cenik B, Herz J, Yu G (2012) Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem 287:32298–32306. 10.1074/jbc.R112.399170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK (2007) JMJD6 is a histone arginine demethylase. Science 318:444–447. 10.1126/science.1145801 [DOI] [PubMed] [Google Scholar]
- Chang MC, Srinivasan K, Friedman BA, Suto E, Modrusan Z, Lee WP, Kaminker JS, Hansen DV, Sheng M (2017) Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J Exp Med 214:2611–2628. 10.1084/jem.20160999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li S, Su L, Sheng J, Lv W, Chen G, Xu Z (2015) Association of progranulin polymorphism rs5848 with neurodegenerative diseases: a meta-analysis. J Neurol 262:814–822. 10.1007/s00415-014-7630-2 [DOI] [PubMed] [Google Scholar]
- Chitramuthu BP, Baranowski DC, Kay DG, Bateman A, Bennett HP (2010) Progranulin modulates zebrafish motoneuron development in vivo and rescues truncation defects associated with knock-down of survival motor neuron 1. Mol Neurodegener 5:41. 10.1186/1750-1326-5-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, Raposo G (2013) Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 126:5553–5565. 10.1242/jcs.128868 [DOI] [PubMed] [Google Scholar]
- Elrick MJ, Lieberman AP (2013) Autophagic dysfunction in a lysosomal storage disorder due to impaired proteolysis. Autophagy 9:234–235. 10.4161/auto.22501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrick MJ, Yu T, Chung C, Lieberman AP (2012) Impaired proteolysis underlies autophagic dysfunction in niemann-pick type C disease. Hum Mol Genet 21:4876–4887. 10.1093/hmg/dds324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers BM, Rodriguez-Navas C, Tesla RJ, Prange-Kiel J, Wasser CR, Yoo KS, McDonald J, Cenik B, Ravenscroft TA, Plattner F, Rademakers R, Yu G, White CL 3rd, Herz J (2017) Lipidomic and transcriptomic basis of lysosomal dysfunction in progranulin deficiency. Cell Rep 20:2565–2574. 10.1016/j.celrep.2017.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J, Lee WC, Cook C, Finch N, Stetler C, Jansen-West K, Lewis J, Link CD, Rademakers R, Nykjær A, Petrucelli L (2012) Progranulin regulates neuronal outgrowth independent of sortilin. Mol Neurodegener 7:33. 10.1186/1750-1326-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, Janssens J, van der Zee J, Lang CM, Kremmer E, Martin JJ, Engelborghs S, Kretzschmar HA, Arzberger T, Van Broeckhoven C, Haass C, Capell A (2014) Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol 127:845–860. 10.1007/s00401-014-1262-6 [DOI] [PubMed] [Google Scholar]
- Gross C, Dubois-Pot H, Wasylyk B (2008) The ternary complex factor Net/Elk-3 participates in the transcriptional response to hypoxia and regulates HIF-1 alpha. Oncogene 27:1333–1341. 10.1038/sj.onc.1210736 [DOI] [PubMed] [Google Scholar]
- Heim A, Grimm C, Müller U, Häußler S, Mackeen MM, Merl J, Hauck SM, Kessler BM, Schofield CJ, Wolf A, Böttger A (2014) Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine-serine-rich (RS) domains of SR- and SR-like proteins. Nucleic Acids Res 42:7833–7850. 10.1093/nar/gku488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo SH, Cho JY (2014) ELK3 suppresses angiogenesis by inhibiting the transcriptional activity of ETS-1 on MT1-MMP. Int J Biol Sci 10:438–447. 10.7150/ijbs.8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DS, Banerji U, Tavana B, George GC, Aaron J, Kurzrock R (2013) Targeting the molecular chaperone heat shock protein 90 (HSP90): lessons learned and future directions. Cancer Treat Rev 39:375–387. 10.1016/j.ctrv.2012.10.001 [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Arai T, Masuda-Suzukake M, Kondo H, Matsuwaki T, Nishihara M, Hasegawa M, Akiyama H (2015) Progranulin reduction is associated with increased tau phosphorylation in P301L tau transgenic mice. J Neuropathol Exp Neurol 74:158–165. 10.1097/NEN.0000000000000158 [DOI] [PubMed] [Google Scholar]
- Hsiung GY, Fok A, Feldman HH, Rademakers R, Mackenzie IR (2011) rs5848 polymorphism and serum progranulin level. J Neurol Sci 300:28–32. 10.1016/j.jns.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Padukkavidana T, Vægter CB, Brady OA, Zheng Y, Mackenzie IR, Feldman HH, Nykjaer A, Strittmatter SM (2010) Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron 68:654–667. 10.1016/j.neuron.2010.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenzer C, Manil-Ségalen M, Lefebvre C, Largeau C, Glatigny A, Legouis R (2014) Human GABARAP can restore autophagosome biogenesis in a C. elegans lgg-1 mutant. Autophagy 10:1868–1872. 10.4161/auto.29745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämäläinen A, Viswanathan J, Natunen T, Helisalmi S, Kauppinen T, Pikkarainen M, Pursiheimo JP, Alafuzoff I, Kivipelto M, Haapasalo A, Soininen H, Herukka SK, Hiltunen M (2013) GRN variant rs5848 reduces plasma and brain levels of granulin in Alzheimer's disease patients. J Alzheimers Dis 33:23–27. 10.3233/JAD-2012-120946 [DOI] [PubMed] [Google Scholar]
- Liu J, Li H, Zhou B, Xu L, Kang X, Yang W, Wu S, Sun H (2015) PGRN induces impaired insulin sensitivity and defective autophagy in hepatic insulin resistance. Mol Endocrinol 29:528–541. 10.1210/me.2014-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Long YH, Wang SQ, Zhang YY, Li YF, Mi JS, Yu CH, Li DY, Zhang JH, Zhang XJ (2019) JMJD6 regulates histone H2A.X phosphorylation and promotes autophagy in triple-negative breast cancer cells via a novel tyrosine kinase activity. Oncogene 38:980–997. [DOI] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, Coppola G, Sloan SA, Hsieh CL, Kim CC, Bigio EH, Weintraub S, Mesulam MM, Rademakers R, Mackenzie IR, Seeley WW, et al. (2016) Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell 165:921–935. 10.1016/j.cell.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Lü L, Lu Y, Zhang L, Li B, Guo K, Chen L, Wang Y, Shao Y, Xu J (2014) Effects of hypoxia on progranulin expression in HT22 mouse hippocampal cells. Mol Med Rep 9:1675–1680. 10.3892/mmr.2014.2016 [DOI] [PubMed] [Google Scholar]
- Mantri M, Loik ND, Hamed RB, Claridge TD, McCullagh JS, Schofield CJ (2011) The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. Chembiochem 12:531–534. 10.1002/cbic.201000641 [DOI] [PubMed] [Google Scholar]
- Martens LH, Zhang J, Barmada SJ, Zhou P, Kamiya S, Sun B, Min SW, Gan L, Finkbeiner S, Huang EJ, Farese RV Jr (2012) Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J Clin Invest 122:3955–3959. 10.1172/JCI63113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S. (2016) No ATG8s, no problem? How LC3/GABARAP proteins contribute to autophagy. J Cell Biol 215:761–763. 10.1083/jcb.201611116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassa DS, Amoroso MR, Maddalena F, Landriscina M, Esposito F (2012) New insights into TRAP1 pathway. Am J Cancer Res 2:235–248. [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, Kirchof A, Gatti E, Helfrich MH, Wakatsuki S, Behrends C, Pierre P, Dikic I (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Minami SS, Min SW, Krabbe G, Wang C, Zhou Y, Asgarov R, Li Y, Martens LH, Elia LP, Ward ME, Mucke L, Farese RV Jr, Gan L (2014) Progranulin protects against amyloid beta deposition and toxicity in Alzheimer's disease mouse models. Nat Med 20:1157–1164. 10.1038/nm.3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojsilovic-Petrovic J, Nedelsky N, Boccitto M, Mano I, Georgiades SN, Zhou W, Liu Y, Neve RL, Taylor JP, Driscoll M, Clardy J, Merry D, Kalb RG (2009) FOXO3a is broadly neuroprotective in vitro and in vivo against insults implicated in motor neuron diseases. J Neurosci 29:8236–8247. 10.1523/JNEUROSCI.1805-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima T, Shigematsu N, Maruki R, Urano Y, Tanaka H, Shimaya A, Shimokawa T, Shibasaki M (2010) Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol 78:961–970. 10.1124/mol.110.065714 [DOI] [PubMed] [Google Scholar]
- Nguyen AD, Nguyen TA, Zhang J, Devireddy S, Zhou P, Karydas AM, Xu X, Miller BL, Rigo F, Ferguson SM, Huang EJ, Walther TC, Farese RV Jr (2018) Murine knockin model for progranulin-deficient frontotemporal dementia with nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A 115:E2849–E2858. 10.1073/pnas.1722344115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M (2016) Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol 215:857–874. 10.1083/jcb.201607039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka M, Ito D, Yagi T, Nihei Y, Suzuki N (2015) Evidence of a link between ubiquilin 2 and optineurin in amyotrophic lateral sclerosis. Hum Mol Genet 24:1617–1629. 10.1093/hmg/ddu575 [DOI] [PubMed] [Google Scholar]
- Pacheco CD, Lieberman AP (2007) Lipid trafficking defects increase beclin-1 and activate autophagy in niemann-pick type C disease. Autophagy 3:487–489. 10.4161/auto.4586 [DOI] [PubMed] [Google Scholar]
- Pacheco CD, Kunkel R, Lieberman AP (2007) Autophagy in niemann-pick C disease is dependent upon beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet 16:1495–1503. 10.1093/hmg/ddm100 [DOI] [PubMed] [Google Scholar]
- Park JH, Kim KP, Ko JJ, Park KS (2016) PI3K/Akt/mTOR activation by suppression of ELK3 mediates chemosensitivity of MDA-MB-231 cells to doxorubicin by inhibiting autophagy. Biochem Biophys Res Commun 477:277–282. 10.1016/j.bbrc.2016.06.057 [DOI] [PubMed] [Google Scholar]
- Pereson S, Wils H, Kleinberger G, McGowan E, Vandewoestyne M, Van Broeck B, Joris G, Cuijt I, Deforce D, Hutton M, Van Broeckhoven C, Kumar-Singh S (2009) Progranulin expression correlates with dense-core amyloid plaque burden in alzheimer disease mouse models. J Pathol 219:173–181. 10.1002/path.2580 [DOI] [PubMed] [Google Scholar]
- Perry DC, Lehmann M, Yokoyama JS, Karydas A, Lee JJ, Coppola G, Grinberg LT, Geschwind D, Seeley WW, Miller BL, Rosen H, Rabinovici G (2013) Progranulin mutations as risk factors for Alzheimer disease. JAMA Neurol 70:774–778. 10.1001/2013.jamaneurol.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau TL, Leavitt BR (2014) Progranulin in neurodegenerative disease. Trends Neurosci 37:388–398. 10.1016/j.tins.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Petkau TL, Neal SJ, Milnerwood A, Mew A, Hill AM, Orban P, Gregg J, Lu G, Feldman HH, Mackenzie IR, Raymond LA, Leavitt BR (2012) Synaptic dysfunction in progranulin-deficient mice. Neurobiol Dis 45:711–722. 10.1016/j.nbd.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Petoukhov E, Fernando S, Mills F, Shivji F, Hunter D, Krieger C, Silverman MA, Bamji SX (2013) Activity-dependent secretion of progranulin from synapses. J Cell Sci 126:5412–5421. 10.1242/jcs.132076 [DOI] [PubMed] [Google Scholar]
- Picher-Martel V, Dutta K, Phaneuf D, Sobue G, Julien JP (2015) Ubiquilin-2 drives NF-kappaB activity and cytosolic TDP-43 aggregation in neuronal cells. Mol Brain 8:71. 10.1186/s13041-015-0162-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscopo P, Rivabene R, Adduci A, Mallozzi C, Malvezzi-Campeggi L, Crestini A, Confaloni A (2010) Hypoxia induces up-regulation of progranulin in neuroblastoma cell lines. Neurochem Int 57:893–898. 10.1016/j.neuint.2010.09.008 [DOI] [PubMed] [Google Scholar]
- Poulard C, Rambaud J, Hussein N, Corbo L, Le Romancer M (2014) JMJD6 regulates ERalpha methylation on arginine. PLoS One 9:e87982. 10.1371/journal.pone.0087982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putcha P, Danzer KM, Kranich LR, Scott A, Silinski M, Mabbett S, Hicks CD, Veal JM, Steed PM, Hyman BT, McLean PJ (2010) Brain-permeable small-molecule inhibitors of Hsp90 prevent alpha-synuclein oligomer formation and rescue alpha-synuclein-induced toxicity. J Pharmacol Exp Ther 332:849–857. 10.1124/jpet.109.158436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovici GD, Miller BL (2010) Frontotemporal lobar degeneration: epidemiology, pathophysiology, diagnosis and management. CNS Drugs 24:375–398. 10.2165/11533100-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Neumann M, Mackenzie IR (2012) Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol 8:423–434. 10.1038/nrneurol.2012.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl L, Mackenzie IR, Förstl H, Kurz A, Diehl-Schmid J (2014) Frontotemporal lobar degeneration: current perspectives. Neuropsychiatr Dis Treat 10:297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan TA, Tumbarello DA (2018) Optineurin: a coordinator of membrane-associated cargo trafficking and autophagy. Front Immunol 9:1024. 10.3389/fimmu.2018.01024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenchenko K, Wasylyk C, Cheung H, Tourrette Y, Maas P, Schalken JA, van der Pluijm G, Wasylyk B (2016) XRP44X, an inhibitor of Ras/Erk activation of the transcription factor Elk3, inhibits tumour growth and metastasis in mice. PLoS One 11:e0159531. 10.1371/journal.pone.0159531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D, Mole SE, Staropoli JF, Sims KB, Lewis J, Lin WL, Dickson DW, Dahl HH, Bahlo M, Berkovic SF (2012) Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet 90:1102–1107. 10.1016/j.ajhg.2012.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh HS, Choi N, Tarassishin L, Lee SC (2012) Regulation of progranulin expression in human microglia and proteolysis of progranulin by matrix metalloproteinase-12 (MMP-12). PLoS One 7:e35115. 10.1371/journal.pone.0035115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, Miyata S, Takamura H, Katayama T, Tohyama M (2011) Mitochondrial TRAP1 regulates the unfolded protein response in the endoplasmic reticulum. Neurochem Int 58:880–887. 10.1016/j.neuint.2011.02.015 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Matsuwaki T, Yamanouchi K, Nishihara M (2013a) Exacerbated inflammatory responses related to activated microglia after traumatic brain injury in progranulin-deficient mice. Neuroscience 231:49–60. 10.1016/j.neuroscience.2012.11.032 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Matsuwaki T, Yamanouchi K, Nishihara M (2013b) Increased lysosomal biogenesis in activated microglia and exacerbated neuronal damage after traumatic brain injury in progranulin-deficient mice. Neuroscience 250:8–19. 10.1016/j.neuroscience.2013.06.049 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Chambers JK, Matsuwaki T, Yamanouchi K, Nishihara M (2014) Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta Neuropathol Commun 2:78. 10.1186/s40478-014-0078-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Suzuki G, Matsuwaki T, Hosokawa M, Serrano G, Beach TG, Yamanouchi K, Hasegawa M, Nishihara M (2017) Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum Mol Genet 26:969–988. 10.1093/hmg/ddx011 [DOI] [PubMed] [Google Scholar]
- Tanimoto R, Palladino C, Xu SQ, Buraschi S, Neill T, Gomella LG, Peiper SC, Belfiore A, Iozzo RV, Morrione A (2017) The perlecan-interacting growth factor progranulin regulates ubiquitination, sorting, and lysosomal degradation of sortilin. Matrix Biol 64:27–39. 10.1016/j.matbio.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Reineke LC, Jain A, Jung SY, Lloyd RE (2017) Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J Biol Chem 292:18886–18896. 10.1074/jbc.M117.800706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov AS, Miller J, Arrasate M, Wong JS, Pleiss MA, Finkbeiner S (2010) A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model. Proc Natl Acad Sci U S A 107:16982–16987. 10.1073/pnas.1004498107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, Shaby BA, Finkbeiner S (2013) Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol 9:586–592. 10.1038/nchembio.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Waxse BJ, Arden SD, Bright NA, Kendrick-Jones J, Buss F (2012) Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol 14:1024–1035. 10.1038/ncb2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbarello DA, Kendrick-Jones J, Buss F (2013) Myosin VI and its cargo adaptors: linking endocytosis and autophagy. J Cell Sci 126:2561–2570. 10.1242/jcs.095554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M, Masuda A, Dohmae N, Arita K, Yoshimatsu M, Iwai Y, Fukui Y, Ueda K, Hamamoto R, Shirakawa M, Sasaki H, Nakamura Y (2013) Lysyl 5-hydroxylation, a novel histone modification, by jumonji domain containing 6 (JMJD6). J Biol Chem 288:6053–6062. 10.1074/jbc.M112.433284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Chowdhury R, Schiller R, Ge W, Kawamura A, Schofield CJ (2016) Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun 7:11974. 10.1038/ncomms11974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sun HQ, Zhu X, Zhang L, Albanesi J, Levine B, Yin H (2015) GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc Natl Acad Sci U S A 112:7015–7020. 10.1073/pnas.1507263112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Liu NS, Seet LF, Hong W (2010) The emerging role of VHS domain-containing Tom1, Tom1L1 and Tom1L2 in membrane trafficking. Traffic 11:1119–1128. 10.1111/j.1600-0854.2010.01098.x [DOI] [PubMed] [Google Scholar]
- Ward ME, Chen R, Huang HY, Ludwig C, Telpoukhovskaia M, Taubes A, Boudin H, Minami SS, Reichert M, Albrecht P, Gelfand JM, Cruz-Herranz A, Cordano C, Alavi MV, Leslie S, Seeley WW, Miller BL, Bigio E, Mesulam MM, Bogyo MS, et al. (2017) Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med 9:eaah5642. 10.1126/scitranslmed.aah5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C, Zheng H, Castell C, Debussche L, Multon MC, Wasylyk B (2008) Inhibition of the ras-net (Elk-3) pathway by a novel pyrazole that affects microtubules. Cancer Res 68:1275–1283. 10.1158/0008-5472.CAN-07-2674 [DOI] [PubMed] [Google Scholar]
- Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR 3rd, Delahunty CM, Hahn P, Lengeling A, Mann M, Proudfoot NJ, Schofield CJ, Böttger A (2009) Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325:90–93. 10.1126/science.1175865 [DOI] [PubMed] [Google Scholar]
- Xu HM, Tan L, Wan Y, Tan MS, Zhang W, Zheng ZJ, Kong LL, Wang ZX, Jiang T, Tan L, Yu JT (2017) PGRN is associated with late-onset Alzheimer's disease: a case-control replication study and meta-analysis. Mol Neurobiol 54:1187–1195. 10.1007/s12035-016-9698-4 [DOI] [PubMed] [Google Scholar]
- Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, Zhang J, Xu P, Xu T (2012) Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 9:727–729. 10.1038/nmeth.2021 [DOI] [PubMed] [Google Scholar]
- Zhang X, Gao Y, Lu L, Zhang Z, Gan S, Xu L, Lei A, Cao Y (2015) JmjC domain-containing protein 6 (Jmjd6) derepresses the transcriptional repressor transcription factor 7-like 1 (Tcf7l1) and is required for body axis patterning during xenopus embryogenesis. J Biol Chem 290:20273–20283. 10.1074/jbc.M115.646554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG (2010) Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol 12:665–675. 10.1038/ncb2069 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Brady OA, Meng PS, Mao Y, Hu F (2011) C-terminus of progranulin interacts with the beta-propeller region of sortilin to regulate progranulin trafficking. PLoS One 6:e21023. 10.1371/journal.pone.0021023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun L, Bastos de Oliveira F, Qi X, Brown WJ, Smolka MB, Sun Y, Hu F (2015) Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol 210:991–1002. 10.1083/jcb.201502029 [DOI] [PMC free article] [PubMed] [Google Scholar]