Abstract
The nonsense-mediated mRNA decay pathway selects and degrades its targets using a dense network of RNA-protein and protein-protein interactions. Together, these interactions allow the pathway to collect copious information about the translating mRNA, including translation termination status, splice junction positions, mRNP composition, and 3’UTR length and structure. The core NMD machinery, centered on the RNA helicase UPF1, integrates this information to determine the efficiency of decay. A picture of NMD is emerging in which many factors contribute to the dynamics of decay complex assembly and disassembly, thereby influencing the probability of decay. The ability of the NMD pathway to recognize mRNP features of diverse potential substrates allows it to simultaneously perform quality control and regulatory functions. In vertebrates, increased transcriptome complexity requires balance between these two functions since high NMD efficiency is desirable for maintenance of quality control fidelity but may impair expression of normal mRNAs. NMD has adapted to this challenge by employing mechanisms to enhance identification of certain potential substrates, while using sequence-specific RNA-binding proteins to shield others from detection. These elaborations on the conserved NMD mechanism permit more sensitive post-transcriptional gene regulation but can have severe deleterious consequences, including the failure to degrade pathogenic aberrant mRNAs in many B cell lymphomas.
Graphical/Visual Abstract and Caption
The nonsense-mediated mRNA decay pathway, using the RNA helicase UPF1 as a focal point, collects many types of information about RNA-protein complexes to select its targets.
Introduction
Nonsense-mediated mRNA decay (NMD) was originally identified as a system to recognize and degrade mRNAs containing premature termination codons caused by genetic lesions (Maquat et al. 1981; Losson and Lacroute 1979), but this designation only begins to describe the various roles of this highly conserved pathway. Over the succeeding decades, it has become clear that the NMD machinery is responsible for a broad range of both quality control and regulatory functions, together affecting the expression of 5–10% of genes in diverse eukaryotes (Mühlemann and Jensen 2012). It cannot be conclusively determined whether NMD originally evolved because of pressure to develop regulatory or quality control capabilities, although it is widely hypothesized that suppression of aberrant transcript expression was an important driving force (Lloyd, 2018). The quality control functions of NMD are generally considered to involve elimination of transcripts with premature termination codons (PTCs) arising from mutations or mRNA processing errors, along with suppression of repetitive element and pseudogene expression (Rebbapragada and Lykke-Andersen 2009). In addition, a widespread role for NMD in degrading transcripts that are prone to low translation fidelity or frameshifting has also been proposed, and several viral RNA targets of NMD have been identified (May et al. 2018; Fontaine et al. 2018; Wada et al. 2018; Fiorini et al. 2018; Weil and Beemon 2006; Hogg 2016; Ashton T. Belew, Advani, and Dinman 2011; Ashton Trey Belew et al. 2014; Garcia-Moreno et al. 2019; Balistreri et al. 2014). All other NMD substrates that are not obviously aberrant or extracellular in nature are considered to be potential regulatory targets. The idea that degradation of such substrates as regulatory in nature is bolstered by several lines of evidence, including evolutionarily conserved targeting of specific mRNAs and pathways by NMD, as discussed below (Lloyd 2018).
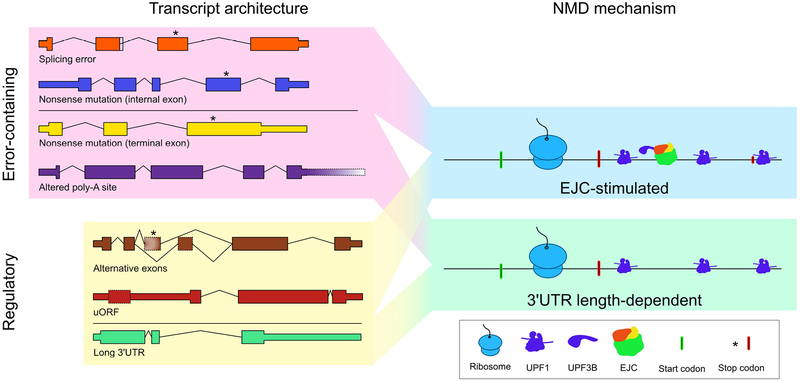
Distinguishing quality control and regulatory roles of NMD provides a conceptual framework for understanding the functional outcomes of decay, but the biochemical mechanism of NMD relies on sensing mRNP features rather than functional “intent.” The NMD machinery itself cannot assess the ultimate consequences (error elimination versus tuning the transcriptome) of mRNA decay events. Instead, the NMD pathway detects mRNP properties associated with premature translation termination events (Doma and Parker 2007), including features of translation, mRNA architecture, and mRNP composition [as discussed at length below and reviewed in (Karousis and Mühlemann 2018)]. Figure 1 captures various mRNA architectures associated with NMD susceptibility and the corresponding mRNP properties that drive decay. Notably, similar decay-promoting mRNP states can arise from either errors or regulated events. For example, exon junction complex (EJC)-stimulated decay can be caused by highly conserved alternative splicing events, while NMD of long 3’UTRs can be used for regulatory purposes but may also result from defective transcription termination. The biochemical similarity between mRNPs arising from errors and conserved regulatory processes suggests that the barrier to adaptation of a quality control mechanism for regulatory aims (or vice versa) is low.
Figure 1: The ribonucleoprotein (RNP) configuration of the 3’UTR of a message influences substrate selection for NMD.
Transcripts identified as decay substrates by the NMD machinery may contain errors or may be a part of a cellular regulatory program. The trigger for decay, however, is not determined by the process that generates a transcript but rather its 3’UTR RNP composition. Left, colored boxes indicate exons where narrow sections represent the 5’ and 3’ UTRs, while wide sections represent the coding sequence (CDS). Typical termination codons are at the point where a wide box transitions to a narrow box. Chevrons indicate introns. Right, various input transcripts have been distilled to the common RNP elements directing decay. Transcripts that have exon junction complexes (EJCs) downstream of a termination codon are efficiently degraded by NMD. This configuration may arise when the termination codon is positioned upstream (asterisk) from the typical termination codon or is in its native position but is still upstream of an EJC. A second signal that could activate the machinery is a long 3’UTR. The 3’UTR could be defined by a termination codon in its typical position, or a premature termination codon (PTC) in the last exon. In either case, UPF1 molecules accumulate downstream of the terminating ribosome in a length-dependent manner, enhancing the probability of decay. Specific molecules are labeled in the figure key.
Forty years of research into the functions of the pathway in humans and various model organisms have generated a body of data illuminating mechanisms and biological implications of NMD. Work from numerous labs has been highly successful in identifying factors required for NMD and describing a multitude of protein-protein and protein-RNA interactions that contribute to target selection and decay. From these efforts, it is clear that NMD requires a large number of pathway-specific factors, multifunctional RNA binding proteins, degradative enzymes, ribosomes, and translation factors to select and degrade its targets. These factors combine to perform essential roles in numerous cellular and developmental processes, such that many eukaryotes require NMD for viability (Anastasaki et al. 2011; McIlwain et al. 2010; Wittkopp et al. 2009; Metzstein and Krasnow 2006; Medghalchi et al. 2001; Weischenfeldt et al. 2008).
Biochemical analysis of interactions required for decay has been complemented by extensive structural analysis of the NMD machinery, generating a rich picture of RNP complexes formed en route to decay. Still, major questions remain. We do not understand how spatial and temporal organization of protein-protein and protein-RNA interactions affect the fates of potential NMD target mRNAs, or at what point in the process mRNAs are committed to decay. Critically, the gaps in our knowledge of how NMD protein interactions are orchestrated on mRNPs include how the NMD pathway works with the translation termination machinery to detect and analyze translation termination events. Further, as the field expands to examine the physiological functions of NMD, much remains to be learned about how NMD mechanisms are shaped by the competing needs for quality control and regulation in complex vertebrate transcriptomes.
BALANCING EFFICIENCY AND SPECIFICITY
With increasing biological complexity, the range of what should be considered normal expands and the task of accurately selecting decay targets by detecting deviations from normal mRNA properties and behaviors becomes more difficult. By way of analogy, a T-shirt factory producing only plain, white shirts can perform quality control by identifying any non-white shirts, while production of a range of colors requires verification that each individual shade is correct. This principle represents a problem for vertebrate NMD pathways, which are responsible for surveilling transcriptomes that are vastly more complex than those of the lower eukaryotes in which NMD is thought originally to have emerged (Causier et al. 2017; Lloyd 2018). Vertebrates have evolved expanded uses of many mechanisms that permit diversification of mRNA functions and regulation, including upstream open reading frames (uORFs), alternative pre-mRNA splicing (AS), alternative polyadenylation (APA), and 3’UTR-based regulation of mRNA translation and stability (e.g. binding sites for microRNAs and regulatory proteins) (Schaefke et al. 2018; Mayr 2016). Each of these mechanisms allows heightened control over gene expression but also increases the chance that a gene will produce mRNAs with features that are recognized by the NMD pathway (Figure 1).
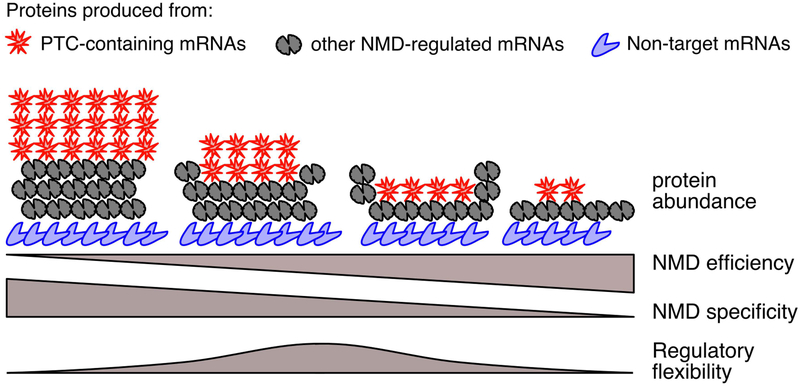
Degradation of mRNA is an irreversible event, and the decision to decay must therefore be approached with care. Success in identifying appropriate targets relies on maintaining the correct balance between decay efficiency and specificity (Figure 2). In other words, cells must concurrently work to minimize false-positives (inappropriate degradation of “normal” mRNAs) and false-negatives (failure to degrade aberrant mRNAs). Both types of errors can be costly: false negatives may result in retention of aberrant mRNAs that can cause expression of nonfunctional and/or dominant-negative proteins, and false positives can cause inappropriate degradation of transcripts encoding functional protein, thereby reducing or eliminating expression of important cellular factors. Particularly for rare transcripts, the cost of individual errors in quality control is expected to be high, as a substantial alteration of protein abundance can result from mis-regulation of individual mRNA molecules. As highlighted in Figure 2, overall NMD pathway efficiency may represent a compromise between aggressive clearance of transcripts containing true errors and preservation of apparently normal mRNAs with potential NMD-inducing features.
Figure 2: In order to simultaneously regulate and safeguard the transcriptome, NMD pathway functionality must balance multiple parameters.
As NMD efficiency increases, the protein products from PTC-containing and other NMD-regulated mRNAs will be strongly down-regulated, but mRNAs that are not normally NMD targets may also become susceptible to decay. Thus, aggressive clearance of aberrant transcripts may come at the cost of sacrificing beneficial mRNAs. Conversely, increased specificity could prevent targeting of all but mRNAs with the strongest NMD-promoting features. Further, an intermediate level of NMD efficiency may be optimal to simultaneously influence the function of multiple cellular pathways, as illustrated by hypothetical values of “regulatory flexibility” across the spectrum of NMD efficiency. Optimal NMD efficiencies to maintain regulatory flexibility are expected to vary according to cell type and physiological conditions.
BALANCING QUALITY CONTROL AND REGULATION
The potential susceptibility of apparently normal mRNAs to NMD can cause a loss of quality control specificity, but it can also be exploited by cells for regulatory purposes. We currently understand the physiological consequences of only a small subset of NMD-based regulatory events, but several uses of NMD to tune important regulatory pathways have been uncovered. In one provocative and well-developed example, numerous stress response genes in humans, flies, and plants are targeted by NMD through recognition of stop codons associated with upstream open reading frames (uORFs), 3’UTR introns, and long 3’UTRs (Goetz and Wilkinson 2017). Reciprocally, stress response pathways have been implicated in repression of NMD (Gardner 2008, 2010). As part of this regulatory circuit, Gadd45, a uORF-containing transcript and one of the few NMD targets conserved from flies to humans, promotes apoptosis in response to stress signaling. Remarkably, deletion of Gadd45 reverses the embryonic lethality of NMD deficiency in Drosophila (Nelson et al. 2016). Against the expectation that the lethal outcome of NMD knockout would be due to its ability to target a broad range of transcripts, including those encoding aberrant proteins, this finding demonstrates that misregulation of a single, important gene can have dramatic consequences. As suggested by the conservation of NMD-based Gadd45 regulation across great evolutionary distance, depletion of human paralogs of the protein was also capable of partially rescuing viability in human cells deficient for NMD activity. Conversely, increasing the efficiency of NMD, thereby causing dramatic down-regulation of Gadd45 and other stress response genes, could result in a failure to effectively mount responses to dangerous cellular conditions (Karam et al. 2015).
In plants, animals, and fungi, alternative splicing events linked to NMD (AS-NMD) are a commonly used mechanism to control gene expression (McGlincy and Smith 2008; Hamid and Makeyev 2014). The most extensively studied AS-NMD events involve skipping or inclusion of an alternatively spliced exon to produce an mRNA containing a premature termination codon, but AS-NMD can also involve alternative 5’ or 3’ splice site usage or intron retention (de Lima Morais and Harrison 2010). AS-NMD is particularly prevalent among genes involved in alternative splicing, many of which bind their own pre-mRNAs to promote the use of NMD-sensitive transcript isoforms (McGlincy and Smith 2008). Because this regulatory strategy does not require generation or evolutionary maintenance of particular coding sequences, it is straightforward for genes to add and subtract exons and/or retained introns to modulate NMD sensitivity. Perhaps for this reason, AS-NMD has independently arisen many times throughout eukaryotes; in fact, even for homologous genes regulated by AS-NMD, distinct RNA elements are often used to confer NMD sensitivity in different species (Lareau and Brenner 2015; de Lima Morais and Harrison 2010). Paradoxically, this highly flexible mode of gene regulation is, in several cases, associated with extreme levels of sequence conservation (Lareau and Brenner 2015). For example, members of the serine/arginine (SR) and heterogeneous nuclear ribonucleoprotein (hnRNP) families of RNA-binding proteins are frequently auto- and cross- regulated by alternative splicing of exons found within ultraconserved elements, defined as consisting of 200 or more continuous base pairs of perfect conservation among human, mouse, and rat (Bejerano et al. 2004; Lareau et al. 2007; Ni et al. 2007). The absolute sequence requirements for AS-NMD are minimal, making it unclear why the regulatory exons of these important splicing factors are so intolerant to sequence alteration.
The NMD pathway itself provides a third well-characterized example of NMD-based regulation. In human cells, NMD pathway components UPF1, UPF2, UPF3B, SMG1, SMG5, and SMG6 mRNAs have all been shown to be targets of NMD (Mendell et al. 2004; Yepiskoposyan et al. 2011; Lulu Huang et al. 2011; Singh, Rebbapragada, and Lykke-Andersen 2008). Introduction of several 3’UTRs from NMD factor mRNAs to reporter transcripts has confirmed that they are sufficient to induce recognition and decay by the NMD pathway (Yepiskoposyan et al. 2011; Singh, Rebbapragada, and Lykke-Andersen 2008). The concurrent auto-regulation of multiple NMD factors through long 3’UTR-directed NMD offers an elegant means of buffering changes in concentration or activity of individual pathway components. The utility of this regulatory mechanism is again underlined by its recurrent use in distinct kingdoms of life: NMD of UPF3 mRNAs is important for normal pathway function in Arabidopsis (Degtiar et al. 2015; Vexler et al. 2016), and Drosophila, like mammalian cells, control expression of SMG5 and SMG6 through NMD (Rehwinkel et al. 2005; Chapin et al. 2014).
In each of the cases discussed here, cells use NMD as a way to modulate expression of multiple components of important pathways. The use of feedback is pervasive, occurring between stress responses and NMD, between productive and non-productive splicing, and between synthesis and decay of NMD factors. Further, these examples illustrate that NMD is not necessarily optimized for efficient clearance of mRNAs containing NMD-sensitive stop codons. Instead, the evolutionarily conserved roles of NMD outlined here suggest that pathway activities are not only subject to potential competing needs of regulation and quality control but are also shaped by diverse regulatory aims that may favor more or less efficient decay. In Figure 2, we refer to this concept as “regulatory flexibility,” which we envision as the ability of NMD to simultaneously contribute to many regulatory programs. The optimal level of NMD activity will vary according to organism, cell type, and condition, but the diversity of known NMD functions and the demonstrated autoregulatory control of NMD through long 3’UTRs imply that regulatory flexibility may often be maximized at an intermediate level of NMD activity (Figure 2).
We propose that the NMD machinery and the transcriptome at large have co-evolved to permit expansion of post-transcriptional regulation while maintaining robust quality control of mRNAs encoding premature termination codons. In order to optimize the various outcomes of NMD, calibration of NMD substrate recognition is essential. Thus, NMD and transcriptome adaptations involve both enhancement and inhibition of multiple steps in decay target selection, balancing efficiency with accuracy, regulation with quality control, and diverse regulatory programs with each other (Figure 2). Below, we will discuss how the NMD machinery components interact to influence these outcomes.
NMD PROTEIN FACTORS AND MECHANISTIC FEATURES
The suite of proteins involved in the decay of specific NMD substrates is large and extensively studied. As several recent reviews have given comprehensive overviews of NMD pathway components and interactions, (Karousis and Mühlemann 2018; Popp and Maquat 2013; Smith and Baker 2015; Jaffrey and Wilkinson 2018; Raimondeau, Bufton, and Schaffitzel 2018; Hug, Longman, and Cáceres 2016), this review will focus on how factors involved in the pathway might collectively act to achieve quality control and regulation, as well as the trade-offs inherent in maintaining the balance between these functions of NMD. To establish the groundwork for this discussion, we briefly summarize key properties and functions of the core NMD proteins below.
The UPF proteins
The three most widely conserved NMD pathway components are the up-frameshift proteins UPF1, UPF2, and UPF3, so named because they were originally discovered in a S. cerevisiae genetic screen for proteins involved in ribosomal frameshifting (Culbertson, Underbrink, and Fink 1980). Together, the UPF proteins form the core machinery responsible for substrate discrimination and recruitment of decay enzymes throughout eukaryotes.
UPF1
An RNA helicase, UPF1 contains tandem RecA domains that together bind and hydrolyze ATP to drive 5’−3’ translocation (Fairman-Williams, Guenther, and Jankowsky 2010; Weng, Czaplinski, and Peltz 1998). In addition to providing the ATPase active site, the UPF1 RecA domains form an extended channel for non-specific RNA binding, mediated almost entirely by contacts with the sugar-phosphate backbone (Chakrabarti et al. 2011). This extended surface allows for tight RNA binding and processive translocation in vitro (Fiorini et al. 2015; Kanaan et al. 2018). RNA binding brings the ATP binding pocket into an active conformation, strongly enhancing ATP hydrolysis. Reciprocally, as discussed at length below, conformational changes in the protein upon ATP binding and hydrolysis regulate the affinity of UPF1 for RNA (Gowravaram et al. 2018; Cheng et al. 2007). The UPF1 enzymatic core is specialized for function in NMD through accessory protein features that regulate the helicase activity of UPF1 and permit it to engage in numerous protein-protein interactions. These include an N-terminal cysteine-histidine rich domain (CH domain), which recruits decapping factors and docks on the RecA2 domain to repress UPF1 helicase activity (Swisher and Parker 2011; He and Jacobson 2015; Chakrabarti et al. 2011), and N- and C-terminal extensions that directly engage decay enzymes (Ohnishi et al. 2003; Okada-Katsuhata et al. 2011; Loh, Jonas, and Izaurralde 2013; Dehecq et al. 2018).
UPF2
UPF2 harbors three phylogenetically conserved MIF4G domains, protein modules often found to mediate protein-protein interactions in mRNA processing, translation, and decay (Kadlec, Izaurralde, and Cusack 2004; Serin et al. 2001; Lykke-Andersen, Shu, and Steitz 2000). The C-terminus of UPF2 adopts an ordered conformation upon interacting with UPF1, forming a bipartite α-helix and β-hairpin interaction module that clamps onto the CH domain of UPF1 (Clerici et al. 2009). For an RNA-bound UPF1, this mode of interaction competes with the association of the UPF1 CH domain with its own RecA2 in the helicase core (Chakrabarti et al. 2011). As a result, binding of UPF2 to UPF1 causes the CH domain to release from RecA2, stimulating UPF1 ATPase and helicase activity (Chakrabarti et al. 2011). Thus, UPF2 is considered an activator of UPF1 that promotes NMD by relieving the autoinhibition of the UPF1 CH domain on its helicase core, stimulating ATP hydrolysis. In higher eukaryotes, UPF2 also functions to promote UPF1 phosphorylation, a decay-promoting event likely mediated by direct interaction between UPF2 and SMG1 (Kashima et al. 2006; Clerici et al. 2013).
UPF3
UPF3, a shuttling protein enriched in the nucleus at steady-state, binds UPF2 through an N-terminal RNP domain (also referred to as a RRM-like domain) (Kadlec, Izaurralde, and Cusack 2004; Serin et al. 2001; Lykke-Andersen, Shu, and Steitz 2000). Two paralogs of UPF3 are expressed throughout vertebrates, UPF3A and UPF3B (also known as UPF3X), and are considered to have opposing roles in NMD, which will be discussed at length in further sections (Serin et al. 2001; Shum et al. 2016). The discovery that UPF3B directly binds the EJC, a multi-protein complex deposited upstream of splice junctions shortly after the completion of splicing, provided a molecular explanation for the ability of the EJC to stimulate decay (V. N. Kim, Kataoka, and Dreyfuss 2001; Gehring et al. 2003, 2005; Chamieh et al. 2008; H. Le Hir et al. 2001). The EJC minimally consists of eIF4A3 held in an inactive conformation by the RBM8A/MAGOH heterodimer (Andersen et al. 2006; H. Le Hir et al. 2000; H. Le Hir, Moore, and Maquat 2000; Kataoka et al. 2000; V. N. Kim et al. 2001; Ballut et al. 2005), which in turn directly interacts with the C-terminal domain of UPF3B (Chamieh et al. 2008; Buchwald et al. 2010; Melero et al. 2012; Gehring et al. 2003). UPF3B alone does not significantly enhance UPF1 ATPase activity but does greatly promote UPF1 RNA helicase activity in the presence of UPF2 (Chamieh et al. 2008). UPF3B also promotes the phosphorylation of UPF1 in higher eukaryotes, an event that is attributed to its ability to bind UPF2 (Kashima et al. 2006). Importantly, UPF3 proteins are also required in organisms such as yeast that do not use the EJC for NMD and for at least some long 3’UTR-mediated decay events in vertebrates (Leeds et al. 1992; Cui et al. 1995; B. S. Lee and Culbertson 1995; Singh, Rebbapragada, and Lykke-Andersen 2008; Eberle et al. 2008; Boehm et al. 2014), suggesting that UPF3 can participate in conserved interactions with UPF1/2 without being directly recruited to NMD substrates through the EJC.
Mutations in the UPF3B gene are associated with intellectual disability and other neurodevelopmental disorders in humans, and Upf3b knockout mice demonstrate learning deficits (Laumonnier et al. 2010; Lynch et al. 2012; Tarpey et al. 2007; Xu et al. 2013; Addington et al. 2011; Szyszka et al. 2012; Jaffrey and Wilkinson 2018; L. Huang et al. 2017). In the frontal cortex of Upf3b knockout mice, hundreds of transcripts are upregulated, consistent with the possibility of NMD compromise, and several of these were shown to overlap with transcripts upregulated in UPF1 knockdown embryonic stem cells (L. Huang et al. 2017). The role of UPF3B in disease is attributed to its canonical role in promoting NMD through EJC-stimulated decay since transcripts with exon junctions downstream of termination codons are enriched among those upregulated in Upf3b knockout mice (Huang et al. 2017).
Factors linking the UPF proteins to decay
In metazoans, NMD induction requires phosphorylation of UPF1 by the PI3K-like kinase SMG1 (Kashima et al. 2006). Phosphorylation of multiple sites at the N- and C-termini provide binding platforms for the 14-3-3 domains of NMD proteins SMG5, SMG6, and SMG7 (Ohnishi et al. 2003; Okada-Katsuhata et al. 2011; Loh, Jonas, and Izaurralde 2013). SMG6, which engages in additional phosphorylation-independent interactions with UPF1, is a specialized PIN domain-containing endonuclease that has been proposed to carry out the bulk of the NMD degradative activities in metazoans (Chakrabarti et al. 2014; Nicholson et al. 2014; Eberle et al. 2009; Huntzinger et al. 2008; Gatfield and Izaurralde 2004). Alternatively, SMG5 and SMG7 form a sub-complex capable of recruiting the CCR4-Not deadenylase complex and decapping factors (Loh, Jonas, and Izaurralde 2013; Nicholson et al. 2018; Unterholzner and Izaurralde 2004).
While these observations have led to the model that SMG6 and SMG5/7 specify independent decay processes that each favor different substrates (Ottens et al. 2017), other RNA-Seq studies have found that a very similar profile of mRNAs is up-regulated in the absence of either SMG5/7 or SMG6 (Colombo et al. 2016). One possible explanation for the requirement for SMG5/7 and SMG6 for most or all NMD events is that SMG5/7 binding to phosphorylated UPF1 may be important for stabilizing UPF1 binding to target 3’UTRs, an activity that may help to promote both SMG6-dependent and SMG5/7-dependent decay (Kurosaki et al. 2014). Interestingly, the function of the C-terminus of UPF1 in recruiting degradative enzymes appears to be conserved even among organisms such as S. cerevisiae that have lost SMG1 (Dehecq et al. 2018).
INVESTIGATING STOP CODON POSITION THROUGH mRNP COMPOSITION
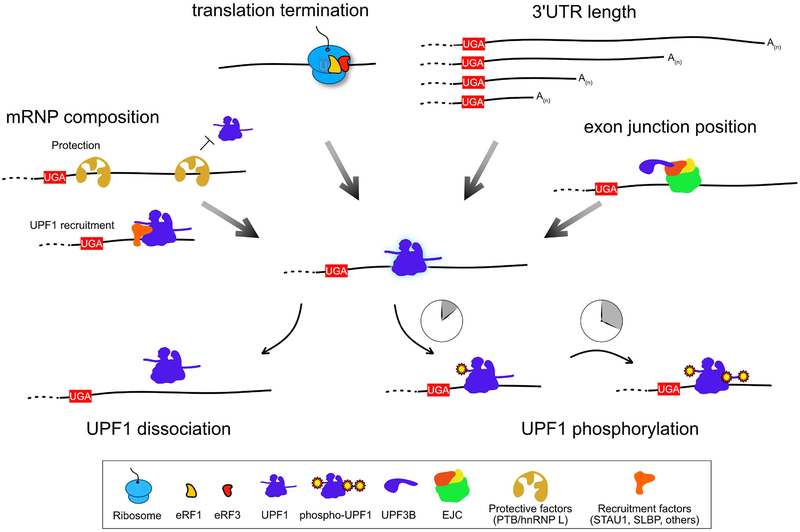
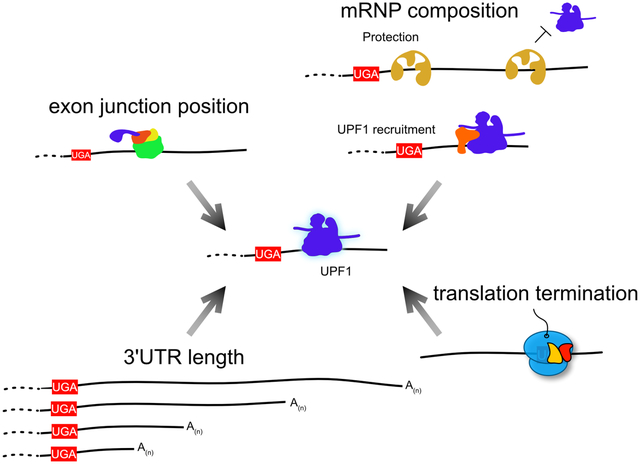
In order to correctly select its targets, the NMD pathway collects a large amount of information about each individual mRNP it surveys. The clues it collects to investigate an mRNA can include 3’UTR length, structure, and sequence; where, when, and perhaps how efficiently translation termination takes place; which proteins comprise the mRNP; and the position of exon junctions (Figure 3). Each mRNP feature surveyed by NMD contributes to a molecular analysis of the probability that a transcript should be cleared, allowing the cell to make the decision either to permit ongoing translation or to degrade the mRNA. Directly or indirectly, all of this information passes through the RNA helicase UPF1, the one universally recognized indispensable NMD factor. Integrating the accumulated knowledge of the mRNP, UPF1 can either engage and activate the degradative machinery or dissociate from the mRNP, preventing NMD. UPF1 thus serves as a molecular link between information-gathering and action.
Figure 3: UPF1 integrates information about mRNP configuration to define NMD substrates.
UPF1 occupies a central position in the NMD pathway due to its role in directly or indirectly sensing features of the mRNP. These signals include the length of the 3’UTR, the positions of EJCs, the presence of protective or recruitment factors, and the timing of translation termination. Integration of these signals will influence UPF1 phosphorylation, an important mark for recruitment and activation of decay factors. UPF1 molecules persistently associated with mRNPs are more likely to be highly phosphorylated. Specific molecules in the figure are labeled in the figure key.
Sensing Translation Termination
Translation termination is initiated when release factor eRF1, in complex with eRF3, recognizes a stop codon in the A site of the ribosome. Incorporation of eRF1 into the A site leads eRF3 to hydrolyze GTP and dissociate from the complex (L. Frolova et al. 1996; Alkalaeva et al. 2006; Eyler, Wehner, and Green 2013; Salas-Marco and Bedwell 2004). A conserved GGQ motif in eRF1 then extends to the peptidyl transferase center of the ribosome, where it mediates peptide hydrolysis (L. Y. Frolova et al. 1999). This activity can be aided by the AAA+ ATPase protein ABCE1, which promotes the active conformation of eRF1 and subsequently drives ribosome subunit splitting (Pisarev et al. 2010; Pisareva et al. 2011; Shoemaker and Green 2011). The individual ribosomal subunits are then recycled to participate in subsequent rounds of translation. The window of opportunity for the NMD pathway to recognize termination events is thus presumably between ribosome arrival at a termination codon and dissociation of the ribosomal subunits from the site of termination. The identity of the key player in the recognition of termination by the NMD machinery within this critical window is currently the subject of debate (Gao and Wilkinson 2017).
Evidence for recognition of termination by UPF1
The prevailing model in the field has long been that UPF1 directly senses termination events by interacting with release factors eRF1 and eRF3 in the A site. This model was originally based on the ability of UPF1 to co-immunoprecipitate with both eRF1 and eRF3 from yeast extracts (Czaplinski et al. 1998; W. Wang et al. 2001), an association later found to be conserved in mammalian cells (Ivanov et al. 2008; Singh, Rebbapragada, and Lykke-Andersen 2008; Kashima et al. 2006). Interaction studies with purified components indicated that UPF1 can independently bind to eRF1 and eRF3 (W. Wang et al. 2001; Kervestin et al. 2012). Further, purified recombinant eRF1 and eRF3 inhibited the RNA-dependent ATPase activity of UPF1 (Czaplinski et al. 1998; S. R. Lee et al. 2015), consistent with functionally significant, direct protein-protein interactions between UPF1 and the release factors.
In metazoans, the specific signal for decay has been proposed to depend on phosphorylation of UPF1 in a complex containing at least SMG-1, UPF1, and the release factors (SURF) (Kashima et al. 2006). However, only limited biochemical characterization of the interactions among UPF1, SMG1 and release factors has been performed (Neu-Yilik et al. 2017; Kervestin et al. 2012; Czaplinski et al. 1998; W. Wang et al. 2001; Kashima et al. 2006; Ivanov et al. 2008; Singh, Rebbapragada, and Lykke-Andersen 2008), and it remains to be directly demonstrated that SURF complex formation is required for UPF1 phosphorylation. In support of SMG1 activity outside of the translation termination context, it has been reported that UPF1 is more highly phosphorylated in cells treated with translation inhibitors cycloheximide or puromycin or that encounter other blocks to NMD (Dang et al. 2009; Durand, Franks, and Lykke-Andersen 2016). Further consistent with the possibility that UPF1 can be phosphorylated outside of the immediate context of translation termination, a transcriptome-wide study detected phospho-UPF1 binding throughout 3’UTRs rather than concentrated near the termination codon (Kurosaki et al. 2014).
While it is clear that translation termination is a required signal for NMD activation, the evidence for a reciprocal effect of NMD components on translation termination is mixed: deletion of UPF1 in yeast decreases termination efficiency, but depletion of UPF1 from mammalian cells has the opposite effect on termination in the context of a dual-luciferase reporter system (Ivanov et al. 2008; Keeling et al. 2004; W. Wang et al. 2001). Resolving this discrepancy hinges on determining whether UPF1 depletion affects the ribosome directly or indirectly. For example, the result of UPF1 deletion on yeast translation termination appears to be, at least in part, indirect: a defect in magnesium homeostasis occurs upon inactivation of the yeast NMD pathway, which in turn causes defective termination (Johansson and Jacobson 2010). In addition to this indirect effect of NMD on termination, more recent experiments in yeast also point to a direct role for UPF1 ATPase activity at the terminating ribosome. Expression of UPF1 variants defective for ATP hydrolysis was found to cause accumulation of 3’ decay intermediates derived from NMD target mRNAs (Serdar, Whiteside, and Baker 2016), potentially similar to those previously observed in human cells upon overexpression of analogous UPF1 mutants (Franks, Singh, and Lykke-Andersen 2010). These fragments induced by ATPase-dead yeast UPF1 expression appeared to be protected from exonucleolytic degradation by stably bound ribosomes, suggesting that UPF1 ATPase activity may be required for termination and/or ribosome recycling at NMD-inducing termination codons (Serdar, Whiteside, and Baker 2016).
An alternative model: UPF3 at the terminating ribosome
Recently, studies using purified cellular components have generated evidence for an alternative model of NMD factor engagement with the translation termination machinery (Neu-Yilik et al. 2017). In contrast to previous in vitro binding and functional assays indicating that UPF1 could directly interact with both eRF1 and eRF3 and that UPF2 and UPF3 were each independently capable of directly interacting with eRF3 (Kervestin et al. 2012; W. Wang et al. 2001; López-Perrote et al. 2016; Czaplinski et al. 1998; S. R. Lee et al. 2015), Neu-Yilik et al., found no evidence for a direct interaction between release factors and UPF1 or UPF2 (Neu-Yilik et al. 2017). Under conditions of limiting release factor abundance and in the absence of ABCE1, UPF3B was found to inhibit termination in a reconstituted human translation system. Further, post-termination complexes were more efficiently dissociated from model mRNAs in the presence of UPF3B. While UPF1 could still directly or indirectly affect these processes in the context of the cellular NMD pathway, it had no influence on these or other aspects of the reconstituted human translation termination system, or a similar system employing yeast components (Neu-Yilik et al. 2017; Schuller et al. 2018). These findings raise the possibility that UPF3 proteins communicate the status of the translation termination event to UPF1 and the remainder of the NMD pathway.
Are the kinetics of premature termination sensed by the NMD pathway?
Regardless of which factor or factors are responsible for linking NMD to translation termination, a central question remains: is the pathway capable of selectively recognizing some aspect of “premature” termination (i.e. a termination event associated with NMD) as distinct from normal termination events? Based on ribosome toeprinting studies in cell-free translation systems, it has been suggested that termination at a PTC is less efficient than termination at a normal stop codon due to a reduced pro-termination interaction between poly-A binding protein (PABP) and eRF3 (Amrani et al. 2004; Peixeiro et al. 2012). To explain these results, a “faux 3’UTR” model posits that increased distance between the terminating ribosome and the poly-A tail disfavors the interaction between PABP and eRF3, impairing termination efficiency (Amrani et al. 2004). In combination with enhanced recognition of long 3’UTRs by UPF1, this model may help explain why the probability of decay increases with 3’UTR length. Tethering of PABP in the vicinity of stop codons has been shown to stabilize NMD-sensitive mRNAs in yeast, flies, and mammals. These results are consistent with the idea that reduced PABP-eRF interactions permit UPF1-eRF binding and decay (Amrani et al. 2004; Ivanov et al. 2008; Singh, Rebbapragada, and Lykke-Andersen 2008; Eberle et al. 2008; Behm-Ansmant et al. 2007), but the stabilizing effect of PABP recruitment could also be explained by reduced UPF1 association with mRNPs (S. R. Lee et al. 2015). The hypothesis that NMD recognizes termination events due to the absence of a functional PABP-eRF3 interaction has been challenged by efforts in yeast to directly test the model: elimination of PABP-eRF3 binding, transient depletion of PABP, and use of reporter mRNAs lacking poly-A tails all failed to generate the predicted NMD defects (Meaux, van Hoof, and Baker 2008; Kervestin et al. 2012; Roque et al. 2015). As elucidating the events at the terminating ribosome is of central importance to understanding NMD mechanisms, further studies of termination and/or ribosome recycling at NMD-inducing stop codons in cells and reconstituted systems are needed.
Enhancing decay specificity by sensing splice junctions
In complex vertebrate transcriptomes, the EJC is a molecular marker that enhances the discrimination of NMD substrates (Woodward et al. 2016). Transcriptomes in eukaryotes that use the EJC for NMD have co-evolved with the pathway such that it is relatively rare for stop codons to occur more than 50 nt upstream of the final exon-exon junction (Bicknell et al. 2012). Given that EJCs are deposited ~20 nucleotides upstream of an exon-exon boundary, EJCs remaining more than ~30 nucleotides downstream of the TC can thus be used as a marker of premature termination, providing a rationale for degradation by the NMD pathway (Nagy and Maquat 1998).
A role for EJC protein partners in modulating NMD
The core EJC can associate with an array of peripheral EJC components, through which it promotes mRNA splicing fidelity, export, translation, and decay (Singh et al. 2015; Hervé Le Hir, Saulière, and Wang 2016). Of particular importance for NMD, UPF3B associates with the EJC in the nucleus and is exported on the spliced mRNP (Gehring, Lamprinaki, Hentze, et al. 2009; Baird et al. 2018). Following export of the EJC-UPF3B-bound mRNP, a combination of elongating ribosomes and the PYM protein displaces EJCs from the 5’UTR, ORF, and immediate vicinity of the TC, leaving only those EJCs deposited at junctions more than 50 nt downstream of the termination codon (Gehring, Lamprinaki, Kulozik, et al. 2009; Diem et al. 2007). UPF3B retained on the 3’UTR-associated EJCs is thought to drive recruitment of UPF2 to the mRNP, where it can activate UPF1 helicase activity and phosphorylation (Buchwald et al. 2010; Kashima et al. 2006; Chamieh et al. 2008; Chakrabarti et al. 2011). Additionally, the specific protein composition of the EJC presents opportunities for regulating and fine-tuning the NMD pathway. As the EJC matures, exchange of the multifunctional RNA binding protein RNPS1 for CASC3/MLN51/Barentz, a protein found to mark canonical sites of EJC deposition, has been proposed to cause a reduction in the NMD-enhancing potential of the EJC (Mabin et al. 2018; Hauer et al. 2016). If differential EJC composition modulates NMD efficiency, this may have far-reaching implications for substrate selection. A number of questions arise from this possibility, including whether EJCs downstream of PTCs undergo a distinct maturation process and whether transcripts with CASC3-containing EJCs may evade quality control. The mechanism by which NMD efficiency is altered by differential EJC composition remains to be elucidated, but could be due to higher levels of UPF2 and/or SR-protein recruitment to RNPS1-bound EJCs (Mabin et al. 2018; Aznarez et al. 2018; Gehring et al. 2005).
Competition between UPF3A and UPF3B
A second way in which EJC activity in NMD can be regulated is by modulating the link between the EJC and the NMD machinery. While UPF3B has been observed to be important for degradation of many NMD target mRNAs, UPF3A has been found to have at best weak activity in NMD (Chan et al. 2007; Kunz et al. 2006; Lykke-Andersen, Shu, and Steitz 2000). Explaining UPF3A’s conserved role in NMD, Shum and colleagues have recently presented evidence that UPF3A can act as an inhibitor of NMD, antagonizing UPF3B (Shum et al. 2016). In this work, cell lines and mice deficient for UPF3A exhibited more rapid decay of NMD targets, suggesting that the protein acts globally to inhibit NMD in multiple contexts. This activity of UPF3A is linked to its ability to decouple the EJC from NMD by engaging in interactions with UPF2 but failing to form a decay-promoting complex with the EJC (Shum et al. 2016; V. N. Kim et al. 2001; Kunz et al. 2006; Buchwald et al. 2010). UPF3A expression can be an important point of regulatory control. For example,UPF3A is highly upregulated in spermatocytes and is required for gametogenesis. Offering further opportunity for regulation, tissue-specific alternative splicing of UPF3A mRNAs generates proteins with differential ability to interact with UPF2 and inhibit NMD (Shum et al. 2016).
ICE1: promoting the link between splicing and decay
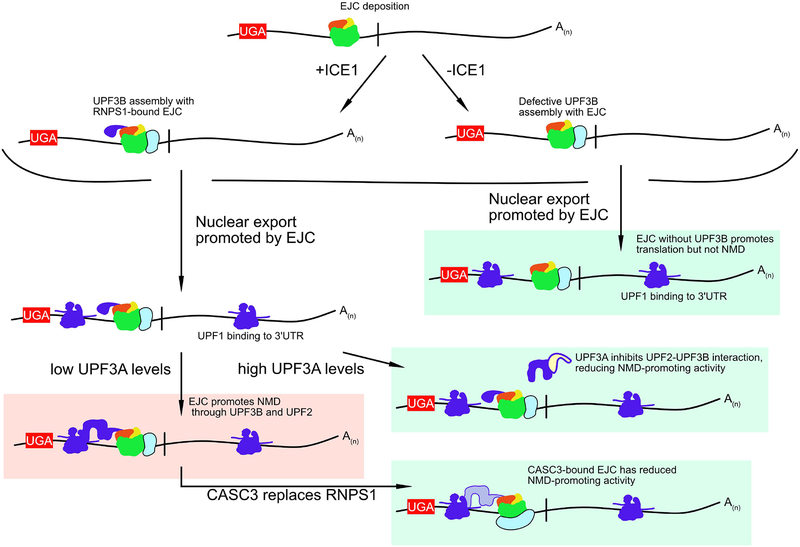
While UPF3A breaks the connection between the EJC and the NMD machinery by competing with UPF3B for UPF2 binding, a newly identified protein required for EJC-enhanced decay suggests that a similar effect can be accomplished by preventing the stable assembly of UPF3B with EJCs (Baird et al. 2018). Identified by whole-genome siRNA screening as promoting decay of a well-characterized EJC-stimulated NMD substrate, ICE1 uses a putative MIF4G domain to bind fully assembled EJCs (Baird et al. 2018). In the absence of ICE1, the interaction between the EJC and UPF3B is disrupted, causing reduced export of UPF3B from the nucleus (Figure 4). Supporting a model in which ICE1 transiently interacts with EJCs to promote UPF3B association, UPF3B overexpression was sufficient to partially rescue ICE1 knockdown. The requirement for ICE1 to promote stable linkage between the EJC and NMD implies the existence of an additional step in EJC-UPF3B assembly that may allow independent control of mRNA biogenesis and decay functions of the EJC.
Figure 4: Peripheral EJC factors influence NMD substrate selection.
ICE1-mediated assembly of the EJC with UPF3B leads to transcripts that are more likely to be targeted for decay. EJC-enhanced decay can be disrupted by UPF3A, which effectively competes with UPF3B for UPF2 binding. Additionally, replacement of RNPS1 with CASC3 may reduce the likelihood of NMD targeting, through a mechanism that remains to be elucidated but may involve altered association of NMD factors. Transcripts more likely to be degraded are over a red background while transcripts less likely to be degraded are over a green background.
Sensing 3’UTR length
Another physical consequence of increasing the distance between the termination codon and the poly-A tail (i.e. a longer 3’UTR) is an increase in mRNA sequence not traversed by the ribosome. In numerous experiments spanning the full range of model organisms used to investigate NMD, 3’UTR extension has been observed to increase the probability that an mRNA will undergo decay (Bühler et al. 2006; Singh, Rebbapragada, and Lykke-Andersen 2008; Behm-Ansmant and Izaurralde 2006; Hogg and Goff 2010; Matsuda et al. 2007; Baker and Hogg 2017). Using its high-affinity, sequence non-specific RNA binding activity, UPF1 can directly recognize this common feature of NMD substrates.
Dynamic, non-specific binding drives 3’UTR length sensing
Several distinct experimental contexts have uncovered a striking relationship between 3’UTR length and UPF1 binding. This principle has been demonstrated through the use of RNA tag-based mRNP purifications and UPF1 immunoprecipitations of several reporters with unrelated 3’UTR sequences (Hogg and Goff 2010; Kurosaki and Maquat 2013; Baker and Hogg 2017). Widespread sequence-independent association of UPF1 with 3’UTRs is supported by UV cross-linking and immunoprecipitation (CLIP) experiments (Hurt, Robertson, and Burge 2013; Zünd et al. 2013; Sundararaman et al. 2016), and both CLIP and RIP-Seq have identified a genome-wide correlation between UPF1 binding and 3’UTR length (Hurt, Robertson, and Burge 2013; S. R. Lee et al. 2015; Kishor, Ge, and Hogg 2019). As suggested by the ability of translational readthrough events to disrupt UPF1 binding to long 3’UTRs containing ORF extensions (Hogg and Goff 2010), UPF1 CLIP reads, usually concentrated on 3’UTRs, were broadly distributed across ORFs and 3’UTRs under conditions of global translational repression (Zünd et al. 2013). Together, these data support a model in which UPF1 is enriched on long 3’UTRs by a combination of non-specific mRNA binding events and clearance from ORFs by translocating ribosomes.
The apparent displacement of UPF1 by translation elongation presents an opportunity to probe UPF1 dynamics and activity on mRNPs by using readthrough-promoting RNA elements to modulate elongation through a region of 3’UTR lacking additional in-frame stop codons (Hogg and Goff 2010; Baker and Hogg 2017; Tang et al. 2016; Hogg 2011). Tuning translational readthrough efficiency with variants of the Moloney murine leukemia virus readthrough-promoting pseudoknot revealed that 5% readthrough of a ~700 nt 3’UTR caused displacement of UPF1 from the conditionally translated ORF, while less efficient readthrough driven by compromised pseudoknot variants allowed UPF1 to re-populate the mRNP (Hogg and Goff 2010). These findings suggested that steady state levels of UPF1 occupancy on 3’UTRs were determined by continuous binding and dissociation events.
A loophole: shielding genuine long 3’UTRs with protective proteins
The ability of UPF1 to discriminate potential NMD targets through sequence non-specific binding endangers the expression of messages that have evolved long 3’UTRs for important regulatory purposes. Vertebrate mRNAs have undergone a dramatic expansion of 3’UTR length, at least in part to allow binding of factors that regulate mRNA localization, decay, and translation (Mayr 2016). To safeguard expression of messages with long 3’UTRs and permit 3’UTR-based regulation, vertebrates have evolved a protective mechanism to inhibit decay by antagonizing UPF1 binding (Ge et al. 2016; Chester et al. 2003; Kishor, Ge, and Hogg 2019; Balagopal and Beemon 2017). The two most extensively characterized protective proteins, PTBP1 and hnRNP L, have a common evolutionary origin and both use four RNA recognition motifs (RRMs) to recognize their target RNAs with nanomolar affinity (Blatter et al. 2015). PTBP1 binds sequences high in pyrimidines (CU-rich tracts), while hnRNP L prefers CA-rich sequences (Pérez, McAfee, and Patton 1997; Hui et al. 2005). Knockdown and RNA-Seq studies indicate that they are independently responsible for shielding hundreds of human mRNAs from NMD in a manner correlated with their ability to inhibit UPF1 association (Ge et al. 2016; Kishor, Ge, and Hogg 2019). Importantly, mRNAs with long 3’UTRs and a high density of protective protein binding sites recruit similar levels of UPF1 as unprotected short 3’UTRs. Therefore, the protective proteins appear to function by preventing accurate 3’UTR length sensing. These findings suggest that the transcriptome and the NMD pathway have co-evolved to use PTBP1 and/or hnRNP L binding sites in the vicinity of stop codons to allow diversification of 3’UTR functions without NMD-imposed constraints on 3’UTR length.
The perils of removing mRNAs from surveillance
Increasing organismal complexity demands tradeoffs, and the use of protective proteins to shield physiological long 3’UTRs from NMD can have severe deleterious consequences due to a failure to properly recognize pathogenic aberrant mRNAs (Kishor, Ge, and Hogg 2019). Over 30 years ago, it was first recognized that VDJ recombination in B cells can lead to frequent translocations between an IgH locus on chromosome 14 and a BCL2 locus on chromosome 18 (Cleary, Smith, and Sklar 1986), causing over-expression of the anti-apoptotic BCL2 protein. These t(14:18) translocations are found in ~90% of follicular B cell lymphomas and ~15% of diffuse large B cell lymphomas, the two most common forms of adult lymphoma (Schuetz et al. 2012; Leich, Ott, and Rosenwald 2011). Because BCL2 overexpression promotes cell survival, t(14:18) translocations are thought to be important initiating events in the development of these neoplasms (Czabotar et al. 2014). In about half of the t(14:18) translocations found in B cell lymphoma patients, the translocation occurs in a small (~270 bp) major breakpoint region in the middle of the BCL2 3’UTR, resulting in hybrid BCL2:IGH mRNAs containing the BCL2 coding sequence, ~2500 nucleotides of BCL2 3’UTR, and several efficiently spliced IgH-derived exons (Seto 2002; Deng et al. 2007; Cleary, Smith, and Sklar 1986). Because the BCL2:IGH fusion mRNA 3’UTRs are long and contain multiple introns, they represent prototypical NMD substrates. However, instead of being decayed, these transcripts cause overexpression of BCL2 protein.
Discovery of hnRNP L as a protective protein led to the identification of the BCL2 mRNA as a transcript that could evade NMD by recruiting hnRNP L. The BCL2 mRNA engages hnRNP L through an extended CA-rich tract near its termination codon (D.-H. Lee et al. 2009), preserving the ~5000 nt BCL2 3’UTR for regulation by numerous miRNAs and RBPs (Wei, Li, and Gupta 2014; Chen et al. 2010; P. Huang et al. 2015; Sabirzhanov et al. 2014; H. Wang et al. 2015; Ouyang et al. 2012; Zhong et al. 2016; Cimmino et al. 2005; Klein et al. 2010; Calin et al. 2005, 2002). In the case of the BCL2:IgH fusion transcript, hnRNP L engages the TC-proximal CA-tract to override the NMD-promoting signals of the aberrant 3’UTR architecture. Mutation of the hnRNP L binding sites in reporter mRNAs restored the expected NMD sensitivity of the fusion mRNAs, and disruption of the tract by CRISPR/Cas9 in a B cell lymphoma line carrying the t(14:18) translocation caused reduced BCL2 expression and increased apoptosis (Kishor, Ge, and Hogg 2019). These findings suggest that the protective mechanism causes a critical failure to eliminate aberrant BCL2:IGH fusion transcripts in B cell lymphomas and may also contribute to the survival of the many cancers that rely on BCL2 overexpression (Czabotar et al. 2014).
UPF1 functional modulation: ATPase activity and phosphorylation
A major factor influencing UPF1 occupancy and ability to correctly distinguish substrates is regulation of RNA binding through ATP binding and hydrolysis. UPF1 has long been known to exhibit reduced RNA binding activity in the presence of ATP (Weng, Czaplinski, and Peltz 1998; Czaplinski et al. 1995). UPF1 mutants deficient for ATPase activity exhibit enhanced RNA binding, causing them to be indiscriminately and non-productively locked onto substrate and non-substrate 3’UTRs alike (S. R. Lee et al. 2015; Kurosaki et al. 2014). Conversely, a point mutation that relieves the autoinhibition of UPF1 ATPase activity imposed by the CH domain (F192E in human UPF1) greatly reduces steady-state RNA binding and prevents target selection (Chakrabarti et al. 2011; S. R. Lee et al. 2015). Thus, regulation of UPF1 association with mRNAs by ATP binding and hydrolysis provides a mechanism for continuous cycling of the protein throughout the transcriptome. These observations imply that any disruption of normal UPF1 equilibrium binding, either to favor association or dissociation, can serve as a mechanism to regulate NMD efficiency. Consistent with this idea, increasing recruitment of UPF1, either through artificial means such as MS2 coat protein fusions or with endogenous factors like Staufen1, SRSF1, the nuclear cap binding complex, Regnase, the glucocorticoid receptor, or the histone stem loop binding protein promotes mRNA decay (Y. K. Kim et al. 2005; Kaygun and Marzluff 2005; Aznarez et al. 2018; Mino et al. 2015; Y. K. Kim and Maquat 2019; Hwang et al. 2010; Lykke-Andersen, Shu, and Steitz 2000).
Continued residence of UPF1 on mRNAs appears to favor UPF1 phosphorylation, cumulatively increasing the potential for decay factor recruitment and activation (S. R. Lee et al. 2015; Kurosaki et al. 2014; Durand, Franks, and Lykke-Andersen 2016; Pal et al. 2001). However, there are apparent limits on the NMD-promoting effects of increased UPF1 phosphorylation. Evidence that ATPase-deficient mutants of UPF1 are hyper-phosphorylated (presumably due to their long residence time on mRNAs) but do not support efficient decay suggests that UPF1 phosphorylation does not commit an mRNP to decay (Isken et al. 2008; Kashima et al. 2006; Durand, Franks, and Lykke-Andersen 2016). Phosphorylated UPF1 appears to be stabilized on mRNPs through interactions with the SMG5/7 complex, but the effects of phosphorylation may be reversed either by phosphatases such as PP2A while UPF1 is still engaged with target or by the dissociation of phospho-UPF1 from the target mRNP (with UPF1 dephosphorylation occurring subsequently) (Anders, Grimson, and Anderson 2003; Ohnishi et al. 2003; Kurosaki et al. 2014).
HOW DOES SENSING BECOME DOING?
NMD models often depict a linear pathway, involving an ordered, stepwise, irreversible progression to decay. Alternatively, we envision that the decision to decay is due to an ensemble of potential interactions and enzymatic reactions that are not strictly ordered and are often reversible in nature (Figure 5). This view gains inspiration from detailed single-molecule analyses of pre-mRNA splicing performed by the Moore and Gelles labs (A. A. H. Hoskins et al. 2011; A. A. Hoskins et al. 2016). Those studies revealed ordered but frequently reversible steps in spliceosome assembly that progressively increase the probability that the pre-mRNA will be spliced, none of which individually represent a commitment to splicing. Likewise, it is possible that no individual events prior to decapping or SMG6-mediated cleavage commit a message to NMD. Instead, increased assembly of pro-decay factors on an mRNA may cumulatively make mRNA degradation more likely. In the case of NMD, many events are known to increase the probability of decay but not be strictly required for decay of all substrates. For example, the EJC is a potent stimulator of NMD in vertebrates, but a large number of transcripts constitutively undergo decay in a 3’UTR EJC-independent manner (Metze et al. 2013). Moreover, several groups have presented evidence for NMD pathway “branching”, in which even core NMD factors such as UPF2 and UPF3B are dispensable for decay of certain substrates (Metze et al. 2013; Lulu Huang et al. 2011; Gehring et al. 2005; Chan et al. 2007). These findings suggest that there are multiple possible assemblies of NMD components that can achieve decay, up to and including initiation of decay by entirely distinct degradative enzymes. Mechanistic flexibility may be another strategy evolved by the NMD pathway to function effectively in complex transcriptomes, since the streamlined yeast pathway shows equal dependence on UPF1, UPF2, and UPF3 for degradation of NMD target mRNAs (Celik et al. 2017).
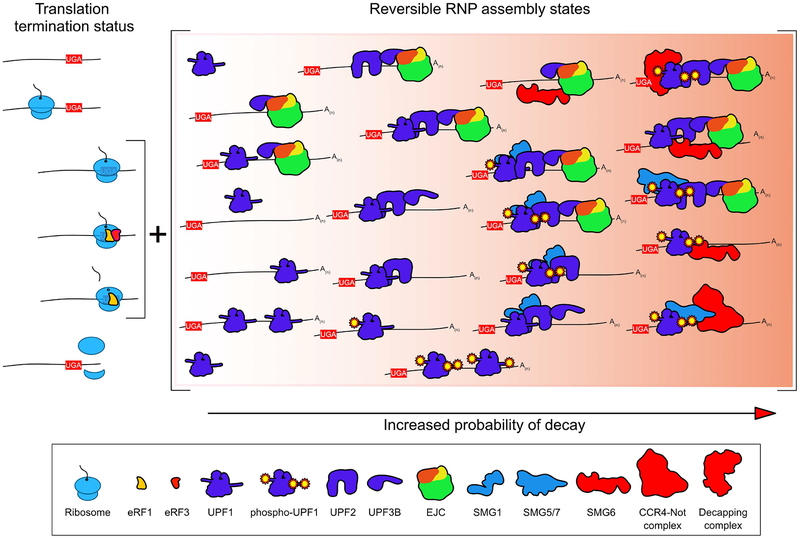
Figure 5: Reversible RNP assembly and modification states may determine the probability of NMD.
NMD requires concurrent translation termination (the left side of figure indicates the state of the terminating ribosome, and the bracket represents the critical window of opportunity for NMD activation) and occupancy of NMD proteins on the transcript. Assembly of these components, however, does not have to be a linear process. Instead, many binding events may be reversible, and modifications such as UPF1 phosphorylation may be undone by the activity of phosphatases. This model highlights the heterogeneity of possible RNP configurations that may ultimately lead to decay. Specific molecules in the figure are labeled in the figure key.
We have described in this review several aspects of mRNP function, structure, and composition that are evaluated by UPF1 and the rest of the NMD machinery in the process of decay target selection. Importantly, many of the inputs to the NMD decision are not static properties of the mRNP but are instead highly dynamic, meaning that the decision to decay can be thought of as a complex kinetic competition between decay promoting and decay inhibiting events (i.e. assembly and disassembly of NMD factors). The precise parameters of this competition remain unclear, but it is highly likely that a minimal requirement for NMD is that some component of the pathway must be mRNP-associated and in communication with a terminating ribosome. In the absence of in vivo evidence that ribosomes are particularly long-lived at NMD-inducing stop codons, the window of opportunity between eRF1 incorporation and ribosome recycling could be fleeting, albeit recurrent. Increased association of NMD-promoting factors with an mRNP would therefore increase the probability of recognition of a potential NMD-inducing termination event. This is the probable mechanism by which the EJC stimulates decay--the local concentration of UPF3B and UPF2 is increased on the message, promoting phosphorylation of 3’UTR-bound UPF1 and/or facilitating interactions with the terminating ribosome. Enhanced association of any individual pro-NMD factor could similarly increase the probability of decay, as evidenced by the ability of many NMD factors to promote decay when artificially tethered to mRNAs (Nicholson et al. 2018; Lykke-Andersen, Shu, and Steitz 2000; Unterholzner and Izaurralde 2004; Nicholson, Joncourt, and Mühlemann 2012; Cho, Kim, and Kim 2009; Loh, Jonas, and Izaurralde 2013; Ivanov et al. 2008; Boehm et al. 2016).
Conclusion
In this review, we have outlined a picture of NMD as a pathway that is mechanistically and functionally flexible, yet constrained in important ways by the need to maintain evolutionarily conserved regulatory and surveillance functions. The flexibility of the pathway allows different organisms and cell types to adapt the core quality control machinery to distinct regulatory aims in ways that are only beginning to be appreciated. Future efforts to explore NMD in diverse systems will certainly offer new surprises about the functional consequences of NMD, both beneficial and deleterious. Along with expansion of studies of NMD to new biological and disease contexts, better experimental and bioinformatic approaches are needed to unambiguously identify direct and indirect targets of NMD, including the use of metabolic labeling techniques to assess transcript stability (Tani et al. 2012). Developing a better understanding of the manifold functions of NMD will in turn aid the elucidation of NMD mechanisms.
There is much to be discovered, but the known diversity of NMD targets and mechanisms has already given rise to a many models for NMD target selection and decay. In part, this likely reflects a complex biological reality in which NMD is the product of dynamic, reversible, and frequently redundant interactions rather than a strictly ordered pathway. The view of NMD as an ensemble of possible mRNP states with a gradient of decay probabilities outlined here implies that many of the outstanding mechanistic questions in the field require approaches that allow analysis at the level of the RNP. It is of particular importance to understand how specific RNA features, the translation apparatus, and NMD factors combine as components of large RNA-protein assemblies to generate coherent regulatory outputs. Efforts to approach these questions will require methods capable of accounting for the activities of multiple factors in time and space, as the RNP makes the journey from translation to decay.
ACKNOWLEDGEMENTS
We thank Nazmul Haque for critical reading of the manuscript.
FUNDING INFORMATION
This work was supported by the Intramural Research Program, National Institutes of Health, National Heart, Lung, and Blood Institute
Contributor Information
Aparna Kishor, Biochemistry and Biophysics Center, National Heart, Lung, and Blood Institute, National Institutes of Health,.
Sarah E. Fritz, Biochemistry and Biophysics Center, National Heart, Lung, and Blood Institute, National Institutes of Health,.
J. Robert Hogg, Biochemistry and Biophysics Center, National Heart, Lung, and Blood Institute, National Institutes of Health,.
References
- Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, et al. 2011. “A Novel Frameshift Mutation in UPF3B Identified in Brothers Affected with Childhood Onset Schizophrenia and Autism Spectrum Disorders.” Molecular Psychiatry 16 (3): 238–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva Elena Z., Pisarev Andrey V., Frolova Lyudmila Y., Kisselev Lev L., and Pestova Tatyana V.. 2006. “In Vitro Reconstitution of Eukaryotic Translation Reveals Cooperativity between Release Factors eRF1 and eRF3.” Cell 125 (6): 1125–36. [DOI] [PubMed] [Google Scholar]
- Amrani Nadia, Ganesan Robin, Kervestin Stephanie, Mangus David A., Ghosh Shubhendu, and Jacobson Allan. 2004. “A Faux 3’-UTR Promotes Aberrant Termination and Triggers Nonsense-Mediated mRNA Decay.” Nature 432 (7013): 112–18. [DOI] [PubMed] [Google Scholar]
- Anastasaki Corina, Longman Dasa, Capper Amy, Patton E. Elizabeth, and Cáceres Javier F.. 2011. “Dhx34 and Nbas Function in the NMD Pathway and Are Required for Embryonic Development in Zebrafish.” Nucleic Acids Research 39 (9): 3686–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen Christian B. F., Ballut Lionel, Johansen Jesper S., Chamieh Hala, Nielsen Klaus H., Oliveira Cristiano L. P., Jan Skov Pedersen, Bertrand Séraphin, Hervé Le Hir, and Gregers Rom Andersen. 2006. “Structure of the Exon Junction Core Complex with a Trapped DEAD-Box ATPase Bound to RNA.” Science 313 (5795): 1968–72. [DOI] [PubMed] [Google Scholar]
- Anders Kirk R., Grimson Andrew, and Anderson Philip. 2003. “SMG-5, Required for C.elegans Nonsense-Mediated mRNA Decay, Associates with SMG-2 and Protein Phosphatase 2A.” The EMBO Journal 22 (3): 641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznarez Isabel, Nomakuchi Tomoki T., Jaclyn Tetenbaum-Novatt Rahman Mohammad Alinoor, Fregoso Oliver, Rees Holly, and Krainer Adrian R.. 2018. “Mechanism of Nonsense-Mediated mRNA Decay Stimulation by Splicing Factor SRSF1.” Cell Reports 23 (7): 2186–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird Thomas D., Ken Chih-Chien Cheng Yu-Chi Chen, Buehler Eugen, Martin Scott E., Inglese James, and Hogg J. Robert. 2018. “ICE1 Promotes the Link between Splicing and Nonsense-Mediated mRNA Decay.” eLife 7 (March). 10.7554/eLife.33178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Stacey L., and Hogg J. Robert. 2017. “A System for Coordinated Analysis of Translational Readthrough and Nonsense-Mediated mRNA Decay.” PloS One 12 (3): e0173980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal Vidya, and Beemon Karen L.. 2017. “Rous Sarcoma Virus RNA Stability Element Inhibits Deadenylation of mRNAs with Long 3’UTRs.” Viruses 9 (8). 10.3390/v9080204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balistreri Giuseppe, Horvath Peter, Schweingruber Christoph, David Zünd Gerald McInerney, Merits Andres, Oliver Mühlemann Claus Azzalin, and Helenius Ari. 2014. “The Host Nonsense-Mediated mRNA Decay Pathway Restricts Mammalian RNA Virus Replication.” Cell Host & Microbe 16 (3): 403–11. [DOI] [PubMed] [Google Scholar]
- Ballut Lionel, Marchadier Brice, Baguet Aurélie, Tomasetto Catherine, Bertrand Séraphin, and Hervé Le Hir. 2005. “The Exon Junction Core Complex Is Locked onto RNA by Inhibition of eIF4AIII ATPase Activity.” Nature Structural & Molecular Biology 12 (10): 861–69. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant Isabelle, Gatfield David, Rehwinkel Jan, Hilgers Valérie, and Izaurralde Elisa. 2007. “A Conserved Role for Cytoplasmic poly(A)-Binding Protein 1 (PABPC1) in Nonsense-Mediated mRNA Decay.” The EMBO Journal 26 (6): 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant Isabelle, and Izaurralde Elisa. 2006. “Quality Control of Gene Expression: A Stepwise Assembly Pathway for the Surveillance Complex That Triggers Nonsense-Mediated mRNA Decay.” Genes & Development 20 (4): 391–98. [DOI] [PubMed] [Google Scholar]
- Bejerano Gill, Pheasant Michael, Makunin Igor, Stephen Stuart, Kent W. James, Mattick John S., and Haussler David. 2004. “Ultraconserved Elements in the Human Genome.” Science 304 (5675): 1321–25. [DOI] [PubMed] [Google Scholar]
- Belew Ashton T., Advani Vivek M., and Dinman Jonathan D.. 2011. “Endogenous Ribosomal Frameshift Signals Operate as mRNA Destabilizing Elements through at Least Two Molecular Pathways in Yeast.” Nucleic Acids Research 39 (7): 2799–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belew Ashton Trey, Meskauskas Arturas, Musalgaonkar Sharmishtha, Advani Vivek M., Sulima Sergey O., Kasprzak Wojciech K., Shapiro Bruce A., and Dinman Jonathan D.. 2014. “Ribosomal Frameshifting in the CCR5 mRNA Is Regulated by miRNAs and the NMD Pathway.” Nature 512 (7514): 265–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell Alicia A., Cenik Can, Chua Hon N., Roth Frederick P., and Moore Melissa J.. 2012. “Introns in UTRs: Why We Should Stop Ignoring Them.” BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology 34 (12): 1025–34. [DOI] [PubMed] [Google Scholar]
- Blatter Markus, Stanislaw Dunin-Horkawicz Inna Grishina, Maris Christophe, Thore Stephane, Maier Timm, Bindereif Albrecht, Bujnicki Janusz M., and Frédéric H-T Allain. 2015. “The Signature of the Five-Stranded vRRM Fold Defined by Functional, Structural and Computational Analysis of the hnRNP L Protein.” Journal of Molecular Biology 427 (19): 3001–22. [DOI] [PubMed] [Google Scholar]
- Boehm Volker, Gerbracht Jennifer V., Marx Marie-Charlotte, and Gehring Niels H.. 2016. “Interrogating the Degradation Pathways of Unstable mRNAs with XRN1-Resistant Sequences.” Nature Communications 7 (December): 13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm Volker, Haberman Nejc, Ottens Franziska, Ule Jernej, and Gehring Niels H.. 2014. “3’ UTR Length and Messenger Ribonucleoprotein Composition Determine Endocleavage Efficiencies at Termination Codons.” Cell Reports 9 (2): 555–68. [DOI] [PubMed] [Google Scholar]
- Buchwald Gretel, Ebert Judith, Basquin Claire, Sauliere Jerome, Jayachandran Uma, Bono Fulvia, Le Hir Hervé, and Elena Conti. 2010. “Insights into the Recruitment of the NMD Machinery from the Crystal Structure of a Core EJC-UPF3b Complex.” Proceedings of the National Academy of Sciences of the United States of America 107 (22): 10050–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler Marc, Steiner Silvia, Mohn Fabio, Paillusson Alexandra, and Oliver Mühlemann. 2006. “EJC-Independent Degradation of Nonsense Immunoglobulin-Mu mRNA Depends on 3’ UTR Length.” Nature Structural & Molecular Biology 13 (5): 462–64. [DOI] [PubMed] [Google Scholar]
- Calin George Adrian, Calin Dan Dumitru Masayoshi Shimizu, Bichi Roberta, Zupo Simona, Noch Evan, Aldler Hansjuerg, et al. 2002. “Frequent Deletions and down-Regulation of Micro- RNA Genes miR15 and miR16 at 13q14 in Chronic Lymphocytic Leukemia.” Proceedings of the National Academy of Sciences of the United States of America 99 (24): 15524–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin George Adrian, Ferracin Manuela, Cimmino Amelia, Gianpiero Di Leva Masayoshi Shimizu, Wojcik Sylwia E., Iorio Marilena V., et al. 2005. “A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia.” The New England Journal of Medicine 353 (17): 1793–1801. [DOI] [PubMed] [Google Scholar]
- Causier Barry, Li Zhen, De Smet Riet, James P. B. Lloyd, Van de Peer Yves, and Davies Brendan. 2017. “Conservation of Nonsense-Mediated mRNA Decay Complex Components Throughout Eukaryotic Evolution.” Scientific Reports 7 (1): 16692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik Alper, Baker Richard, He Feng, and Jacobson Allan. 2017. “High Resolution Profiling of NMD Targets in Yeast Reveals Translational Fidelity as a Basis for Substrate Selection.” RNA, February 10.1261/rna.060541.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti Sutapa, Bonneau Fabien, Steffen Schüssler Elfriede Eppinger, and Conti Elena. 2014. “Phospho-Dependent and Phospho-Independent Interactions of the Helicase UPF1 with the NMD Factors SMG5-SMG7 and SMG6.” Nucleic Acids Research 42 (14): 9447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti Sutapa, Jayachandran Uma, Bonneau Fabien, Fiorini Francesca, Basquin Claire, Domcke Silvia, Hervé Le Hir, and Elena Conti. 2011. “Molecular Mechanisms for the RNA-Dependent ATPase Activity of Upf1 and Its Regulation by Upf2.” Molecular Cell 41 (6): 693–703. [DOI] [PubMed] [Google Scholar]
- Chamieh Hala, Ballut Lionel, Bonneau Fabien, and Le Hir Hervé. 2008. “NMD Factors UPF2 and UPF3 Bridge UPF1 to the Exon Junction Complex and Stimulate Its RNA Helicase Activity.” Nature Structural & Molecular Biology 15 (1): 85–93. [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, and Chang YF. 2007. “An Alternative Branch of the Nonsense-mediated Decay Pathway.” The EMBO Journal. http://emboj.embopress.org/content/26/7/1820.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin Alex, Hu Hao, Rynearson Shawn G., Hollien Julie, Yandell Mark, and Metzstein Mark M.. 2014. “In Vivo Determination of Direct Targets of the Nonsense-Mediated Decay Pathway in Drosophila.” G3 4 (3): 485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Gong, Zhu Wei, Shi Dezhi, Lv Li, Zhang Chun, Liu Ping, and Hu Weixing. 2010. “MicroRNA-181a Sensitizes Human Malignant Glioma U87MG Cells to Radiation by Targeting Bcl-2.” Oncology Reports 23 (4): 997–1003. [DOI] [PubMed] [Google Scholar]
- Cheng Zhihong, Muhlrad Denise, Meng Kiat Lim Roy Parker, and Song Haiwei. 2007. “Structural and Functional Insights into the Human Upf1 Helicase Core.” The EMBO Journal 26 (1): 253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester Ann, Somasekaram Angelika, Tzimina Maria, Jarmuz Adam, Gisbourne Jane, Raymond O’Keefe James Scott, and Navaratnam Naveenan. 2003. “The Apolipoprotein B mRNA Editing Complex Performs a Multifunctional Cycle and Suppresses Nonsense-Mediated Decay.” The EMBO Journal 22 (15): 3971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Hana, Kyoung Mi Kim, and Yoon Ki Kim. 2009. “Human Proline-Rich Nuclear Receptor Coregulatory Protein 2 Mediates an Interaction between mRNA Surveillance Machinery and Decapping Complex.” Molecular Cell 33 (1): 75–86. [DOI] [PubMed] [Google Scholar]
- Cimmino Amelia, George Adrian Calin Muller Fabbri, Iorio Marilena V., Ferracin Manuela, Shimizu Masayoshi, Wojcik Sylwia E., et al. 2005. “miR-15 and miR-16 Induce Apoptosis by Targeting BCL2.” Proceedings of the National Academy of Sciences of the United States of America 102 (39): 13944–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary Michael L., Smith Stephen D., and Sklar Jeffrey. 1986. “Cloning and Structural Analysis of cDNAs for Bcl-2 and a Hybrid Bcl-2/immunoglobulin Transcript Resulting from the T (14; 18) Translocation.” Cell 47 (1): 19–28. [DOI] [PubMed] [Google Scholar]
- Clerici Marcello, Deniaud Aurélien, Boehm Volker, Gehring Niels H., Schaffitzel Christiane, and Cusack Stephen. 2013. “Structural and Functional Analysis of the Three MIF4G Domains of Nonsense-Mediated Decay Factor UPF2.” Nucleic Acids Research. 10.1093/nar/gkt1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici Marcello, André Mourão Irina Gutsche, Gehring Niels H., Hentze Matthias W., Kulozik Andreas, Kadlec Jan, Sattler Michael, and Cusack Stephen. 2009. “Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, UPF1 and UPF2.” The EMBO Journal 28 (15): 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo Martino, Karousis Evangelos D., Bourquin Joël, Bruggmann Rémy, and Mühlemann Oliver. 2016. “Transcriptome-Wide Identification of NMD-Targeted Human mRNAs Reveals Extensive Redundancy between SMG6- and SMG7-Mediated Degradation Pathways.” RNA, November 10.1261/rna.059055.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Hagan KW, Zhang S, and Peltz SW. 1995. “Identification and Characterization of Genes That Are Required for the Accelerated Degradation of mRNAs Containing a Premature Translational Termination Codon.” Genes & Development 9 (4): 423–36. [DOI] [PubMed] [Google Scholar]
- Culbertson MR, Underbrink KM, and Fink GR. 1980. “Frameshift Suppression Saccharomyces Cerevisiae. II. Genetic Properties of Group II Suppressors.” Genetics 95 (4): 833–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar Peter E., Lessene Guillaume, Strasser Andreas, and Adams Jerry M.. 2014. “Control of Apoptosis by the BCL-2 Protein Family: Implications for Physiology and Therapy.” Nature Reviews. Molecular Cell Biology 15 (1): 49–63. [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, and Peltz SW. 1998. “The Surveillance Complex Interacts with the Translation Release Factors to Enhance Termination and Degrade Aberrant mRNAs.” Genes & Development 12 (11): 1665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, and Peltz SW. 1995. “Purification and Characterization of the Upf1 Protein: A Factor Involved in Translation and mRNA Degradation.” RNA 1 (6): 610–23. [PMC free article] [PubMed] [Google Scholar]
- Dang Yongjun, Low Woon-Kai, Xu Jing, Gehring Niels H., Dietz Harry C., Romo Daniel, and Liu Jun O.. 2009. “Inhibition of Nonsense-Mediated mRNA Decay by the Natural Product Pateamine A through Eukaryotic Initiation Factor 4AIII.” The Journal of Biological Chemistry 284 (35): 23613–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtiar Evgeniya, Fridman Adi, Gottlieb Dror, Vexler Karina, Berezin Irina, Farhi Ronit, Golani Linoy, and Shaul Orit. 2015. “The Feedback Control of UPF3 Is Crucial for RNA Surveillance in Plants.” Nucleic Acids Research 43 (8): 4219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehecq Marine, Decourty Laurence, Namane Abdelkader, Proux Caroline, Kanaan Joanne, Hervé Le Hir Alain Jacquier, and Saveanu Cosmin. 2018. “Nonsense-Mediated mRNA Decay Involves Two Distinct Upf1-Bound Complexes.” The EMBO Journal, October 10.15252/embj.201899278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Jing, Carlson Nicole, Takeyama Kunihiko, Paola Dal Cin Margaret Shipp, and Letai Anthony. 2007. “BH3 Profiling Identifies Three Distinct Classes of Apoptotic Blocks to Predict Response to ABT-737 and Conventional Chemotherapeutic Agents.” Cancer Cell 12 (2): 171–85. [DOI] [PubMed] [Google Scholar]
- Diem Michael D., Chan Chia C., Younis Ihab, and Dreyfuss Gideon. 2007. “PYM Binds the Cytoplasmic Exon-Junction Complex and Ribosomes to Enhance Translation of Spliced mRNAs.” Nature Structural & Molecular Biology 14 (12): 1173–79. [DOI] [PubMed] [Google Scholar]
- Doma Meenakshi K., and Parker Roy. 2007. “RNA Quality Control in Eukaryotes.” Cell 131 (4): 660–68. [DOI] [PubMed] [Google Scholar]
- Durand Sébastien, Franks Tobias M., and Lykke-Andersen Jens. 2016. “Hyperphosphorylation Amplifies UPF1 Activity to Resolve Stalls in Nonsense-Mediated mRNA Decay.” Nature Communications 7 (August): 12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle Andrea B., Søren Lykke-Andersen, Oliver Mühlemann, and Torben Heick Jensen. 2009. “SMG6 Promotes Endonucleolytic Cleavage of Nonsense mRNA in Human Cells.” Nature Structural & Molecular Biology 16 (1): 49–55. [DOI] [PubMed] [Google Scholar]
- Eberle Andrea B., Stalder Lukas, Mathys Hansruedi, Rodolfo Zamudio Orozco, and Oliver Mühlemann. 2008. “Posttranscriptional Gene Regulation by Spatial Rearrangement of the 3′ Untranslated Region.” PLoS Biology 6 (4): e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler Daniel E., Wehner Karen A., and Green Rachel. 2013. “Eukaryotic Release Factor 3 Is Required for Multiple Turnovers of Peptide Release Catalysis by Eukaryotic Release Factor 1.” The Journal of Biological Chemistry 288 (41): 29530–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams Margaret E., Guenther Ulf-Peter, and Jankowsky Eckhard. 2010. “SF1 and SF2 Helicases: Family Matters.” Current Opinion in Structural Biology 20 (3): 313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini Francesca, Bagchi Debjani, Hervé Le Hir, and Vincent Croquette. 2015. “Human Upf1 Is a Highly Processive RNA Helicase and Translocase with RNP Remodelling Activities.” Nature Communications 6 10.1038/ncomms8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini Francesca, Robin Jean-Philippe, Kanaan Joanne, Borowiak Malgorzata, Croquette Vincent, Hervé Le Hir Pierre Jalinot, and Mocquet Vincent. 2018. “HTLV-1 Tax Plugs and Freezes UPF1 Helicase Leading to Nonsense-Mediated mRNA Decay Inhibition.” Nature Communications 9 (1): 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine Krystal A., Leon Kristoffer E., Khalid Mir M., Tomar Sakshi, David Jimenez-Morales Mariah Dunlap, Kaye Julia A., et al. 2018. “The Cellular NMD Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein.” mBio 9 (6). 10.1128/mBio.02126-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks Tobias M., Singh Guramrit, and Jens Lykke-Andersen. 2010. “Upf1 ATPase-Dependent mRNP Disassembly Is Required for Completion of Nonsense- Mediated mRNA Decay.” Cell 143 (6): 938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova L, Le Goff X, Zhouravleva G, Davydova E, Philippe M, and Kisselev L. 1996. “Eukaryotic Polypeptide Chain Release Factor eRF3 Is an eRF1- and Ribosome-Dependent Guanosine Triphosphatase.” RNA 2 (4): 334–41. [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, and Kisselev LL. 1999. “Mutations in the Highly Conserved GGQ Motif of Class 1 Polypeptide Release Factors Abolish Ability of Human eRF1 to Trigger Peptidyl-tRNA Hydrolysis.” RNA 5 (8): 1014–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Zhaofeng, and Wilkinson Miles. 2017. “An RNA Decay Factor Wears a New Coat: UPF3B Modulates Translation Termination.” F1000Research 6 (December): 2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno Manuel, Noerenberg Marko, Ni Shuai, Järvelin Aino I., González-Almela Esther, Lenz Caroline E., Bach-Pages Marcel, et al. 2019. “System-Wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection.” Molecular Cell, February 10.1016/j.molcel.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner Lawrence B. 2008. “Hypoxic Inhibition of Nonsense-Mediated RNA Decay Regulates Gene Expression and the Integrated Stress Response.” Molecular and Cellular Biology 28 (11): 3729–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner Lawrence B.. 2010. “Nonsense-Mediated RNA Decay Regulation by Cellular Stress: Implications for Tumorigenesis.” Molecular Cancer Research: MCR 8 (3): 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield David, and Izaurralde Elisa. 2004. “Nonsense-Mediated Messenger RNA Decay Is Initiated by Endonucleolytic Cleavage in Drosophila.” Nature 429 (6991): 575–78. [DOI] [PubMed] [Google Scholar]
- Gehring Niels H., Kunz Joachim B., Gabriele Neu-Yilik Stephen Breit, Viegas Marcelo H., Hentze Matthias W., and Kulozik Andreas E.. 2005. “Exon-Junction Complex Components Specify Distinct Routes of Nonsense-Mediated mRNA Decay with Differential Cofactor Requirements.” Molecular Cell 20 (1): 65–75. [DOI] [PubMed] [Google Scholar]
- Gehring Niels H., Lamprinaki Styliani, Hentze Matthias W., and Kulozik Andreas E.. 2009. “The Hierarchy of Exon-Junction Complex Assembly by the Spliceosome Explains Key Features of Mammalian Nonsense-Mediated mRNA Decay.” PLoS Biology 7 (5): e1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring Niels H., Lamprinaki Styliani, Kulozik Andreas E., and Hentze Matthias W.. 2009. “Disassembly of Exon Junction Complexes by PYM.” Cell 137 (3): 536–48. [DOI] [PubMed] [Google Scholar]
- Gehring Niels H., Gabriele Neu-Yilik, Thomas Schell, Hentze Matthias W., and Kulozik Andreas E.. 2003. “Y14 and hUpf3b Form an NMD-Activating Complex.” Molecular Cell 11 (4): 939–49. [DOI] [PubMed] [Google Scholar]
- Ge Zhiyun, Bao Lin Quek Karen L. Beemon, and Hogg J. Robert. 2016. “Polypyrimidine Tract Binding Protein 1 Protects mRNAs from Recognition by the Nonsense-Mediated mRNA Decay Pathway.” eLife 5 (January). 10.7554/eLife.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz Alexandra E., and Wilkinson Miles. 2017. “Stress and the Nonsense-Mediated RNA Decay Pathway.” Cellular and Molecular Life Sciences: CMLS 74 (19): 3509–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowravaram Manjeera, Bonneau Fabien, Kanaan Joanne, Maciej Vincent D., Fiorini Francesca, Raj Saurabh, Croquette Vincent, Hervé Le Hir, and Sutapa Chakrabarti. 2018. “A Conserved Structural Element in the RNA Helicase UPF1 Regulates Its Catalytic Activity in an Isoform-Specific Manner.” Nucleic Acids Research, January 10.1093/nar/gky040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid Fursham M., and Makeyev Eugene V.. 2014. “Emerging Functions of Alternative Splicing Coupled with Nonsense-Mediated Decay.” Biochemical Society Transactions 42 (4): 1168–73. [DOI] [PubMed] [Google Scholar]
- Hauer Christian, Sieber Jana, Schwarzl Thomas, Hollerer Ina, Curk Tomaz, Alleaume Anne-Marie, Hentze Matthias W., and Kulozik Andreas E.. 2016. “Exon Junction Complexes Show a Distributional Bias toward Alternatively Spliced mRNAs and against mRNAs Coding for Ribosomal Proteins.” Cell Reports 16 (6): 1588–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Feng, and Jacobson Allan. 2015. “Control of mRNA Decapping by Positive and Negative Regulatory Elements in the Dcp2 C-Terminal Domain.” RNA 21 (9): 1633–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg J. Robert. 2011. “This Message Was Inspected by Upf1: 3’UTR Length Sensing in mRNA Quality Control.” Cell Cycle 10 (3): 372–73. [DOI] [PubMed] [Google Scholar]
- Hogg J. Robert. 2016. “Viral Evasion and Manipulation of Host RNA Quality Control Pathways.” Journal of Virology, May 10.1128/JVI.00607-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg J. Robert, and Stephen P. Goff. 2010. “Upf1 Senses 3′ UTR Length to Potentiate mRNA Decay.” Cell 143 (3): 379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AAH, Friedman LJF, Gallagher SSG, Crawford DJC, Anderson EGA, Wombacher RW, Ramirez NR, Cornish VWC, Gelles JG, and Moore MJ. 2011. “Ordered and Dynamic Assembly of Single Spliceosomes.” Science 331 (6022): 1289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins Aaron A., Rodgers Margaret L., Friedman Larry J., Gelles Jeff, and Moore Melissa J.. 2016. “Single Molecule Analysis Reveals Reversible and Irreversible Steps during Spliceosome Activation.” eLife 5 (May). 10.7554/eLife.14166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Shum EY, Jones SH, Lou C-H, Dumdie J, Kim H, Roberts AJ, et al. 2017. “A Upf3b-Mutant Mouse Model with Behavioral and Neurogenesis Defects.” Molecular Psychiatry, September 10.1038/mp.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Lulu, Lou Chih-Hong, Chan Waikin, Shum Eleen Y., Shao Ada, Stone Erica, Karam Rachid, Song Hye-Won, and Wilkinson Miles F.. 2011. “RNA Homeostasis Governed by Cell Type-Specific and Branched Feedback Loops Acting on NMD.” Molecular Cell 43 (6): 950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Ping, Ye Bo, Yang Yu, Shi Jianxin, and Zhao Heng. 2015. “MicroRNA-181 Functions as a Tumor Suppressor in Non-Small Cell Lung Cancer (NSCLC) by Targeting Bcl-2.” Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine 36 (5): 3381–87. [DOI] [PubMed] [Google Scholar]
- Hug Nele, Longman Dasa, and Cáceres Javier F.. 2016. “Mechanism and Regulation of the Nonsense-Mediated Decay Pathway.” Nucleic Acids Research, January 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui Jingyi, Hung Lee-Hsueh, Heiner Monika, Schreiner Silke, Norma Neumüller Gregor Reither, Haas Stefan A., and Bindereif Albrecht. 2005. “Intronic CA-Repeat and CA-Rich Elements: A New Class of Regulators of Mammalian Alternative Splicing.” The EMBO Journal 24 (11): 1988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger Eric, Kashima Isao, Fauser Maria, Jérôme Saulière, and Elisa Izaurralde. 2008. “SMG6 Is the Catalytic Endonuclease That Cleaves mRNAs Containing Nonsense Codons in Metazoan.” RNA 14 (12): 2609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt Jessica A., Robertson Alex D., and Burge Christopher B.. 2013. “Global Analyses of UPF1 Binding and Function Reveal Expanded Scope of Nonsense-Mediated mRNA Decay.” Genome Research 23 (10): 1636–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Jungwook H., Sato Hanae S., Tang Yalan T., Matsuda Daiki M., and Maquat Lynne E. M.. 2010. “UPF1 Association with the Cap-Binding Protein, CBP80, Promotes Nonsense-Mediated mRNA Decay at Two Distinct Steps.” Molecular Cell 39 (3): 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken Olaf, Yoon Ki Kim Nao Hosoda, Mayeur Greg L., Hershey John W. B., and Maquat Lynne E.. 2008. “Upf1 Phosphorylation Triggers Translational Repression during Nonsense-Mediated mRNA Decay.” Cell 133 (2): 314–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov Pavel V., Gehring Niels H., Kunz Joachim B., Hentze Matthias W., and Kulozik Andreas E.. 2008. “Interactions between UPF1, eRFs, PABP and the Exon Junction Complex Suggest an Integrated Model for Mammalian NMD Pathways.” The EMBO Journal 27 (5): 736–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey Samie R., and Wilkinson Miles F.. 2018. “Nonsense-Mediated RNA Decay in the Brain: Emerging Modulator of Neural Development and Disease.” Nature Reviews. Neuroscience, November 10.1038/s41583-018-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson Marcus J. O., and Jacobson Allan. 2010. “Nonsense-Mediated mRNA Decay Maintains Translational Fidelity by Limiting Magnesium Uptake.” Genes & Development 24 (14): 1491–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec Jan, Izaurralde Elisa, and Cusack Stephen. 2004. “The Structural Basis for the Interaction between Nonsense-Mediated mRNA Decay Factors UPF2 and UPF3.” Nature Structural & Molecular Biology 11 (4): 330–37. [DOI] [PubMed] [Google Scholar]
- Kanaan Joanne, Raj Saurabh, Decourty Laurence, Saveanu Cosmin, Croquette Vincent, and Le Hir Hervé. 2018. “UPF1-like Helicase Grip on Nucleic Acids Dictates Processivity.” Nature Communications 9 (1): 3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam Rachid, Lou Chih-Hong H., Kroeger Heike, Huang Lulu, Lin Jonathan H., and Wilkinson Miles F.. 2015. “The Unfolded Protein Response Is Shaped by the NMD Pathway.” EMBO Reports 16 (5): 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karousis Evangelos D., and Oliver Mühlemann. 2018. “Nonsense-Mediated mRNA Decay Begins Where Translation Ends.” Cold Spring Harbor Perspectives in Biology, June 10.1101/cshperspect.a032862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima Isao, Yamashita Akio, Izumi Natsuko, Kataoka Naoyuki, Morishita Ryo, Hoshino Shinichi, Ohno Mutsuhito, Dreyfuss Gideon, and Ohno Shigeo. 2006. “Binding of a Novel SMG-1-Upf1-eRF1-eRF3 Complex (SURF) to the Exon Junction Complex Triggers Upf1 Phosphorylation and Nonsense-Mediated mRNA Decay.” Genes & Development 20 (3): 355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka NK, Yong JY, Kim VNK, Velazquez FV, Perkinson RAP, Wang FW, and Dreyfuss GD. 2000. “Pre-mRNA Splicing Imprints mRNA in the Nucleus with a Novel RNA-Binding Protein That Persists in the Cytoplasm.” Molecular Cell 6 (3): 673–82. [DOI] [PubMed] [Google Scholar]
- Kaygun Handan, and Marzluff William F.. 2005. “Regulated Degradation of Replication-Dependent Histone mRNAs Requires Both ATR and Upf1.” Nature Structural & Molecular Biology 12 (9): 794–800. [DOI] [PubMed] [Google Scholar]
- Keeling Kim M., Lanier Jessica, Du Ming, Joe Salas-Marco, Lin Gao, Kaenjak-Angeletti Anisa, and Bedwell David M.. 2004. “Leaky Termination at Premature Stop Codons Antagonizes Nonsense-Mediated mRNA Decay in S. Cerevisiae.” RNA 10 (4): 691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin Stephanie, Li Chunfang, Buckingham Richard, and Jacobson Allan. 2012. “Testing the Faux-UTR Model for NMD: Analysis of Upf1p and Pab1p Competition for Binding to eRF3/Sup35p.” Biochimie, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, and Dreyfuss G. 2001. “Role of the Nonsense-Mediated Decay Factor hUpf3 in the Splicing-Dependent Exon-Exon Junction Complex.” Science 293 (5536): 1832–36. [DOI] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, and Dreyfuss G. 2001. “The Y14 Protein Communicates to the Cytoplasm the Position of Exon-Exon Junctions.” The EMBO Journal 20 (8): 2062–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Yoon Ki, Furic Luc, Luc DesGroseillers, and Lynne E. Maquat. 2005. “Mammalian Staufen1 Recruits Upf1 to Specific mRNA 3′ UTRs so as to Elicit mRNA Decay.” Cell 120 (2): 195–208. [DOI] [PubMed] [Google Scholar]
- Kim Yoon Ki, and Maquat Lynne E.. 2019. “UPFront and Center in RNA Decay: UPF1 in Nonsense-Mediated mRNA Decay and beyond.” RNA, January 10.1261/rna.070136.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishor Aparna, Ge Zhiyun, and Hogg J. Robert. 2019. “hnRNP L-Dependent Protection of Normal mRNAs from NMD Subverts Quality Control in B Cell Lymphoma.” The EMBO Journal 38 (3): e99128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Ulf, Lia Marie, Crespo Marta, Siegel Rachael, Shen Qiong, Mo Tongwei, Alberto Ambesi-Impiombato, et al. 2010. “The DLEU2/miR-15a/16–1 Cluster Controls B Cell Proliferation and Its Deletion Leads to Chronic Lymphocytic Leukemia.” Cancer Cell 17 (1): 28–40. [DOI] [PubMed] [Google Scholar]
- Kunz Joachim B., Gabriele Neu-Yilik Matthias W. Hentze, Kulozik Andreas E., and Gehring Niels H.. 2006. “Functions of hUpf3a and hUpf3b in Nonsense-Mediated mRNA Decay and Translation.” RNA 12 (6): 1015–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki Tatsuaki, Li Wencheng, Hoque Mainul, Maximilian W-L Popp Dmitri N. Ermolenko, Tian Bin, and Maquat Lynne E.. 2014. “A Post-Translational Regulatory Switch on UPF1 Controls Targeted mRNA Degradation.” Genes & Development 28 (17): 1900–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki Tatsuaki, and Maquat Lynne E.. 2013. “Rules That Govern UPF1 Binding to mRNA 3’ UTRs.” Proceedings of the National Academy of Sciences of the United States of America 110 (9): 3357–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau Liana F., and Brenner Steven E.. 2015. “Regulation of Splicing Factors by Alternative Splicing and NMD Is Conserved between Kingdoms yet Evolutionarily Flexible.” Molecular Biology and Evolution 32 (4): 1072–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau Liana F., Inada Maki, Green Richard E., Wengrod Jordan C., and Brenner Steven E.. 2007. “Unproductive Splicing of SR Genes Associated with Highly Conserved and Ultraconserved DNA Elements.” Nature 446 (7138): 926–29. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, Briault S, et al. 2010. “Mutations of the UPF3B Gene, Which Encodes a Protein Widely Expressed in Neurons, Are Associated with Nonspecific Mental Retardation with or without Autism.” Molecular Psychiatry 15 (7): 767–76. [DOI] [PubMed] [Google Scholar]
- Lee BS, and Culbertson MR. 1995. “Identification of an Additional Gene Required for Eukaryotic Nonsense mRNA Turnover.” Proceedings of the National Academy of Sciences of the United States of America 92 (22): 10354–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Dong-Hyoung, Lim Mi-Hyun, Youn Dong-Ye, Seung Eun Jung Young Soo Ahn, Tsujimoto Yoshihide, and Lee Jeong-Hwa. 2009. “hnRNP L Binds to CA Repeats in the 3′ UTR of Bcl-2 mRNA.” Biochemical and Biophysical Research Communications 382 (3): 583–87. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, and Culbertson MR. 1992. “Gene Products That Promote mRNA Turnover in Saccharomyces Cerevisiae.” Molecular and Cellular Biology 12 (5): 2165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Suzanne R., Pratt Gabriel A., Martinez Fernando J., Yeo Gene W., and Jens Lykke-Andersen. 2015. “Target Discrimination in Nonsense-Mediated mRNA Decay Requires Upf1 ATPase Activity.” Molecular Cell 59 (3): 413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hir Le, Hervé Jérôme Saulière, and Wang Zhen. 2016. “The Exon Junction Complex as a Node of Post-Transcriptional Networks.” Nature Reviews. Molecular Cell Biology 17 (1): 41–54. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, and Moore MJ. 2001. “The Exon-Exon Junction Complex Provides a Binding Platform for Factors Involved in mRNA Export and Nonsense-Mediated mRNA Decay.” The EMBO Journal 20 (17): 4987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, and Moore MJ. 2000. “The Spliceosome Deposits Multiple Proteins 20–24 Nucleotides Upstream of mRNA Exon-Exon Junctions.” The EMBO Journal 19 (24): 6860–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, and Maquat LEM. 2000. “Pre-mRNA Splicing Alters mRNP Composition: Evidence for Stable Association of Proteins at Exon-Exon Junctions.” Genes & Development 14 (9): 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Leich E, Ott G, and Rosenwald A. 2011. “Pathology, Pathogenesis and Molecular Genetics of Follicular NHL.” Best Practice & Research. Clinical Haematology 24 (2): 95–109. [DOI] [PubMed] [Google Scholar]
- Morais Lima, de David A, and Harrison Paul M.. 2010. “Large-Scale Evidence for Conservation of NMD Candidature across Mammals.” PloS One 5 (7): e11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd James P. B. 2018. “The Evolution and Diversity of the Nonsense-Mediated mRNA Decay Pathway.” F1000Research 7 (August). 10.12688/f1000research.15872.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Belinda, Jonas Stefanie, and Izaurralde Elisa. 2013. “The SMG5-SMG7 Heterodimer Directly Recruits the CCR4-NOT Deadenylase Complex to mRNAs Containing Nonsense Codons via Interaction with POP2.” Genes & Development 27 (19): 2125–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Perrote Andrés, Raquel Castaño Roberto Melero, Zamarro Teresa, Kurosawa Hitomi, Ohnishi Tetsuo, Uchiyama Akiko, et al. 2016. “Human Nonsense-Mediated mRNA Decay Factor UPF2 Interacts Directly with eRF3 and the SURF Complex.” Nucleic Acids Research, January 10.1093/nar/gkv1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R, and Lacroute F. 1979. “Interference of Nonsense Mutations with Eukaryotic Messenger RNA Stability.” Proceedings of the National Academy of Sciences of the United States of America 76 (10): 5134–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, and Steitz JA. 2000. “Human Upf Proteins Target an mRNA for Nonsense-Mediated Decay When Bound Downstream of a Termination Codon.” Cell 103 (7): 1121–31. [DOI] [PubMed] [Google Scholar]
- Lynch Sally Ann, Lam Son Nguyen Li Yen Ng, Waldron Mary, Denise McDonald, and Jozef Gecz. 2012. “Broadening the Phenotype Associated with Mutations in UPF3B: Two Further Cases with Renal Dysplasia and Variable Developmental Delay.” European Journal of Medical Genetics 55 (8–9): 476–79. [DOI] [PubMed] [Google Scholar]
- Mabin Justin W., Woodward Lauren A., Patton Robert D., Yi Zhongxia, Jia Mengxuan, Wysocki Vicki H., Bundschuh Ralf, and Singh Guramrit. 2018. “The Exon Junction Complex Undergoes a Compositional Switch That Alters mRNP Structure and Nonsense-Mediated mRNA Decay Activity.” Cell Reports 25 (9): 2431–46.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Kinniburgh AJ, Rachmilewitz EA, and Ross J. 1981. “Unstable Beta-Globin mRNA in mRNA-Deficient Beta O Thalassemia.” Cell 27 (3 Pt 2): 543–53. [DOI] [PubMed] [Google Scholar]
- Matsuda Daiki, Hosoda Nao, Kim Yoon Ki, and Maquat Lynne E.. 2007. “Failsafe Nonsense-Mediated mRNA Decay Does Not Detectably Target eIF4E-Bound mRNA.” Nature Structural & Molecular Biology 14 (10): 974–79. [DOI] [PubMed] [Google Scholar]
- May Jared P., Yuan Xuefeng, Sawicki Erika, and Simon Anne E.. 2018. “RNA Virus Evasion of Nonsense-Mediated Decay.” PLoS Pathogens 14 (11): e1007459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr Christine. 2016. “Evolution and Biological Roles of Alternative 3’UTRs.” Trends in Cell Biology 26 (3): 227–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy Nicholas J., and Smith Christopher W. J.. 2008. “Alternative Splicing Resulting in Nonsense-Mediated mRNA Decay: What Is the Meaning of Nonsense?” Trends in Biochemical Sciences 33 (8): 385–93. [DOI] [PubMed] [Google Scholar]
- McIlwain David R., Pan Qun, Reilly Patrick T., Elia Andrew J., Susan McCracken Andrew C. Wakeham, Annick Itie-Youten Benjamin J. Blencowe, and Mak Tak W.. 2010. “Smg1 Is Required for Embryogenesis and Regulates Diverse Genes via Alternative Splicing Coupled to Nonsense-Mediated mRNA Decay.” Proceedings of the National Academy of Sciences of the United States of America 107 (27): 12186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux Stacie, van Hoof Ambro, and Baker Kristian E.. 2008. “Nonsense-Mediated mRNA Decay in Yeast Does Not Require PAB1 or a poly(A) Tail.” Molecular Cell 29 (1): 134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, and Dietz HC. 2001. “Rent1, a Trans-Effector of Nonsense-Mediated mRNA Decay, Is Essential for Mammalian Embryonic Viability.” Human Molecular Genetics 10 (2): 99–105. [DOI] [PubMed] [Google Scholar]
- Melero Roberto, Buchwald Gretel, Raquel Castaño Monika Raabe, Gil David, Melisa Lázaro Henning Urlaub, Conti Elena, and Llorca Oscar. 2012. “The Cryo-EM Structure of the UPF–EJC Complex Shows UPF1 Poised toward the RNA 3′ End.” Nature Structural & Molecular Biology 19 (April): 498. [DOI] [PubMed] [Google Scholar]
- Mendell Joshua T., Sharifi Neda A., Meyers Jennifer L., Francisco Martinez-Murillo, and Dietz Harry C.. 2004. “Nonsense Surveillance Regulates Expression of Diverse Classes of Mammalian Transcripts and Mutes Genomic Noise.” Nature Genetics 36 (10): 1073–78. [DOI] [PubMed] [Google Scholar]
- Metze Stefanie, Herzog Veronika A., Ruepp Marc-David, and Mühlemann Oliver. 2013. “Comparison of EJC-Enhanced and EJC-Independent NMD in Human Cells Reveals Two Partially Redundant Degradation Pathways.” RNA 19 (10): 1432–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzstein Mark M., and Krasnow Mark A.. 2006. “Functions of the Nonsense-Mediated mRNA Decay Pathway in Drosophila Development.” PLoS Genetics 2 (12): e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino Takashi, Murakawa Yasuhiro, Fukao Akira, Vandenbon Alexis, Wessels Hans-Hermann, Ori Daisuke, Uehata Takuya, et al. 2015. “Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms.” Cell 161 (5): 1058–73. [DOI] [PubMed] [Google Scholar]
- Mühlemann Oliver, and Torben Heick Jensen. 2012. “mRNP Quality Control Goes Regulatory.” Trends in Genetics: TIG 28 (2): 70–77. [DOI] [PubMed] [Google Scholar]
- Nagy E, and Maquat LE. 1998. “A Rule for Termination-Codon Position within Intron-Containing Genes: When Nonsense Affects RNA Abundance.” Trends in Biochemical Sciences 23 (6): 198–99. [DOI] [PubMed] [Google Scholar]
- Nelson Jonathan O., Moore Kristin A., Chapin Alex, Hollien Julie, and Metzstein Mark M.. 2016. “Degradation of Gadd45 mRNA by Nonsense-Mediated Decay Is Essential for Viability.” eLife 5 (March). 10.7554/eLife.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu-Yilik Gabriele, Raimondeau Etienne, Eliseev Boris, Yeramala Lahari, Amthor Beate, Deniaud Aurélien, Huard Karine, et al. 2017. “Dual Function of UPF3B in Early and Late Translation Termination.” The EMBO Journal, September 10.15252/embj.201797079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson Pamela, Gkratsou Asimina, Josi Christoph, Colombo Martino, and Oliver Mühlemann. 2018. “Dissecting the Functions of SMG5, SMG7 and PNRC2 in Nonsense-Mediated mRNA Decay of Human Cells.” RNA, January 10.1261/rna.063719.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson Pamela, Joncourt Raphael, and Oliver Mühlemann. 2012. “Analysis of Nonsense-Mediated mRNA Decay in Mammalian Cells.” Current Protocols in Cell Biology. 10.1002/0471143030.cb2704s55. [DOI] [PubMed] [Google Scholar]
- Nicholson Pamela, Josi Christoph, Kurosawa Hitomi, Yamashita Akio, and Mühlemann Oliver. 2014. “A Novel Phosphorylation-Independent Interaction between SMG6 and UPF1 Is Essential for Human NMD.” Nucleic Acids Research. 10.1093/nar/gku645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Julie Z., Grate Leslie, John Paul Donohue Christine Preston, Nobida Naomi, Georgeann O’Brien Lily Shiue, Clark Tyson A., Blume John E., and Ares Manuel Jr. 2007. “Ultraconserved Elements Are Associated with Homeostatic Control of Splicing Regulators by Alternative Splicing and Nonsense-Mediated Decay.” Genes & Development 21 (6): 708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Tetsuo, Yamashita Akio, Kashima Isao, Schell Thomas, Anders Kirk R., Grimson Andrew, Hachiya Takahisa, Hentze Matthias W., Anderson Philip, and Ohno Shigeo. 2003. “Phosphorylation of hUPF1 Induces Formation of mRNA Surveillance Complexes Containing hSMG-5 and hSMG-7.” Molecular Cell 12 (5): 1187–1200. [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, and Ohno S. 2011. “N- and C-Terminal Upf1 Phosphorylations Create Binding Platforms for SMG-6 and SMG-5:SMG-7 during NMD.” Nucleic Acids Research, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottens Franziska, Boehm Volker, Sibley Christopher R., Ule Jernej, and Gehring Niels H.. 2017. “Transcript-Specific Characteristics Determine the Contribution of Endo- and Exonucleolytic Decay Pathways during the Degradation of Nonsense-Mediated Decay Substrates.” RNA 23 (8): 1224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Yi-Bing, Lu Yu, Yue Sibiao, and Giffard Rona G.. 2012. “miR-181 Targets Multiple Bcl-2 Family Members and Influences Apoptosis and Mitochondrial Function in Astrocytes.” Mitochondrion 12 (2): 213–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M, Ishigaki Y, Nagy E, and Maquat LE. 2001. “Evidence That Phosphorylation of Human Upfl Protein Varies with Intracellular Location and Is Mediated by a Wortmannin-Sensitive and Rapamycin-Sensitive PI 3-Kinase-Related Kinase Signaling Pathway.” RNA 7 (1): 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixeiro Isabel, Ângela Inácio, Cristina Barbosa, Silva Ana Luísa, Liebhaber Stephen A., and Romão Luísa. 2012. “Interaction of PABPC1 with the Translation Initiation Complex Is Critical to the NMD Resistance of AUG-Proximal Nonsense Mutations.” Nucleic Acids Research 40 (3): 1160–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez I, McAfee JG, and Patton JG. 1997. “Multiple RRMs Contribute to RNA Binding Specificity and Affinity for Polypyrimidine Tract Binding Protein.” Biochemistry 36 (39): 11881–90. [DOI] [PubMed] [Google Scholar]
- Pisarev Andrey V., Skabkin Maxim A., Pisareva Vera P., Skabkina Olga V., Rakotondrafara Aurélie M., Hentze Matthias W., Hellen Christopher U. T., and Pestova Tatyana V.. 2010. “The Role of ABCE1 in Eukaryotic Posttermination Ribosomal Recycling.” Molecular Cell 37 (2): 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva Vera P., Skabkin Maxim A., Hellen Christopher U. T., Pestova Tatyana V., and Pisarev Andrey V.. 2011. “Dissociation by Pelota, Hbs1 and ABCE1 of Mammalian Vacant 80S Ribosomes and Stalled Elongation Complexes.” The EMBO Journal 30 (9): 1804–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp Maximilian Wei-Lin, and Maquat Lynne E.. 2013. “Organizing Principles of Mammalian Nonsense-Mediated mRNA Decay.” Annual Review of Genetics 47: 139–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondeau Etienne, Bufton Joshua C., and Schaffitzel Christiane. 2018. “New Insights into the Interplay between the Translation Machinery and Nonsense-Mediated mRNA Decay Factors.” Biochemical Society Transactions, April 10.1042/BST20170427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada Indrani, and Jens Lykke-Andersen. 2009. “Execution of Nonsense-Mediated mRNA Decay: What Defines a Substrate?” Current Opinion in Cell Biology 21 (3): 394–402. [DOI] [PubMed] [Google Scholar]
- Rehwinkel Jan, Letunic Ivica, Raes Jeroen, Bork Peer, and Izaurralde Elisa. 2005. “Nonsense-Mediated mRNA Decay Factors Act in Concert to Regulate Common mRNA Targets.” RNA 11 (10): 1530–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque Sylvain, Cerciat Marie, Isabelle Gaugué Liliana Mora, Floch Aurélie G., de Zamaroczy Miklos Valérie Heurgué-Hamard, and Kervestin Stephanie. 2015. “Interaction between the poly(A)-Binding Protein Pab1 and the Eukaryotic Release Factor eRF3 Regulates Translation Termination but Not mRNA Decay in Saccharomyces Cerevisiae.” RNA 21 (1): 124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabirzhanov Boris, Zhao Zaorui, Stoica Bogdan A., Loane David J., Wu Junfang, Borroto Carlos, Dorsey Susan G., and Faden Alan I.. 2014. “Downregulation of miR-23a and miR-27a Following Experimental Traumatic Brain Injury Induces Neuronal Cell Death through Activation of Proapoptotic Bcl-2 Proteins.” The Journal of Neuroscience: The Official Journal of the Society for Neuroscience 34 (30): 10055–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco Joe, and Bedwell David M.. 2004. “GTP Hydrolysis by eRF3 Facilitates Stop Codon Decoding during Eukaryotic Translation Termination.” Molecular and Cellular Biology 24 (17): 7769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefke Bernhard, Sun Wei, Li Yi-Sheng, Fang Liang, and Chen Wei. 2018. “The Evolution of Posttranscriptional Regulation.” Wiley Interdisciplinary Reviews. RNA, May, e1485. [DOI] [PubMed] [Google Scholar]
- Schuetz JM, Johnson NA, Morin RD, Scott DW, Tan K, Ben-Nierah S, Boyle M, et al. 2012. “BCL2 Mutations in Diffuse Large B-Cell Lymphoma.” Leukemia 26 (6): 1383–90. [DOI] [PubMed] [Google Scholar]
- Schuller Anthony P., Zinshteyn Boris, Enam Syed Usman, and Green Rachel. 2018. “Directed Hydroxyl Radical Probing Reveals Upf1 Binding to the 80S Ribosomal E Site rRNA at the L1 Stalk.” Nucleic Acids Research 46 (4): 2060–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serdar Lucas D., Whiteside Dajuan L., and Baker Kristian E.. 2016. “ATP Hydrolysis by UPF1 Is Required for Efficient Translation Termination at Premature Stop Codons.” Nature Communications 7 (December): 14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, and Maquat LE. 2001. “Identification and Characterization of Human Orthologues to Saccharomyces Cerevisiae Upf2 Protein and Upf3 Protein (Caenorhabditis Elegans SMG-4).” Molecular and Cellular Biology 21 (1): 209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Masao. 2002. “Molecular Mechanisms of Lymphomagenesis through Transcriptional Disregulation by Chromosome Translocation.” International Journal of Hematology 76 Suppl 1 (August): 323–26. [DOI] [PubMed] [Google Scholar]
- Shoemaker Christopher J., and Green Rachel. 2011. “Kinetic Analysis Reveals the Ordered Coupling of Translation Termination and Ribosome Recycling in Yeast.” Proceedings of the National Academy of Sciences of the United States of America 108 (51): E1392–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum Eleen Y., Jones Samantha H., Shao Ada, Dumdie Jennifer, Krause Matthew D., Chan Wai-Kin, Lou Chih-Hong, et al. 2016. “The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay.” Cell, March 10.1016/j.cell.2016.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Guramrit, Pratt Gabriel, Yeo Gene W., and Moore Melissa J.. 2015. “The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion.” Annual Review of Biochemistry 84 (March): 325–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Guramrit, Rebbapragada Indrani, and Lykke-Andersen Jens. 2008. “A Competition between Stimulators and Antagonists of Upf Complex Recruitment Governs Human Nonsense-Mediated mRNA Decay.” PLoS Biology 6 (4): e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Jenna E., and Baker Kristian E.. 2015. “Nonsense-Mediated RNA Decay--a Switch and Dial for Regulating Gene Expression.” BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology 37 (6): 612–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararaman Balaji, Zhan Lijun, Blue Steven M., Stanton Rebecca, Elkins Keri, Olson Sara, Wei Xintao, et al. 2016. “Resources for the Comprehensive Discovery of Functional RNA Elements.” Molecular Cell 61 (6): 903–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher Kylie D., and Parker Roy. 2011. “Interactions between Upf1 and the Decapping Factors Edc3 and Pat1 in Saccharomyces Cerevisiae.” PloS One 6 (10): e26547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyszka Paulina, Sharp Sally I., Dedman Alexandra, Gurling Hugh M. D., and McQuillin Andrew. 2012. “A Nonconservative Amino Acid Change in the UPF3B Gene in a Patient with Schizophrenia.” Psychiatric Genetics 22 (3): 150–51. [DOI] [PubMed] [Google Scholar]
- Tang Xuhua, Zhu Yiping, Baker Stacey L., Bowler Matthew W., Benjamin Jieming Chen Chen Chen, Hogg J. Robert, Goff Stephen P., and Song Haiwei. 2016. “Structural Basis of Suppression of Host Translation Termination by Moloney Murine Leukemia Virus.” Nature Communications 7 (June): 12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani Hidenori, Imamachi Naoto, Kazi Abdus Salam Rena Mizutani, Ijiri Kenichi, Irie Takuma, Yada Tetsushi, Suzuki Yutaka, and Akimitsu Nobuyoshi. 2012. “Identification of Hundreds of Novel UPF1 Target Transcripts by Direct Determination of Whole Transcriptome Stability.” RNA Biology 9 (11): 1370–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey Patrick S., Raymond F. Lucy, Nguyen Lam S., Rodriguez Jayson, Hackett Anna, Vandeleur Lucianne, Smith Raffaella, et al. 2007. “Mutations in UPF3B, a Member of the Nonsense-Mediated mRNA Decay Complex, Cause Syndromic and Nonsyndromic Mental Retardation.” Nature Genetics 39 (9): 1127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner Leonie, and Izaurralde Elisa. 2004. “SMG7 Acts as a Molecular Link between mRNA Surveillance and mRNA Decay.” Molecular Cell 16 (4): 587–96. [DOI] [PubMed] [Google Scholar]
- Vexler Karina, Cymerman Miryam A., Berezin Irina, Fridman Adi, Golani Linoy, Lasnoy Michal, Saul Helen, and Shaul Orit. 2016. “The Arabidopsis NMD Factor UPF3 Is Feedback-Regulated at Multiple Levels and Plays a Role in Plant Response to Salt Stress.” Frontiers in Plant Science 7 (September): 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Masami, Lokugamage Kumari G., Nakagawa Keisuke, Narayanan Krishna, and Makino Shinji. 2018. “Interplay between Coronavirus, a Cytoplasmic RNA Virus, and Nonsense-Mediated mRNA Decay Pathway.” Proceedings of the National Academy of Sciences of the United States of America, October 10.1073/pnas.1811675115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Hongjiang, Li Jing, Chi Hongjie, Zhang Fan, Zhu Xiaoming, Cai Jun, and Yang Xinchun. 2015. “MicroRNA-181c Targets Bcl-2 and Regulates Mitochondrial Morphology in Myocardial Cells.” Journal of Cellular and Molecular Medicine 19 (9): 2084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, and Peltz SW. 2001. “The Role of Upf Proteins in Modulating the Translation Read-through of Nonsense-Containing Transcripts.” The EMBO Journal 20 (4): 880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Chuanyu, Li Li, and Gupta Sudhiranjan. 2014. “NF-κB-Mediated miR-30b Regulation in Cardiomyocytes Cell Death by Targeting Bcl-2.” Molecular and Cellular Biochemistry 387 (1–2): 135–41. [DOI] [PubMed] [Google Scholar]
- Weil Jason E., and Beemon Karen L.. 2006. “A 3’ UTR Sequence Stabilizes Termination Codons in the Unspliced RNA of Rous Sarcoma Virus.” RNA 12 (1): 102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt Joachim, Damgaard Inge, Bryder David, Kim Theilgaard-Mönch Thoren Lina A., Finn Cilius Nielsen Jacobsen Sten Eirik W., Nerlov Claus, and Porse Bo Torben. 2008. “NMD Is Essential for Hematopoietic Stem and Progenitor Cells and for Eliminating by-Products of Programmed DNA Rearrangements.” Genes & Development 22 (10): 1381–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, and Peltz SW. 1998. “ATP Is a Cofactor of the Upf1 Protein That Modulates Its Translation Termination and RNA Binding Activities.” RNA 4 (2): 205–14. [PMC free article] [PubMed] [Google Scholar]
- Wittkopp Nadine, Huntzinger Eric, Weiler Catrin, Jérôme Saulière Steffen Schmidt, Sonawane Mahendra, and Izaurralde Elisa. 2009. “Nonsense-Mediated mRNA Decay Effectors Are Essential for Zebrafish Embryonic Development and Survival.” Molecular and Cellular Biology 29 (13): 3517–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward Lauren A., Mabin Justin W., Gangras Pooja, and Singh Guramrit. 2016. “The Exon Junction Complex: A Lifelong Guardian of mRNA Fate.” Wiley Interdisciplinary Reviews. RNA, December 10.1002/wrna.1411. [DOI] [PubMed] [Google Scholar]
- Xu X, Zhang L, Tong P, Xun G, Su W, Xiong Z, Zhu T, et al. 2013. “Exome Sequencing Identifies UPF3B as the Causative Gene for a Chinese Non-Syndrome Mental Retardation Pedigree.” Clinical Genetics 83 (6): 560–64. [DOI] [PubMed] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, and Muhlemann O. 2011. “Autoregulation of the Nonsense-Mediated mRNA Decay Pathway in Human Cells.” RNA, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Xiaomin, Zheng Lan, Shen Jianfeng, Zhang Dongmei, Xiong Minmin, Zhang Youyou, He Xinhong, et al. 2016. “Suppression of MicroRNA 200 Family Expression by Oncogenic KRAS Activation Promotes Cell Survival and Epithelial-Mesenchymal Transition in KRAS-Driven Cancer.” Molecular and Cellular Biology 36 (21): 2742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zünd David, Gruber Andreas R., Zavolan Mihaela, and Oliver Mühlemann. 2013. “Translation-Dependent Displacement of UPF1 from Coding Sequences Causes Its Enrichment in 3’ UTRs.” Nature Structural & Molecular Biology 20 (8): 936–43. [DOI] [PubMed] [Google Scholar]