Abstract
Chronic Hepatitis B virus (HBV) infection remains a worldwide concern and public health problem. Two key aspects of the HBV life cycle are essential for viral replication and thus the development of chronic infections: the establishment of the viral minichromosome, covalently closed circular (ccc) DNA, within the nucleus of infected hepatocytes, and the expression of the regulatory Hepatitis B virus X protein (HBx). Interestingly, nuclear HBx redirects host epigenetic machinery to activate cccDNA transcription. In this Perspective, we provide an overview of recent advances in understanding the regulation of cccDNA and the mechanistic and functional roles of HBx. We also describe the progress toward targeting both cccDNA and HBx for therapeutic purposes. Finally, we outline standing questions in the field and propose complementary chemical biology approaches to address them.
Keywords: Hepatitis B virus, cccDNA, HBx, chemical biology
Graphical Abstract
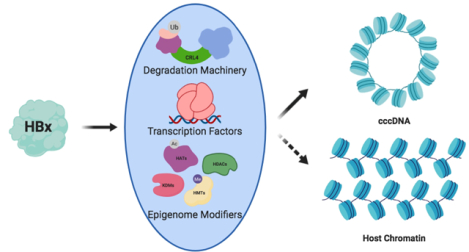
Chronic Hepatitis B virus (HBV) infection afflicts over 250 million people and is responsible for almost half of all cases of hepatocellular carcinoma (HCC), the fourth leading cause of cancer mortality.1,2 The key replicative intermediate and transcriptional template for HBV is a viral minichromosome of covalently closed circular (ccc)DNA, which supports chronic infection. While existing HBV DNA polymerase inhibitors are able to reduce other HBV replicative intermediates, cccDNA levels remain remarkably stable to such treatments. Consequently, upon interruption or termination of therapy, HBV replication is able to resume with the transcription of unaffected cccDNA. Thus, directly targeting the transcription or clearance of cccDNA remains a key step toward a complete cure for chronic infections. Furthermore, the master regulatory Hepatitis B virus X protein (HBx) has been shown not only to be recruited to cccDNA in vivo to promote viral replication,3,4 but also to contribute to the progression of hepatocellular carcinogenesis through abrogation of several host signaling pathways.5,6 Despite this, a mechanistic understanding of HBx function remains strikingly elusive. Clearly, a deeper understanding of both HBx and cccDNA at the molecular level is required to elucidate the malignant transformation of hepatocytes during infection and to identify novel targets for the disruption of cccDNA.
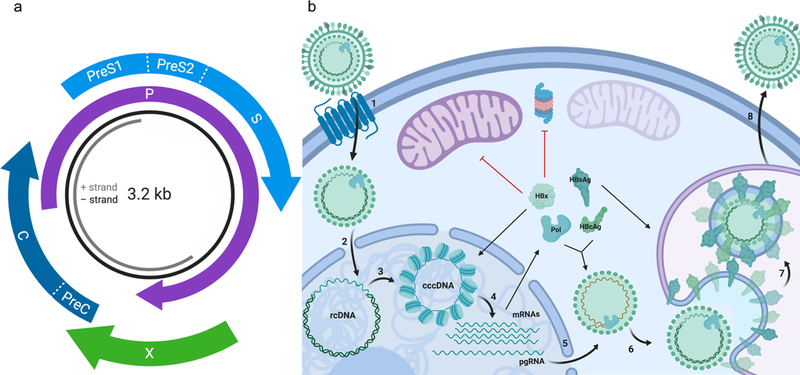
HBV is a member of the Hepadnaviridae family, members of which have characteristically small, partially double-stranded DNA genomes and a particularly unique life cycle compared to other DNA viruses. The 3.2 kb HBV genome encodes four overlapping open reading frames (ORFs), dubbed C, P, S, and X (Figure 1a). The PreC/C frame encodes both the core antigen (HBcAg) that composes the viral nucleocapsid and a PreCore polypeptide, which is the precursor of secreted Hepatitis B e-antigen (HBeAg). Translation of the P frame yields a single polypeptide and the only enzyme encoded within the HBV genome, the viral polymerase and reverse transcriptase (Pol). The PreS1/PreS2/S reading frame contains three different surface antigen proteins (HBsAg), which decorate the exterior of the mature virion and mediate entry and exit from hepatocytes. Finally, the X frame encodes the multifunctional regulatory protein HBx. Each of these elements contributes to the HBV lifecycle, which has been thoroughly reviewed previously (Figure 1b).7 Briefly, infection is mediated by the attachment of the virion to a hepatocyte-specific bile acid receptor, the sodium taurocholate cotransporting polypeptide (SLC10A1 or NTCP),8 followed by endocytosis, after which the viral nucleocapsid escapes the endosome into the cytosol.9 The viral capsid proteins mediate passage through the nuclear pore. Upon nuclear translocation, the capsid disassembles to release the partially double-stranded relaxed circular (rc) viral DNA. Host cell DNA repair mechanisms then remove the covalently attached viral polymerase and complete the synthesis of both strands of the DNA. Finally, this covalently closed circular (ccc) DNA is populated with host histones to form a viral minichromosome. Upon the establishment of cccDNA, which serves as the primary transcriptional template for HBV, the genome is transcribed into the replicative intermediate pregenomic RNA (pgRNA) and the five viral transcripts described earlier.10 The pgRNA is packaged along with Pol into the viral capsid (composed of HBcAg), after which pgRNA is reverse-transcribed into rcDNA and subsequently digested. Finally, the nucleocapsid associates with surface proteins at late-endosomal multivesicular bodies, becomes enveloped, and is subsequently secreted by the ESCRT (endosomal sorting complex required for transport) pathway.11 Furthermore, mature nucleocapsids can also re-enter the nucleus to recycle the HBV genome, thus maintaining a stable intracellular cccDNA pool.12
Figure 1. Overview of HBV genome structure and life cycle.
(a) HBV has a partially double-stranded DNA genome which encodes four overlapping open reading frames, termed C, P, S, and X. (b) The virion enters hepatocytes via the the sodium taurocholate cotransporting polypeptide (NTCP) (1). The relaxed circular DNA (rcDNA) viral genome is released from the capsid upon nuclear translocation (2) and undergoes DNA repair to establish cccDNA (3). HBx mediates cccDNA maintenance and transcription (4). The pregenomic RNA (pgRNA) is encapsidated together with the viral polymerase (Pol) (5), and reverse transcribed within the nucleocapsid (6). Finally, the nucleocapsid is enveloped at late-endosomal multivesicular bodies (7), and secreted via the endosomal sorting complex required for transport (ESCRT) pathway pathway (8).
Effective treatments are able to reduce virion production, either with existing drugs targeting Pol or a promising new class of small molecules that target nucleocapsid assembly.13,14 However, no therapy has been developed to target cccDNA, the template of HBV transcription responsible for chronic infection, although some promising reports suggest this may be possible by upregulating a class of DNA editing enzymes through IFN-α treatment.15 Still fewer encouraging leads exist to therapeutically target HBx, which both promotes cccDNA transcription and is implicated in hepatocellular carcinogenesis. Here, we discuss recent advances in the biochemical understanding of cccDNA physiology and HBx function in cccDNA and host chromatin (mis)regulation, as well as chemical biology approaches that may provide new insights to these crucial elements of HBV pathogenesis.
The cccDNA Minichromosome
Host DNA Repair and cccDNA Establishment
Successful establishment of cccDNA is critical for HBV replication. However, the specific host mechanisms that convert rcDNA into cccDNA remain poorly understood. Furthermore, due to the complex life cycle of the virus, cccDNA is not the only HBV-derived DNA species present in cells, though it is considered the most stable. Aberrant reverse transcription of pgRNA has been shown to occasionally yield double-stranded linear DNA (dslDNA) in the virion.16 In that context, both homologous repair and non-homologous end joining pathways have been shown to circularize dslDNA to yield an incompetent cccDNA-like species termed Ψ-cccDNA.17 Furthermore, both dslDNA and cccDNA can be occasionally integrated into the host genome.16 HBV integration is correlated with HCC, though it occurs early in HBV infection prior to HCC development.18 Integration was recently shown to occur most often at fragile genomic sites and functional genomic regions.19 However, a more detailed understanding of the distinct processes underlying genomic integration of HBV remains lacking.
Recent advances have begun to identify the roles of various host enzymes in cccDNA generation, either by rationally testing enzymes involved in DNA metabolism whose substrates resemble aspects of rcDNA or by RNA silencing (RNAi) screens. For example, the first enzyme shown to be involved in rcDNA repair was tyrosyl-DNA-phosphodiesterase 2 (TDP2), which was identified due to its physiological role in resolving abortive topoisomerase cleavage complexes, similar to the covalent link between Pol and rcDNA.20 However, other reports have shown that while TDP2 is important in vitro, it may not be necessary for productive infection in vivo.21,22 Another recent study also found similarities between the antisense strand of rcDNA structure and 5’ flap structures formed during Okazaki fragment maturation to identify flap structure-specific endonuclease 1 (FEN1) as a contributor to cccDNA formation.23 Moreover, RNAi screening studies identified several DNA polymerases (α, η, κ, and λ) that mediate cccDNA establishment from either de novo infection (η, κ, λ) or intracellular amplification (α).24,25 Newly developed inhibitors specific to some of these error-prone polymerases may prove useful to further define their roles in cccDNA formation.26 An shRNA screen targeting host DNA repair enzymes identified DNA ligases I and III as having a role in the conversion of rcDNA into cccDNA, while DNA ligase IV was shown to mediate the so-called “illegitimate” conversion of dslDNA into cccDNA.27 Furthermore, a chemical compound screen targeting DNA replication and repair enzymes demonstrated a nonredundant requirement for both topoisomerase 1 (TOP1) and TOP2 in cccDNA synthesis.28 Taken together, targeting these newly identified host DNA repair factors may represent a novel therapeutic avenue for the treatment of chronic HBV infection, either alone or in combination with existing treatments.
Chromatinized cccDNA: Features and Regulation
Kinetic experiments studying initial cccDNA establishment showed that cells maintain a relatively low copy number of the minichromosome, with some estimates ranging between 1 and 5 copies per cell on average.29,30 Perhaps due to its low abundance, the components and structure of cccDNA have only recently begun to be characterized in depth. Early studies showed that cccDNA is rapidly populated with host histones and chromatinized in the nucleus.31,32 However, it is unclear whether only the canonical histones form part of this minichromosome or if histone variants play a role in cccDNA biology. A recent Hi-C-based study examined the three-dimensional localization of cccDNA within higher-order chromatin architecture and found it contacts the host genome at CpG islands and highly expressed genomic regions.33 The same study identified an interaction between cccDNA and the CpG- binding protein CXXC finger protein 1 (Cfp1), suggesting a role for Cfp1 in cccDNA transcriptional regulation. These findings are in accordance with earlier results identifying up to three CpG islands within cccDNA itself.34
More recent studies have shown that HBcAg remains associated with cccDNA by binding to CpG islands within the viral genome and promotes HBV transcription,35,36 in contrast with earlier reports that suggested that HBcAg compacts the minichromosome, which would have a repressive effect.32 Further biochemical and biophysical studies are still required to better define the role of cccDNA-bound HBcAg. Such a platform would prove invaluable for the study not only of cccDNA alone, but also of HBx, which has been suggested to also localize to cccDNA in the nucleus and redirect a vast array of host enzymes involved in chromatin regulation.
Nuclear HBx and cccDNA Regulation
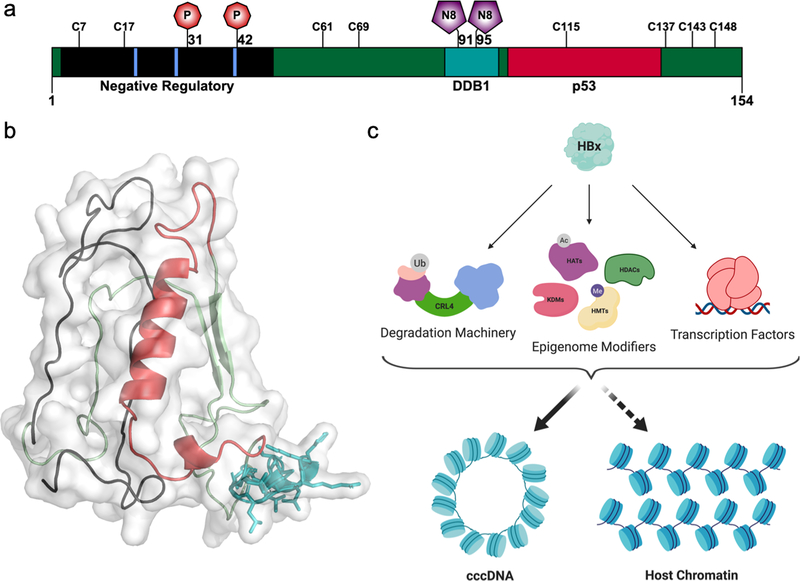
HBx carries out many functions within the infected hepatocyte and is the primary effector protein encoded by HBV (Figure 2). Numerous human proteins have been identified as potential HBx interactors, though relatively few of these interactions have known functional outcomes.37–39 Reflective of the ambiguity surrounding the mechanistic functions of HBx is the protein’s poorly understood structure. Although decades of study have focused on HBx, remarkably little experimental data has shed any light on its three-dimensional architecture. As known elements of HBx structure and their functional implications have been reviewed recently,40 in this review we will highlight those elements that relate to HBx’s functions in the nucleus. Various regions within HBx have been associated with certain functional outputs including transactivation activity, relevance to protein stability, and binding to interacting partners (Figure 2a). The amino-terminus of HBx (aa 1–50) is predicted to be largely unstructured and described as a negative regulatory region since mutational studies have shown it to be dispensable for HBx’s transactivation function.40 The proposed disorder of this region may contribute to HBx’s ability to bind diverse interactors, though this remains to be proven. The remainder of the protein is expected to have a more defined structure based on spectroscopic measurements of secondary structure.41 Nevertheless, only small fragments of HBx have been structurally characterized, typically in complex with a binding partner.42,43 Despite the unavailability of any experimental structure, several groups have reported computationally modeled structures of HBx and used them to predict or explain interactions (Figure 2b).44–46
Figure 2. Snapshot of HBx structure and function in chromatin signaling.
(a) Overview of key domains, residues, and PTMs in HBx. The negative regulatory region is marked in black, the DDB1-binding helix is in teal, the p53 interaction domain is in red, and sites of proline isomerization are in blue. Red octagons mark phosphorylation sites, and purple pentagons mark sites of neddylation. (b) Computationally modeled structure of HBx, generated using the intensive mode of the Phyre2 web application.112 (c) Schematic model of HBx functions in chromatin signaling.
In the nucleus, HBx carries out the dual roles of 1) redirecting host transcription factors (including p53, NF-κB, and CREB, among others) to change expression of a wide variety of gene families, and 2) initiating virus replication by stimulating transcription of cccDNA (Figure 2c).4,40 Its role in transcription also presents a unique paradox. HBx appears to be critical for cccDNA transcription and for the development of viraemia in vivo,4,47 yet the protein itself is not packaged in the mature virion. Thus, the question arises of how during initial infection HBx can be expressed without already being present in the cell. Recent findings that HBx specifically hijacks host machinery to induce the degradation of an antiviral restriction complex further illustrate this seeming incongruity.48 Current theories propose that HBx may be transcribed from rcDNA or dslDNA before or during conversion into cccDNA, though one recent report identified HBx mRNA in both cell culture derived virus preparations and HBV patient plasma, suggesting that it may indeed be packaged into the virion.4,49 Below, we describe some of the more well-known functions and regulatory mechanisms of HBx.
HBx and CRL4
An interaction between HBx and damaged DNA-binding protein 1 (DDB1) was identified decades ago, yet neither the structural basis for this interaction nor its functional significance was known until recently. DDB1 serves as an obligate adaptor protein for the cullin-RING ligase 4 (CRL4) E3 ligase complex. The other minimal components of CRL4 are one of two highly similar scaffold proteins, Cul4A or Cul4B, and the E3 ligase Rbx1 (Figure 2c).50 Within this complex, DDB1 serves as an adaptor that recruits a class of substrate receptors, named DDB1-Cul4 associated factors (DCAFs), which bring ubiquitination substrates into proximity with Rbx1. A crystal structure of DDB1 bound to an α-helical motif of HBx found that this interaction greatly resembles that of known DCAFs, suggesting that HBx could have a similar function.42 Later proteomic studies corroborated this observation and identified the HBx-mediated degradation of the structural maintenance of chromosomes (Smc)5/6 complex.48,51 Furthermore, this degradation was shown to be crucial for cccDNA transcription, since cells with either DDB1-binding deficient HBx or Cul4-binding deficient DDB1 exhibited no cccDNA transcription.42,48 In the absence of HBx, Smc5/6 tethers cccDNA to ND10 bodies, nuclear regions of transcriptional repression, in contrast to cccDNA in the presence of HBx, which localizes to regions of active transcription.33,49
Viral hijacking of the host ubiquitin system is not uncommon – several viruses either encode their own deubiquitinase or E3 ligase enzymes, or proteins that redirect host machinery.52 In fact, other mammalian viruses (for example, Simian Virus 5 or HIV) either hijack or express DCAFs that lead to degradation of several antiviral signaling factors.53,54 Therefore, it may be unlikely that Smc5/6 is the only new target for ubiquitination upon HBx binding to Cul4-DDB1. Indeed, both studies reporting this role also detected core histone ubiquitination, which has been shown to mediate a variety of signaling cascades.55–57 Moreover, both HBx and the CRL4 complex are present in both the cytosol and nucleus, raising the possibility that HBx-CRL4 has additional cellular targets. When this is taken into consideration along with earlier studies that show that HBx has a wide variety of well-documented binding partners throughout the cell, it seems more likely that HBx could be a DCAF for other novel substrates. More targeted in vitro and cell-based studies will be required to identify any such activities and determine their functional significance.
HBx and Chromatin Modifications
Despite the importance of HBx’s role in redirecting CRL4 to degrade Smc5/6, this is far from its only role in cccDNA transcriptional and chromatin regulation. Decades of study have shown that HBx is able to redirects cellular machinery responsible for the deposition or erasure of histone post-translational modifications (PTMs) to produce an active chromatin landscape on cccDNA.58–60 For example, cccDNA was found to be hyperacetylated in vivo in an HBx-dependent manner.61,62 Furthermore, chromatin immunoprecipitation (ChIP)-based studies found an increased occupancy of the histone deacetylases Sirt1 and HDAC1 on cccDNA in HBx-deficient infections, suggesting that HBx may outcompete for binding sites or mediate their degradation.62 Detailed explanations for these observations still remain undetermined. Aside from acetylation, HBx may also perturb histone methylation through interactions with lysine and arginine methylases, though this needs to be investigated further.60,63 Interestingly, studies have shown that HBx colocalizes with cccDNA in cells, yet the mechanisms underlying this interaction remain unclear since HBx lacks any known DNA-binding motifs, suggesting the involvement of another factor. It was also observed that the recruitment of HBx to cccDNA paralleled with cccDNA H3 hyperacetylation, implying that the recruitment may be mediated by the epigenetic machinery.62
In addition to histone PTMs, HBx also modifies the epigenetic landscape of cccDNA and host chromatin through interactions with DNA methyltransferases (DNMTs). HBx upregulates the expression of DNMT1, DNMT3A, and DNMT3B, and was shown to directly interact with DNMT3A.64 Thus, HBx could be responsible for DNA methylation within the CpG islands of cccDNA, which is correlated with the development of HCC in infected patients.65 The mechanism by which HBx is able to specifically direct DNA methylation to certain areas of both the host and viral genomes is unclear but remains open for future study.
In addition to the above-mentioned canonical “epigenetic” regulators that HBx is thought to recruit to cccDNA, recent studies have also implicated HBx in mediating interactions between newly identified enzymes in chromatin regulation and cccDNA. For example, an interaction between HBx, cccDNA, and two peptidyl-prolyl cis/trans isomerases (PPIases) was recently described. While the nuclear PPIase Pin1 was shown previously to bind to HBx and enhance hepatocarcinogenesis, a direct effect on cccDNA was not reported.66 Moreover, an interaction between HBx and two isoforms of the PPIase-encoding PIN4 gene: parvulin 14 (Par14) and parvulin 17 (Par17) was described .67 Briefly, inhibition of the parvulin family or knockdown led to downregulation of cccDNA formation and HBV replication in an HBx-dependent manner. Direct interaction of Par14 and Par17 with HBx was shown by co-immunoprecipitation and immunofluorescence colocalization, and their interaction with cccDNA was confirmed by ChIP. Together, these data implicate prolyl isomerization of HBx in the regulation of cccDNA. Though proline isomerization has been previously implicated in transcriptional and epigenetic regulation,68,69 its functional role in cccDNA biology requires further study. Moreover, the ability of HBx to recruit such diverse proteins to cccDNA, in terms of both their enzymatic functions and range of molecular weights, raises questions about the mechanism by which it mediates such interactions.
Post-translational Modifications of HBx
Another layer of complexity to HBx biology and regulation is that HBx itself is post-translationally modified in order to modulate its many functions. Some HBx PTMs have been known for several years, but many of their functions remained enigmatic. Recent studies have begun to describe biochemical roles for HBx PTMs and the enzymes that deposit them, potentially providing insight into novel pathways that indirectly target HBx function. Perhaps unsurprising due to its proximity and involvement with host machinery regulating ubiquitination, HBx itself is subject to dynamic modification by both ubiquitin and a ubiquitin-like protein. Similar to many cellular proteins, HBx is degraded by the ubiquitin-proteasome system. It has been shown to directly interact with the deubiquitinating enzyme Usp15. The presence of Usp15 increased steady-state levels of HBx as well as its half-life within cells.70 The other recently-described ubiquitin-like modification of HBx is NEDDylation, which is most typically associated with cullin activation.71,72 As seen with ubiquitination, HBx’s proximity to Cul4 makes it an unsurprising substrate for NEDDylation, shown to occur on lysine resides 91 and 95. This mark was found not only to increase HBx stability (potentially by obscuring a ubiquitination site), but also to enhance HBx chromatin localization, and consequently transactivation activity.71 While these larger PTMs were shown to correspond to HBx stability, others have been shown to have more unique functions. For example, it was found that the kinase Akt1 specifically phosphorylates S31 of HBx in vitro and in cells, and that this phosphorylation increases the oncogenic potential of HBx.73 Notably, residue 31 is not conserved across all major HBV strains, leading to the suggestion that HBx PTMs may potentially be responsible for the observation that some genotypes correlate with more severe clinical outcomes than others.74 However, S31 is potentially not the only phosphorylation site of HBx. A computational study of HBx searching for conserved residues and motifs across all viral genotypes identified three residues (S25, S41, and T81) that are predicted to be phosphorylated. The same sites were also predicted to be O-GlcNAcylated in competition with phosphorylation in what is known as a “Yin Yang” relationship.45
HBx contains 9 or 10 cysteine residues, depending on the viral genotype, which add another layer of complexity as disulfide bonds are potentially an important element of HBx structure and function. One early study purified HBx from insect cells and identified 5 intramolecular disulfide linkages.75 Similarly, a more recent mass spectrometry study of bacterially purified recombinant HBx (from a viral strain with 9 Cys residues) found 4 intramolecular disulfide linkages and posited that the remaining free Cys residue may participate in an intermolecular disulfide bond to dimerize HBx.76 While there is some data supporting that HBx may exist as a dimer in solution,77 it is uncommon for a cytosolic or nuclear protein to contain stable disulfide bonds. Moreover, cysteine to serine alteration resulted in negligible changes to HBx secondary-structure as measured by circular dichroism (CD) spectroscopy.41 These mutations seemed not to effect HBx activity, as measured in vitro using p53 transactivation assays.41,78 Conversely, disulfide-containing HBx exhibited stronger stimulatory effects on the kinase Cdk2 in vitro and higher transactivation in a cellular reporter assay as compared to a cysteine-free mutant.76 Clearly, more detailed analysis of HBx structure and PTMs in vivo is required, not least so that future in vitro studies with recombinant HBx can use more physiologically relevant conditions.
Advances in Targeting HBx and cccDNA
As described in this review, a great body of work has highlighted the importance of cccDNA and HBx to HBV replication and for the persistence of chronic infection.79 To this end, several recent studies have reported a variety of approaches to target these viral elements using small molecule or biologic tools. These methods range from rationally targeted approaches with established and recently developed methodologies to target-agnostic high-throughput screens, which we will describe below.
Rational Approaches Targeting cccDNA
Genome editing methods allow for the relatively facile, locus-specific generation of double-stranded DNA breaks (DSBs), which led to the theory that such an approach could be used to target cccDNA for degradation. Several independent groups reported relative success in this approach, with findings generally showing suppression of viral gene expression and reduction of cccDNA levels.80,81 However, a few barriers still remain preventing such an approach from proceeding into the clinic. Perhaps most obvious of these concerns is that gene editing therapy has not gone through clinical trials. Furthermore, due to its replicative RNA intermediate, HBV has a 10-fold higher mutation rate than conventional DNA viruses.82 As such, resistance mutations may rapidly arise under such a strong selective pressure. Additionally, development of a therapy capable of targeting each of the documented HBV genotypes may prove difficult due to the ~8 % sequence divergence at the DNA level.83 Finally, and perhaps most importantly, the generation of a DSB in cccDNA would generate dslDNA, which could lead to genome integration by host DNA repair mechanisms and consequently promote oncogenesis.19 Nevertheless, such an approach remains attractive for its potential uses in both research and the clinic, and ongoing efforts to address these shortcomings may prove fruitful.
Several studies have identified host factors that contribute to the restriction of HBV and may have therapeutic potential if explored further.84 Some of these factors include zinc finger antiviral protein (ZAP), Tat-interacting protein 60 kDa (TIP60), and the APOBEC3 family of proteins.85–88 Of these, perhaps the most interesting and well-described factor is the APOBEC3 enzyme family, which is composed of seven C-to-U deaminases.89,90 Intriguingly, studies have shown that HBV itself seems to have evolved mechanisms to defend itself from these enzymes. HBx indirectly depletes APOBEC3B through the upregulation of the E3 ligase MSL2 and depletes APOBEC3G by promoting its export.88,91 Such antagonism may be evolutionarily favorable, as several enzymes in this family have been shown to target cccDNA (and other viral episomes) for depurination and subsequent degradation.15,92,93 Thus, induction of APOBEC3 expression is another promising route toward the development of cccDNA-targeting drugs.
Screening Strategies to Target HBV Functions
The complexity of the HBV life cycle undoubtedly makes the virus challenging to study, but also presents several different pathways or actions that can be adapted for use in high-throughput screening (HTS) approaches. Consequently, several studies have recently reported hits from screening approaches that show promise. While this topic has been reviewed in greater detail elsewhere,94 we here highlight several screens which specifically sought to target HBx functions or restrict cccDNA expression (Figure 3).
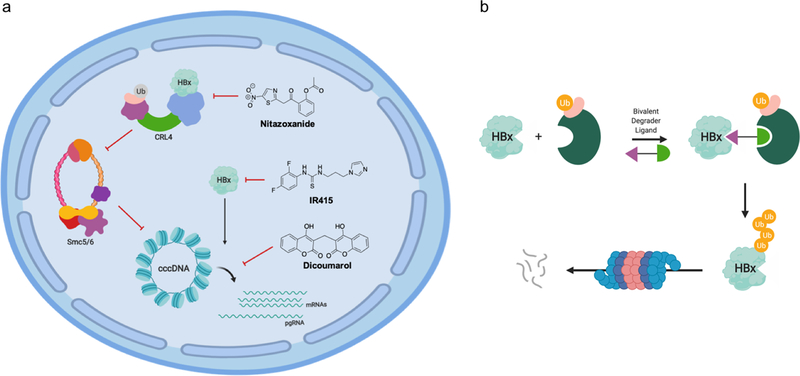
Figure 3. Small molecule modulators of HBx and cccDNA.
(a) Recently identified small molecules that disrupt HBx or cccDNA. (b) Potential development of a catalytic small molecule “degrader” of HBx based on recently identified chemical ligands.
One such example reported recently was identified in a HTS which took advantage of HBx’s documented role inhibiting certain RNAi pathways.95 Briefly, computational prediction was used to generate a three-dimensional structural model of HBx, and virtual docking simulation was used to identify potential binders from a 14400 compound library. Multiple iterations of the screen were used to identify 100 compounds to be experimentally evaluated. These leading hits were used to screen for reversal of HBx-mediated RNAi suppression, and generated a lead compound dubbed IR415.46 Further in vitro assays with recombinant HBx and analyses with infection models verified that IR415 is a bona fide HBx-targeting small molecule that inhibits its role in RNAi suppression. Additional studies will be required in order to determine whether IR415 disrupts any other aspects of HBx function, such as its interactions with epigenome modifiers or transcription factors. These analyses will prove necessary to define the utility of IR415 for research purposes, as well as any therapeutic potential.
More recently, another HBx-targeting molecule was identified by a split luciferase screening assay. Due to the critical role of HBx’s DCAF function in HBV gene expression, this screen appended the two halves of the split luciferase to HBx and DDB1 to screen for molecules that would disrupt this interaction.96 Screening a library of FDA-approved drugs for decreased luciferase signal identified nitazoxanide (NTZ), a broad-spectrum anti-infective agent typically used to treat parasitic infections. This interaction was further validated using recombinant proteins and in cells. However, whether NTZ binds to HBx or DDB1 individually, or only the proteins in complex together, remains unknown and should be further examined in future studies.
Finally, a chemical screen aiming to identify inhibitors of episomal DNA expression was reported recently. Briefly, an integration-deficient lentivirus expressing a luciferase reporter was used to screen a chemical library for inhibitors of luciferase signal and the small molecule dicoumarol was shown to decrease episomal DNA expression.97 This was subsequently validated using HBV-infected primary human hepatocytes, where a dose-dependent depletion of cccDNA was observed. Although dicoumarol has been shown to target the vitamin K epoxide reductase (VKOR), inhibition of this target with a different compound failed to reduce cccDNA levels, indicating that dicoumarol must inhibit a different host factor to do so. Still, dicoumarol is the first reported small molecule to specifically deplete cccDNA rather than targeting downstream elements of the HBV life cycle. As such, it may serve as the basis for future drug development to optimize anti-cccDNA efficacy.
Future Perspectives
Methodological developments and the application of interdisciplinary techniques to study HBV have led to significant advances in our understanding of cccDNA establishment and regulation, as well as certain mechanisms behind HBx function. However, more questions remain for both of these key components of HBV. In this section, we describe outstanding questions and propose possible “chemical virology” approaches to address them that can complement the existing genetic and virological methods.
Chemical Biology and Protein Engineering Strategies to Study HBx and cccDNA
A persistent difficulty in the study of HBx has been the scarcity of methods to manipulate or modulate native HBx in the context of an active HBV infection. It has largely been studied alone by genetic manipulation in cell culture, often at greater expression levels than may be present in actual infections. The recently reported HBx-targeting small molecules discussed in this review present a unique opportunity to chemically address this challenge. By harnessing targeted protein degradation methodologies, chemical ligands of HBx could be adapted to study its function, as has been done for other viral proteins recently (Figure 3b).98,99 Such a molecular tool could even have therapeutic potential for chronic HBV infection.
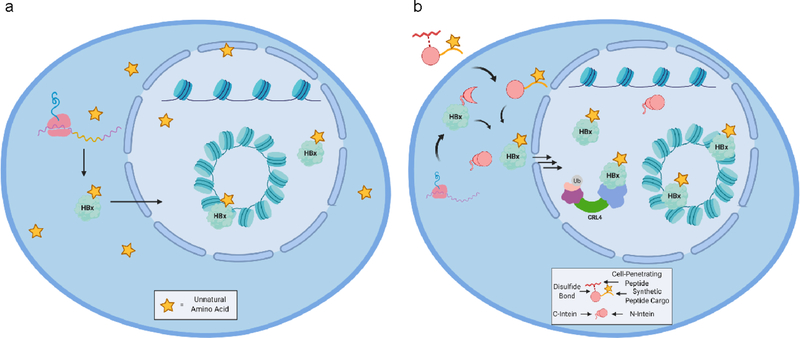
In parallel, protein engineering strategies to better study HBx in vitro and in vivo should also be developed. Due to its poor yield and solubility when recombinantly expressed in E. coli, HBx has rarely been the focus of in vitro biochemical experiments, as mentioned above. Protein fusion methods to improve yield have already been reported in the literature, though continued efforts may build on this progress.76 Engineering approaches to site-specifically incorporate PTMs or chemical handles would contribute greatly to our mechanistic understanding of HBx by enabling more targeted studies to be performed.100,101 The ability to prepare high-quality, chemically homogeneous recombinant HBx by either protein splicing or unnatural amino acid incorporation methods would greatly expand the breadth of questions that could be answered about its many functions (Figure 4). The incorporation of synthetic moieties like fluorophores or cross-linking agents into HBx could interrogate its dynamics or direct interactors, as has been done previously in other systems.102,103 Such methods could also be used to study the effects of individual PTMs on HBx function without perturbing the host cell by manipulating the endogenous enzymes responsible for depositing or erasing them.
Figure 4. Protein engineering approaches to study HBx.
(a) Genetic code expansion (GCE) could be used to site-specifically incorporate unnatural amino acids (UAAs) into HBx. (b) Protein trans-splicing methods could be utilized in live cells to attach UAAs, PTMs, or synthetic cargoes to HBx and study their functions independently.
With respect to cccDNA, despite progress in identifying the host elements responsible for its repair and establishment, many details about its structural and regulatory components remain enigmatic. In particular, recent efforts have been made to characterize the histone PTM landscape of cccDNA.58 However, most such endeavors rely heavily on ChIP coupled with next-generation DNA sequencing (ChIP-seq), which has several shortcomings, including inherent target-bias due to antibody quality and availability as well as having relatively low throughput. Although rational, targeted studies in this area have proved useful to understand cccDNA regulation, it is quite likely that they fail to capture the full complement of factors involved. As such, unbiased high-throughput methods could prove fruitful to generate leads for further mechanistic studies. Recently, different approaches toward genomic locus-specific proteomics have been reported.104–106 These methods generally employ a “reverse-ChIP” workflow, whereby a particular chromatin locus is enriched and then subjected to proteomic analyses. Application of such techniques toward the study of cccDNA could further illuminate the host elements involved in the transcriptional regulation, in part by allowing for proteomic analysis of the histone PTM landscape on the minichromosome.
The development of ever more sophisticated methods to study chromatin biology also has implications in the study of cccDNA. Long-standing protein engineering techniques such as genetic code expansion and expressed protein ligation have been adapted in the past few years for the study of chromatin-associated processes.101,107–109 The implementation of such techniques in HBV-susceptible cell lines could establish a platform for the interrogation of key biochemical questions in cccDNA. Complementing such an approach, the development of a method to reconstitute a cccDNA model for in vitro biochemical and biophysical studies could potentially provide crucial insight into the mechanisms behind recognition and recruitment of host factors onto cccDNA. Although model chromatin fragments have long been used in biochemistry, few studies have been performed on circularized chromatin fragments akin to cccDNA. The few recent reports that have done so analyzed nucleosome spacing and DNA supercoiling,110,111 but a reconstituted cccDNA model could be used for a wide array of biophysical and biochemical studies.
Outlook
Continued efforts in recent years have revealed critical details about the cccDNA life cycle and HBx function that may potentially serve as the basis for novel therapeutic approaches. Indeed, both rational targeted approaches and high-throughput screening methods have identified novel components of the DDR involved in cccDNA establishment. The chromatin landscape of cccDNA, particularly its histone PTMs and regulatory proteins, continues to be further elaborated. Importantly, new roles have been described for HBx, both as it relates to cccDNA expression and with respect to its own stability and regulation. All of these advances have informed the design of assays amenable to high-throughput screens, which have begun to yield small molecule modulators of HBx and cccDNA. Even still, ongoing research is needed to further build upon these advances. Chemical biology and protein engineering techniques are perfectly poised to fully illuminate these two cryptic yet indispensable components of chronic HBV infection.
Acknowledgements
Elements of all figures were prepared with BioRender. N. A. P. is supported by the National Science Foundation Graduate Research Fellowship Grant Number 2017239554 and the NIH T32 GM115327-Tan chemistry-biology interface training grant. Y.B. is supported by NIH Grants R03DK117252, and R01CA234614. R.E.S is supported by NIH grants K08DK101754, R03DK117252, and R01CA234614 and by the Irma Hirschl Trust Research Award Scholar program. Y.D. is supported by the Josie Robertson Foundation, the Pershing Square Sohn Cancer Research Alliance, the NIH (CCSG core grant P30 CA008748, MSK SPORE P50 CA192937, and R21 DA044767), and Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center.
References
- (1). Yuen M-F; Chen D-S; Dusheiko GM; Janssen HLA; Lau DTY; Locarnini SA; Peters MG; Lai C-L Hepatitis B Virus Infection. Nat. Rev. Dis. Prim. 2018, 4, 18035. [DOI] [PubMed] [Google Scholar]
- (2). WHO. (2017) Cancer: World Health Organization Fact Sheet 204, https://www.who.int/news-room/fact-sheets/detail/cancer (accessed Aug 18, 2019). [Google Scholar]
- (3). Allweiss L; Dandri M The Role of cccDNA in HBV Maintenance. Viruses 2017, 9 (6), 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4). Lucifora J; Arzberger S; Durantel D; Belloni L; Strubin M; Levrero M; Zoulim F; Hantz O; Protzer U Hepatitis B Virus X Protein Is Essential to Initiate and Maintain Virus Replication after Infection. J. Hepatol 2011, 55 (5), 996–1003. [DOI] [PubMed] [Google Scholar]
- (5). Neuveut C; Wei Y; Buendia MA Mechanisms of HBV-Related Hepatocarcinogenesis. J. Hepatol 2010, 52 (4), 594–604. [DOI] [PubMed] [Google Scholar]
- (6). Levrero M; Zucman-Rossi J Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatol 2016, 64 (1), S84–S101. [DOI] [PubMed] [Google Scholar]
- (7). Hu J; Cheng J; Tang L; Hu Z; Luo Y; Li Y; Zhou T; Chang J; Guo J-T Virological Basis for the Cure of Chronic Hepatitis B. ACS Infect. Dis 2019, 5 (5), 659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8). Yan H; Zhong G; Xu G; He W; Jing Z; Gao Z; Huang Y; Qi Y; Peng B; Wang H;Fu L; Song M; Chen P; Gao W; Ren B; Sun Y; Cai T; Feng X; Sui J; Li W Sodium Taurocholate Cotransporting Polypeptide Is a Functional Receptor for Human Hepatitis B and D Virus. eLife 2012, 2012 (1), 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9). Macovei A; Petrareanu C; Lazar C; Florian P; Branza-Nichita N Regulation of Hepatitis B Virus Infection by Rab5, Rab7, and the Endolysosomal Compartment. J. Virol 2013, 87 (11), 6415–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10). Lamontagne RJ; Bagga S; Bouchard MJ Hepatitis B Virus Molecular Biology and Pathogenesis. Hepatoma Res 2016, 2 (7), 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11). Selzer L; Zlotnick A Assembly and Release of Hepatitis B Virus. Cold Spring Harb. Perspect. Med 2015, a021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12). Ko C; Chakraborty A; Chou W-M; Hasreiter J; Wettengel JM; Stadler D; Bester R; Asen T; Zhang K; Wisskirchen K; McKeating JA; Ryu W-S; Proter U Hepatitis B Virus Genome Recycling and de Novo Secondary Infection Events Maintain Stable cccDNA Levels. J. Hepatol 2018, 69 (6), 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13). Guo F; Zhao Q; Sheraz M; Cheng J; Qi Y; Su Q; Cuconati A; Wei L; Du Y; Li W; Chang J; Guo J-T. HBV Core Protein Allosteric Modulators Differentially Alter cccDNA Biosynthesis from de Novo Infection and Intracellular Amplification Pathways. PLoS Pathog. 2017, 13 (9), e1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14). Yang L; Liu F; Tong X; Hoffmann D; Zuo J; Lu M Treatment of Chronic Hepatitis B Virus Infection Using Small Molecule Modulators of Nucleocapsid Assembly: Recent Advances and Perspectives. ACS Infect. Dis 2019, 5 (5), 713–724. [DOI] [PubMed] [Google Scholar]
- (15). Lucifora J; Xia Y; Reisinger F; Zhang K; Stadler D; Cheng X; Sprinzl MF; Koppensteiner H; Makowska Z; Volz T; Remouchamps C; Chou W-M; Thasler WE; Hüser N; Durantel D; Liang TJ; Münk C; Heim MH; Browning JL; Dejardin E; Dandri M; Schindler M; Heikenwalder M; Protzer U Specific and Nonhepatotoxic Degradation of Nuclear Hepatitis B Virus cccDNA. Science (80-. ) 2014, 343 (6176), 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16). Zhao X-L; Yang J-R; Lin S-Z; Ma H; Guo F; Yang R-F; Zhang H-H; Han J-C; Wei L; Pan X-B Serum Viral Duplex-Linear DNA Proportion Increases with the Progression of Liver Disease in Patients Infected with HBV. Gut 2016, 65 (3), 502–511. [DOI] [PubMed] [Google Scholar]
- (17). Schreiner S; Nassal M A Role for the Host DNA Damage Response in Hepatitis B Virus cccDNA Formation—and Beyond? Viruses 2017, 9 (5), 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18). Tu T; Budzinska MA; Vondran FWR; Shackel NA; Urban S Hepatitis B Virus DNA Integration Occurs Early in the Viral Life Cycle in an In Vitro Infection Model via Sodium Taurocholate Cotransporting Polypeptide-Dependent Uptake of Enveloped Virus Particles. J. Virol 2018, 92 (11), e02007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19). Zhao L-H; Liu X; Yan H-X; Li W-Y; Zeng X; Yang Y; Zhao J; Liu S-P; Zhuang X-H; Lin C; Qin C-J; Zhao Y; Pan Z-Y; Huang G; Liu H; Zhang J; Wang R-Y; Yang Y; Wen W; Lv G-S; Zhang H-L; Wu H; Huang S; WAng M-D; Tang L; Cao H-Z; Wang L; Lee T-L; Jiang H; Tan Y-X; Yuan S-X; Hou G-J; Tao Q-F; Xu Q-G; Zhang X-Q; Wu M-C; Xu X; Wang J; Yang H-M; Zhou W-P; Wang H-Y Genomic and Oncogenic Preference of HBV Integration in Hepatocellular Carcinoma. Nat. Commun 2016, 7 (1), 12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20). Königer C; Wingert I; Marsmann M; Rösler C; Beck J; Nassal M Involvement of the Host DNA-Repair Enzyme TDP2 in Formation of the Covalently Closed Circular DNA Persistence Reservoir of Hepatitis B Viruses. Proc. Natl. Acad. Sci 2014, 111 (40), E4244–E4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21). Cui X; McAllister R; Boregowda R; Sohn JA; Ledesma FC; Caldecott KW; Seeger C; Hu J Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS One 2015, 10 (6), e0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22). Winer BY; Huang TS; Pludwinski E; Heller B; Wojcik F; Lipkowitz GE; Parekh A; Cho C; Shrirao A; Muir TW; Novik E; Ploss A Long-Term Hepatitis B Infection in a Scalable Hepatic Co-Culture System. Nat. Commun 2017, 8 (1), 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23). Kitamura K; Que L; Shimadu M; Koura M; Ishihara Y; Wakae K; Nakamura T; Watashi K; Wakita T; Muramatsu M Flap Endonuclease 1 Is Involved in cccDNA Formation in the Hepatitis B Virus. PLoS Pathog. 2018, 14 (6), e1007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24). Qi Y; Gao Z; Xu G; Peng B; Liu C; Yan H; Yao Q; Sun G; Liu Y; Tang D; Song Z; He W; Sun Y; Guo J-T; Li W DNA Polymerase κ Is a Key Cellular Factor for the Formation of Covalently Closed Circular DNA of Hepatitis B Virus. PLoS Pathog. 2016, 12 (10), e1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25). Tang L; Sheraz M; McGrane M; Chang J; Guo J-T DNA Polymerase Alpha Is Essential for Intracellular Amplification of Hepatitis B Virus Covalently Closed Circular DNA. PLoS Pathog. 2019, 15 (4), e1007742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26). Ketkar A; Maddukuri L; Penthala NR; Reed MR; Zafar MK; Crooks PA; Eoff RL Inhibition of Human DNA Polymerases Eta and Kappa by Indole-Derived Molecules Occurs through Distinct Mechanisms. ACS Chem. Biol 2019, 14 (6), 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27). Long Q; Yan R; Hu J; Cai D; Mitra B; Kim ES; Marchetti A; Zhang H; Wang S; Liu Y; Huang A; Guo H The Role of Host DNA Ligases in Hepadnavirus Covalently Closed Circular DNA Formation. PLoS Pathog. 2017, 13 (12), e1006784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28). Sheraz M; Cheng J; Tang L; Chang J; Guo J-T Cellular DNA Topoisomerases Are Required for the Synthesis of Hepatitis B Virus Covalently Closed Circular DNA. J. Virol 2019, 93 (11), 176–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29). Li M; Sohn JA; Seeger C Distribution of Hepatitis B Virus Nuclear DNA. J. Virol 2017, 92 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30). Qu B; Ni Y; Lempp FA; Vondran FWR; Urban S T5 Exonuclease Hydrolysis of Hepatitis B Virus Replicative Intermediates Allows Reliable Quantification and Fast Drug Efficacy Testing of Covalently Closed Circular DNA by PCR. J. Virol 2018, 92 (23), e01117–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31). Newbold JE; Xin H; Tencza M; Sherman G; Dean J; Bowden S; Locarnini S The Covalently Closed Duplex Form of the Hepadnavirus Genome Exists in Situ as a Heterogeneous Population of Viral Minichromosomes. J. Virol 1995, 69 (6), 3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32). Bock CT; Schwinn S; Locarnini S; Fyfe J; Manns MP; Trautwein C; Zentgraf H Structural Organization of the Hepatitis B Virus Minichromosome. J. Mol. Biol 2001, 307 (1), 183–196. [DOI] [PubMed] [Google Scholar]
- (33). Moreau P; Cournac A; Palumbo GA; Marbouty M; Mortaza S; Thierry A; Cairo S; Lavigne M; Koszul R; Neuveut C Tridimensional Infiltration of DNA Viruses into the Host Genome Shows Preferential Contact with Active Chromatin. Nat. Commun 2018, 9 (1), 4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34). Zhang Y; Li C; Zhang Y; Zhu H; Kang Y; Liu H; Wang J; Qin Y; Mao R; Xie Y; Huang Y; Zhang J Comparative Analysis of CpG Islands among HBV Genotypes. PLoS One 2013, 8 (2), e56711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35). Guo YH; Li YN; Zhao JR; Zhang J; Yan Z HBc Binds to the CpG Islands of HBV cccDNA and Promotes an Epigenetic Permissive State. Epigenetics 2011, 6 (6), 720–726. [DOI] [PubMed] [Google Scholar]
- (36). Chong CK; Cheng CYS; Tsoi SYJ; Huang FY; Liu F; Seto WK; Lai CL; Yuen MF; Wong DKH Role of Hepatitis B Core Protein in HBV Transcription and Recruitment of Histone Acetyltransferases to cccDNA Minichromosome. Antiviral Res. 2017, 144, 1–7. [DOI] [PubMed] [Google Scholar]
- (37). Wu Z-J; Zhu Y; Huang D-R; Wang Z-Q Constructing the HBV-Human Protein Interaction Network to Understand the Relationship between HBV and Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res 2010, 29 (1), 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38). Zhang T; Xie N; He W; Liu R; Lei Y; Chen Y; Tang H; Liu B; Huang C; Wei Y An Integrated Proteomics and Bioinformatics Analyses of Hepatitis B Virus X Interacting Proteins and Identification of a Novel Interactor ApoA-I. J. Proteomics 2013, 84, 92–105. [DOI] [PubMed] [Google Scholar]
- (39). Xie N; Chen X; Zhang T; Liu B; Huang C Using Proteomics to Identify the HBx Interactome in Hepatitis B Virus: How Can This Inform the Clinic? Expert Rev. Proteomics 2014, 11 (1), 59–74. [DOI] [PubMed] [Google Scholar]
- (40). Slagle BL; Bouchard MJ Hepatitis B Virus X and Regulation of Viral Gene Expression. Cold Spring Harb. Perspect. Med 2016, 6 (3), a021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41). Lee S-HH; Cha E-JJ; Lim J-EE; Kwon S-HH; Kim D-HH; Cho H; Han K-HH Structural Characterization of an Intrinsically Unfolded Mini-HBX Protein from Hepatitis B Virus. Mol. Cells 2012, 34 (2), 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42). Li T; Robert EI; van Breugel PC; Strubin M; Zheng N A Promiscuous α-Helical Motif Anchors Viral Hijackers and Substrate Receptors to the CUL4–DDB1 Ubiquitin Ligase Machinery. Nat. Struct. Mol. Biol 2010, 17 (1), 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43). Jiang T; Liu M; Wu J; Shi Y Structural and Biochemical Analysis of Bcl-2 Interaction with the Hepatitis B Virus Protein HBx. Proc. Natl. Acad. Sci 2016, 113 (8), 2074–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44). van Hemert FJ; van de Klundert MAA; Lukashov VV; Kootstra NA; Berkhout B; Zaaijer HL Protein X of Hepatitis B Virus: Origin and Structure Similarity with the Central Domain of DNA Glycosylase. PLoS One 2011, 6 (8), e23392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45). Hernández S; Venegas M; Brahm J; Villanueva RA The Viral Transactivator HBx Protein Exhibits a High Potential for Regulation via Phosphorylation through an Evolutionarily Conserved Mechanism. Infect. Agent. Cancer 2012, 7 (1), 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46). Ghosh S; Kaushik A; Khurana S; Varshney A; Singh AK; Dahiya P; Thakur JK; Sarin SK; Gupta D; Malhotra P; Mukherjee SK; Bhatnagar RK An RNAi-Based High-Throughput Screening Assay to Identify Small Molecule Inhibitors of Hepatitis B Virus Replication. J. Biol. Chem 2017, 292 (30), 12577–12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47). Tsuge M; Hiraga N; Akiyama R; Tanaka S; Matsushita M; Mitsui F; Abe H; Kitamura S; Hatakeyama T; Kimura T; Miki D; Mori N; Imamura M; Tskahashi S; Hayes CN; Chayama K HBx Protein Is Indispensable for Development of Viraemia in Human Hepatocyte Chimeric Mice. J. Gen. Virol 2010, 91 (7), 1854–1864. [DOI] [PubMed] [Google Scholar]
- (48). Decorsière A; Mueller H; van Breugel PC; Abdul F; Gerossier L; Beran RK; Livingston CM; Niu C; Fletcher SP; Hantz O; Strubin M Hepatitis B Virus X Protein Identifies the Smc5/6 Complex as a Host Restriction Factor. Nature 2016, 531 (7594), 386–380. [DOI] [PubMed] [Google Scholar]
- (49). Niu C; Livingston CM; Li L; Beran RK; Daffis S; Ramakrishnan D; Burdette D; Peiser L; Salas E; Ramos H; Yu M; Cheng G; Strubin M; Delaney IV WE; Fletcher SP The Smc5/6 Complex Restricts HBV When Localized to ND10 without Inducing an Innate Immune Response and Is Counteracted by the HBV X Protein Shortly after Infection. PLoS One 2017, 12 (1), e0169648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50). Jackson S; Xiong Y CRL4s: The CUL4-RING E3 Ubiquitin Ligases. Trends Biochem. Sci 2009, pp 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51). Murphy CM; Xu Y; Li F; Nio K; Reszka-Blanco N; Li X; Wu Y; Yu Y; Xiong Y; Su L Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep. 2016, 16 (11), 2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52). Gustin JK; Moses AV; Früh K; Douglas JL Viral Takeover of the Host Ubiquitin System. Front. Microbiol 2011, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53). Li T; Chen X; Garbutt KC; Zhou P; Zheng N Structure of DDB1 in Complex with a Paramyxovirus V Protein: Viral Hijack of a Propeller Cluster in Ubiquitin Ligase. Cell 2006, 124 (1), 105–117. [DOI] [PubMed] [Google Scholar]
- (54). Le Rouzic E; Belaïdouni N; Estrabaud E; Morel M; Rain J-C; Transy C; Margottin-Goguet F HIV1 Vpr Arrests the Cell Cycle by Recruiting DCAF1/VprBP, a Receptor of the Cul4-DDB1 Ubiquitin Ligase. Cell Cycle 2007, 6 (2), 182–188. [DOI] [PubMed] [Google Scholar]
- (55). Fuchs G; Oren M Writing and Reading H2B Monoubiquitylation. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2014, pp 694–701. [DOI] [PubMed] [Google Scholar]
- (56). Grazini U; Zanardi F; Citterio E; Casola S; Goding CR; McBlane F The RING Domain of RAG1 Ubiquitylates Histone H3: A Novel Activity in Chromatin-Mediated Regulation of V(D)J Joining. Mol. Cell 2010, 37 (2), 282–293. [DOI] [PubMed] [Google Scholar]
- (57). Mandemaker IK; van Cuijk L; Janssens RC; Lans H; Bezstarosti K; Hoeijmakers JH; Demmers JA; Vermeulen W; Marteijn JA DNA Damage-Induced Histone H1 Ubiquitylation Is Mediated by HUWE1 and Stimulates the RNF8-RNF168 Pathway. Sci. Rep 2017, 7 (1), 15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58). Tropberger P; Mercier A; Robinson M; Zhong W; Ganem DE; Holdorf M Mapping of Histone Modifications in Episomal HBV cccDNA Uncovers an Unusual Chromatin Organization Amenable to Epigenetic Manipulation. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (42), E5715–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59). Andrisani O Deregulation of Epigenetic Mechanisms by the Hepatitis B Virus X Protein in Hepatocarcinogenesis. Viruses 2013, 5 (3), 858–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60). Hong X; Kim ES; Guo H Epigenetic Regulation of Hepatitis B Virus Covalently Closed Circular DNA: Implications for Epigenetic Therapy against Chronic Hepatitis B. Hepatology 2017, 66 (6), 2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61). Pollicino T; Belloni L; Raffa G; Pediconi N; Squadrito G; Raimondo G; Levrero M Hepatitis B Virus Replication Is Regulated by the Acetylation Status of Hepatitis B Virus cccDNA-Bound H3 and H4 Histones. Gastroenterology 2006, 130 (3), 823–837. [DOI] [PubMed] [Google Scholar]
- (62). Belloni L; Pollicino T; De Nicola F; Guerrieri F; Raffa G; Fanciulli M; Raimondo G; Levrero M Nuclear HBx Binds the HBV Minichromosome and Modifies the Epigenetic Regulation of cccDNA Function. Proc. Natl. Acad. Sci 2009, 106 (47), 19975–19979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63). Rivière L; Gerossier L; Ducroux A; Dion S; Deng Q; Michel M-L; Buendia MA; Hantz O; Neuveut C HBx Relieves Chromatin-Mediated Transcriptional Repression of Hepatitis B Viral cccDNA Involving SETDB1 Histone Methyltransferase. J. Hepatol 2015, 63 (5), 1093–1102. [DOI] [PubMed] [Google Scholar]
- (64). Zheng D-L; Zhang L; Cheng N; Xu X; Deng Q; Teng X-M; Wang K-S; Zhang X; Huang J; Han Z-G Epigenetic Modification Induced by Hepatitis B Virus X Protein via Interaction with de Novo DNA Methyltransferase DNMT3A. J. Hepatol 2009, 50 (2), 377–387. [DOI] [PubMed] [Google Scholar]
- (65). Koumbi L; Karayiannis P The Epigenetic Control of Hepatitis B Virus Modulates the Outcome of Infection. Front. Microbiol 2016, 6 (JAN), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66). Pang R; Lee TKW; Poon RTP; Fan ST; Wong KB; Kwong YL; Tse E Pin1 Interacts With a Specific Serine-Proline Motif of Hepatitis B Virus X-Protein to Enhance Hepatocarcinogenesis. Gastroenterology 2007, 132 (3), 1088–1103. [DOI] [PubMed] [Google Scholar]
- (67). Saeed U; Kim J; Piracha ZZ; Kwon H; Jung J; Chwae Y-J; Park S; Shin H-J; Kim K Parvulin 14 and Parvulin 17 Bind to HBx and cccDNA and Upregulate Hepatitis B Virus Replication from cccDNA to Virion in an HBx-Dependent Manner. J. Virol 2018, 93 (6). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (68). Mayfield JE; Fan S; Wei S; Zhang M; Li B; Ellington AD; Etzkorn FA; Zhang YJ Chemical Tools To Decipher Regulation of Phosphatases by Proline Isomerization on Eukaryotic RNA Polymerase II. ACS Chem. Biol 2015, 10 (10), 2405–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69). Howe FS; Boubriak I; Sale MJ; Nair A; Clynes D; Grijzenhout A; Murray SC; Woloszczuk R; Mellor J Lysine Acetylation Controls Local Protein Conformation by Influencing Proline Isomerization. Mol. Cell 2014, 55 (5), 733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70). Su ZJ; Cao JS; Wu YF; Chen WN; Lin X; Wu YL; Lin X Deubiquitylation of Hepatitis B Virus X Protein (HBx) by Ubiquitin-Specific Peptidase 15 (USP15) Increases HBx Stability and Its Transactivation Activity. Sci. Rep 2017, 7 (August 2016), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71). Liu N; Zhang J; Yang X; Jiao T; Zhao X; Li W; Zhu J; Yang P; Jin J; Peng J; Li Z; Ye X HDM2 Promotes NEDDylation of Hepatitis B Virus HBx To Enhance Its Stability and Function. J. Virol 2017, 91 (16), e00340–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72). Minor M; Slagle B Hepatitis B Virus HBx Protein Interactions with the Ubiquitin Proteasome System. Viruses 2014, 6 (11), 4683–4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73). Khattar E; Mukherji A; Kumar V Akt Augments the Oncogenic Potential of the HBx Protein of Hepatitis B Virus by Phosphorylation. FEBS J 2012, 279 (7), 1220–1230. [DOI] [PubMed] [Google Scholar]
- (74). Sunbul M Hepatitis B Virus Genotypes: Global Distribution and Clinical Importance. World J. Gastroenterol. 2014, 20 (18), 5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75). Urban S; Hildt E; Eckerskorn C; Sirma H; Kekulé A; Hofschneider PH Isolation and Molecular Characterization of Hepatitis B Virus X-Protein from a Baculovirus Expression System. Hepatology 1997, 26 (4), 1045–1053. [DOI] [PubMed] [Google Scholar]
- (76). Sidhu K; Kumar S; Reddy VS; Kumar V Mass Spectrometric Determination of Disulfide Bonds in the Biologically Active Recombinant HBx Protein of Hepatitis B Virus. Biochemistry 2014, 53 (28), 4685–4695. [DOI] [PubMed] [Google Scholar]
- (77). Lin M-H; Lo SJ Dimerization of Hepatitis B Viral X Protein Synthesized in a Cell-Free System. Biochem. Biophys. Res. Commun 1989, 164 (1), 14–21. [DOI] [PubMed] [Google Scholar]
- (78). de Moura PR; Rui E; de Almeida Gonçalves K; Kobarg J The Cysteine Residues of the Hepatitis B Virus Onco-Protein HBx Are Not Required for Its Interaction with RNA or with Human P53. Virus Res. 2005, 108 (1–2), 121–131. [DOI] [PubMed] [Google Scholar]
- (79). Alter H; Block T; Brown N; Brownstein A; Brosgart C; Chang K-M; Chen P-J; Chisari FV; Cohen C; El-Serag H; Feld J; Gish R; Glenn J; Greten T; Guo H; Guo J-T; Hoshida Y; Hu J; Kowdley KV; Li W; Liang J; Locarnini S; Lok AS; Mason W; McMahon B; Mehta A; Perrillo R; Revill P; Rice CM; Rinaudo J; Schinazi R; Seeger C; Shetty K; Tavis J; Zoulim F A Research Agenda for Curing Chronic Hepatitis B Virus Infection. Hepatology 2018, 67 (3), 1127–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80). Dong C; Qu L; Wang H; Wei L; Dong Y; Xiong S Targeting Hepatitis B Virus cccDNA by CRISPR/Cas9 Nuclease Efficiently Inhibits Viral Replication. Antiviral Res. 2015, 118, 110–117. [DOI] [PubMed] [Google Scholar]
- (81). Ramanan V; Shlomai A; Cox DBT; Schwartz RE; Michailidis E; Bhatta A; Scott DA; Zhang F; Rice CM; Bhatia SN CRISPR/Cas9 Cleavage of Viral DNA Efficiently Suppresses Hepatitis B Virus. Sci. Rep 2015, 5 (1), 10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82). De Silva Feelixge HS; Stone D; Roychoudhury P; Aubert M; Jerome KR CRISPR/Cas9 and Genome Editing for Viral Disease—Is Resistance Futile? ACS Infect. Dis 2018, 4 (6), 871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83). McNaughton AL; D’Arienzo V; Ansari MA; Lumley SF; Littlejohn M; Revill P; McKeating JA; Matthews PC Insights From Deep Sequencing of the HBV Genome—Unique, Tiny, and Misunderstood. Gastroenterology 2019, 156 (2), 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84). Ortega-Prieto AM; Dorner M Immune Evasion Strategies during Chronic Hepatitis B and C Virus Infection. Vaccines 2017, 5 (3), 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85). Mao R; Nie H; Cai D; Zhang J; Liu H; Yan R; Cuconati A; Block TM; Guo J-T; Guo H Inhibition of Hepatitis B Virus Replication by the Host Zinc Finger Antiviral Protein. PLoS Pathog. 2013, 9 (7), e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86). Nishitsuji H; Ujino S; Harada K; Shimotohno K TIP60 Complex Inhibits Hepatitis B Virus Transcription. J. Virol 2018, 92 (6), 176–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87). Luo X; Huang Y; Chen Y; Tu Z; Hu J; Tavis JE; Huang A; Hu Y Association of Hepatitis B Virus Covalently Closed Circular DNA and Human APOBEC3B in Hepatitis B Virus-Related Hepatocellular Carcinoma. PLoS One 2016, 11 (6), e0157708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88). Chen R; Zhao X; Wang Y; Xie Y; Liu J Hepatitis B Virus X Protein Is Capable of Down-Regulating Protein Level of Host Antiviral Protein APOBEC3G. Sci. Rep 2017, 7, 40783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89). Olson ME; Harris RS; Harki DA APOBEC Enzymes as Targets for Virus and Cancer Therapy. Cell Chem. Biol 2018, 25, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90). Adolph MB; Love RP; Chelico L Biochemical Basis of APOBEC3 Deoxycytidine Deaminase Activity on Diverse DNA Substrates. ACS Infect. Dis 2018, 4 (3), 224–238. [DOI] [PubMed] [Google Scholar]
- (91). Gao Y; Feng J; Yang G; Zhang S; Liu Y; Bu Y; Sun M; Zhao M; Chen F; Zhang W; Ye L; Zhang X Hepatitis B Virus X Protein-Elevated MSL2 Modulates Hepatitis B Virus Covalently Closed Circular DNA by Inducing Degradation of APOBEC3B to Enhance Hepatocarcinogenesis. Hepatology 2017, 66 (5), 1413–1429. [DOI] [PubMed] [Google Scholar]
- (92). Seeger C; Mason WS Molecular Biology of Hepatitis B Virus Infection. Virology 2015, 479–480, 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93). Chen Y; Hu J; Cai X; Huang Y; Zhou X; Tu Z; Hu J; Tavis JE; Tang N; Huang A; Hu Y APOBEC3B Edits HBV DNA and Inhibits HBV Replication during Reverse Transcription. Antiviral Res. 2018, 149, 16–25. [DOI] [PubMed] [Google Scholar]
- (94). Feng S; Gao L; Han X; Hu T; Hu Y; Liu H; Thomas AW; Yan Z; Yang S; Young JAT; Yun H; Zhu W; Shen HC Discovery of Small Molecule Therapeutics for Treatment of Chronic HBV Infection. ACS Infect. Dis 2018, 4 (3), 257–277. [DOI] [PubMed] [Google Scholar]
- (95). Chinnappan M; Singh AK; Kakumani PK; Kumar G; Rooge SB; Kumari A; Varshney A; Rastogi A; Singh AK; Sarin SK; Malhotra P; Mukherjee SK; Bhatnagar RK Key Elements of the RNAi Pathway Are Regulated by Hepatitis B Virus Replication and HBx Acts as a Viral Suppressor of RNA Silencing. Biochem. J 2014, 462 (2), 347–358. [DOI] [PubMed] [Google Scholar]
- (96). Sekiba K; Otsuka M; Ohno M; Yamagami M; Kishikawa T; Suzuki T; Ishibashi R; Seimiya T; Tanaka E; Koike K Inhibition of HBV Transcription From cccDNA With Nitazoxanide by Targeting the HBx–DDB1 Interaction. Cell. Mol. Gastroenterol. Hepatol 2019, 7 (2), 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97). Takeuchi F; Ikeda S; Tsukamoto Y; Iwasawa Y; Qihao C; Otakaki Y; Ryota O; Yao W-L; Narita R; Makoto H; Watashi K; Wakita T; Takeuchi K; Chayama K; Kogure A; Kato H; Fujita T Screening for Inhibitor of Episomal DNA Identified Dicumarol as a Hepatitis B Virus Inhibitor. PLoS One 2019, 14 (2), e0212233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98). Bondeson DP; Mares A; Smith IED; Ko E; Campos S; Miah AH; Mulholland KE; Routly N; Buckley DL; Gustafson JL; Zinn N; Grandi P; Shimamura S; Bergamini G; Faelth-Savitski M; Bantscheff M; Cox C; Gordon DA; Willard RR; Flanagan JJ; Casillas LN; Votta BJ; den Besten W; Famm K; Kruidenier L; Carter PS; Harling JD; Churcher I; Crews CM Catalytic in Vivo Protein Knockdown by Small-Molecule PROTACs. Nat. Chem. Biol 2015, 11 (8), 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99). de Wispelaere M; Du G; Donovan KA; Zhang T; Eleuteri NA; Yuan JC; Kalabathula J; Nowak RP; Fischer ES; Gray NS; Yang PL Small Molecule Degraders of the Hepatitis C Virus Protease Reduce Susceptibility to Resistance Mutations. Nat. Commun 2019, 10 (1), 3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100). Osunsade A; Prescott NA; Hebert JM; Ray DM; Jmeian Y; Lorenz IC; David Y A Robust Method for the Purification and Characterization of Recombinant Human Histone H1 Variants. Biochemistry 2019, 58 (3), 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101). David Y; Muir TW Emerging Chemistry Strategies for Engineering Native Chromatin. J. Am. Chem. Soc 2017, 139 (27), 9090–9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102). Uttamapinant C; Howe JD; Lang K; Beránek V; Davis L; Mahesh M; Barry NP; Chin JW Genetic Code Expansion Enables Live-Cell and Super-Resolution Imaging of Site-Specifically Labeled Cellular Proteins. J. Am. Chem. Soc 2015, 137 (14), 4602–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103). Kleiner RE; Hang LE; Molloy KR; Chait BT; Kapoor TM A Chemical Proteomics Approach to Reveal Direct Protein-Protein Interactions in Living Cells. Cell Chem. Biol 2017, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104). Tsui C; Inouye C; Levy M; Lu A; Florens L; Washburn MP; Tjian R DCas9-Targeted Locus-Specific Protein Isolation Method Identifies Histone Gene Regulators. Proc. Natl. Acad. Sci 2018, 201718844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105). Korthout T; Poramba-Liyanage DW; van Kruijsbergen I; Verzijlbergen KF; van Gemert FPA; van Welsem T; van Leeuwen F Decoding the Chromatin Proteome of a Single Genomic Locus by DNA Sequencing. PLoS Biol. 2018, 16 (7), e2005542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106). Fujita T; Yuno M; Fujii H EnChIP Systems Using Different CRISPR Orthologues and Epitope Tags. BMC Res. Notes 2018, 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107). David Y; Vila-Perelló M; Verma S; Muir TW Chemical Tagging and Customizing of Cellular Chromatin States Using Ultrafast Trans-Splicing Inteins. Nat. Chem 2015, 7 (5), 394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108). Elsässer SJ; Ernst RJ; Walker OS; Chin JW Genetic Code Expansion in Stable Cell Lines Enables Encoded Chromatin Modification. Nat. Methods 2016, 13 (2), 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109). Liszczak GP; Brown ZZ; Kim SH; Oslund RC; David Y; Muir TW Genomic Targeting of Epigenetic Probes Using a Chemically Tailored Cas9 System. Proc. Natl. Acad. Sci 2017, 114 (4), 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110). Nikitina T; Norouzi D; Grigoryev SA; Zhurkin VB DNA Topology in Chromatin Is Defined by Nucleosome Spacing. Sci. Adv 2017, 3 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111). Bass MV; Nikitina T; Norouzi D; Zhurkin VB; Grigoryev SA Nucleosome Spacing Periodically Modulates Nucleosome Chain Folding and DNA Topology in Circular Nucleosome Arrays. J. Biol. Chem 2019, 294 (11), 4233–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112). Kelley LA; Mezulis S; Yates CM; Wass MN; Sternberg MJE The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc 2015, 10 (6), 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]