Abstract
Objectives.
Cervical cancer (CC) remains a major health problem worldwide. Poly (adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitors (PARPi) have emerged as a promising class of chemotherapeutics in ovarian cancer. We explored the preclinical in vitro and in vivo activity of olaparib against multiple primary whole exome sequenced (WES) CC cells lines and xenografts.
Methods.
Olaparib cell-cycle, apoptosis, homologous-recombination-deficiency (HRD), PARP trapping and cytotoxicity activity was evaluated against 9 primary CC cell lines in vitro. PARP and PAR expression were analyzed by western blot assays. Finally, olaparib in vivo antitumor activity was tested against CC xenografts.
Results.
While none of the cell lines demonstrated HRD, three out of 9 (33.3%) primary CC cell lines showed strong PARylation activity and demonstrated high sensitivity to olaparib in vitro treatment (cutoff IC50 values < 2μM, p=0.0012). Olaparib suppressed CC cell growth through cell cycle arrest in the G2/M phase and caused apoptosis (p<0.0001). Olaparib activity in CC involved both PARP enzyme inhibition and trapping. In vivo, olaparib significantly impaired CC xenografts tumor growth (p=0.0017) and increased overall animal survival (p=0.008).
Conclusions.
A subset of CC primary cell lines is highly responsive to olaparib treatment in vitro and in vivo. High level of PARylation correlated with olaparib preclinical activity and may represent a useful biomarker for the identification of CC patients benefitting the most from PARPi.
BACKGROUND
Despite the implementation of prophylactic vaccination strategies against Human Papillomavirus (HPV) infection and advances in chemoradiation and immunotherapy, cervical cancer (CC) remains a major health problem in the United States with 13,240 new cases and 4,170 related deaths in 2018 [1]. Chemoradiation represents the standard of care for patients with locally advanced disease not suitable for curative surgery [2] while the usual treatment for recurrent/metastatic CC is a combination of paclitaxel and cisplatin or paclitaxel, cisplatin and bevacizumab. These chemotherapy treatments, although not curative, result in median survival times of approximately one to 1.5 years [3–5]. Once patients progress after this initial therapy for recurrent or metastatic disease, options are limited (there are no FDA approved or NCCN level 1 or 2A therapies available). Identification of novel, effective therapies for CC patients with disease resistant to standard treatment modalities remains an unmet medical need.
In recent years, Poly (adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitors (PARPi) have emerged as a promising class of chemotherapeutic agents for ovarian cancer associated with defects in homologous recombination DNA repair (HRR) system [6–10]. PARP1 is one of the most abundant proteins among several members of the PARP family and multiple studies implicated PARP1 as having pleiotropic cellular functions, such as maintenance of genomic integrity, DNA repair and regulation of apoptotic and survival balance in cells [11–14]. Furthermore, the enzyme is involved in the PARylation of nuclear proteins (i.e., the post-translational modification process by which polymers of ADP-ribose (poly(adenosine diphosphate-ribose)) are covalently attached to proteins by PAR polymerase enzymes), recruitment of DNA repair factors and stabilization of chromatin for transcriptional regulation [15]. Importantly, since PARPi prevents repair of single strand breaks, causing DNA destabilization and eventual double strand breaks, cancer cells with deficient double strand repair (HRR) are particularly sensitive to PARPi [16]. Accordingly, based on preclinical and clinical results, in 2014 the US Food and Drug Administration (FDA) approved the first PARPi (i.e., olaparib) for treatment of patients with germline BRCA-mutated advanced ovarian cancer, who have been treated with three or more prior lines of chemotherapy. Since 2017, three PARP inhibitors (i.e., olaparib, rucaparib and niraparib), have received FDA approval in the ovarian cancer recurrent setting as maintenance therapy following platinum-based therapy [17–19].
Although several clinical trials are currently underway investigating the clinical efficacy and safety of PARPi for various human malignancies, limited preclinical and clinical information is currently available on the potential activity of olaparib in CC patients [20]. Accordingly, in this study, we evaluated the preclinical activity of olaparib against multiple homologous recombination competent (HRD) primary CC cell lines (i.e., both squamous and adenocarcinoma) and xenografts. Furthermore, we also investigated possible mechanisms behind CC sensitivity to PARPi and elucidated the correlation between sensitivity to olaparib and PARylation activity.
MATERIAL AND METHODS
Establishment of CC cell lines
Study approval was obtained from the Institutional Review Board (IRB), and all patients signed consent prior to tissue collection according to the institutional guidelines. Nine primary CC cell lines (Table 1) were established from fresh tumor biopsy samples and maintained at 37 °C, 5% CO2 in Keratinocytes-SFM (Gibco®, Life Technologies™), supplemented with prequalified human recombinant Epidermal Growth Factor 1–53 (EGF 1–53), Bovine Pituitary Extract (BPE), 10%, 1% penicillin/streptomycin (Mediatech, Manassas, VA), and 1% Fungizone (Life Technologies, Carlsbad, CA). Briefly, cervical tumor biopsies were obtained from all patients and viable tumor tissue was mechanically minced under sterile conditions in enzyme solution [0.14% Collagenase Type I (Sigma St. Louis, MO) and 0.01% DNAse (Sigma, 2000 KU/mg)] in RPMI 1640, and incubated on a magnetic stirring apparatus 40’ at room temperature. The resultant cell suspension was washed in RPMI 1640 plus 10% FBS and then washed in PBS. Tumors were staged according to the International Federation of Gynecology and Obstetrics (FIGO) staging system. Patient characteristics are noted in Table 1
Table 1.
Characteristics and demographic data of cervical cancer cell lines.
| Cell line | Age | RACE | FIGO stage | Histology | HPV |
|---|---|---|---|---|---|
| CVX3 | 35 | B | IB2 | SCC | 16 |
| CVX4 | 40 | W | IIA | SCC | 16 |
| CVX5 | 42 | W | IB2 | SCC | 18 |
| CVX7 | 22 | H | IB2 | SCC | 16 |
| CVX8 | 29 | W | IB1 | SCC | 16 |
| ADX1 | 33 | W | IB | ADSQ | 18 |
| ADX2 | 33 | B | IB | ACA | 18 |
| ADX3 | 25 | W | IB | ACA | 18 |
| ADX4 | 47 | B | IB | SCC | 45 |
Homologous recombination deficiency (HRD) evaluation in CC cell lines
Log2-ratios of read counts in exonic intervals in whole exome sequenced (WES) tumor and normal samples [21], were tabulated (Figure 1S). Intervals were determined from high coverage regions in the normal samples, and intervals that did not overlap with RefSeq annotations were removed, to ensure remaining intervals corresponded to known genic loci. SNP allele frequencies were calculated in these exonic intervals, using SNPs defined in the phase 3 1000 Genomes dataset (Figure 2S). The log2-ratios and allele frequencies were used to assess HRD status for each sample using an ad hoc scoring algorithm, similar to the one used in the ARIEL2 trial [22].
Immunoblotting and antibodies
Cells were washed twice in ice-cold PBS and harvested with radioimmunoprecipitation assay buffer (RIPA) (50 mmol/L Tris–HCl, pH 8, 150 mmol/L NaCl, Triton X-100 1%, Na deoxycholate 0.5%, SDS 0.1%, MgCl 5 mmol/L in H2O) supplemented with Protease and Phosphatase Inhibitor (cat#78430, Thermo Fisher Scientific). Protein concentrations were measured by BCA Protein Assay Kit (Pierce™ #23225) to ensure equal loading. Proteins were denatured at 95°C for 5 minutes in Laemmli sample buffer (S3401; Sigma-Aldrich) and then resolved in SDS-PAGE electrophoresis, transferred on nitrocellulose, and blotted with corresponding antibodies. The antibodies used for western blotting were as follows: PAR (#4336, Trevigen), PARP (#9532, Cell Signaling Technology, Inc.), and GAPDH (#2118, Cell Signaling Technology, Inc.).
Cell viability assay
CC cell lines were plated at log phase of growth in 6-well tissue culture plates at a density of 80,000–100,000 cells/well. After 24 hours of incubation, cells were treated with Olaparib (AZD2281, LYNPARZA™, AstraZeneca) at a concentration of 0, 0.15, 1.5, 3, 12 μM. 72 hours after drug treatment, cells were harvested in their entirety, centrifuged and stained with propidium iodide (2 μl of 500 μg/ml stock solution in PBS). Count was performed using a flow-cytometry based assay to quantify percent viable cells as a mean ± SEM relative to untreated cells as 100% viable control. A minimum of three independent experiments per cell line was performed.
Cell-cycle analysis
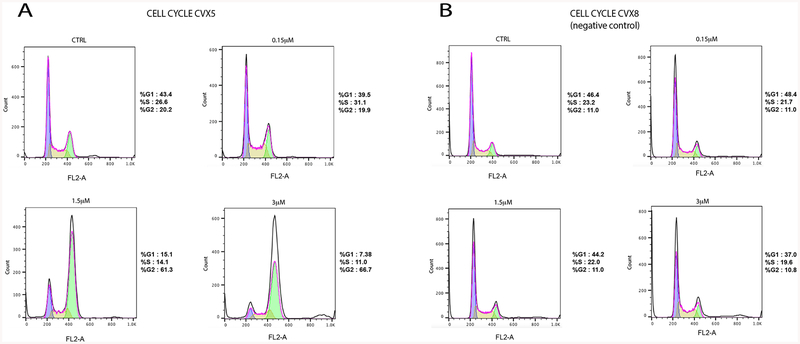
After 48h incubation at the conditions described in Figure 4, cells were harvested and washed with ice-cold PBS, fixed in ice-cold 70% ethanol at −20°C for a minimum of 30 minutes to overnight. Subsequently, cells were washed in PBS, incubated with ribonuclease A (100 μg/ml) for 5 minutes at room temperature and stained with propidium iodide (20 μg/ml) in PBS. Cell-cycle phase distributions were analyzed with Flow-Jo software program (v. 8.7).
Fig. 4.
A) Cell cycle assay on CVX5 after 48h Olaparib treatment at the following concentrations: 0.15, 1.5, 3 μM (p=0.00005) B) Cell cycle assay on CVX8 (representative resistant cell line) after 48h Olaparib treatment at the following concentrations: 0.15, 1.5, 3 μM (p>0.05).
Annexin V-FITC/PI double staining
Annexin V-fluorescein isothiocyanate/propidium iodide (Annexin V-FITC/PI) double staining was used to quantify apoptosis. Adherent cells were incubated with 0, 0.15, 1.5, 3 μM of olaparib for 72 hours, then harvested and collected. Cells were washed twice with ice-cold PBS and resuspended in 1× Binding Buffer at a concentration of 1×106 cells/ml. 5 μl of Annexin V-FITC and 5 μl of propidium iodide were added to 100 μl of the cell suspension. After 15 minutes of incubation, 400 μl of Binding Buffer were added to each cell suspension. Cells were analyzed by flow cytometry within 1 hour.
siRNA transfection
Cells were plated in 6 well plate in Keratinocytes-SFM (Gibco®, Life Technologies™), supplemented with prequalified human recombinant Epidermal Growth Factor 1–53 (EGF 1–53), Bovine Pituitary Extract (BPE), 1% penicillin/streptomycin (Mediatech, Manassas, VA), and 1% Fungizone (Life Technologies, Carlsbad, CA). 70–80% confluent cells were subjected to transfection. PARP1 siRNA and negative control siRNA were purchased from Ambion®, Life Technologies™. Briefly, the siRNA was incubated with Lipofectamine™ RNAiMAX reagent (Invitrogen, CA, USA) in OptiMEM™ medium for 20 minutes, then added to a monolayer of cells in Keratinocytes-SFM without antibiotics. Twenty-four hours after the transfection, cells were treated with scalar amounts of olaparib ranging from 1.5 μM to 400 μM. Cells were then counted by flow cytometry.
Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
RNA was obtained from cells after 48 hours of incubation with olaparib (Table 1S) using AllPrep DNA/RNA/Protein Mini Kit (Qiagen) according to the manufacturer’s instructions. Total RNA (5 μg) was reverse-transcribed using Superscript III (Invitrogen). Quantitative PCR was carried out to evaluate the expression level of PARP-1 (PARP-1, Assay ID: Hs00242302_m1, Applied Biosystems) in all samples with a 7500 Real-Time PCR System (Applied Biosystems) following the manufacturer’s protocol. Each reaction was run in duplicate. The internal control GAPDH (Assay ID: Hs99999905_ml, Applied Biosystems) was used to normalize variations in cDNA quantities from different samples. The comparative threshold cycle (Ct) method was used for the calculation of amplification fold as specified by the manufacturer. Analyses were performed using SDS software 2.2.2 (Applied Biosystems/Life Technologies).
In vivo treatment
The in vivo antitumor activity of olaparib was tested in xenograft models. Briefly, four to six-week-old CB17/SCID mice were given a single subcutaneous injection in the abdominal region of 7 × 106 CVX5 cells in approximately 300 μl of a 1:1 suspension of sterile PBS containing cells and Matrigel® (BD Biosciences). Xenografted mice were randomized into treatment groups (6 mice each group) when mean tumor burden was 0.15–0.25 cm3, and dosing (vehicle PO or olaparib 10 mg/kg BID, PO) was delivered to the CVX5 xenografts for 4 weeks (7 days/week). Drug dosage was chosen according to previous studies [23, 24]. Tumor and weight measurements of each mouse were recorded twice weekly. Mice were humanely euthanized when tumor volume reached 1.5 cm3 using the formula (width2 × height)/2. Animal care and euthanasia were carried out according to the rules and regulations as set forth by the Institutional Animal Care and Use Committee (IACUC).
Statistical analysis
Statistical analysis was performed using Graph Pad Prism version 8 (Graph Pad Software, Inc. San Diego, CA). The inhibition of proliferation in the CC cell lines after exposure to olaparib was evaluated by the two-tailed unpaired student t-test. Unpaired t-test was used to evaluate significant differences in the tumor volumes at specific time points in the in vivo experiments. Overall survival data was analyzed and plotted using the Kaplan-Meier method. Survival curves were compared using the log-rank test. Differences in all comparisons were considered statistically significant at p-values < 0.05.
RESULTS
Olaparib suppresses CC cell lines growth
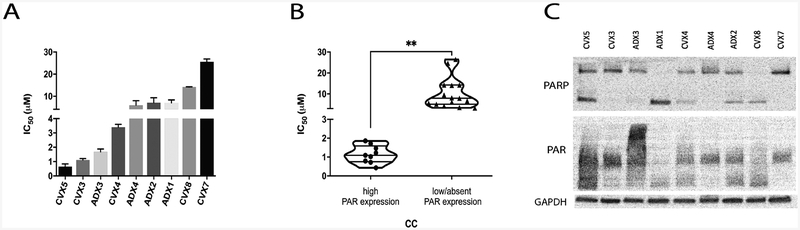
To evaluate the potential of PARP inhibitors on CC, we investigated the in vitro effects of olaparib on the growth of 9 primary CC cell lines using flow cytometric-based assay as described in the methods. As shown in Figure 1A, 1B, after 72 hours of incubation with increasing concentrations of olaparib, we found a progressive, dose-response decrease in cell proliferation in 33% of CC lines tested, with a significant difference in IC50 values between the sensitive and resistant group (p= 0.0012).
Fig. 1.
A) In vitro proliferation assay overview of the established primary CC cell lines (n=9) B) Violin scatter dot plot representing grouped sensitive cell lines and resistant cell lines (p=0.0012) C) Western blot analysis displaying basal expression of PARP, PAR, and GAPDH in all nine CC cell lines.
Sensitivity to olaparib is strongly correlated to PARP activity
To better understand the mechanisms behind the sensitivity to olaparib in a subset of primary CC, we analyzed PARP and PAR basal expression in all nine CC cell lines as well as their mutation spectrum (i.e., HRD), as defined in the methods section. None of the tested CC cell lines demonstrated HRD. Indeed, within the nine CC cell lines, genomic loss of heterozygosity (LOH) results ranged from 0–12.3% (Table 2S), which falls short of the initial ARIEL2 cutoff of 14% (and the current revised cutoff of 16%) used to classify a tumor as HRD [22]. In contrast, as demonstrated in Figure 1C, using immunoblot (i.e., cells lysates were loaded in order from the most sensitive to the most resistant CC based on IC50 values previously obtained by flow cytometric-based assay) we found a direct correlation between basal expression level of PARP activity (PAR) and sensitivity to olaparib treatment. Indeed, CVX5, CVX1 and CVX3 (i.e., the 3 CC primary cell lines with the higher PARP expression of both PARP isoforms 116 and 89 kDa), consistently demonstrated the higher sensitivity to olaparib exposure in the in vitro experiments.
Silencing of PARP-1 elicits resistance to olaparib
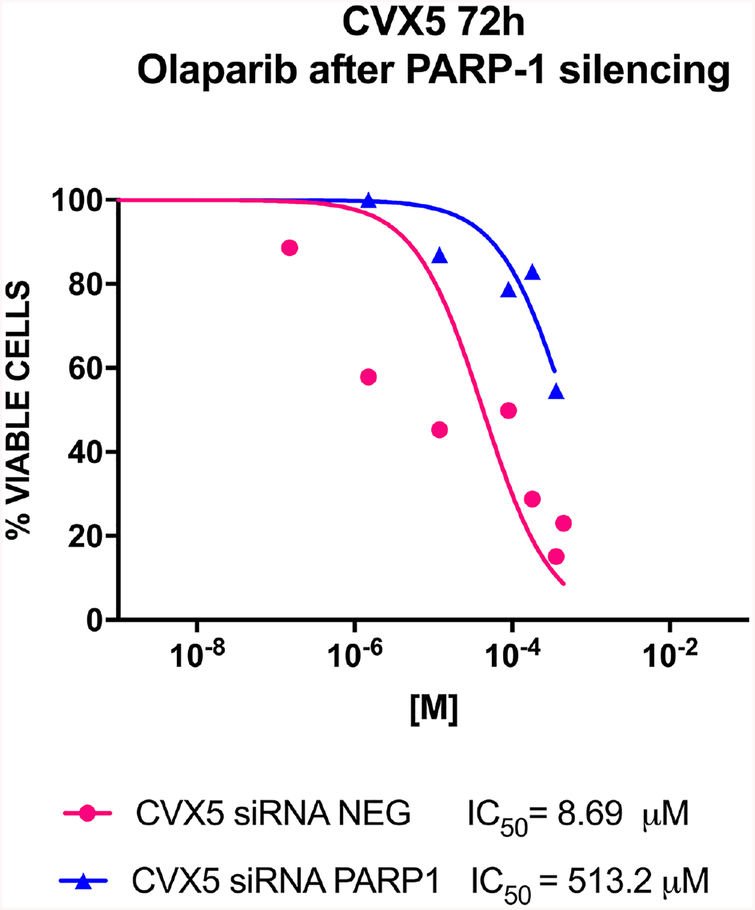
To evaluate further the correlation between PARP-1 activity and sensitivity of CC to olaparib we transiently transfected CVX5 cells with PARP-1 siRNA and negative siRNA control as described in materials and methods section. After 72 hours of olaparib treatment, IC50 values of either PARP-1 siRNA and negative control siRNA transfected CVX5 cells were evaluated through flow cytometric-based assay as described in Methods. Validation of PARP-1 mRNA silencing in tumor cells was confirmed with q-real time PCR (Table S1). As shown in Figure 2, CVX5 cells transfected with PARP-1 siRNA from sensitive become highly resistant (i.e., IC50 from 8.69 μM to 513.2 μM) to olaparib treatment (p=0.0063).
Fig. 2.
In vitro proliferation assay in PARP-1 silenced CVX5 cell line versus non-silenced control (p=0.0063).
Olaparib triggers apoptosis of CC in a dose-dependent manner
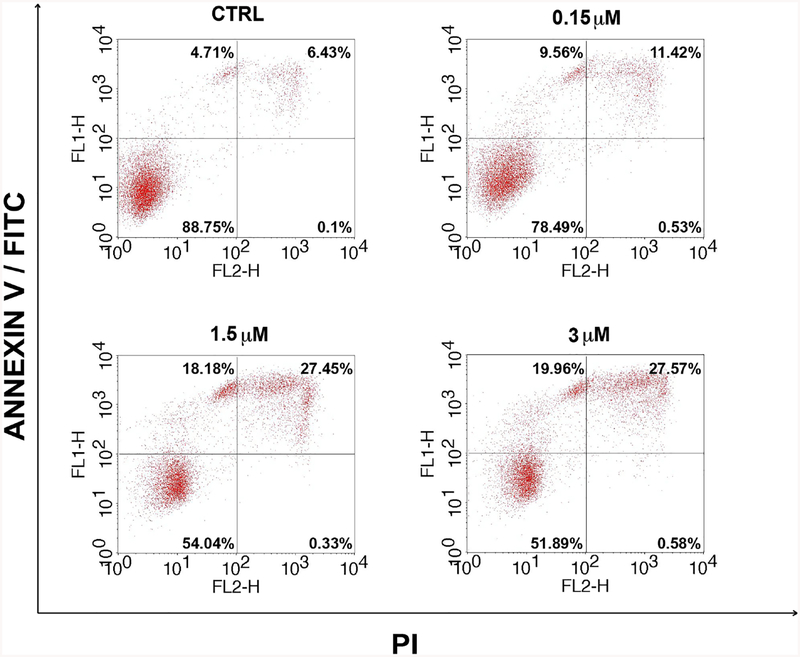
To gain better insight into the mechanism of PARPi activity, CVX5 was exposed to increasing concentration of olaparib (0.15, 1.5, 3 μM) for 48 hours before being harvested for Annexin V/PI staining. As shown in Figure 3, we demonstrated that olaparib at the dose of 1.5 μM and 3 μM induced apoptosis in 18% to 20% of cells, respectively, and tardive apoptosis in an additional 27.5% of cells (p<0.0001).
Fig. 3.
Up Left (UL) and Up Right (UR) quadrants show single positive events for FL1-H (ANNEXIN V-FITC) and double positive events for FL1-H and FL2-H, respectively. Double positive events stand for tardive apoptosis, corroborated by the absence of events in Down Right (DR) quadrant (single positive for FL2-H representing cell necrosis) (p<0.0001).
Olaparib sensitivity is associated with G2/M cell cycle arrest
We next examined the cell cycle profiles of CVX5 (i.e., a representative olaparib-sensitive CC cell lines) and CVX8 (a representative olaparib-resistant CC cell line) after 24 hours of olaparib treatment. As shown in Figure 4A, starting at 1.5 μM of olaparib, 67.7% of CVX5 cells demonstrated a G2/M cell cycle arrest (in comparison to non-treated cells (i.e., 22.3%) (p=0.000061). This percentage increased at the dose of 3 μM olaparib (78.3%) (p=0.00005). In contrast, as demonstrated in Figure 4B, CVX8 cell cycle was not affected by olaparib treatment at any dose tested (16.2% cells in G2-M non-treated cells vs 13.5% cells in G2-M after 3 μM olaparib treatment) (p>0.05).
Olaparib PARP inhibition and PARP trapping on sensitive CC
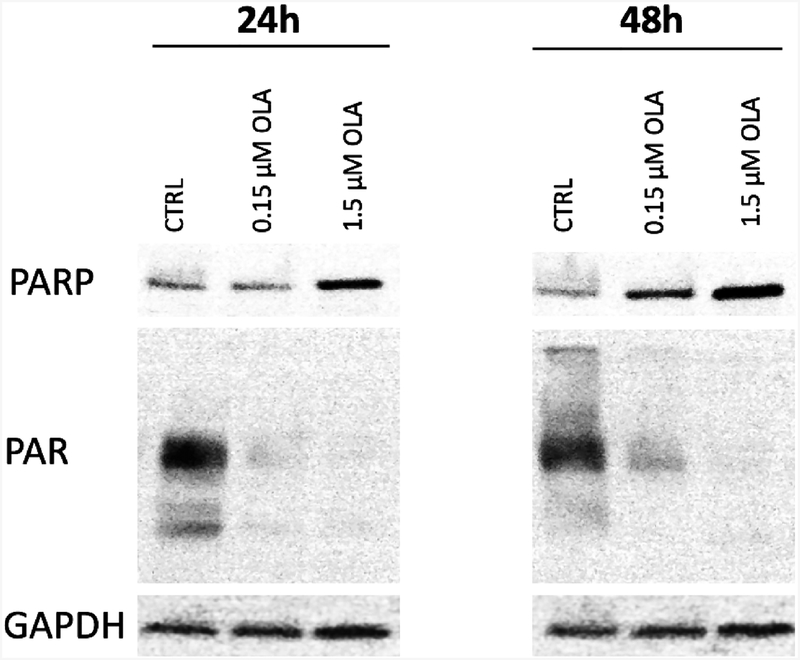
Next, we analyzed PARP-1 and PAR expression in CVX5 cells by immunoblotting assay after exposure to different doses of olaparib (0.15 μM – 1.5 μM) at two different time points (24–48 hours). As shown in Figure 5, PARP expression increased after exposure to 1.5 μM olaparib at both time points while no significant variation was detected in PARP-1 mRNA expression level at 24 or 48 hours (Table S1). A dramatic reduction in PAR levels was detected at both doses of olaparib (0.15 and 1.5 μM) (Figure 5).
Fig. 5.
Western blot analysis displaying expression of PARP, PAR, and GAPDH in CVX5 cells after 24–48 hours of treatment with 0.15 and 1.5 μM Olaparib.
Olaparib impairs CVX5 xenograft tumor growth in vivo
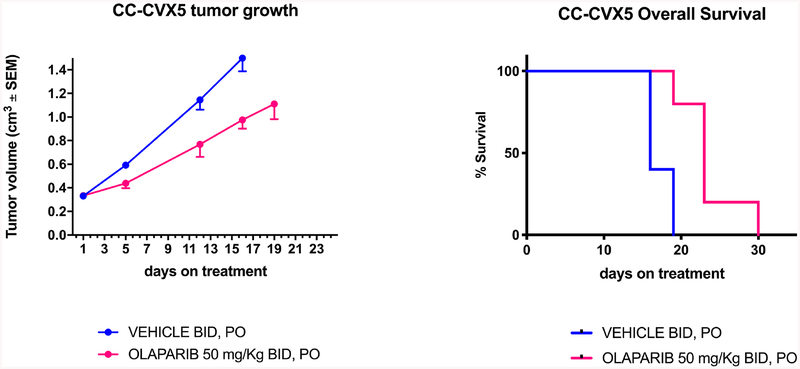
The in vivo effects of olaparib was determined by establishing xenografts from the primary CVX5 CC cell line. Briefly, after the tumors had reached the goal size, animals were randomized into treatment groups and treated as described in Materials and Methods. Tumor size was assessed weekly and mice were sacrificed if tumors became necrotic, reached a volume of 1.5 cm3, or mice appeared to be in poor health. Twice daily oral dose of olaparib 50 mg/kg was well tolerated with no clear impact on body weight compared with vehicle control (data not shown). As shown in Figure 6, mice undergoing olaparib treatment exhibited a significantly slower rate of tumor growth, compared to vehicle control starting at day 12 (p=0.0017). Furthermore, the overall survival was significantly prolonged in the treated group (Log Rank Mantel-Cox test p=0.008).
Fig. 6.
A) In vivo tumor growth inhibition following 19 days dosing olaparib or vehicle of CVX5 injected xenografts (p=0.0017) B) overall survival (p=0.008).
DISCUSSION
The inhibition of PARP was initially demonstrated to determine ‘synthetic lethality’ in cancer patients harboring specific DNA repair defects, (i.e., BRCA1 or BRCA2 (BRCA1/2) mutations) causing deficiency in the cell homologous recombination (HR) repair system [25, 26]. Accordingly, initial FDA approval was restricted to the treatment of patients harboring deleterious or suspected deleterious germline BRCA-mutated (gBRCAm) advanced ovarian cancer who have been treated with three or more prior lines of chemotherapy. More recently, however, PARPi approval was expanded to maintenance therapy for patients with platinum-sensitive relapsed ovarian cancer, who responded to their second line regimen, regardless of BRCA1 or BRCA2 (BRCA1/2) mutation status [27] [28]. This broader use of PARPi stems from the evidence that tumors that share molecular features with BRCA-mutant tumors (i.e., BRCAness) also exhibit different levels of defective homologous recombination DNA repair, and therefore will respond to PARP inhibition [29]. Importantly however, recent results from large prospective randomized clinical trials have demonstrated significant PARPi clinical activity also against patients harboring HR-competent/BRCA wild-type tumors [30].
Unfortunately, while olaparib, rucaparib and niraparib are currently FDA-approved in ovarian cancer and multiple clinical trials are currently evaluating PARPi as single agents or in combination against multiple human tumors, limited information is currently available on the role of olaparib in CC patients. Accordingly, in this study, we thoroughly investigated the preclinically activity of olaparib against multiple primary CC cell lines in vitro and in vivo.
We found three of the nine primary CC cell lines to be highly sensitive to olaparib exposure with a cutoff IC50 value < 2μM [31]. To gain further insight into the molecular characteristics making these CC cell lines sensitive to olaparib treatment we evaluated their mutation spectrum (i.e., HRD), as well as their level of PARP1 expression [20, 32], and the potential role played by PARylation. Using the ARIEL2 study cutoff of 14% (and the current revised cutoff of 16%) used to classify a tumor as HRD [22], we found none of the tested CC cell lines to demonstrate HRD. Importantly, we found the level of PARylation but not PARP1 expression in the tumors to consistently correlate with CC cell line sensitivity to olaparib. To prove the correlation between PARylation overexpression and olaparib sensitivity was causative, we downregulated PARP1 mRNA through PARP1 siRNA transfection in a representative cell line (i.e., CVX5 cell line) and analyzed the IC50 values in comparison to transfected CVX5 with a universal negative control siRNA. We found CVX5 transfected with PARP1 siRNA to gain high resistance to olaparib treatment (p=0.0063), confirming that PARP activity (PAR) is of utmost importance in determining olaparib sensitivity in CC cell lines. These results are similar to the results obtained by Michels et al., who also found a positive correlation between cellular PARylation levels and sensitivity to PARP inhibitors in non-small cell lung carcinoma cell lines [33]. Moreover, in agreement with our results, other groups demonstrated that in the absence of functional HR, PARP1 or PARP2 knockout cells are resistant to PARP inhibitors [34, 35]. Taken together, these data combined with our findings in CC strongly suggest that determination of the level of PARP1 protein activity (i.e., PAR expression), may represent a biomarker potentially able to identify the most sensitive CC patients for treatment with PARPi. Accordingly, testing the possible link between PARP expression/activity and sensitivity to PARP inhibitors in the clinical setting may be warranted in future CC studies.
To better understand the functional mechanisms of olaparib in inhibiting CC cell growth, we performed cell cycle analysis experiments. We found olaparib, in a dose-dependent manner, to consistently arrest cell cycle in G2/M phase in all sensitive cell lines, ultimately preventing cells to going through the G1 phase. In contrast, no detectable alteration was found in the cell cycle of olaparib-resistant CC cell lines (i.e., CVX8). This effect of olaparib, as previously demonstrated in ovarian cancer, is explained by the PARP trapping mechanism, by which PARP inhibitors induce the formation of cytotoxic PARP–DNA complexes, preventing DNA replication [34]. When we investigated the mechanism of cell death in the CC cell lines exposed to olaparib, we found that only less than 1% of total cells demonstrated necrosis, corroborating the result that olaparib triggers and induces apoptosis in olaparib-sensitive CC cell lines.
To further elucidate the mechanism of action of olaparib against PARP, we analyzed PARP-1 and PAR protein expression in a representative cell line (i.e., CVX5) during olaparib treatment. Our immunoblot experiments clearly demonstrated a dose dependent increase of PARP1, as main consequence to olaparib exposure, further supporting an olaparib-induced PARP trapping phenomenon. In agreement with this interpretation, PARP-1 mRNA levels were not increased in any of the condition tested in any CC cell line. Taken together, our results support the notion of PARP-1 accumulation in cells treated with increasing concentrations of olaparib as main mechanism of action in CC. Importantly, when we evaluated the activity of olaparib in vivo in xenografted animals injected with CVX5, our result were confirmatory of the in vitro results with significant impairment of CVX5 tumor growth, and a significant increase in animal overall survival (p=0.008).
In conclusion, we demonstrated in vitro and in vivo activity of olaparib in a significant subset of CC primary cell lines and suggest that PAR expression may represent a novel biomarker for the potential prediction of PARPi response in patients with CC. Future studies with PARPi used alone or in combination with other targeted agents in patients with CC resistant to standard treatment modalities are warranted.
Supplementary Material
HIGHLIGHTS.
A subset of primary CC cell lines is highly sensitive to olaparib in vitro and in vivo
High PARylation activity correlates with sensitivity to olaparib in CC cell lines
Silencing of PARP-1 reverses CC cell line sensitivity to olaparib and induce resistance
Financial support:
This work was supported in part by grants from NIH U01 CA176067–01A1, the Deborah Bunn Alley Foundation, the Tina Brozman Foundation, the Discovery to Cure Foundation and the Guido Berlucchi Foundation to A.D.S., and Gilead Sciences Inc., Foster City, CA. This investigation was also supported by NIH Research Grant CA-16359 from NCI and Standup-to-cancer (SU2C) convergence grant 2.0 to Alessandro Santin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Koh WJ, Abu-Rustum NR, Bean S, Bradley K, Campos SM, Cho KR, et al. Cervical Cancer, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17:64–84. [DOI] [PubMed] [Google Scholar]
- [3].Tewari KS, Sill MW, Penson RT, Huang H, Ramondetta LM, Landrum LM, et al. Bevacizumab for advanced cervical cancer: final overall survival and adverse event analysis of a randomised, controlled, open-label, phase 3 trial (Gynecologic Oncology Group 240). Lancet. 2017;390:1654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cadron I, Van Gorp T, Amant F, Leunen K, Neven P, Vergote I. Chemotherapy for recurrent cervical cancer. Gynecol Oncol. 2007;107:S113–8. [DOI] [PubMed] [Google Scholar]
- [5].Tewari KS, Sill MW, Long HJ 3rd, Penson RT, Huang H, Ramondetta LM, et al. Improved survival with bevacizumab in advanced cervical cancer. N Engl J Med. 2014;370:734–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weil MK, Chen AP. PARP inhibitor treatment in ovarian and breast cancer. Curr Probl Cancer. 2011;35:7–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zimmer AS, Gillard M, Lipkowitz S, Lee JM. Update on PARP Inhibitors in Breast Cancer. Curr Treat Options Oncol. 2018;19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pulliam N, Fang F, Ozes AR, Tang J, Adewuyi A, Keer H, et al. An Effective Epigenetic-PARP Inhibitor Combination Therapy for Breast and Ovarian Cancers Independent of BRCA Mutations. Clin Cancer Res. 2018;24:3163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–9. [DOI] [PubMed] [Google Scholar]
- [10].Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1274–84. [DOI] [PubMed] [Google Scholar]
- [11].D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342 (Pt 2):249–68. [PMC free article] [PubMed] [Google Scholar]
- [12].Ossovskaya V, Koo IC, Kaldjian EP, Alvares C, Sherman BM. Upregulation of Poly (ADP-Ribose) Polymerase-1 (PARP1) in Triple-Negative Breast Cancer and Other Primary Human Tumor Types. Genes Cancer. 2010;1:812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ba X, Garg NJ. Signaling mechanism of poly(ADP-ribose) polymerase-1 (PARP-1) in inflammatory diseases. Am J Pathol. 2011;178:946–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10:1192–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim G, Ison G, McKee AE, Zhang H, Tang S, Gwise T, et al. FDA Approval Summary: Olaparib Monotherapy in Patients with Deleterious Germline BRCA-Mutated Advanced Ovarian Cancer Treated with Three or More Lines of Chemotherapy. Clin Cancer Res. 2015;21:4257–61. [DOI] [PubMed] [Google Scholar]
- [18].Le D, Gelmon KA. Olaparib tablets for the treatment of germ line BRCA-mutated metastatic breast cancer. Expert Rev Clin Pharmacol. 2018;11:833–9. [DOI] [PubMed] [Google Scholar]
- [19].Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med. 2018;379:2495–505. [DOI] [PubMed] [Google Scholar]
- [20].Prasad CB, Prasad SB, Yadav SS, Pandey LK, Singh S, Pradhan S, et al. Olaparib modulates DNA repair efficiency, sensitizes cervical cancer cells to cisplatin and exhibits anti-metastatic property. Sci Rep. 2017;7:12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zammataro L, Lopez S, Bellone S, Pettinella F, Bonazzoli E, Perrone E, et al. Whole exome sequencing of cervical carcinomas identifies activating ERBB2/PIK3CA mutations as targets for combination therapy. PNAS (in press). 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. [DOI] [PubMed] [Google Scholar]
- [23].Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tahara M, Inoue T, Sato F, Miyakura Y, Horie H, Yasuda Y, et al. The use of Olaparib (AZD2281) potentiates SN-38 cytotoxicity in colon cancer cells by indirect inhibition of Rad51-mediated repair of DNA double-strand breaks. Mol Cancer Ther. 2014;13:1170–80. [DOI] [PubMed] [Google Scholar]
- [25].Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. [DOI] [PubMed] [Google Scholar]
- [26].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. [DOI] [PubMed] [Google Scholar]
- [27].Miller RE, Ledermann JA. The status of poly(adenosine diphosphate-ribose) polymerase (PARP) inhibitors in ovarian cancer, part 2: extending the scope beyond olaparib and BRCA1/2 mutations. Clin Adv Hematol Oncol. 2016;14:704–11. [PubMed] [Google Scholar]
- [28].Sonnenblick A, de Azambuja E, Azim HA Jr., Piccart M. An update on PARP inhibitors--moving to the adjuvant setting. Nat Rev Clin Oncol. 2015;12:27–41. [DOI] [PubMed] [Google Scholar]
- [29].Gadducci A, Guerrieri ME. PARP inhibitors alone and in combination with other biological agents in homologous recombination deficient epithelial ovarian cancer: From the basic research to the clinic. Crit Rev Oncol Hematol. 2017;114:153–65. [DOI] [PubMed] [Google Scholar]
- [30].Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med. 2016;375:2154–64. [DOI] [PubMed] [Google Scholar]
- [31].Min A, Im SA, Yoon YK, Song SH, Nam HJ, Hur HS, et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol Cancer Ther. 2013;12:865–77. [DOI] [PubMed] [Google Scholar]
- [32].Thomas A, Murai J, Pommier Y. The evolving landscape of predictive biomarkers of response to PARP inhibitors. J Clin Invest. 2018;128:1727–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Michels J, Vitale I, Galluzzi L, Adam J, Olaussen KA, Kepp O, et al. Cisplatin resistance associated with PARP hyperactivation. Cancer Res. 2013;73:2271–80. [DOI] [PubMed] [Google Scholar]
- [34].Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.