Abstract
Objective
To develop an automated vessel wall segmentation method using convolutional neural networks (CNN) to facilitate the quantification on magnetic resonance (MR) vessel wall images of patients with intracranial atherosclerotic disease (ICAD).
Methods
Vessel wall images of 56 subjects were acquired with our recently developed whole-brain 3D MR vessel wall imaging (VWI) technique. An intracranial vessel analysis (IVA) framework was presented to extract, straighten, and resample the interested vessel segment into 2D slices. A U-net-like fully convolutional networks (FCN) method was proposed for automated vessel wall segmentation by hierarchical extraction of low- and high-order convolutional features.
Results
The network was trained and validated on 1160 slices and tested on 545 slices. The proposed segmentation method demonstrated satisfactory agreement with manual segmentations with Dice coefficient of 0.89 for the lumen and 0.77 for the vessel wall. The method was further applied to a clinical study of additional 12 symptomatic and 12 asymptomatic patients with >50% ICAD stenosis at the middle cerebral artery (MCA). Normalized wall index (NWI) at the focal MCA ICAD lesions was found significantly larger in symptomatic patients compared to asymptomatic patients.
Conclusion
We have presented an automated vessel wall segmentation method based on FCN as well as the IVA framework for 3D intracranial MR VWI.
Significance
This approach would make large-scale quantitative plaque analysis more realistic and promote the adoption of MR VWI in ICAD management.
Keywords: Vessel wall imaging, deep learning, segmentation, quantification, Intracranial Atherosclerotic Disease
I. Introduction
Intracranial atherosclerotic disease (ICAD) is an important cause of ischemic stroke worldwide [1–4]. The disease is characterized by the development, progression, and complication of atherosclerotic plaques in the vessel wall of the intracranial arteries [4]. Luminography imaging, routinely used in the diagnostic workup of ICAD, is restricted to the detection of luminal stenosis, which is, however, not a specific marker for confirming and risk-stratifying atherosclerotic plaques [5]. Magnetic resonance (MR) vessel wall imaging (VWI) is an emerging noninvasive imaging modality that can directly visualize the intracranial vessel wall and characterize plaque features due to excellent soft-tissue contrast [6–12]. Several VWI-derived morphological features, in particular quantitative measures such as wall area, remodeling ratio, and normalized wall index (NWI), have been investigated in ICAD patients and demonstrated the great potential for the identification of high-risk ICAD lesions [11, 13–20].
Morphological quantification of ICAD lesions based on MR VWI is currently of limited use in clinical practice. ICAD lesions are typically assessed by neuroradiologists in a qualitative fashion [21–23]. Dichotomized classification or ordinal scores are used to describe morphological features, which is reader experience-dependent and potentially unsatisfactory in accuracy and reproducibility. Thus, quantitative assessment of plaque morphology is highly desirable. Its feasibility has previously been demonstrated for two-dimensional (2D) VWI whereby cross-sectional images were used for manually tracing the lumen and outer vessel wall boundaries at each isolated plaque [11]. This segmentation step then allows for subsequent quantitative analyses. However, when moving into the era of three-dimensional (3D) VWI [24], this manual approach to vessel segmentation becomes prohibitively time-consuming because of the need for analyzing a largely increased number of imaging slices from multiple plaques and/or arterial segments. An automated vessel wall segmentation method would make quantitative plaque analysis more realistic, which would in turn promote the adoption of 3D MR VWI in the clinical management of ICAD.
To date, there is a paucity of literature on automated segmentation of the vessel wall from intracranial MR VWI. A challenge unique to the intracranial vessel wall is that the ultra-thin, tortuous structure is immediately surrounded by brain parenchyma or cerebrospinal fluid (CSF) that may show similar contrast. Recent technological advancements, such as isotropic, high spatial resolution imaging [6, 8, 25, 26] and CSF signal suppression [8, 26–28], have achieved enhanced quality of vessel delineation, potentially facilitating automated segmentation. Qiao et al. has demonstrated the feasibility by using the software LAVA that employs a method of nonuniform rational B-spline surface modeling [14]. However, the software was originally developed for carotid vessels, more rigorous evaluations on intracranial vessels are warranted.
Convolutional neural network (CNN) is a recently emerged deep-learning technique that can be used for image segmentation [29]. Inspired by neurons that make up the human brain, CNN organizes neural networks as a layered and hierarchical architecture with sufficient flexibility to represent complex multisource data, reveal hidden patterns, and build models for prediction. As a type of CNN, fully convolutional networks (FCN) have been successfully used in segmentation tasks [30]. In addition to convolutional layers for feature extraction and pooling layers for feature contraction, FCN introduces upsampling layers to expand the contracted features back to the same size of the input image, which is ideal for segmentation. Following this idea, a U-shaped deep convolutional network called U-net was proposed that employs multiple upsampling layers along with skip connections between the downsampling and upsampling paths to help recover fine-grained information [31].
In this work, we developed a U-net-like CNN segmentation method that can identify vessel wall boundaries in cross-sectional vessel wall images as pre-reformatted from whole-brain 3D MR VWI data [8, 32]. As a preliminary application demonstration, the method was then applied in a prospective VWI study to determine the difference in a plaque burden metric, NWI, between symptomatic and asymptomatic ICAD patients.
II. Materials and Methods
A. Human Subjects
We retrospectively reviewed our institutional MR VWI database, and prepared a dataset for the segmentation network development. Briefly, a total of 56 patients with diagnosed brain ischemia (i.e. ischemic stroke or transient ischemic attack) secondary to ICAD were selected. Enrollment criteria included: at least one >50% intracranial diameter stenosis on MR angiography or CT angiography within the ischemic territory, ⩾ 1 atherosclerotic risk factor (hypertension, diabetes mellitus, hyperlipidemia, and cigarette smoking), and exclusion of other etiologic causes such as coexistent >50% ipsilateral extracranial stenosis, nonatherosclerosis vasculopathy (e.g. dissection, vasculitis, or Moyamoya disease), and cardioembolism based on thorough evaluations.
We prepared a second dataset to further demonstrate the clinical use of the proposed framework. For that purpose, we prospectively recruited 12 symptomatic and 12 asymptomatic patients with age matched to investigate the difference in the NWI of ICAD lesions. All subjects had one >50% stenosis at the M1 segment of a unilateral middle cerebral artery (MCA) on MR angiography or CT angiography and ⩾ 1 atherosclerotic risk factor. Symptomatic subjects qualified if there was an ischemic stroke or transient ischemic attack in the territory of the stenotic MCA within the past 8 weeks and had received thorough evaluations to exclude other etiologic causes. Asymptomatic subjects were considered for inclusion if no ischemic events had occurred in the territory of the stenotic MCA and no nonatherosclerosis vasculopathy had been documented.
The above studies were approved by the local ethics committee and the informed consent were waived for the retrospective study and obtained from prospectively recruited subjects.
B. Imaging Protocol
Intracranial vessel wall images were acquired on a 3-Tesla whole-body MR system (MAGNETOM Verio; Siemens Healthcare, Erlangen, Germany) equipped with a 32-channel head coil (Siemens Healthcare) using our recently developed whole-brain 3D VWI sequence [8]. The imaging parameters include: 3D sagittal orientation; TR/TE=900/15 ms; field of view=70×170 mm2; 240 slices with spatial resolution=0.53 mm isotropic without any interpolation. With this, VWI of entire intracranial vasculatures can be completed in 8 minutes with enhanced T1 contrast weighting and CSF-signal suppression.
C. Image Preprocessing
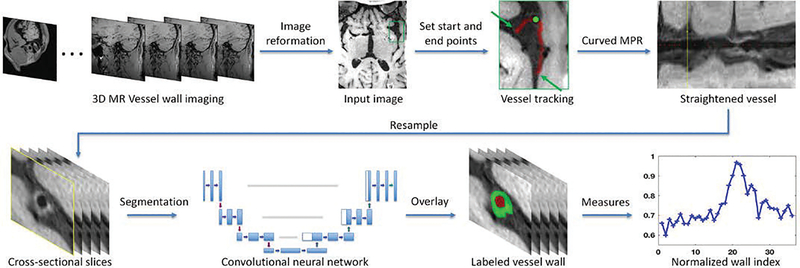
We propose an intracranial vessel analysis (IVA) framework that provides integrated vessel wall image analysis (Fig. 1). All images were preprocessed by a neuroradiologist (over 10 years of experience) using an in-lab intracranial vessel analysis software plug-in built on Horos, a free, open source medical image viewer (http://www.horosproject.org). Briefly, a 3D image set acquired using an MR VWI sequence was first reformatted for optimal visualization of a vessel segment of interest (e.g. pathology involved). For each patient, a vessel segment implicated in ICAD was selected. The start and end points of the vessel segment were then manually designated. Vessel centerline tracking was then performed in Horos with manual modifications if necessary. After that, 3D curved multiplanar reconstruction (MPR) was used to straighten the segment, and consecutive 2D cross-sectional slices with 0.5-mm slice thickness were thus reconstructed and exported in DICOM format. Note that the in-plane resolution was upsampled to 0.1 mm for the resulting 2D slices. Depending on the interrogated vessel segment (i.e. the terminal internal carotid artery, the M1 of the middle cerebral artery, the vertebral artery, or the basilar artery) and the long-axis diffuseness of the interest of ICAD lesions, 17 to 50 contiguous cross-sectional images, covering both normal and thickened vessel wall parts, were generated from each subject. These 2D slices were manually delineated for the lumen and vessel wall by an expert rater using ITK-Snap software (http://itksnap.org), providing the ground truth for the training and validation of segmentation model.
Fig. 1.
Image processing pipeline proposed in IVA framework. Top row shows the image preparing steps and bottom row demonstrates the image segmentation and quantification steps. The originally acquired 3D VWI image set is first reviewed to identify the interested intracranial vessel (the green box). Then this region is zoomed up where the start and end points of the vessel could be manually designated. It is followed by vessel centerline tracking and vessel straighten using curved MPR (the yellow line in the “Straightened vessel” panel corresponds to the location marked by a greed dot in the “vessel tracking” panel), and sliced into contiguous cross-sectional 2D slices that will in turn undergo vessel segmentation and quantification analysis.
D. FCN-based Vessel Wall Segmentation
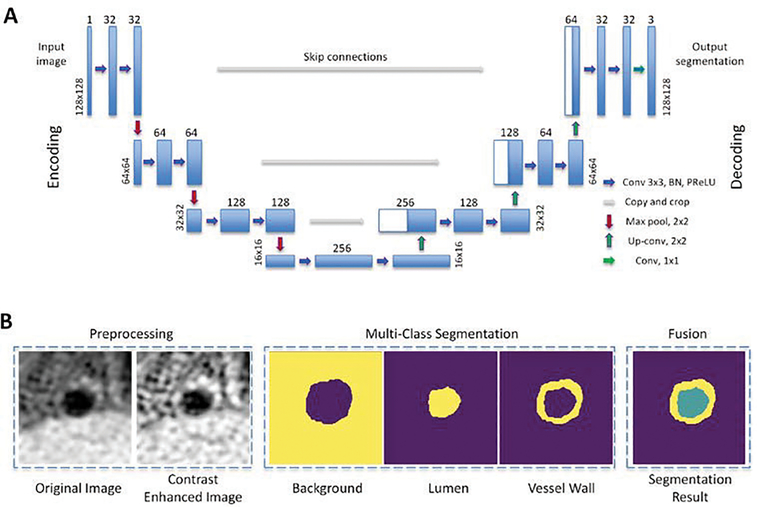
We propose a U-net-like FCN architecture to segment the vessel wall from 2D cross-sectional image slices, as shown in Fig. 2A. The network architecture consists of an encoding path followed by a decoding path. The encoding path includes 3 convolutional units, where each of them is composed of a convolutional layer, batch normalization layer, parametric rectified linear unit (PReLU) activation layer [33], and another convolution layer. Adjacent convolutional units are connected by a max-pooling layer for downsampling the image to half its size. An input 2D image sized 128×128, corresponding to a field of view of (12.8mm)2, is first processed to improve the local contrast using contrast-limited adaptive histogram equalization (CLAHE). CLAHE performs histogram equalization in local regions, and neighborhood regions are combined to eliminate artificially induced boundaries. The contrast, especially in homogeneous areas, will be limited to avoid amplifying any noise that might be present in the image. The contrast-enhanced images are then sent to the first convolutional unit in which a total of 32 learnable filters of size K = 3 are used to convolute with the input image, respectively, producing 32 feature maps. The size of resulting feature maps O is the same as the input image size W according to the equation O = (W – K + 2P)/S + 1, where we set the stride S = 1 and padding P = 1. The following batch normalization, PReLU activation, and convolution layers also have the same filter size and feature map number. The max-pooling layer downsamples the image to 64×64. Subsequent convolutional units further increase feature map numbers while reducing image size, resulting in hierarchical extraction of low- and high-order convolutional features.
Fig. 2.
Illustration of the (A) architecture of the proposed FCN segmentation and (B) examples of input image, intermediate result, and final segmentation.
The features derived from the above encoding path later undergo a decoding path that reverses the convolution process. An upsampled segmentation result that eventually has the same dimension as the input image is generated. Similarly, 3 convolutional units with the same settings as mentioned above are used. Each of them is concatenated with a corresponding unit in the encoding path through a skip connection to help the decoding path better recover spatially detailed information by reusing feature maps. The skip connection has previously proved useful to alleviate the gradient vanish problems in network optimization [34].
The proposed FCN network architecture is modified from the originally introduced U-net [31] in several aspects. In the decoding path, instead of direct upsampling, we employ a transposed convolution layer that achieves transformation in the opposite direction of a normal convolution [35]. This layer is trainable and promises better performance. Moreover, PReLU is used for the activation function after each convolution layer instead of the commonly used rectified linear unit (ReLU) [33]. This is because the PReLU does not have the saturate problem and thus avoids the “dead features” caused by zero gradients in ReLU.
Given the input is a 2D image slice, the output is designed as a multi-class image in a form of a 128×128×3 matrix, where the three channels are for the background, lumen, and vessel wall, respectively. Comparing to the approach that only segment the vessel wall, this multi-class segmentation may achieve a better segmentation performance in both the lumen and vessel wall since the relative position of them could potentially be utilized. Thus, the final activation function is set as softmax instead of sigmoid, where the former one guarantees the sum of three channels is 1 in all pixels while the latter one estimates each channel separately. Fig. 2B demonstrates an example of the input image, intermediate segmentation, and final results.
Dice coefficient is used to evaluate the overlapping ratio between automated segmentation and manual ground truth. Conventionally, Dice is defined as: D = 2(A ⋂ M)/(|A| + |M|), where the nominator represents twice the number of pixels with the same label in automated segmentation A and manual result M and the denominator represents the total number of pixels. Note that in our work the output segmentation from FCN is a probability map. To take advantage of this information instead of using the Dice on a thresholded map, we define a soft Dice coefficient: D′ = 2 ∗ sum(A ∗ M)/(sum(A ∗ A) + sum(M ∗ M)), where the A is a probability map, A and M are reshaped as vectors, and the (∗) is a dot operation. Dice ranges from 0 to 1, where 1 means perfect match between automated and manual segmentations. The Dice is computed separately for the lumen and vessel wall so that they have equal importance and the result is not dominated by the one with more voxels. Moreover, to accommodate multi-class results, the loss function is defined as: . The background channel is not included as it is the counterpart of other two channels.
E. Implementation
The neural network was implemented in Keras with backend of Tensorflow (https://keras.io). The loss function is the mean Dice coefficient of the lumen and vessel wall. Network optimization is realized with Adam gradient descent. The learning rate is 1e-5 with a batch size of 64. The training time on 1030 slices took around 1 hour on a NVIDIA GeForce GTX 1080Ti 8GB GPU, including 7 seconds for training an epoch and we set the maximum epoch number as 500 and keep the best model determined by the performance in validation data, although the algorithm training generally converges around 200 epochs. For testing, it takes around 0.1 ms on an unseen slice.
F. Experimental Settings
The first dataset was used for the training, validation, and testing of the segmentation model. We randomly split the imaging data of 56 patients into 30 patients for training (1030 slices), 6 patients for validation (130 slices), and 20 patients for testing (545 slices). For data augmentation, the 2D slices in training data were randomly flipped in vertical and horizontal directions. Using the Dice metric, the baseline U-net from [31] was compared in testing subjects with the approaches involving additional components including batch normalization, PReLU, CLAHE, and dropout for further evaluations. Following vessel segmentation, morphological measurements were also derived for consecutive cross sectional images in the testing group, including wall area Awall, lumen area Alumen, and NWI defined as NWI = Awall/(Awall + Alumen), where the index ranges from 0 to 1, and the closer to 1 the heavier plaque burden). For each of these metrics, all measurements along the analyzed vessel segment were compared between the automatic and manual segmentation approaches using the Pearson correlation analysis.
In the second dataset, the proposed segmentation method was applied to the VWI data of the M1 arterial segment in age-matched symptomatic and asymptomatic ICAD patients. Wall and lumen areas as well as NWI were generated following automated segmentation. The three metrics at the slice with the peak NWI (by definition corresponding to the highest plaque burden) were analyzed for the differences between the two groups. Moreover, after including neighbouring 2 and 4 slices, these metrics were respectively averaged over totally 3 and 5 slices and then undergo comparisons. Two-sample t-tests were used for the comparison of quantification metrics and Chi-square tests used for comparison of patient demographics between the two patient groups. A p-value less than 0.05 indicated statistical significance.
III. Results
A. Segmentation network evaluation
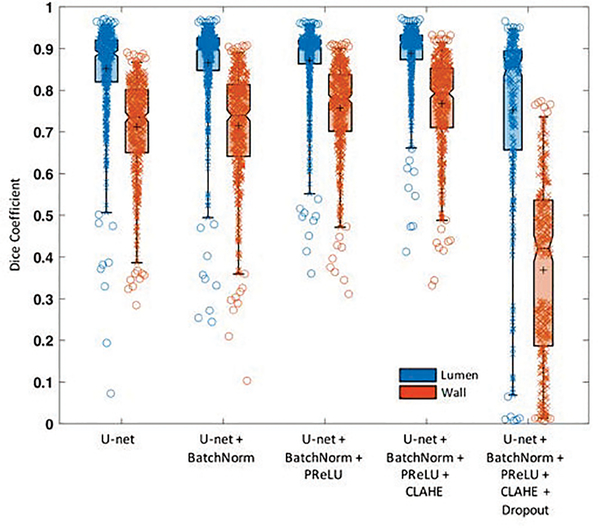
As shown in Fig. 3, on the testing subjects, the baseline U-net achieved a Dice coefficient of 0.852 for the lumen and 0.712 for the vessel wall when comparing automated and manual results. The performance kept improving when successively adding batch normalization (0.866 and 0.715, respectively), PReLU (0.872 and 0.757, respectively), and CLAHE (0.889 and 0.767, respectively). The addition of dropout, however, showed the lowest performance (0.752 and 0.369, respectively). As a result, our finalized network included batch normalization, PReLU activation, and CLAHE for image preprocessing and excluded the dropout procedure.
Fig. 3.
Segmentation performance evaluation on different methods. Boxplot shows the median with 25th and 75th percentiles. The “+” symbol points the mean and “o” shows the outliers. In this work, we adopted the U-net+BatchNorm+PReLU+CLAHE method that has Dice coefficient of 0.889±0.041 and 0.767±0.104 for lumen and vessel wall, respectively.
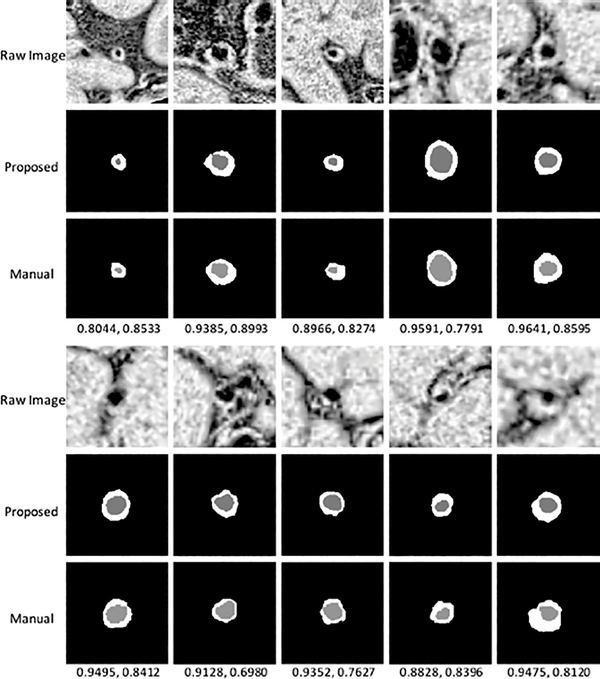
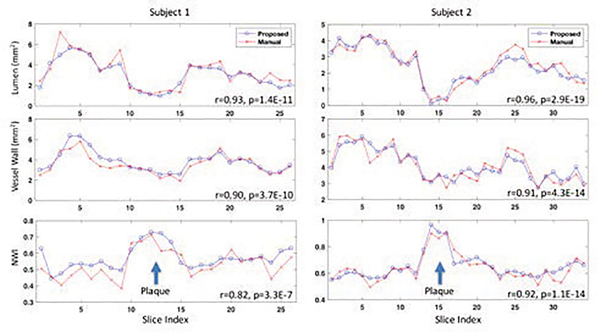
Fig. 4 shows the segmentation results from 10 subjects as well as their manual segmentations for references. Our method was able to locate the vessel region and provide reasonable segmentation, even when the boundaries in neighboring tissues were essentially invisible to naked eyes. Example curves of lumen area, wall area, and NWI (defined as the wall area divided by the summation of wall area and lumen area) for automated and manual segmentations, respectively, along a vessel segment of 2 test subjects were shown in Fig. 5. The average Pearson correlations between the curves on all test subjects were 0.89, 0.84, 0.88, respectively, on lumen area, wall area, and NWI.
Fig. 4.
Illustration of segmentation results from 10 random subjects. The numbers below the manual results show Dice coefficient of lumen and vessel wall, respectively.
Fig. 5.
Results of lumen area, wall area, and NWI from 2 testing subjects, showing comparable patterns for determining plaque existence and measuring its characteristics.
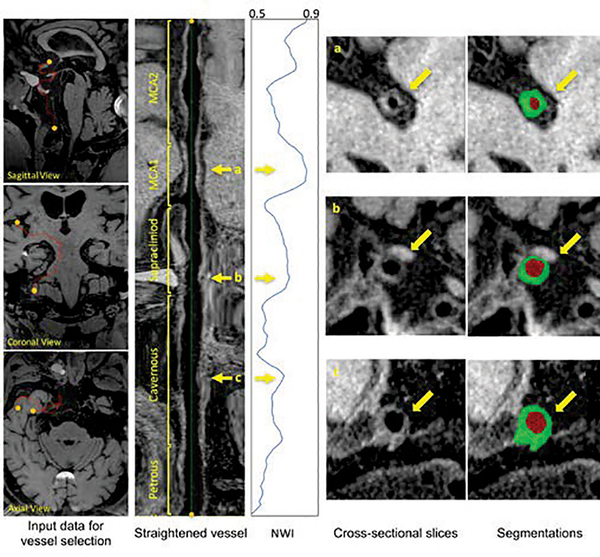
Fig. 6 demonstrates the feasibility of NWI quantification in large-range anterior circulation vasculatures (from the intracranial internal carotid artery to the MCA M2) on a subject using the IVA framework and proposed segmentation approach. The vessels of interest underwent centerline tracking, straighten, and slicing. Following automated vessel wall segmentation, a NWI versus slice curve was derived.
Fig. 6.
Illustrations of the anterior circulation vessel wall analysis for a subject. From left to right, after defining the start and ending points, the intracranial internal carotid artery and middle cerebral artery are centerline tracked and straightened. The sliced images are then processed with the proposed segmentation and used to calculate NWI. Three interested locations in the vessel, as marked by yellow arrows, could be further checked for the NWI values, cross-sectional slices and their corresponding segmentations.
B. Clinical Applications in ICAD patients
We also demonstrate the use of IVA in clinical application of ICAD patients. MCA stenosis is a major component of ICAD, which could be symptomatic or asymptomatic. Recent studies suggest they may belong to distinct groups associated with different stroke risks [11]. Table I shows the demographic data of the recruited 24 patients. VWI images were acquired and the proposed framework was used for image processing. The consecutive 2D slices from the arterial segments of interest underwent segmentation analysis and quantification including lumen area, wall area, and NWI.
TABLE I.
Demographic data of 24 patients
| Symptomatic | Asymptomatic | p-value | |
|---|---|---|---|
| Number | 12 | 12 | - |
| Age (yrs) | 48.1±10.9 | 49.2±13.1 | 0.83 |
| Male | 6 (50%) | 9 (75%) | 0.23 |
| Stenosis located in the left MCA (others are at right MCA) | 6 (50%) | 6 (50%) | 1 |
The slice with a peak NWI, corresponding to the location with the most severe plaque burden, was chosen for comparisons (Table II). To test the robustness, we further included the neighboring 2 and 4 slices, along with itself. The lumen area and wall area in symptomatic patients were slightly smaller and larger, respectively, than those of asymptomatic patients, but the differences did not reach a statistical significance (p > 0.05 from two-sample t-tests). However, the peak NWI in symptomatic patients were significantly higher than that of asymptomatic patients (p = 0.0312), and the difference became more prominent when including neighboring slices (p = 0.0073 for 3 slices and p = 0.0045 for 5 slices).
TABLE II.
Morphology analysis of vessel in 24 patients with a high-grade (>50% diameter stenosis) intracranial atherosclerotic plaques at the middle cerebral artery M1 segment.
| Symptomatic (n=12) | Asymptomatic (n=12) | t | p | ||||
|---|---|---|---|---|---|---|---|
| Mean | Std | Mean | Std | ||||
| Lumen area (mm2) | Peak slice | 1.31 | 0.62 | 1.63 | 0.76 | −1.14 | 0.2647 |
| 3 slices | 1.59 | 0.54 | 1.90 | 0.71 | −1.18 | 0.2494 | |
| 5 slices | 1.96 | 0.57 | 2.29 | 0.74 | −1.22 | 0.2369 | |
| Wall area (mm2) | Peak slice | 7.53 | 1.60 | 6.62 | 1.92 | 1.26 | 0.2218 |
| 3 slices | 7.41 | 1.15 | 6.36 | 1.84 | 1.66 | 0.1104 | |
| 5 slices | 7.35 | 1.19 | 6.27 | 1.73 | 1.77 | 0.0904 | |
| NWI | Peak slice | 0.86 | 0.04 | 0.81 | 0.06 | 2.30 | 0.0312* |
| 3 slices | 0.83 | 0.04 | 0.77 | 0.05 | 2.96 | 0.0073* | |
| 5 slices | 0.79 | 0.04 | 0.73 | 0.05 | 3.16 | 0.0045* | |
Note: “Std” denotes standard deviation. Two-sample t-tests were used for group comparison, where t and p were provided.
denotes significance reached (p<0.05).
IV. Discussion
In this work, we developed an automated vessel wall segmentation method for the intracranial arteries based on convolutional neural networks. The proposed method achieved good agreements with manual segmentation. With this method, a significant difference in lesion NWI between symptomatic patients and asymptomatic patients was detected, although lumen area and raw wall area were comparable in the two groups.
MR VWI is increasingly being adopted in ICAD studies. However, current plaque analysis heavily relies on qualitative assessment of a few plaque features through expert image review. This is in part due to the fact that a proper computer-aided tool for quantitative plaque analysis is unavailable. Vessel wall segmentation is a challenging task but prerequisite for automating quantitative analysis. For medical images of anatomic structures other than intracranial vessels, deep learning has shown its advantages in object localization and segmentation over conventional methods such as histogram thresholding, shape filters, and surface models. Our study, the first report to our knowledge, presented a deep-learning based approach for automated segmentation of intracranial vessels.
There are only few semi-automatic segmentation methods previously proposed for extracranial vessels [17, 36–39]. These methods may not be robust when applying to smaller vessels, for example intracranial arteries, due to the possible leak at low-contrast outer wall boundary. Advantageous over conventional machine-learning approaches where features are hand-engineered, FCN can automatically learn the comprehensive data-driven features from training data. In this work, specifically, we proposed a U-net-like FCN to strengthen the capacity of resolving the outer wall boundary. Our results suggest that the combination of baseline U-net with three other components, including batch normalization PReLU, and CLAHE, are necessary for a high performance in segmentation. Also, we performed a multi-class segmentation to extract both lumen and wall instead of only segmenting wall, which is a superior strategy as the latter approach may result into a C-shape segmentation for the wall that is difficult to restore lumen or wall through post-processing such as morphological opening (see Fig. S1 in the supplementary material). As we continue to accumulate the image and label data, the neural network weights could be further tuned for improved segmentation performance.
The utility of the automated vessel wall segmentation method was demonstrated in this study. When integrated into an intracranial vessel analysis framework, the proposed method permitted efficient analysis for large-range anterior circulation vasculatures, which is otherwise tremendously time-consuming in a manual fashion (see Fig. S2 in the supplementary material). In addition, a significantly higher NWI was observed in symptomatic ICAD lesions compared to asymptomatic ones despite their similar severity of stenosis. More importantly, NWI exhibited a stronger association with symptomatic lesions compared to wall and lumen areas. Hence, it is anticipated that this marker may have significant clinical value for early identifying event-prone plaques or informing the response of medical management in ICAD patients. Our results also highly suggest that the most stenotic location of a focal ICAD lesion is associated with a locally peak NWI. Whether this finding would hold true and could be utilized to automatically detect focal ICAD lesions is worthy of investigations in the future.
In this study, we proposed to simplify the segmentation task by straightening the vessel and using 2D cross-sectional slices. The reason is that the original vessels are tortuous in 3D space that is difficult for manual rater to delineate. Also, automated segmentation of 3D images would need a lot more training data to learn the segmentation in that complex environment. Result checking find most failure cases are at the vessel branching area, where one vessel is splitting to two or more vessels and so the vessel wall boundary in this 2D slice is no longer the general circle shape (see Fig. S3 in the supplementary material). To improve this, we could include more vessel branching slices in the training sample and so the program could learn this shape better. Second, neighboring cross-sectional slices from the same vessel may provide additional regularization for the segmentation results. The program could be extended to work on multiple 2D slices or even 3D images to get better overall structure information. Third, we measured the NWI as well as the areas of lumen and wall in this study. In future work, we would further evaluate vessel wall thickness, or eccentricity index (EI) [40] when considering the effects of stenosis, as relevant clinical parameters in a multi-center large-scale clinical study.
V. Conclusion
We have presented an automated vessel wall segmentation method for intracranial arteries based on convolutional neural networks. The proposed method achieved good agreements with manual rater in terms of Dice coefficient, lumen area, wall area, and NWI. With the method, a significant difference in lesion NWI between symptomatic patients and asymptomatic patients was detected, although lumen area and raw wall area were comparable in the two groups. Such an automated vessel segmentation approach would permit large-scale quantitative plaque analysis and promote the adoption of MR VWI in ICAD management.
Supplementary Material
Acknowledgments
This work was supported in part by American Heart Association (15SDG25710441), National Institutes of Health (NHLBI 2R01HL096119), and Natural Science Foundation of China (NSFC No.61375112).
Contributor Information
Feng Shi, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Qi Yang, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Radiology, Xuanwu Hospital, Beijing, China.
Xiuhai Guo, Department of Neurology, Xuanwu Hospital, Beijing, China.
Touseef Ahmad Qureshi, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Zixiao Tian, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Huijuan Miao, Department of Neurology, Xuanwu Hospital, Beijing, China.
Damini Dey, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Medicine, University of California, Los Angeles, CA, USA.
Debiao Li, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Medicine, University of California, Los Angeles, CA, USA; Department of Bioengineering, University of California, Los Angeles, CA, USA.
Zhaoyang Fan, Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, USA; Department of Medicine, University of California, Los Angeles, CA, USA; Department of Bioengineering, University of California, Los Angeles, CA, USA.
References
- [1].Turan TN, Derdeyn CP, Fiorella D, and Chimowitz MI, “Treatment of atherosclerotic intracranial arterial stenosis,” Stroke, vol. 40, no. 6, pp. 2257–61, Jun 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Holmstedt CA, Turan TN, and Chimowitz MI, “Atherosclerotic intracranial arterial stenosis: risk factors, diagnosis, and treatment,” Lancet Neurol, vol. 12, no. 11, pp. 1106–14, Nov 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gorelick PB, Wong KS, Bae HJ, and Pandey DK, “Large artery intracranial occlusive disease: a large worldwide burden but a relatively neglected frontier,” Stroke, vol. 39, no. 8, pp. 2396–9, Aug 2008. [DOI] [PubMed] [Google Scholar]
- [4].Arenillas JF, “Intracranial atherosclerosis: current concepts,” Stroke, vol. 42, no. 1 Suppl, pp. S20–3, Jan 2011. [DOI] [PubMed] [Google Scholar]
- [5].Alexander MD et al. , “High-resolution intracranial vessel wall imaging: imaging beyond the lumen,” J Neurol Neurosurg Psychiatry, vol. 87, no. 6, pp. 589–97, Jun 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Qiao Y et al. , “Intracranial arterial wall imaging using three-dimensional high isotropic resolution black blood MRI at 3.0 Tesla,” Journal of Magnetic Resonance Imaging, vol. 34, no. 1, pp. 22–30, 2011. [DOI] [PubMed] [Google Scholar]
- [7].van der Kolk AG, Hendrikse J, Zwanenburg JJ, Visser F, and Luijten PR, “Clinical applications of 7T MRI in the brain,” European journal of radiology, vol. 82, no. 5, pp. 708–718, 2013. [DOI] [PubMed] [Google Scholar]
- [8].Fan Z et al. , “Whole-brain intracranial vessel wall imaging at 3 Tesla using cerebrospinal fluid-attenuated T1-weighted 3D turbo spin echo,” Magn Reson Med, vol. 77, no. 3, pp. 1142–1150, Mar 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Klein IF, Lavallee PC, Touboul PJ, Schouman-Claeys E, and Amarenco P, “In vivo middle cerebral artery plaque imaging by high-resolution MRI,” Neurology, vol. 67, no. 2, pp. 327–9, Jul 25 2006. [DOI] [PubMed] [Google Scholar]
- [10].Swartz R et al. , “Intracranial arterial wall imaging using high-resolution 3-tesla contrast-enhanced MRI,” Neurology, vol. 72, no. 7, pp. 627–634, 2009. [DOI] [PubMed] [Google Scholar]
- [11].Xu WH et al. , “In vivo high-resolution MR imaging of symptomatic and asymptomatic middle cerebral artery atherosclerotic stenosis,” Atherosclerosis, vol. 212, no. 2, pp. 507–11, Oct 2010. [DOI] [PubMed] [Google Scholar]
- [12].Ryu CW, Jahng GH, Kim EJ, Choi WS, and Yang DM, “High resolution wall and lumen MRI of the middle cerebral arteries at 3 tesla,” Cerebrovasc Dis, vol. 27, no. 5, pp. 433–42, 2009. [DOI] [PubMed] [Google Scholar]
- [13].Qiao Y et al. , “Intracranial plaque enhancement in patients with cerebrovascular events on high-spatial-resolution MR images,” Radiology, vol. 271, no. 2, pp. 534–42, May 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Qiao Y et al. , “MR Imaging Measures of Intracranial Atherosclerosis in a Population-based Study,” Radiology, vol. 280, no. 3, pp. 860–8, Sep 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qiao Y et al. , “Patterns and Implications of Intracranial Arterial Remodeling in Stroke Patients,” Stroke, vol. 47, no. 2, pp. 434–40, Feb 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Feng C, Hua T, Xu Y, Liu X-Y, and Huang J, “Arterial remodeling of basilar atherosclerosis in isolated pontine infarction,” Neurological Sciences, vol. 36, no. 4, pp. 547–551, 2015. [DOI] [PubMed] [Google Scholar]
- [17].Niu P-P et al. , “Vessel wall differences between middle cerebral artery and basilar artery plaques on magnetic resonance imaging,” Scientific reports, vol. 6, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dieleman N et al. , “Magnetic resonance imaging of plaque morphology, burden, and distribution in patients with symptomatic middle cerebral artery stenosis,” Stroke, vol. 47, no. 7, pp. 1797–1802, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wu F et al. , “Hyperintense Plaque on Intracranial Vessel Wall Magnetic Resonance Imaging as a Predictor of Artery-to-Artery Embolic Infarction,” Stroke, vol. 49, no. 4, pp. 905–911, Apr 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wu F et al. , “Differential Features of Culprit Intracranial Atherosclerotic Lesions: A Whole-Brain Vessel Wall Imaging Study in Patients With Acute Ischemic Stroke,” J Am Heart Assoc, vol. 7, no. 15, Jul 22 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Swartz RH et al. , “Intracranial arterial wall imaging using high-resolution 3-tesla contrast-enhanced MRI,” Neurology, vol. 72, no. 7, pp. 627–34, Feb 17 2009. [DOI] [PubMed] [Google Scholar]
- [22].Yu J, Kwak H, Chung G, Hwang S, Park M, and Park S, “Association of Intraplaque Hemorrhage and Acute Infarction in Patients With Basilar Artery Plaque,” Stroke, vol. 46, no. 10, pp. 2768–2772, 2015. [DOI] [PubMed] [Google Scholar]
- [23].Dieleman N et al. , “Magnetic Resonance Imaging of Plaque Morphology, Burden, and Distribution in Patients With Symptomatic Middle Cerebral Artery Stenosis,” Stroke, vol. 47, no. 7, pp. 1797–1802, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mandell DM et al. , “Intracranial Vessel Wall MRI: Principles and Expert Consensus Recommendations of the American Society of Neuroradiology,” AJNR Am J Neuroradiol, vol. 38, no. 2, pp. 218–229, Feb 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].van der Kolk AG et al. , “Intracranial vessel wall imaging at 7.0-T MRI,” Stroke, vol. 42, no. 9, pp. 2478–2484, 2011. [DOI] [PubMed] [Google Scholar]
- [26].Zhang L et al. , “High resolution three dimensional intracranial arterial wall imaging at 3T using T1 weighted SPACE,” Magnetic resonance imaging, vol. 33, no. 9, pp. 1026–1034, 2015. [DOI] [PubMed] [Google Scholar]
- [27].Wang J, Helle M, Zhou Z, Bornert P, Hatsukami TS, and Yuan C, “Joint blood and cerebrospinal fluid suppression for intracranial vessel wall MRI,” Magn Reson Med, vol. 75, no. 2, pp. 831–8, Feb 2016. [DOI] [PubMed] [Google Scholar]
- [28].Yang H, Zhang X, Qin Q, Liu L, Wasserman BA, and Qiao Y, “Improved cerebrospinal fluid suppression for intracranial vessel wall MRI,” J Magn Reson Imaging, vol. 44, no. 3, pp. 665–72, Sep 2016. [DOI] [PubMed] [Google Scholar]
- [29].Litjens G et al. , “A survey on deep learning in medical image analysis,” Med Image Anal, vol. 42, pp. 60–88, Dec 2017. [DOI] [PubMed] [Google Scholar]
- [30].Shelhamer E, Long J, and Darrell T, “Fully Convolutional Networks for Semantic Segmentation,” IEEE Trans Pattern Anal Mach Intell, vol. 39, no. 4, pp. 640–651, Apr 2017. [DOI] [PubMed] [Google Scholar]
- [31].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015, pp. 234–241: Springer. [Google Scholar]
- [32].Yang Q et al. , “Whole-brain vessel wall MRI: A parameter tune-up solution to improve the scan efficiency of three-dimensional variable flip-angle turbo spin-echo,” J Magn Reson Imaging, vol. 46, no. 3, pp. 751–757, Sep 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].He K, Zhang X, Ren S, and Sun J, “Delving deep into rectifiers: Surpassing human-level performance on imagenet classification,” in The IEEE International Conference on Computer Vision, 2015, pp. 1026–1034. [Google Scholar]
- [34].Huang G, Liu Z, Weinberger KQ, and van der Maaten L, “Densely connected convolutional networks,” arXiv preprint arXiv:1608.06993, 2016. [Google Scholar]
- [35].Dumoulin V and Visin F, “A guide to convolution arithmetic for deep learning,” arXiv preprint arXiv, p. 1603.07285, 2016. [Google Scholar]
- [36].Chiu B, Egger M, Spence JD, Parraga G, and Fenster A, “Quantification of carotid vessel wall and plaque thickness change using 3D ultrasound images,” Med Phys, vol. 35, no. 8, pp. 3691–710, Aug 2008. [DOI] [PubMed] [Google Scholar]
- [37].Chiu B et al. , “Fast plaque burden assessment of the femoral artery using 3D black-blood MRI and automated segmentation,” Med Phys, vol. 38, no. 10, pp. 5370–84, Oct 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Smits LP et al. , “Manual versus Automated Carotid Artery Plaque Component Segmentation in High and Lower Quality 3.0 Tesla MRI Scans,” PLoS One, vol. 11, no. 12, p. e0164267, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Suinesiaputra A, de Koning PJ, Zudilova-Seinstra E, Reiber JH, and van der Geest RJ, “Automated quantification of carotid artery stenosis on contrast-enhanced MRA data using a deformable vascular tube model,” Int J Cardiovasc Imaging, vol. 28, no. 6, pp. 1513–24, Aug 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ohara T et al. , “Eccentric stenosis of the carotid artery associated with ipsilateral cerebrovascular events,” AJNR Am J Neuroradiol, vol. 29, no. 6, pp. 1200–3, Jun 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.