Abstract
Antibodies have long been the main approach used for localizing proteins of interest by light microscopy. In the past 5 yr or so, and with the advent of superresolution microscopy, the diversity of tools for imaging has rapidly expanded. One main area of expansion has been in the area of nanobodies, small single-chain antibodies from camelids or sharks. The other has been the use of artificial scaffold proteins, including Affimers. The small size of nanobodies and Affimers compared with the traditional antibody provides several advantages for superresolution imaging.
BACKGROUND
For many years, the scientific community has been using antibodies for localizing proteins of interest by immunofluorescence microscopy. This approach is well established and works well for standard wide-field and confocal fluorescence imaging. However, more recently, many researchers have begun to use “superresolution” imaging in their research to locate their proteins of interest with better resolution and precision. There are three main approaches to superresolution microscopy: structured illumination microscopy, which provides a twofold improvement in resolution compared with wide-field; stimulated emission depletion (STED), which improves resolution by approximately fivefold (to ∼50 nm); and single-molecule localization microscopy (SMLM) approaches that include photoactivatable light microscopy, stochastic optical reconstruction microscopy (STORM), and DNA points accumulation for imaging in nanoscale topography (PAINT), which provide resolutions of ∼20 nm or better (recently reviewed in Schermelleh et al., 2019). As the resolution increases to ∼20 nm or better, the use of both primary and secondary antibodies starts to become limiting, particularly in SMLM approaches, and new labeling approaches are needed. This Perspective provides a short overview of the different approaches that have begun to be employed to overcome this limitation.
THE TRADITIONAL DUAL-ANTIBODY APPROACH INTRODUCES LOCALIZATION ERROR IN SMLM SUPERRESOLUTION IMAGING
The typical approach of labeling proteins in cells, which uses a combination of primary and secondary antibodies, adds to the localization error in SMLM approaches. Antibodies from most species contain both heavy and light chains and are large in size (molecular mass of ∼150 kDa and ∼12 nm in length; Figure 1A). They can be polyclonal or monoclonal and tend to be immunoglobulin type G (IgG). In immunofluorescence applications, the location of the primary antibody is visualized using a fluorescently labeled secondary antibody that recognizes IgG from the species in which the primary antibody was raised. Secondary antibodies are almost always polyclonal and bind to various epitopes on the primary antibody. The secondary antibody is typically labeled with multiple dyes, usually by ε-amine labeling (e.g., via N-hydroxysuccinimydyl esters; Haugland, 1995), which attaches fluorophores to the lysine residues, of which there are many.
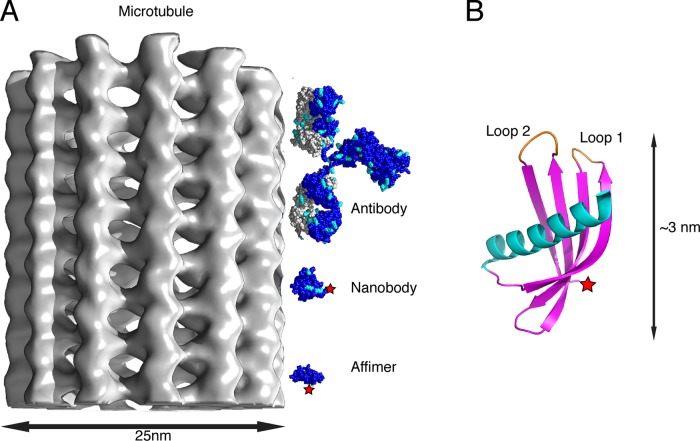
FIGURE 1:
Size comparison between an antibody, a nanobody, and an Affimer. (A) An antibody, a nanobody, and an Affimer (to scale) are shown next to a microtubule. The antibody (IgG: 1RJH) is composed of heavy chains (blue) and light chains (gray). The nanobody (30G0) is composed of a single heavy chain (blue; see text), and the Affimer (4N6T) is also composed of a single polypeptide chain (blue). The location of a dye molecule attached to a C-terminal cysteine residue is indicated by the red star for the Affimer and the nanobody. Otherwise, dye molecules for the antibody and nanobody would be attached to one or more of the lysine residues (shown in cyan). (B) The structure of the Affimer in detail, showing the positions of the two variable loops that interact with its binding partner. The position of the C-terminus, where the dye molecule is attached to a unique cysteine residue, is indicated by the red star.
In wide-field or confocal imaging, with a resolution of ∼250 nm, the small uncertainty (∼20 nm) introduced by using a combination of primary and secondary antibodies is not significant or detectable. However, as the resolution of the imaging methods increases toward that of SMLM, which has a resolution of 20 nm or better, antibody size combined with the polyclonal nature and multiple dye labels of the secondary antibody introduces an additional error in the localization precision for the target protein. Effectively, in SMLM, the location of the protein of interest is determined by imaging several dye labels at some variable but considerable distance (∼20 nm) away from the protein of interest, which leads to a degree of uncertainty about its actual position, the so-called linkage error.
There are various strategies to overcome this problem. If a combination of primary and secondary antibodies has to be used, then the secondary antibody can be labeled in such a way as to make sure it only has one dye label per antibody. This can be achieved by labeling the lysines via ε-amine labeling, and carefully monitoring the extent of the labeling. Alternatively, commercial kits are available to selectively reduce the disulfide bonds in the hinge region of the antibody and couple labels to the resulting free sulfhydryl groups, or to couple dyes to the polysaccharide groups, if present. All of these need care to make sure the antibody only contains one dye per molecule and that its properties are otherwise unaffected. Moreover, this approach does not overcome the polyclonal nature of the secondary antibody and still places the dye molecule at some uncertain distance away from the target protein.
Alternatively, the primary antibody can directly labeled. However, this can expensive, as it usually requires a minimum amount of source antibody to be labeled, and the dye:molecule ratio still needs to be controlled. The unspecific nature of ε-amine labeling can make this difficult to achieve and, in some cases, can eliminate the binding efficacy of the antibody by labeling on/or close to the antigen-binding site (Vira et al., 2010; Szabo et al., 2018). Even when successful, the fluorophore can still be ∼10 nm away from the epitope of interest. Using smaller Fab fragments that have been directly labeled will place the fluorophore closer to the protein of interest, but Fab fragments are often not available commercially and normally have to be made in house. Clearly, alternatives to traditional antibody labeling are needed for SMLM-based superresolution imaging.
NANOBODIES: ALTERNATIVES TO THE TRADITIONAL DUAL ANTIBODY LABELING APPROACH IN SMLM SUPERRESOLUTION IMAGING
One solution to this problem is to use nanobodies. About 50% of the antibodies found in camelid species (Bactrian camels, alpacas, dromedaries, and llamas) are composed of a dimer of heavy chains only (isotypes IgG2 and IgG3). The size of these heavy chains is considerably smaller than those of mammalian antibodies (Hamers-Casterman et al., 1993), and the single N-terminal domain of these antibodies does not need to pair with a second heavy chain in order to bind specifically to its protein target. Similarly, some cartilaginous fish contain a subset of heavy-chain antibodies (Greenberg et al., 1995) that are also able to function via a single monomeric variable antibody domain. These antibodies (or nanobodies) are thus much smaller (∼10–15 kDa) than the traditional monoclonal antibody (Figure 1A). It has become relatively routine to isolate the mRNA from blood lymphocytes from an immunized animal, subclone the VHH sequences into a phage display vector, and then generate small immune libraries (∼106 variants) for screening (Muyldermans, 2001, 2013). Naïve (using nonimmunized animals) and synthetic/semisynthetic libraries have also since been developed (for a review of recent technologies, see Liu et al., 2018), as have humanized nanobodies (Moutel et al., 2016).
The first elegant demonstration that nanobodies are useful for SMLM-based superresolution imaging, was shown through the use of the anti–green fluorescent protein (GFP) nanobody (Ries et al., 2012), for example, to visualize GFP-tagged tubulin in microtubules. A second nanobody against red fluorescent protein has been used similarly (Platonova et al., 2015), and nanobodies that recognize epitope tags (Virant et al., 2018) have since been developed. However, to use these antibodies, the protein of interest must be tagged, which can affect its function and/or location. Nanobodies that recognize mouse or rat primary antibodies and that can be used as secondary antibodies have also been described (Pleiner et al., 2018).
Nanobodies have also been generated to a variety of proteins, including tubulin (Mikhaylova et al., 2015), to allow direct rather than indirect labeling of microtubules and have been used successfully in SMLM approaches, including DNA PAINT (Fabricius et al., 2018; Sograte-Idrissi et al., 2019). They have also been used successfully in correlative light and electron microscopy (EM) approaches (Fang et al., 2018) with better penetration than traditional antibodies. This approach (termed “NATIVE” for nanobody-assisted tissue immunostaining for volumetric EM) is enabled by the better penetration of nanobodies into thick (500-µM) sections, without the need for permeabilization by detergents, which destroys lipid membranes.
As with traditional antibodies, nanobodies can be labeled via the ε-amine (lysine residues), and careful labeling can result in 1–1.5 labels on average per nanobody (Platonova et al., 2015). However, a further innovation demonstrated that nanobodies could be generated with single cysteines for direct labeling at a single site (Pleiner et al., 2015). This approach demonstrated that the nanobodies could be relatively stable without the disulfide bonds they normally contain and could then be purified with high yields from the cytoplasm of Escherichia coli, rather than the periplasm (inclusion bodies), as is more typical (Pleiner et al., 2015).
Importantly, in SMLM-based superresolution imaging, the small size of the nanobody combined with a single unique site for attaching the dye label means that the fluorescent label is now placed ∼2–4 nm away from the protein target, which provides better localization precision.
“NON-ANTIBODY” APPROACHES TO LABELING PROTEINS IN SMLM SUPERRESOLUTION
A number of small molecules and peptides such as phalloidin (Xu et al., 2012) and LifeAct (Kiuchi et al., 2015) to label actin, Taxol derivatives to label tubulin (Lukinavicius et al., 2018), DNA intercalating dyes (Flors et al., 2009), and labels for endoplasmic reticulum and other organelles (Shim et al., 2012) have been used in SMLM-based superresolution microscopy. These place the dye molecule very close to the protein of interest. However, these binders cannot be generated in a systematic way against a range of different target proteins. Moreover, in some instances, they do not appear to work well, an example being the use of phalloidin in DNA-PAINT (Schlichthaerle et al., 2018).
Many different alternatives to antibodies are now being developed that can bind to and label proteins. Typically, these consist of a protein scaffold into which variable binding loops or domains have been engineered to generate a combinatorial phagemid library that can be screened for binding to the protein target of interest (Nygren and Skerra, 2004; Skrlec et al., 2015). These binders can then be expressed and purified from E. coli. One of these is the Affibody (Lofblom et al., 2010), which is derived from the IgG-binding protein A from Staphylococcus and consists of a three-helix bundle (58 amino acids in total). Another is the DARPin (designed ankyrin repeat proteins; Boersma and Pluckthun, 2011). In addition to protein-based alternatives, aptamers (single-stranded DNA or RNA oligonucleotides) have also been reported as useful in SMLM-based superresolution imaging (Opazo et al., 2012). However, so far, many of these antibody alternatives have not been well explored for their potential use in superresolution imaging.
Affimers are an example of another type of non-antibody binding alternative that has been used successfully in SMLM by our group and others (Tiede et al., 2017; Schlichthaerle et al., 2018). Affimers (Figure 1), originally named Adhirons (Tiede et al., 2014), are made up of a scaffold based on the plant phytocystatin protein. The Affimer contains two variable loops containing randomized amino acid sequences, and the combinatorial phagemid library comprises 1.3 × 1010 clones. The sequence is ∼100 amino acids long (10–12 kDa and ∼3 nm in size). The amino acid residue cysteine is excluded, which allows this residue to be cloned in at a later stage to either the N- or C-terminus to allow site-specific labeling. The protein itself is highly thermally stable, making purification of the Affimer (via an introduced His tag) from E. coli very simple, as most of the bacterial proteins can be denatured and precipitated by a short heat treatment before purification. The binding affinity of the Affimers is typically in the nanomolar range, and highly specific reagents can be isolated from the library (Tiede et al., 2014; Hughes et al., 2017; Tang et al., 2017; Robinson et al., 2018). As the Affimers are completely synthetic, they can also be screened against potentially toxic polypeptides.
We reasoned that their small size and the ability to specifically add a fluorescent label via the unique cysteine residue would make Affimers ideal for superresolution imaging. We initially isolated four Affimers to rabbit skeletal F-actin (Lopata et al., 2018) and characterized their ability to image F-actin in live and fixed cells using a variety of approaches. We followed this up by demonstrating that one of these Affimers (Affimer 14) performed well in both the 3D dSTORM and DNA PAINT approaches (Schlichthaerle et al., 2018), outperforming phalloidin. Similarly, we have raised an Affimer to tubulin that also works well in dSTORM (Tiede et al., 2017).
Affimers, like nanobodies, also have the advantage that their smaller size enables them to better penetrate regions of cells from which traditional antibodies are normally excluded. For example, the tubulin Affimer stained up the cytokinetic furrow, a region densely packed with microtubules, which does not stain up with the traditional antibody approach (Tiede et al., 2017). Similarly, nanobodies to SNAP-25 and Syntaxin A1 revealed an additional extrasynaptic localization of these proteins in superresolution (STED) imaging due to their ability to more easily penetrate samples (Maidorn et al., 2019).
Affimers, like antibodies and nanobodies, may well be able to recognize specific posttranslational modifications and/or protein conformations. For example, of the four Affimers raised to F-actin, only one Affimer bound F-actin in paraformaldehyde-fixed cells (Affimer 14; Lopata et al., 2018), whereas all four Affimers bound F-actin in methanol-fixed cells. One potential explanation for this is that methanol fixation has a small effect on F-actin structure, which is enough to prevent the binding of phalloidin (Lopata et al., 2018; Mentes et al., 2018). It is possible that the three F-actin Affimers that only bind F-actin in methanol-fixed cells are also sensitive to this structural change. This suggests that further work and tailoring the screening process could enable the retrieval of Affimers that recognize specific protein conformations. In a further example, we originally recovered 10 Affimers to tubulin, but only one of these recognized and labeled microtubules in interphase cells (Tiede et al., 2017). However, the Affimers were raised against tubulin purified from brain, which contains multiple tubulin isoforms and multiple posttranslational modifications. Therefore, it is possible that some of these Affimers may recognize specific tubulin isoforms or posttranslational modifications that are not commonly present in cultured cells but are present in the brain. Work is currently underway to explore this possibility.
Overall, this is an exciting time for imaging. Technology is driving forward the development of sophisticated imaging techniques that are revealing more detail about protein organization in cells than ever before. The development of nanobodies and small non-antibody binding proteins such as Affimers are allowing us to capitalize on the potential of superresolution imaging by providing probes with very small linkage errors. Together, these approaches provide us with much better certainty as to the location of our favorite proteins of interest.
Acknowledgments
Part of the work in developing Affimers for superresolution imaging was supported by an MRC Next Generation imaging grant (MR/K015613/1) to M.P.
Abbreviations used:
- DARPin
designed ankyrin repeat proteins
- DNA PAINT
DNA points accumulation for imaging in nanoscale topography
- EM
electron microscopy
- GFP
green fluorescent protein
- IgG
immunoglobulin type G
- SMLM
single-molecule localization microscopy
- STED
stimulated emission depletion
- STORM
stochastic optical reconstruction microscopy.
Footnotes
REFERENCES
- Boersma YL, Pluckthun A. (2011). DARPins and other repeat protein scaffolds: advances in engineering and applications. Curr Opin Biotechnol , 849–857. [DOI] [PubMed] [Google Scholar]
- Fabricius F, Lefebre J, Geertsema H, Marino SF, Ewers H. (2018). Rapid and efficient C-terminal labeling of nanobodies for DNA-PAINT. J Phys D: Appl Phys , 474005. [Google Scholar]
- Fang T, Lu X, Berger D, Gmeiner C, Cho J, Schalek R, Ploegh H, Lichtman J. (2018). Nanobody immunostaining for correlated light and electron microscopy with preservation of ultrastructure. Nat Methods , 1029–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors C, Ravarani CN, Dryden DT. (2009). Super-resolution imaging of DNA labelled with intercalating dyes. Chemphyschem , 2201–2204. [DOI] [PubMed] [Google Scholar]
- Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. (1995). A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature , 168–173. [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. (1993). Naturally occurring antibodies devoid of light chains. Nature , 446–448. [DOI] [PubMed] [Google Scholar]
- Haugland RP. (1995). Coupling of monoclonal antibodies with fluorophores. Methods Mol Biol , 205–221. [DOI] [PubMed] [Google Scholar]
- Hughes DJ, Tiede C, Penswick N, Tang AA, Trinh CH, Mandal U, Zajac KZ, Gaule T, Howell G, Edwards TA, et al. (2017). Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology. Sci Signal , eaaj2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T, Higuchi M, Takamura A, Maruoka M, Watanabe N. (2015). Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat Methods , 743–746. [DOI] [PubMed] [Google Scholar]
- Liu W, Song H, Chen Q, Yu J, Xian M, Nian R, Feng D. (2018). Recent advances in the selection and identification of antigen-specific nanobodies. Mol Immunol , 37–47. [DOI] [PubMed] [Google Scholar]
- Lofblom J, Feldwisch J, Tolmachev V, Carlsson J, Stahl S, Frejd FY. (2010). Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett , 2670–2680. [DOI] [PubMed] [Google Scholar]
- Lopata A, Hughes R, Tiede C, Heissler SM, Sellers JR, Knight PJ, Tomlinson D, Peckham M. (2018). Affimer proteins for F-actin: novel affinity reagents that label F-actin in live and fixed cells. Sci Rep , 6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavicius G, Mitronova GY, Schnorrenberg S, Butkevich AN, Barthel H, Belov VN, Hell SW. (2018). Fluorescent dyes and probes for super-resolution microscopy of microtubules and tracheoles in living cells and tissues. Chem Sci , 3324–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidorn M, Olichon A, Rizzoli SO, Opazo F. (2019). Nanobodies reveal an extra-synaptic population of SNAP-25 and Syntaxin 1A in hippocampal neurons. MAbs , 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentes A, Huehn A, Liu X, Zwolak A, Dominguez R, Shuman H, Ostap EM, Sindelar CV. (2018). High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc Natl Acad Sci USA , 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova M, Cloin BM, Finan K, van den Berg R, Teeuw J, Kijanka MM, Sokolowski M, Katrukha EA, Maidorn M, Opazo F, et al. (2015). Resolving bundled microtubules using anti-tubulin nanobodies. Nat Commun , 7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, Ligat L, Rain JC, Favre G, Olichon A, Perez F. (2016). NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife , e16228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. (2001). Single domain camel antibodies: current status. J Biotechnol , 277–302. [DOI] [PubMed] [Google Scholar]
- Muyldermans S. (2013). Nanobodies: natural single-domain antibodies. Annu Rev Biochem , 775–797. [DOI] [PubMed] [Google Scholar]
- Nygren PA, Skerra A. (2004). Binding proteins from alternative scaffolds. J Immunol Methods , 3–28. [DOI] [PubMed] [Google Scholar]
- Opazo F, Levy M, Byrom M, Schafer C, Geisler C, Groemer TW, Ellington AD, Rizzoli SO. (2012). Aptamers as potential tools for super-resolution microscopy. Nat Methods , 938–939. [DOI] [PubMed] [Google Scholar]
- Platonova E, Winterflood CM, Junemann A, Albrecht D, Faix J, Ewers H. (2015). Single-molecule microscopy of molecules tagged with GFP or RFP derivatives in mammalian cells using nanobody binders. Methods , 89–97. [DOI] [PubMed] [Google Scholar]
- Pleiner T, Bates M, Gorlich D. (2018). A toolbox of anti-mouse and anti-rabbit IgG secondary nanobodies. J Cell Biol , 1143–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiner T, Bates M, Trakhanov S, Lee CT, Schliep JE, Chug H, Bohning M, Stark H, Urlaub H, Gorlich D. (2015). Nanobodies: site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. Elife , e11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. (2012). A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods , 582–584. [DOI] [PubMed] [Google Scholar]
- Robinson JI, Baxter EW, Owen RL, Thomsen M, Tomlinson DC, Waterhouse MP, Win SJ, Nettleship JE, Tiede C, Foster RJ, et al. (2018). Affimer proteins inhibit immune complex binding to FcγRIIIa with high specificity through competitive and allosteric modes of action. Proc Natl Acad Sci USA , E72–E81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Ferrand A, Huser T, Eggeling C, Sauer M, Biehlmaier O, Drummen GPC. (2019). Super-resolution microscopy demystified. Nat Cell Biol , 72–84. [DOI] [PubMed] [Google Scholar]
- Schlichthaerle T, Eklund AS, Schueder F, Strauss MT, Tiede C, Curd A, Ries J, Peckham M, Tomlinson DC, Jungmann R. (2018). Site-specific labeling of Affimers for DNA-PAINT microscopy. Angew Chem Int Ed Engl , 11060–11063. [DOI] [PubMed] [Google Scholar]
- Shim SH, Xia C, Zhong G, Babcock HP, Vaughan JC, Huang B, Wang X, Xu C, Bi GQ, Zhuang X. (2012). Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc Natl Acad Sci USA , 13978–13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrlec K, Strukelj B, Berlec A. (2015). Non-immunoglobulin scaffolds: a focus on their targets. Trends Biotechnol , 408–418. [DOI] [PubMed] [Google Scholar]
- Sograte-Idrissi S, Oleksiievets N, Isbaner S, Eggert-Martinez M, Enderlein J, Tsukanov R, Opazo F. (2019). Nanobody detection of standard fluorescent proteins enables multi-target DNA-PAINT with high resolution and minimal displacement errors. Cells , E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Szendi-Szatmari T, Ujlaky-Nagy L, Radi I, Vereb G, Szollosi J, Nagy P. (2018). The effect of fluorophore conjugation on antibody affinity and the photophysical properties of dyes. Biophys J , 688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AA, Tiede C, Hughes DJ, McPherson MJ, Tomlinson DC. (2017). Isolation of isoform-specific binding proteins (Affimers) by phage display using negative selection. Sci Signal , eaan0868. [DOI] [PubMed] [Google Scholar]
- Tiede C, Bedford R, Heseltine SJ, Smith G, Wijetunga I, Ross R, AlQallaf D, Roberts AP, Balls A, Curd A, et al. (2017). Affimer proteins are versatile and renewable affinity reagents. Elife , e24903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede C, Tang AA, Deacon SE, Mandal U, Nettleship JE, Owen RL, George SE, Harrison DJ, Owens RJ, Tomlinson DC, McPherson MJ. (2014). Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications. Protein Eng Des Sel , 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vira S, Mekhedov E, Humphrey G, Blank PS. (2010). Fluorescent-labeled antibodies: balancing functionality and degree of labeling. Anal Biochem , 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant D, Traenkle B, Maier J, Kaiser PD, Bodenhofer M, Schmees C, Vojnovic I, Pisak-Lukats B, Endesfelder U, Rothbauer U. (2018). A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat Commun , 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Babcock HP, Zhuang X. (2012). Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods , 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]