Abstract
Bipolar spindles are organized by motor proteins that generate microtubule-dependent forces to separate the two spindle poles. The fission yeast Cut7 (kinesin-5) is a plus-end-directed motor that generates the outward force to separate the two spindle poles, whereas the minus-end-directed motor Pkl1 (kinesin-14) generates the inward force. Balanced forces by these antagonizing kinesins are essential for bipolar spindle organization in mitosis. Here, we demonstrate that chromosomes generate another outward force that contributes to the bipolar spindle assembly. First, it was noted that the cut7 pkl1 double knockout failed to separate spindle poles in meiosis I, although the mutant is known to succeed it in mitosis. It was assumed that this might be because meiotic kinetochores of bivalent chromosomes joined by cross-overs generate weaker tensions in meiosis I than the strong tensions in mitosis generated by tightly tethered sister kinetochores. In line with this idea, when meiotic mono-oriented kinetochores were artificially converted to a mitotic bioriented layout, the cut7 pkl1 mutant successfully separated spindle poles in meiosis I. Therefore, we propose that spindle pole separation is promoted by outward forces transmitted from kinetochores to spindle poles through microtubules.
INTRODUCTION
Chromosome segregation is driven by a bipolar spindle, which consists of microtubules emanating from spindle poles. Bipolar spindle assembly is regulated by outward pushing forces and inward pulling forces generated by microtubules and microtubule-associated proteins (MAPs). The plus-end-directed motor kinesin-5 (BimC/Eg5/Cut7) cross-links and slides antiparallel microtubules, thereby pushing away the two spindle poles harboring minus ends of the microtubules (Kashina et al., 1996; Kapitein et al., 2005).
The outward force generated by kinesin-5 is essential for bipolar spindle assembly, as its inhibition results in monopolar spindles, defined by a failure in separation of the two spindle poles (Hagan and Yanagida, 1990, 1992; Heck et al., 1993; Mayer et al., 1999; Kapoor et al., 2000). On the contrary, inward forces are generated by minus-end-directed motors such as kinesin-14 and dynein (Sharp et al., 1999; Mitchison et al., 2005). Outward and inward forces are antagonistic to each other during spindle pole separation. The monopolar spindle caused by kinesin-5 inhibition is canceled by the simultaneous depletion of kinesin-14 or dynein in humans, Drosophila, Xenopus eggs, and fission yeast (Pidoux et al., 1996; Sharp et al., 1999; Troxell et al., 2001; Mitchison et al., 2005; Tanenbaum et al., 2008; Ferenz et al., 2009; Olmsted et al., 2014; van Heesbeen et al., 2014). The interplay of the kinesin-5 and kinesin-14 in bipolar spindle assembly was intensively investigated in the fission yeast Schizosaccharomyces pombe.
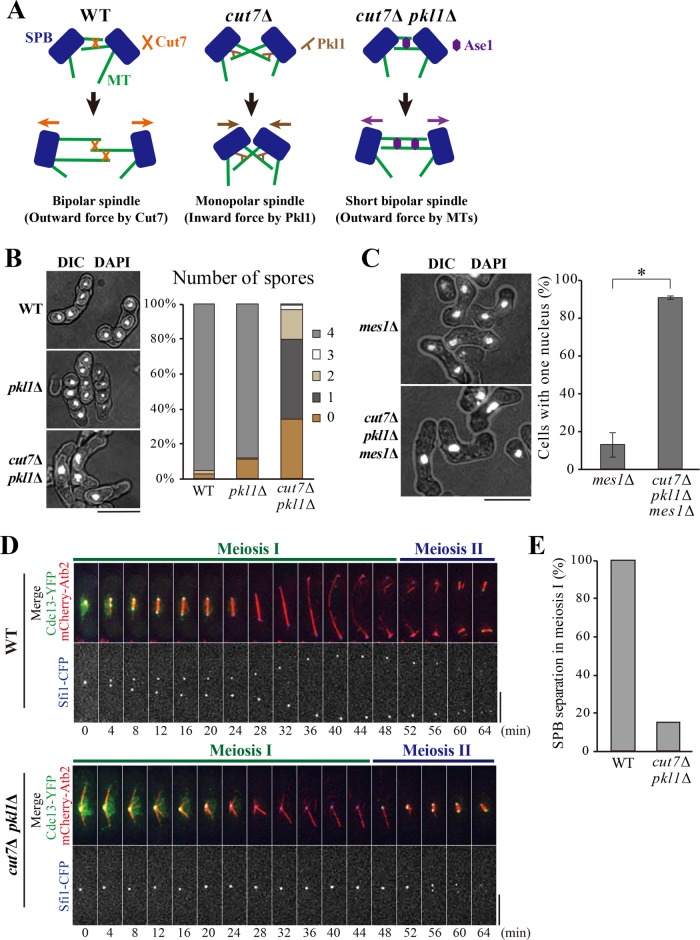
Fission yeast has a kinesin-5 protein Cut7 and two kinesin-14 proteins; Pkl1 and Klp2 (Hagan and Yanagida, 1990; Pidoux et al., 1996; Troxell et al., 2001). Cut7 generates an outward force that slides antiparallel microtubules apart (Hagan and Yanagida, 1990, 1992) (see Figure 1A for a schematic, left). Cut7 is essential for the separation of spindle pole bodies (SPBs, the fungal equivalent of the centrosome) (Hagan and Yanagida, 1990, 1992), and the deletion mutant of Cut7 (cut7∆) is lethal. The conditional (temperature-sensitive) mutant of Cut7 shows the monopolar spindle phenotype at the restrictive temperature, which is a hallmark of SPB separation defects. In contrast, Pkl1 localizes closely to SPBs and generates an inward force that pulls microtubules emanating from opposite poles (Rincon et al., 2017; Yukawa et al., 2017, 2018) (Figure 1A, center).
FIGURE 1:
cut7Δ pkl1Δ cells display the monopolar spindle in meiosis I. (A) Schematics for spindle assembly in the mitosis of WT, cut7Δ, and cut7Δ pkl1Δ cells. (B) The cut7∆ pkl1∆ cells are defective in meiosis and sporulation. Cells of indicated genotypes were stained with DAPI. Images are shown merged with differential interference contrast (DIC) (left). The spores in an ascus were counted (right, WT, n = 111 cells; pkl1Δ, n = 108 cells; cut7∆ pkl1∆, n = 131 cells). Scale bar, 10 μm. (C) Meiosis was induced for mes1Δ and cut7Δ pkl1Δ mes1Δ cells. Cells were stained with DAPI, and the chromosome masses in a zygote were counted (mean ± SD n > 94 cells, N = 2 experiments). Scale bar, 10 μm. *P < 0.005 (Welch’s t test). (D) Living cells expressing Cdc13-YFP (green), mCherry-Atb2 (microtubules; red), and Sfi1-CFP (SPB; blue) were observed from the onset of meiosis I in WT (top) and cut7Δ pkl1Δ (bottom). The value 0 min corresponds to the start of time-lapse imaging. Scale bars, 5 μm. (E) Percentages of WT and cut7Δ pkl1Δ cells that showed SPB separation in meiosis I were calculated from observation in C (WT, n = 11 cells; cut7∆ pkl1∆, n = 13 cells).
The lethality of cut7Δ is suppressed by the deletion of the pkl1 gene; and the cut7∆ pkl1∆ double mutant survives (Olmsted et al., 2014) and manages to eventually separate SPBs by microtubules grown in a later stage than usually seen in wild type (WT) cells (Rincon et al., 2017; Yukawa et al., 2017). The cut7∆ pkl1∆ double mutant utilizes the antiparallel microtubule bundling factor Ase1/PRC1 for SPB separation (Rincon et al., 2017; Yukawa et al., 2017). Ase1 in WT cells generates outward pushing forces in the spindle midzone in anaphase that contributes to elongation of the anaphase spindle (Loïodice et al., 2005; Yamashita et al., 2005) (Figure 1A, right). In silico simulations suggest that the Ase1-dependent mechanism is seemingly sufficient for bipolar spindle assembly in the absence of Cut7 and Pkl1 (Rincon et al., 2017). Therefore, the force balance exerted for the positioning of two SPBs can be explained by Cut7, Pkl1, and additional factors located on interpolar microtubules connecting two SPBs.
In addition to recent knowledge, we here propose an additional outward force that may contribute to the separation of SPBs in the pro-prometaphase of mitosis. Chromosomes play a key role in outward force generation. In mitosis, kinetochores located on sister chromatids are bioriented to associate with microtubules from opposite poles. In the first division of meiosis (meiosis I), homologous chromosomes are paired as bivalent with some cross-overs as a result of meiotic recombination (reviewed in Siomos and Nasmyth, 2003; Yamagishi et al., 2014). Spindle microtubules need to segregate homologues but not sister chromatids. For this purpose, sister kinetochores are tightly tethered throughout meiosis I by the meiotic cohesin Rec8 as well as the monopolin Moa1, so that such mono-oriented sister kinetochores can be associated to microtubules from either of the two SPBs (Yokobayashi and Watanabe, 2005). When spindle microtubules attach to kinetochores to pull them, bivalent chromosomes are stretched by pulling forces generated in the spindle (reviewed in Nicklas, 1974).
In this study, we use mitotic and meiotic cells to investigate how chromosomes affect SPB separation. Through analyses, we propose that kinetochores attached to microtubules provide mechanical support to push SPBs apart during the pro-prometaphase of mitosis.
RESULTS
cut7Δ pkl1Δ cells fail to assemble the bipolar spindle in meiosis I
Recent knowledge regarding the force balance is summarized as a schematic in Figure 1A. Cut7 is essential for SPB separation in fission yeast (Hagan and Yanagida, 1990, 1992). The cut7 deletion mutant (cut7∆) is inviable as it is unable to separate SPBs, but the cut7Δ pkl1Δ double mutant is shown to be viable (Olmsted et al., 2014) (Supplemental Figure S1A). Indeed, cut7Δ pkl1Δ cells separate SPBs, albeit partially, powered by the sliding of interdigitating microtubules with the assistance of the microtubule cross-linker Ase1 (Rincon et al., 2017; Yukawa et al., 2017) (Figure 1A and Supplemental Figure S1, B and C). The spindles in cut7∆ pkl1∆ cells were elongated later in the anaphase, although the timing appeared delayed compared with those in WT cells (Rincon et al., 2017; Yukawa et al., 2017) (Supplemental Figure S1, B and C).
However, it was found that the cut7Δ pkl1Δ mutant showed severe defects in meiotic progression. Although WT and pkl1Δ zygotes underwent normal meiosis and produced four spores, cut7Δ pkl1Δ zygotes mostly produced 0–2 spores per ascus (Figure 1B). The number of nuclei indicated by DAPI (4′,6-diamidino-2-phenylindole) was one or two in most asci, instead of four in WT asci as a result of two rounds of meiotic divisions (meiosis I and II). This result indicates that the cut7Δ pkl1Δ mutant is deficient in chromosome segregation in meiosis.
The mes1∆ mutation, which arrests meiotic progression before meiosis II at the binucleate state, was utilized for further clarification studies (Bresch et al., 1968; Izawa et al., 2005) (Figure 1C). In contrast, ∼90% of cut7Δ pkl1Δ mes1Δ zygotes showed one nucleus (Figure 1C), indicating that cells were unable to undergo meiosis I in the simultaneous absence of Cut7 and Pkl1.
Therefore, the progression of meiosis I by the time-lapse imaging of WT and cut7Δ pkl1Δ cells expressing Cdc13-YFP (cyclin B) (Decottignies et al., 2001), mCherry-Atb2 (α2-tubulin) (Sato et al., 2009), and Sfi1-CFP (a component of the SPB half-bridge) (Kilmartin, 2003) was investigated. In WT cells, SPBs separated and accordingly, the bipolar spindle was formed by the metaphase of meiosis I (4 min onward; note that Cdc13-YFP localized to SPBs; Figure 1D, top). Cdc13 then disappeared, and the spindle began to elongate on entry into the anaphase (24 min). In contrast, SPBs failed to separate in 85% of the cut7Δ pkl1Δ cells, and the resulting monopolar spindle remained until the end of meiosis I when Cdc13-YFP disappeared (Figure 1, D and E, bottom).
Despite the failure in bipolar spindle formation in meiosis I, it appeared that the SPBs separated and the bipolar spindle was assembled later, in the timing of meiosis II (56 min, Figure 1D, bottom), which was indicated by the reaccumulation of Cdc13-YFP on SPBs. Although the spindle for meiosis II started to assemble, it failed to fully elongate and segregate chromosomes. Therefore, most of the mutant cells produced a single spore with a single nucleus (Figure 1, B and D). Taking these results together, it was concluded that the Cut7-independent spindle assembly machinery that operates in mitosis might not properly function in meiosis I.
Deletion of klp2 improves the SPB separation of cut7Δ pkl1Δ cells in meiosis I
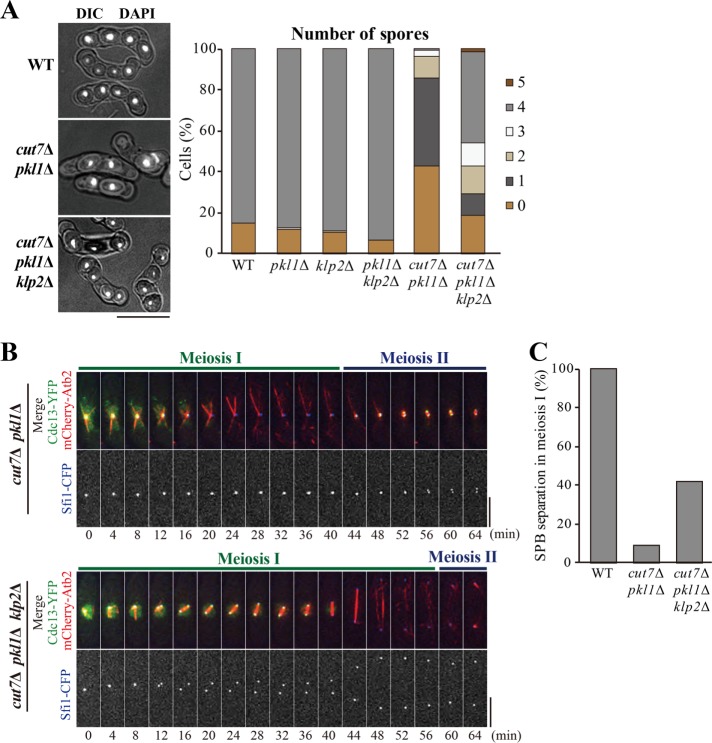
It was then hypothesized that the monopolar spindle phenotype seen in meiosis I of the cut7∆ pkl1∆ mutant was due to the imbalance of spindle forces. The inward forces pulling SPBs might be strong relative to outward pushing forces in the meiosis I spindle. To test this hypothesis, the deletion of the klp2 gene was introduced. While the kinesin-5 Cut7 generates an outward force, two kinesin-14 proteins, Pkl1 and Klp2, generate inward forces in the mitotic spindle (Rincon et al., 2017; Yukawa et al., 2017, 2018, 2019). In line with this, previous observations for mitosis have demonstrated that the bipolar spindle assembled in cut7Δ pkl1Δ cells is ameliorated even by the simultaneous deletion of klp2 (Yukawa et al., 2018, 2019).
Therefore, meiosis I of the cut7Δ pkl1Δ klp2Δ triple knockout cells was observed. First, most of the cut7Δ pkl1Δ cells failed to undergo meiosis I resulting in improper nuclear division, as shown above (Figures 1 and 2A). In contrast, ∼40% of the cut7Δ pkl1Δ klp2Δ cells generated four spores with four nuclei as WT cells did (Figure 2A), indicating that defects in SPB separation were canceled by the additional deletion of klp2. As expected, similar percentages of cut7Δ pkl1Δ klp2Δ cells separated SPBs and assembled the bipolar spindle by metaphase of meiosis I (12 min; Figure 2, B and C, bottom), followed by spindle elongation in the anaphase (40 min).
FIGURE 2:
Deletion of klp2 partly suppresses meiotic defects in cut7∆ pkl1∆. (A) Sporulation was monitored in the cells of indicated genotypes. Cells were stained with DAPI and shown with DIC (left). Proportions of the number of spores are also shown (right, WT, n = 183 cells; pkl1Δ, n = 181; klp2∆, n = 89; pkl1∆ klp2∆, n = 146; cut7∆ pkl1∆, n = 179; cut7∆ pkl1∆ klp2∆, n = 151). Scale bar, 10 μm. (B) Cells expressing Cdc13-YFP (green), mCherry-Atb2 (microtubules; red), and Sfi1-CFP (SPB; blue) were observed from the onset of meiosis I in cut7Δ pkl1Δ (top) and cut7Δ pkl1Δ klp2Δ (bottom). Scale bars, 5 μm. (C) Percentages of WT, cut7Δ pkl1Δ, and cut7Δ pkl1Δ klp2Δ cells that showed SPB separation in meiosis I were calculated from observations in B (WT, n = 12 cells; cut7∆ pkl1∆, n = 11; cut7∆ pkl1∆ klp2∆, n = 12).
These results demonstrate that cut7Δ pkl1Δ cells were unable to separate SPBs in meiosis I because of the imbalance of forces inside the spindle that was not evident during mitotic divisions.
Chromosome configuration affects the balance of forces inside the meiotic spindle
Collectively, these results indicate that in meiosis I, the inward forces are stronger or outward forces are weaker than in mitosis. Therefore, cut7∆ pkl1∆ cells fail to separate SPBs in meiosis I.
As a result, the reasons why the force imbalance was evident, particularly in meiosis I, were examined. This difference could be due to the ploidy of cells used in the experiments, as current knowledge on mitosis has been based on observations using haploid cells, and the meiotic cells used in this study are diploid. The diploid cut7Δ pkl1Δ strain was constructed for the observation of mitosis to test this possibility. Diploid cells of cut7Δ pkl1Δ showed SPB separation and bipolar spindle formation in mitosis (Supplemental Figure S2, A and B), as seen in haploid cells (Olmsted et al., 2014). Hence, ploidy cannot be the reason for the monopolar spindle phenotype seen in meiosis I of the cut7Δ pkl1Δ cells.
Next, it was considered that chromosome configuration in meiosis I may cause the force imbalance. In mitosis and meiosis II, sister chromatids are tied together with cohesin, whereas homologous chromosomes behave independently. On the contrary, homologues are paired, and cross-overs are made prior to meiosis I (reviewed in Moore, 1998; Siomos and Nasmyth, 2003). The bivalent chromosomes with chiasmata are associated with microtubules so that homologues are separated in meiosis I (WT; see Figure 3A for a schematic). In each type of division, chromosome configuration is correlated with the SPB state in cut7∆ pkl1∆ cells. Namely, SPBs separated in mitosis and meiosis II, but not in meiosis I (see Figure 1D).
FIGURE 3:
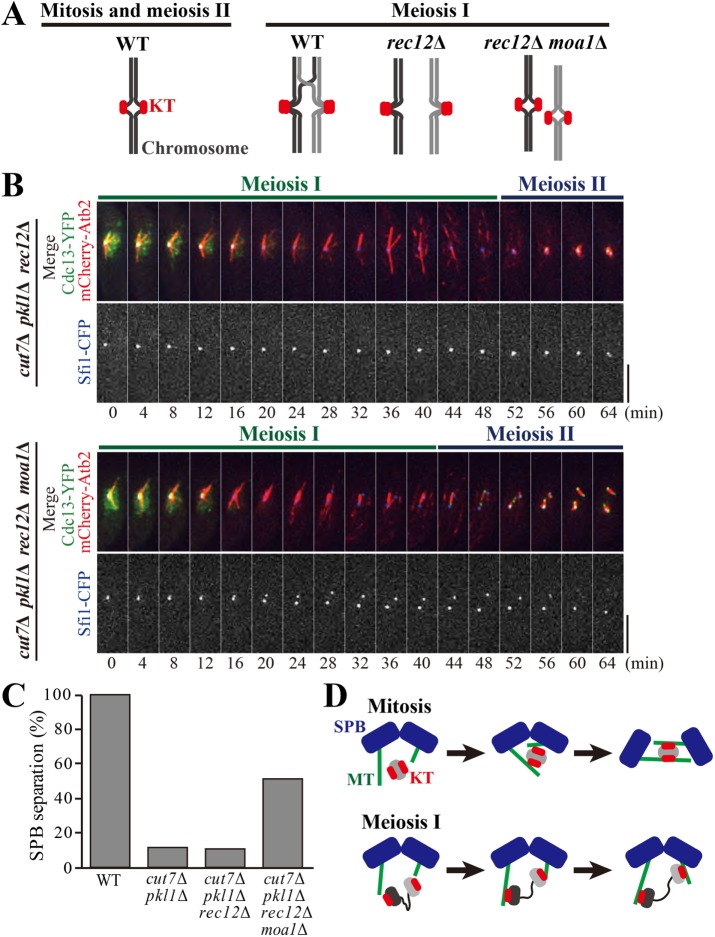
Deletion of Rec12 and Moa1 suppresses defects in the SPB separation of cut7Δ pkl1Δ cells. (A) Schematics for chromosome configuration in mitosis (WT) and meiosis I of WT, rec12Δ, and rec12Δ moa1Δ cells. For meiosis I, a pair of homologous chromosomes (black and gray) are shown. Red, kinetochores (KT) (B) Cells expressing Cdc13-YFP (green), mCherry-Atb2 (microtubules; red), and Sfi1-CFP (SPB; blue) were observed from the onset of meiosis I in cut7Δ pkl1Δ rec12Δ (top) and cut7Δ pkl1Δ rec12Δ moa1Δ (bottom). Scale bars, 5 μm. (C) Percentages of cells that showed the separation of SPBs in meiosis I in indicated strains, as calculated from observations in B (WT, n = 10 cells; cut7∆ pkl1∆, n = 17; cut7∆ pkl1∆ rec12∆, n = 9; cut7∆ pkl1∆ rec12∆ moa1∆, n = 25). (D) Models as to how kinetochores serve as a fulcrum for microtubules emanated from two SPBs. Mitosis, gray ovals indicate sister chromatids of a chromosome. Meiosis I, Gray and black ovals denote homologous chromosomes in a bivalent, the kinetochores of which are loosely connected (denoted by a wavy line). Green filaments denote microtubules (MT).
This indicated that chromosomes associated with the spindle might act as a pivot point for the spindle structure. The fulcrum of the pivot point could be kinetochores, as they form the site for physical attachment to microtubules.
The polarity of kinetochores is key for outward force generation
In mitosis and meiosis II, kinetochores of sister chromatids are oriented in a back-to-back configuration (Mitosis, Figure 3A) called biorientation (reviewed in Yamagishi et al., 2014). In meiosis I, sister kinetochores are united so that microtubules from either of the spindle poles can associate (mono-orientation), and homologues are paired as bivalent in a tetrad connected by cross-overs (Meiosis I, WT, Figure 3A).
SPO11/Rec12 is a protein that introduces double-strand breaks to meiotic chromosomes, thereby facilitating meiotic cross-overs and pairing (Cao et al., 1990; Lin and Smith, 1994; Keeney et al., 1997; Cervantes et al., 2000). In the absence of SPO11/Rec12, homologues are never paired (called univalent), although kinetochores remain mono-oriented (rec12∆, Figure 3A) (Yokobayashi and Watanabe, 2005; Sakuno et al., 2011). The monopolin protein Moa1 is responsible for the mono-orientation of sister kinetochores in meiosis I (Yokobayashi and Watanabe, 2005). Therefore, the removal of Moa1 in rec12∆ cells causes the biorientation of sister kinetochores without forming a bivalent to mimic the chromosome configuration of mitosis (rec12∆ moa1∆, Figure 3A).
To investigate whether chromosome configuration in meiosis I causes the force imbalance in cut7Δ pkl1Δ cells, Rec12 was first removed from the cells and disconnected homologues. The majority of cut7Δ pkl1Δ rec12Δ cells (∼90%) terminated meiosis I without separating SPBs, as in cut7Δ pkl1Δ cells, suggesting that chiasma formation per se is not the reason for force imbalance (cut7Δ pkl1Δ rec12Δ, Figure 3, B and C). When Rec12 and Moa1 were simultaneously removed from cut7∆ pkl1∆ cells, 52% of the cells successfully separated SPBs in meiosis I (16 min, cut7Δ pkl1Δ rec12Δ moa1Δ; Figure 3, B and C). Therefore, the conversion of chromosome configuration from meiosis I to mitosis rebalanced the forces inside the spindle.
In moa1∆ cells, sister kinetochores are converted to the bioriented pattern in meiosis I, although homologous chromosomes are still tied as bivalent (Yokobayashi and Watanabe, 2005) (Supplemental Figure S3A). The cut7∆ pkl1∆ moa1∆ cells frequently recovered SPB separation in meiosis I, which is similar to the case of cut7Δ pkl1Δ rec12Δ moa1Δ cells (Supplemental Figure S3, B and C). This means that the biorientation of kinetochores, no matter whether bivalent or univalent, drives the outward force for SPB separation.
Therefore, artificially bioriented kinetochores restore SPB separation in meiosis I lack both Cut7 and Pkl1. Although cut7Δ pkl1Δ cells without Klp2 and Rec12–Moa1 both showed SPB separation in the prometaphase of meiosis I, they showed distinct behavior in spindle elongation later in the anaphase. The cut7∆ pkl1∆ klp2∆ successfully elongated the spindle (see Figure 2B), whereas cut7Δ pkl1Δ rec12∆ moa1∆ (and cut7Δ pkl1Δ moa1∆) failed to do so, which instead resulted in spindle collapse (28–32 min, Figure 3B, and 24–28 min, Supplemental Figure S3B). As both moa1∆ and rec12∆ moa1∆ cells often displayed lagging chromosomes in the anaphase because of merotelic attachment between microtubules and kinetochores (Yokobayashi and Watanabe, 2005), anaphase spindle elongation might be hampered in those mutants including moa1∆.
In conclusion, in meiosis I without Cut7 and Pkl1, the outward forces that separate SPBs are not fully generated because of the mono-oriented kinetochores of bivalent chromosomes.
In mitosis, sister kinetochores attach to microtubules emanating from two SPBs. Then, kinetochores might be able to serve as a fulcrum for microtubules to generate a repulsive force for SPB separation (Mitosis, Figure 3D). Notably, the bipolar attachment of microtubules to kinetochores is not fully established in the early stages of mitosis. Therefore, the kinetochore-mediated outward force may not be generated through the “end-on” attachment of microtubules to kinetochores. Instead, sister kinetochores first attach to the lateral surfaces of microtubules from two SPBs. Even before conversion to end-on attachment, lateral attachment might be able to interlock the antiparallel microtubules, thereby generating the repulsive force for SPB separation (Figure 3D; see Discussion).
In this context, the force imbalance in cut7∆ pkl1∆ cells during meiosis I may be interpreted as a lack of the repulsive force at kinetochores because homologous kinetochores in a bivalent are relaxed. Figure 3D (Meiosis I) illustrates this situation; gray and black ovals denote homologous chromosomes of a bivalent, and their kinetochores are loosely connected (drawn as a wavy line).
Kinetochore positioning affects the generation of a repulsive force for SPB separation
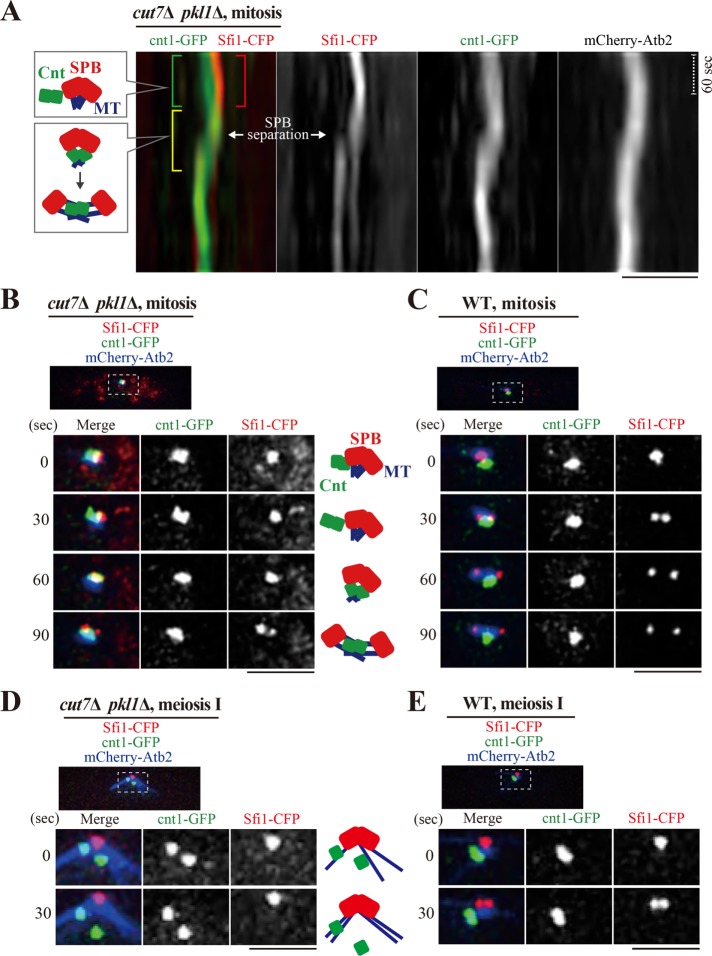
The cnt1-GFP system was employed to investigate whether kinetochores indeed generate the repulsive force to separate SPBs. The central core regions (cnt) of the centromeres in chromosome 1 were selectively visualized with GFP (Sakuno et al., 2009). We monitored the positional relationship between sister kinetochores (cnt1-GFP) and SPBs (Sfi1-CFP) when cut7∆ pkl1∆ (and WT) cells enter mitosis (Figure 4, A–C). Figure 4A displays kymographs derived from images of the cut7∆ pkl1∆ cells shown in Figure 4B. Although cnt1-GFP (green) and SPBs (red) were closely located, on entry into mitosis, the cnt1-GFP foci stood aside two SPBs (0–30 s, Figure 4B). In the corresponding kymographs (Figure 4A), the green and red brackets denote separate side-by-side positioning of sister kinetochores and SPBs, which was observed before SPB separation. This off-site location of the kinetochores implies that sister kinetochores at this stage are unable to serve as a fulcrum to generate a repulsive force through microtubules. After a while, the cnt1-GFP foci overlap with Sfi1-CFP when two SPBs separate (yellow bracket, Figure 4A; 60 s, Figure 4B). This correlation suggests that the location of the sister kinetochores is key for the generation of a repulsive force to trigger SPB separation.
FIGURE 4:
Position of kinetochores and SPBs in mitosis and meiosis I. Location of SPBs (Sfi1-CFP; red) and centromeres of chromosome 1 (cnt1-GFP; green) together with microtubules (mCherry-Atb2; blue) were visualized in the cut7Δ pkl1Δ and WT cells. (A, B) A representative cut7Δ pkl1Δ cell in mitosis. The positional relationship between kinetochores and SPBs over time is shown in kymographs (A) and in time-lapse sequential images (B). In kymographs (A), the mCherry-Atb2 image is not merged. Green and red brackets denote separate positioning of cnt1-GFP and Sfi1-CFP. A yellow bracket denotes overlap of cnt1-GFP and Sfi1-CFP signals. The timing of SPB separation is pinpointed. Corresponding schematics for SPBs, centromeres (Cnt), and microtubules are illustrated (left). The dotted line corresponds to 60 s. In B, the dotted inset is shown below as time-lapse images. Corresponding schematics are shown (right). Time 0 is defined as when nuclear microtubules started to assemble. (C) A mitotic WT cell. (D, E) cut7Δ pkl1Δ (D) and WT (E) cells on entry into meiosis I. Scale bars, 2 μm.
Then, the relaxation of kinetochores in a bivalent during meiosis I was confirmed using the cnt1-GFP system (Figure 4, D and E). On entry into meiosis I, a radial array of microtubules was assembled to capture and reposition centromeres (Kakui et al., 2013) in WT and cut7∆ pkl1∆ cells (Figure 4, D and E). Homologous kinetochores were often observed as two separate dots (for instance, Figure 4D; ∼120 s; Supplemental Figure S4), reflecting that the linkage of homologues is based on chiasmata formed in the arm region of chromosomes, but not on the centromeric cohesion between homologues (Moore, 1998; Siomos and Nasmyth, 2003). In WT cells, SPB separation efficiently occurred using the standard Cut7-dependent machinery (Figure 4E). As the major machinery is absent in cut7∆ pkl1∆ cells, the homologous kinetochores eventually behaved separately and failed to be positioned together at SPBs (Figure 4D and Supplemental Figure S4). This visually demonstrates that homologous kinetochores are unable to serve as a fulcrum to generate a repulsive force to separate SPBs. Therefore, it was concluded that kinetochores generate an outward force in mitosis, but not efficiently in meiosis.
Centromeric cohesion is required for bipolar spindle assembly in cut7Δ pkl1Δ cells
In mitosis, sister kinetochores are tightly tethered by mitotic cohesin at centromeres (Tomonaga et al., 2000; Bernard et al., 2001; Nonaka et al., 2002; Sakuno et al., 2009) (WT, Figure 5A). It was assumed that in mitosis, the centromeric cohesion between sister chromatids contributes to the generation of the outward repulsive force to separate SPBs in cut7Δ pkl1Δ cells.
FIGURE 5:
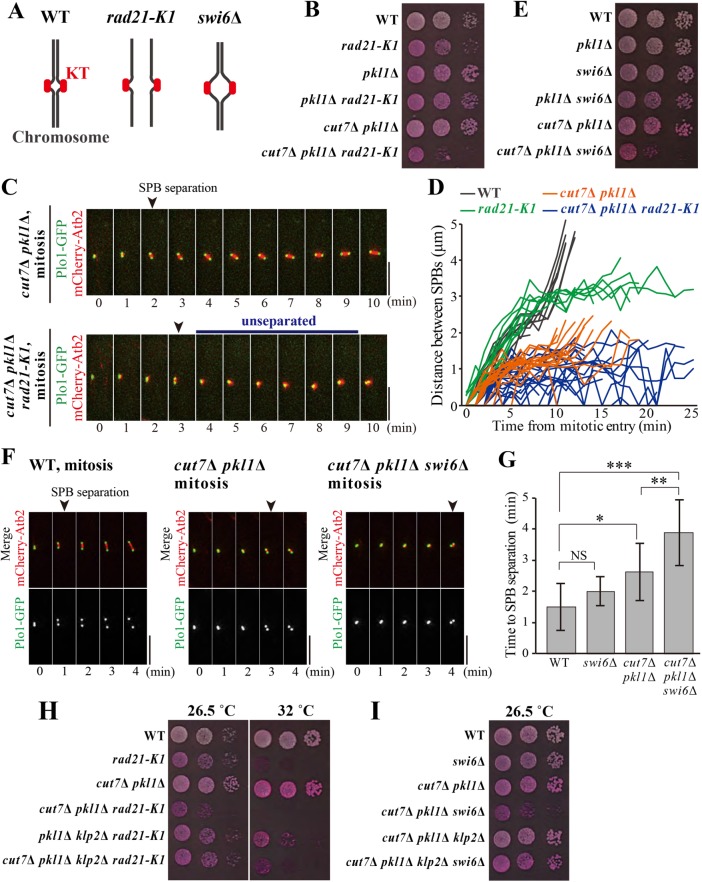
Centromeric cohesion is required for SPB separation in cut7Δ pkl1Δ cells. (A) Schematics for chromosome configurations in WT, rad21-K1, and swi6Δ cells in mitosis. (B) Strains with indicated genotypes were spotted onto agar plates to test their growth. Tenfold serial dilutions were spotted. The plates contained Phloxine B, which stains dead or sick cells. (C) Time-lapse images for mitotic cells of cut7Δ pkl1Δ and cut7Δ pkl1Δ rad21-K1. Microtubules and SPBs were visualized with mCherry-Atb2 and Plo1-GFP, respectively. Time 0 (min) corresponds to when a nuclear punctate signal of mCherry-Atb2 first emerged. The timing of SPB separation is indicated with arrowheads. (D) Temporal kinetics of the distance between two SPBs. Mitotic cells of WT (gray; n = 10 cells), rad21-K1 (green; n = 11), cut7Δ pkl1Δ (orange; n = 16), and cut7Δ pkl1Δ rad21-K1 (blue; n = 24) were observed at 36°C. (E) Plate-based growth assay as shown in B. (F) Time-lapse images of WT (left), cut7Δ pkl1Δ (center), or cut7Δ pkl1Δ swi6Δ cells (right) expressing Plo1-GFP (SPB; green) and mCherry-Atb2 (microtubules; red), as in C. The timing of SPB separation is indicated with arrowheads. (G) Time required for SPB separation since a nuclear punctate signal of mCherry-Atb2 first appeared (mean ± SD WT, n = 8 cells; swi6∆, n = 10; cut7∆ pkl1∆, n = 18; cut7∆ pkl1∆ swi6∆, n = 9). *P < 0.01; **P < 0.005; ***P < 0.001; NS, not significant (Welch’s t test). (H, I) Plate-based growth test using 10-fold serial dilutions of indicated strains as in B and E. Scale bars, 5 μm.
The function of the mitotic cohesin Rad21 connecting the sister chromatids was impaired by introducing the rad21-K1 temperature-sensitive hypomorphic mutation to test this assumption (Tatebayashi et al., 1998) (Figure 5A). Indeed, at the restrictive temperature, the rad21-K1 mutant precociously separated centromeres before the anaphase (Bernard et al., 2001) (Supplemental Figure S5). The rad21-K1 mutation was introduced to the cut7∆ pkl1∆ cells, and the triple mutant cut7Δ pkl1Δ rad21-K1 showed a severe synthetic growth defect even at 26.5°C, whereas both cut7∆ pkl1∆ and rad21-K1 mutants grew (Figure 5B).
The cut7Δ pkl1Δ rad21-K1 mitotic cells visualizing microtubules and mitotic SPBs with mCherry-Atb2 and Plo1-GFP (Polo-like kinase) (Bähler et al., 1998b), respectively, were then observed. In the majority of mitotic cut7Δ pkl1Δ cells, SPB separation occurred, and the distance between the two SPBs remained constant at ∼1.5 µm for 5–10 min (Figure 5, C and D). On the contrary, the distance between SPBs fluctuated in the cut7Δ pkl1Δ rad21-K1 cells. Two SPBs were located alongside each other most of the time during observation, and SPBs were reconnected even if once separated (Figure 5, C and D). These results imply that the impaired cohesion between sister chromatids is a reason for a loss of the outward force in the spindle.
In the rad21-K1 mutant, cohesion between the sister chromatids was lost. To further specify whether cohesion at centromeres generates the outward tension, the swi6Δ strain was introduced to dismiss cohesin from the pericentromeric regions. Swi6/HP1 is a heterochromatin protein, which recruits cohesin to the pericentromeric regions (Ekwall et al., 1995; Bernard et al., 2001; Nonaka et al., 2002). As reported previously, the core centromeres of sister chromatids (indicated by cnt1-GFP) behave in a discrete manner even before the anaphase in swi6Δ cells, confirming that the linkage between core centromeres is relaxed by the removal of Swi6 (Ekwall et al., 1995; Bernard et al., 2001; Nonaka et al., 2002) (Figure 5A and Supplemental Figure S5D). When Swi6 was removed from the cut7∆ pkl1∆ strain, the cut7Δ pkl1Δ swi6Δ triple mutant showed severe growth defects, although neither cut7∆ pkl1∆ nor swi6∆ showed growth defects in these conditions (Figure 5E). The frequency of mitotic cells showing the short spindle (≤1.5 µm) increased when swi6 was removed from cut7∆ pkl1∆ cells, which was comparable to when the rad21-K1 mutation was introduced (Supplemental Figure S6).
In the live-cell imaging of WT cells, two SPBs separated and the bipolar spindle was stably assembled to a size of ∼2 µm immediately after mitotic entry (1 min, WT; Figure 5F). SPB separation took longer in the cut7Δ pkl1Δ cells (3 min, Figure 5F), reflecting the reduction of outward forces for SPB separation. Furthermore, SPBs in cut7Δ pkl1Δ swi6Δ cells remained unseparated for longer than in the cut7Δ pkl1Δ cells (3.9 ± 1.1 min vs. 2.6 ± 0.9 min; Figure 5G), indicating that centromeric cohesion promotes SPB separation in the cut7Δ pkl1Δ mutant. This may mimic the chromosome state in meiosis I of the cut7∆ pkl1∆ cells, in which the outward force is not fully generated due to the relaxed connection between the homologous kinetochores. The defects in SPB separation in cut7Δ pkl1Δ swi6Δ cells were partial, probably because sister-centromeres were only partially loosened by the removal of Swi6.
As the force balance in meiosis I was ameliorated by the removal of the klp2 gene from the cut7Δ pkl1Δ cells (Figure 2), it was reasoned by analogy that the deletion of klp2 could suppress defects in cut7Δ pkl1Δ swi6Δ cells. As expected, the mitotic growth of both cut7Δ pkl1Δ rad21-K1 and cut7∆ pkl1∆ swi6∆ cells was partially restored by the removal of the klp2 gene (Figure 5, H and I). This implies that inward and outward forces inside the spindle are rebalanced there so that cells can assemble the bipolar spindle. These results demonstrate that centromeric cohesion contributes to generating the outward forces and bipolar spindle assembly in the absence of Cut7 and Pkl1.
The functional outer kinetochore is required for efficient SPB separation
Although the results using mutants of cohesin and Swi6 imply that centromeric cohesion could be key for generation of the outward force, it is alternatively possible that the merotelic attachment of microtubules to kinetochores impedes SPB separation, as such attachment is frequently observed in cohesin mutants (Courthéoux et al., 2009). It was thought that the effects of merotelic attachment to SPB separation in pro-prometaphase are minor because merotelic attachment is thought to affect microtubule force balance, particularly in the anaphase when chromosomes with normal attachment have already finished segregation (Cimini et al., 2004; Courthéoux et al., 2009). Indeed, although moa1∆ rec12∆ often causes merotelic attachment, it restored SPB separation in cut7∆ pkl1∆ in pro-prometaphase, and spindle elongation defects could be seen only in the anaphase (see Figure 3B).
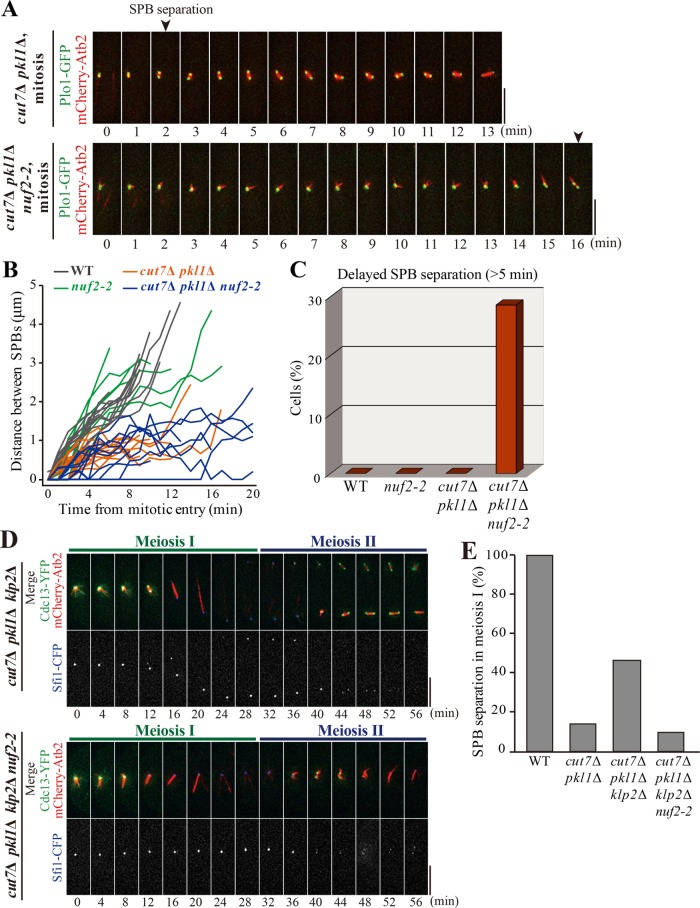
To further clarify this point, it is necessary to investigate more directly whether kinetochore generates the outward force for SPB separation through microtubules. Therefore, the mutant of Nuf2, a component of the Ndc80 complex located in the outer kinetochore, which serves as the interface for kinetochore-microtubule attachment, was employed (Wigge and Kilmartin, 2001). Then, the temperature-sensitive nuf2-2 mutant (Nabetani et al., 2001) was crossed with the cut7∆ pkl1∆ mutant to investigate whether SPB separation is affected by defective outer kinetochores.
The cut7∆ pkl1∆ nuf2-2 cells were delayed in SPB separation at the onset of mitosis (Figure 6A). The cut7∆ pkl1∆ (nuf2+) cells started to separate SPBs within 5 min after the mitotic onset, whereas more than 25% of the cut7∆ pkl1∆ nuf2-2 cells did not separate SPBs within 5 min (Figure 6, B and C). Once SPBs were separated, the spindle was occasionally longer than 1 µm. As the nuf2-2 mutant is not completely null, the effects of the mutation might be partial. Nonetheless, the nuf2-2 mutation caused severe delays in the initial stage of SPB separation in cut7∆ pkl1∆ cells, indicating that the outer kinetochore complex plays a central role in SPB separation.
FIGURE 6:
Functional kinetochore is required for generation of the outward force. (A) Time-lapse images of Plo1-GFP and mCherry-Atb2 in the mitosis of cut7∆ pkl1∆ cells and cut7∆ pkl1∆ nuf2-2 cells at 36°C. Arrowheads indicate the timing of SPB separation. Time 0 corresponds to the onset of mitosis. (B) Kinetics of distance between two SPBs in indicated strains. (C) Percentages of cells with delayed SPB separation in each strain. Mitotic cells in which SPBs did not separate for more than 5 min after the mitotic onset are counted (WT, n = 16 cells; nuf2-2, n = 7; cut7∆ pkl1∆, n = 20; cut7∆ pkl1∆ nuf2-2, n = 14). (D) Cells expressing Cdc13-YFP (green), mCherry-Atb2 (microtubules; red), and Sfi1-CFP (SPB; blue) were observed at 36°C from the onset of meiosis I in cut7Δ pkl1Δ klp2Δ (top) and cut7Δ pkl1Δ klp2Δ nuf2-2 (bottom). (E) Percentages of cells that showed SPB separation in meiosis I in indicated strains, as calculated from observations in D (WT, n = 8 cells; cut7∆ pkl1∆, n = 14; cut7∆ pkl1∆ klp2∆, n = 15; cut7∆ pkl1∆ klp2∆ nuf2-2, n = 10). Scale bars, 5 µm.
This was also tested in meiosis I. As shown above, cut7∆ pkl1∆ klp2∆ cells showed SPB separation in meiosis I (Figures 2, B and C, and 6D), but cut7∆ pkl1∆ klp2∆ nuf2-2 cells were mostly defective in SPB separation at meiosis I (Figure 6, D and E). Therefore, it is concluded that SPB separation in meiosis I in the absence of kinesin-5 (Cut7) and both kinesin-14s (Pkl1 and Klp2) depends on kinetochore-microtubule attachment through the outer kinetochore complex.
DISCUSSION
The outward force mediated by kinetochores
Visualization of kinetochores and the genetic experiments demonstrate that the tight connection between sister kinetochores as well as the functional outer kinetochore complex are the bases for a repulsive outward force for microtubules to separate SPBs. This is particularly evident in cut7Δ pkl1Δ cells lacking two opposing kinesins (Figure 7). It is proposed that kinetochores contribute to the generation of outward forces for SPB separation in mitosis, but kinetochores fail to do so in meiosis I, because the interkinetochore tension in a meiotic bivalent (i.e., between homologous kinetochores but not sister kinetochores) is low, compared with that in a mitotic univalent (Figure 7). SPBs in cut7Δ pkl1Δ cells were separated later in meiosis II, in which kinetochore configuration is reconverted to mitotic style (bioriented kinetochores in a univalent). Moreover, the artificial conversion of kinetochore geometry in meiosis I from the mono-oriented to mitotic bioriented style (by use of moa1∆ and rec12∆ moa1∆) partially restored SPB separation (Figure 3, A–D, and Supplemental Figure S3, A–C). Therefore, it is concluded that the kinetochore-mediated outward force is relatively low in meiosis I due to the loose linkages between homologues (Figure 7).
FIGURE 7:
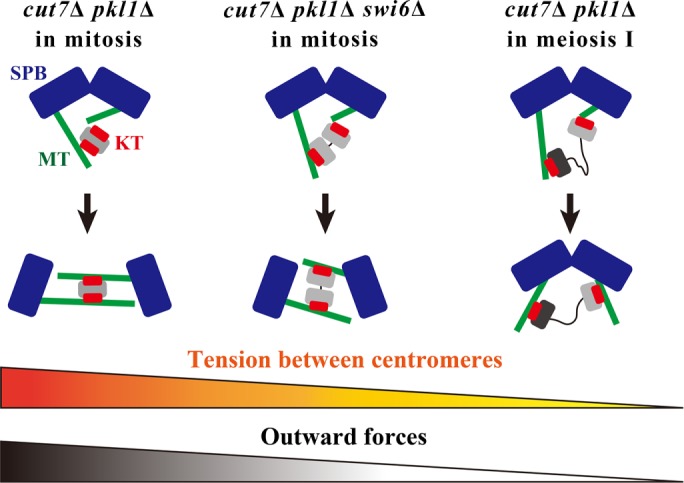
Model for the relationship between centromeric cohesion and the outward force to separate SPBs in cut7Δ pkl1Δ. The behavior of kinetochores and SPBs in cut7∆ pkl1∆ mitotic cells (left), cut7∆ pkl1∆ swi6∆ mitotic cells (center), and cut7∆ pkl1∆ cells in meiosis I (right) are shown. Gray and black ovals denote a pair of homologous chromosomes. Wavy lines indicate that the connection between the chromosomes is relaxed. See text for details.
Balance of inward and outward forces
As previously shown in studies on S. pombe mitosis, SPB separation is orchestrated by a number of outward and inward forces within the spindle (Pidoux et al.,1996; Rincon et al., 2017; Yukawa et al., 2017). In the presence of Cut7, the rad21-K1, swi6∆, and nuf2-2 single mutants separated SPBs almost normally (Figures 5, D and G, and 6B). Therefore, Cut7 appears to be the major outward-force generator that cross-links and slides microtubules in WT cells, and the kinetochore may serve as an additional factor to further assist the outward force.
In the absence of both Cut7 and Pkl1 in mitosis, outward forces dominate and therefore SPBs are separated. Tight cohesion of sister kinetochores contributes to the generation of the outward forces for efficient SPB separation.
However, in meiosis I, the outward forces do not dominate. SPBs do not separate in the double knockout, because kinetochores in a bivalent are loosely connected. Recent studies indicate that the microtubule-bundling factor Ase1 is responsible for the generation of the outward force that separates SPBs in the absence of both Cut7 and Pkl1 (Rincon et al., 2017; Yukawa et al., 2017). Based on this notion, defects in SPB separation in meiosis I could be interpreted as an absence of Ase1 functions at this stage. Inward forces driven by Klp2 (Yukawa et al., 2018) might be stronger than in mitosis to hold back SPB separation, or Ase1 might not fully function in meiosis I, although those possibilities are not mutually exclusive.
Indeed, the removal of Klp2 in cut7∆ pkl1∆ cells restored SPB separation, supporting this notion. Regarding Ase1, it is speculated that the transient bilateral attachment of kinetochores to microtubules generates the interlock of antiparallel microtubules (see Figure 7), thereby providing a base for Ase1 localization to promote SPB separation. In meiosis I of the cut7∆ pkl1∆ cells, flexible cohesion in a bivalent does not produce sufficient interdigitating microtubules. Therefore, Ase1 might not be able to fully function at this stage.
Lack of the kinetochore-mediated outward force in meiosis I
Kinetochore positioning in the nucleus is regulated distinctly in mitosis and meiosis. In the interphase of the mitotic cell cycle, kinetochores are tethered at SPBs in the nucleus (Funabiki et al., 1993). Therefore, it is reasonable to utilize kinetochores as a pivot point for bipolar spindle assembly when spindle microtubules are emanated from SPBs on entry into mitosis.
In contrast, kinetochores locate distal from SPBs and are scattered in the nucleus before entry into meiosis I (Chikashige et al., 1994). They are repositioned at the onset of meiosis I since distal kinetochores are captured and retrieved by a radial array of microtubules assembled from unseparated SPBs (Kakui et al., 2013).
As the radial array is reminiscent of the monopolar “V-shaped” spindle seen in mitosis of the cut7 hypomorphic mutant (Hagan and Yanagida, 1990), the radial array of microtubules in meiosis may need to be assembled from unseparated SPBs that are juxtaposed to each other.
In this study, two possible explanations have been identified as to how the monopolar radial array of microtubules are assembled specifically at the onset of meiosis I. First, it is possible that Cut7 is not fully functional at the onset of meiosis I, although Cut7 is localized to SPBs at this stage (unpublished data). The other possibility, although not mutually exclusive, is that the meiosis-specific configuration of chromosomes prevents SPB separation, thereby assisting the assembly of the monopolar radial array of microtubules. Kinetochores in meiosis I are connected loosely in a bivalent and are distal from SPBs. These circumstances do not allow the generation of kinetochore-mediated outward forces that promote SPB separation. This brings a substantial delay in SPB separation, which promotes the assembly of the monopolar array of microtubules instead, thereby providing an opportunity for kinetochore repositioning. SPBs then start to separate as soon as the radial array diminishes (Kakui et al., 2013).
Moreover, the close proximity of kinetochores and SPBs promotes bipolar spindle assembly in meiosis I (Fennell et al., 2015). These observations indicate that although meiotic kinetochores in a bivalent are relaxed, kinetochores contribute to separate SPBs, albeit partially, as soon as they are retrieved to SPBs. Therefore, the positioning and configuration of kinetochores control the timing of SPB separation.
MATERIALS AND METHODS
Yeast strains, media, and genetics
S. pombe strains used in this study are listed in Supplemental Table S1. For vegetative growth, cells were grown in the rich medium YE5S containing yeast extract, glucose, and five supplements (adenine, leucine, uracil, lysine, and histidine). To induce mating, meiosis, and sporulation, homothallic (h90) cells were grown in YE5S and then spotted onto sporulation agar (SPA) plates. For plate-based growth assays, 10-fold serial dilutions of cell suspensions (ranging from 1 × 104 to 1 × 102 cells) were spotted onto YE5S plates with or without Phloxine B. The plates were incubated at 26.5°C or 32°C. Standard methods for yeast genetics and PCR-based gene targeting for gene deletion and the tagging of fluorescent proteins were used (Moreno et al., 1991; Bähler et al., 1998a; Sato et al., 2005, 2009). For the visualization of microtubules, the fusion gene of mCherry and atb2 (α2-tubulin) placed under the native promoter and terminator of the atb2+ gene was integrated into a chromosome as an extra copy of endogenous atb2+(the resultant strain is referred to as “Z2-mCherry-atb2”) (Ohta et al., 2012).
Microscopy
Cells on SPA (for 24 h) were fixed with ethanol and then mounted on the glass slide using VECTASHIELD mounting medium with DAPI (Vector Laboratories) to visualize DNA in spores. Cells were spotted onto SPA and incubated at 30°C for 18 h and fixed with 3.2% formaldehyde (FUJIFILM Wako Pure Chemical) and mounted similarly to visualize DNA in the mes1Δ mutant. Images were taken using the DeltaVision-SoftWoRx system (Applied Precision).
For the live-cell imaging of meiotic cells, homothallic cells were spotted onto SPA and incubated at 30°C for 7–10 h, except for the nuf2-2 strains incubated on SPA at 30°C for 5 h and shifted to 36°C for 2 h prior to observation. For the observation of mitosis, most of the strains were cultured in the YE5S liquid medium until the mid–log phase at 30°C. Exceptionally, the rad21-K1 and nuf2-2 strains were cultured in the YE5S liquid medium at 25°C and shifted to 36°C for 2.5 h (rad21-K1) or 4 h (nuf2-2) prior to observation. Cells were then mounted on a glass-bottom dish (Iwaki glass) precoated with lectin from Glycine max (Sigma) and filled with a synthetic medium with adenine, leucine, uracil, lysine, and histidine (SD5S) for the observation of mitosis, alternatively with minimal medium without a nitrogen source supplemented with uracil and leucine for meiosis. Filming was performed using the DeltaVision-SoftWoRx system (Applied Precision) as described previously (Sato et al., 2009). The rad21-K1 and nuf2-2 mutants were incubated at 36°C during observation. Images were acquired as serial sections along the z-axis and stacked using the quick projection algorithm in SoftWoRx. Kymographs were generated using the SoftWoRx software, and image contrasts were adjusted with Adobe Photoshop CC 2019.
Immunofluorescence
Cells were cultured in the YE5S liquid medium until the mid–log phase at 25°C and then shifted up to 36°C. After 4 h, cells were fixed with 3.2% formaldehyde (FUJIFILM Wako Pure Chemical). Samples were then stained with the mouse anti–α-tubulin antibody B-5-1-2 (Santa Cruz Biotechnology) as a primary antibody and with Goat anti-mouse immunoglobulin G (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (Thermo Fisher Scientific). The samples were then sealed with VECTASHIELD mounting medium with DAPI (Vector Laboratories).
Statistical analysis
Welch’s t test was used to evaluate the statistical significance of the difference between two values. All data are presented as the mean ± SD.
Supplementary Material
Acknowledgments
We thank Yasushi Hiraoka, Takeshi Sakuno, Yoshinori Watanabe, Yasutaka Kakui, and the National Bioresource Project of Japan for strains. We are grateful to Mika Toya for discussion. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI JP25291041, JP15H01359, JP16H04787, JP16H01317, and JP18K19347 to M.S. This study was also supported by The Uehara Memorial Foundation and by Waseda University grants for Special Research Projects 2017B-242, 2017B-243, 2018B-222, and 2019C-570 to M.S. This work was partly supported by the JSPS Core-to-Core Program, A.Advanced Research Networks.
Abbreviations used:
- cnt
central core region
- DAPI
4′,6-diamidino-2-phenylindole
- GFP/YFP/CFP
green/yellow/cyan fluorescent protein
- MAP
microtubule-associated protein
- SPA
sporulation agar
- SPB
spindle pole body
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0366) on September 18, 2019.
REFERENCES
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. (1998a). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast , 943–951. [DOI] [PubMed] [Google Scholar]
- Bähler J, Steever AB, Wang YL, Pringle JR, Gould KL. (1998b). Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol , 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Javerzat JP. (2001). Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol , 522–526. [DOI] [PubMed] [Google Scholar]
- Bresch C, Muller G, Egel R. (1968). Genes involved in meiosis and sporulation of a yeast. Mol Gen Genet , 301–306. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. (1990). A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell , 1089–1101. [DOI] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. (2000). Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell , 883–888. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. (1994). Telomere-led premeiotic chromosome movement in fission yeast. Science , 270–273. [DOI] [PubMed] [Google Scholar]
- Cimini D, Cameron LA, Salmon ED. (2004). Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol , 2149–2155. [DOI] [PubMed] [Google Scholar]
- Courthéoux T, Gay G, Gachet Y, Tournier S. (2009). Ase1/Prc1-dependent spindle elongation corrects merotely during anaphase in fission yeast. J Cell Biol , 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A, Zarzov P, Nurse P. (2001). In vivo localisation of fission yeast cyclin-dependent kinase cdc2p and cyclin B cdc13p during mitosis and meiosis. J Cell Sci , 2627–2640. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. (1995). The chromodomain protein Swi6: a key component at fission yeast centromeres. Science , 1429–1431. [DOI] [PubMed] [Google Scholar]
- Fennell A, Fernández-Álvarez A, Tomita K, Cooper JP. (2015). Telomeres and centromeres have interchangeable roles in promoting meiotic spindle formation. J Cell Biol , 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P. (2009). Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol , 1833–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. (1993). Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol , 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. (1990). Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature , 563–566. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. (1992). Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature , 74–76. [DOI] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS. (1993). The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol , 665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. (2005). Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature , 529–533. [DOI] [PubMed] [Google Scholar]
- Kakui Y, Sato M, Okada N, Toda T, Yamamoto M. (2013). Microtubules and Alp7-Alp14 (TACC-TOG) reposition chromosomes before meiotic segregation. Nat Cell Biol , 786–796. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. (2005). The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature , 114–118. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol , 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. (1996). A bipolar kinesin. Nature , 270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell , 375–384. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV. (2003). Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol , 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Smith GR. (1994). Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics , 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loïodice I, Staub J, Setty TG, Nguyen N-PT, Paoletti A, Tran PT. (2005). Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell , 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. (1999). Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science , 971–974. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. (2005). Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell , 3064–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DP. (1998). Chromosome segregation during meiosis: building an unambivalent bivalent. Curr Top Dev Biol , 263–299. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol , 795–823. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. (2001). A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma , 322–334. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. (1974). Chromosome segregation mechanisms. Genetics , 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SIS, Watanabe Y. (2002). Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol , 89–93. [DOI] [PubMed] [Google Scholar]
- Ohta M, Sato M, Yamamoto M. (2012). Spindle pole body components are reorganized during fission yeast meiosis. Mol Biol Cell , 1799–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted ZT, Colliver AG, Riehlman TD, Paluh JL. (2014). Kinesin-14 and kinesin-5 antagonistically regulate microtubule nucleation by γ-TuRC in yeast and human cells. Nat Commun , 5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, LeDizet M, Cande WZ. (1996). Fission yeast pkl1 is a kinesin-related protein involved in mitotic spindle function. Mol Biol Cell , 1639–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon SA, Lamson A, Blackwell R, Syrovatkina V, Fraisier V, Tran PT. (2017). Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast. Nat Commun , 15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Tada K, Watanabe Y. (2009). Kinetochore geometry defined by cohesion within the centromere. Nature , 852–858. [DOI] [PubMed] [Google Scholar]
- Sakuno T, Tanaka K, Hauf S, Watanabe Y. (2011). Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev Cell , 534–545. [DOI] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T. (2005). New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast , 583–591. [DOI] [PubMed] [Google Scholar]
- Sato M, Toya M, Toda T. (2009). Visualization of fluorescence-tagged proteins in fission yeast: the analysis of mitotic spindle dynamics using GFP-tubulin under the native promoter. Methods Mol Biol , 185–203. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM. (1999). Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol , 51–54. [DOI] [PubMed] [Google Scholar]
- Siomos MF, Nasmyth K. (2003). Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell , 423–440. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macu˚rek L, Galjart N, Medema RH. (2008). Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. Embo J , 3235–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Kato J, Ikeda H. (1998). Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics , 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, et al (2000). Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev , 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR. (2001). pkl1+and klp2+: Two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell , 3476–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesbeen RGHP, Tanenbaum ME, Medema RH. (2014). Balanced activity of three mitotic motors is required for bipolar spindle assembly and chromosome segregation. Cell Rep , 948–956. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV. (2001). The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol , 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Goto Y, Watanabe Y. (2014). Kinetochore composition and its function: lessons from yeasts. FEMS Microbiol Rev , 185–200. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Fujita A, Yamamoto M. (2005). The roles of fission yeast Ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell , 1378–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. (2005). The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell , 803–817. [DOI] [PubMed] [Google Scholar]
- Yukawa M, Kawakami T, Okazaki M, Kume K, Tang NH, Toda T. (2017). A microtubule polymerase cooperates with the kinesin-6 motor and a microtubule cross-linker to promote bipolar spindle assembly in the absence of kinesin-5 and kinesin-14 in fission yeast. Mol Biol Cell , 3647–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa M, Yamada Y, Toda T. (2019). Suppressor analysis uncovers that MAPs and microtubule dynamics balance with the Cut7/Kinesin-5 motor for mitotic spindle assembly in Schizosaccharomyces pombe. G3 (Bethesda) , 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa M, Yamada Y, Yamauchi T, Toda T. (2018). Two spatially distinct kinesin-14 proteins, Pkl1 and Klp2, generate collaborative inward forces against kinesin-5 Cut7 in S. pombe. J Cell Sci , jcs210740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.