Abstract
Stable localization of the Rheb GTPase to lysosomes is thought to be required for activation of mTOR complex 1 (mTORC1) signaling. However, the lysosome targeting mechanisms for Rheb remain unclear. We therefore investigated the relationship between Rheb subcellular localization and mTORC1 activation. Surprisingly, we found that Rheb was undetectable at lysosomes. Nonetheless, functional assays in knockout human cells revealed that farnesylation of the C-terminal CaaX motif on Rheb was essential for Rheb-dependent mTORC1 activation. Although farnesylated Rheb exhibited partial endoplasmic reticulum (ER) localization, constitutively targeting Rheb to ER membranes did not support mTORC1 activation. Further systematic analysis of Rheb lipidation revealed that weak, nonselective, membrane interactions support Rheb-dependent mTORC1 activation without the need for a specific lysosome targeting motif. Collectively, these results argue against stable interactions of Rheb with lysosomes and instead that transient membrane interactions optimally allow Rheb to activate mTORC1 signaling.
INTRODUCTION
The mTOR complex 1 (mTORC1) signaling pathway plays a major role in matching cell growth and metabolism to ongoing changes in environmental conditions. Multiple signals converge on the surface of lysosomes to regulate the activity of the Rag and Rheb small GTPases that recruit and activate mTORC1, respectively. This has led to a widely accepted two-step model for mTORC1 activation wherein Rags recruit mTORC1 to lysosomes followed by Rheb-dependent activation of mTORC1 kinase activity (Supplemental Figure S1A) (Sancak et al., 2008; Ferguson, 2015; Ben-Sahra and Manning, 2017; Saxton and Sabatini, 2017). However, even though farnesylation at its C-terminus has been widely accepted as a means of localizing Rheb to lysosomes, there is only limited direct support for significant enrichment of Rheb on lysosomes (Sancak et al., 2008, 2010; Menon et al., 2014). Furthermore, as a farnesyl group is only expected to confer transient membrane interactions without selectivity for binding to lysosomes over other organelles (Silvius and l’Heureux, 1994; Silvius et al., 2006), the underlying mechanism of lysosome targeting is unexplained.
Adding to the confusion concerning the relationship between Rheb localization and function, it has also recently been proposed that Rheb instead resides on either the endoplasmic reticulum (ER) or the Golgi and activates mTORC1 via contact sites between these organelles and lysosomes (Hao et al., 2018; Walton et al., 2018). These observations are paralleled by some older studies that also reported enrichment of overexpressed Rheb at the ER (Buerger et al., 2006; Hanker et al., 2010). The conflicting messages in these studies reveal uncertainty about both where Rheb functions within cells and how Rheb is targeted to its specific site of action.
To address these questions, we systematically investigated the relationship between Rheb localization and function in human cells. Surprisingly, our data indicate that Rheb does not require stable enrichment on a specific organelle. Instead, weak, nonselective, membrane interactions are sufficient to support mTORC1 activation. Collectively these data argue against the requirement for a stable and highly selective membrane interaction mechanism for optimal Rheb function. Our new data are not completely inconsistent with the widely accepted role for Rheb in activating mTORC1 at lysosomes. However, instead of stable residence of Rheb at lysosomes, we propose that transient membrane interactions are sufficient to satisfy the need to bring Rheb into proximity with mTORC1 that has been recruited to lysosomes via Rag-dependent mechanisms.
RESULTS AND DISCUSSION
Rheb is not enriched on the surface of lysosomes
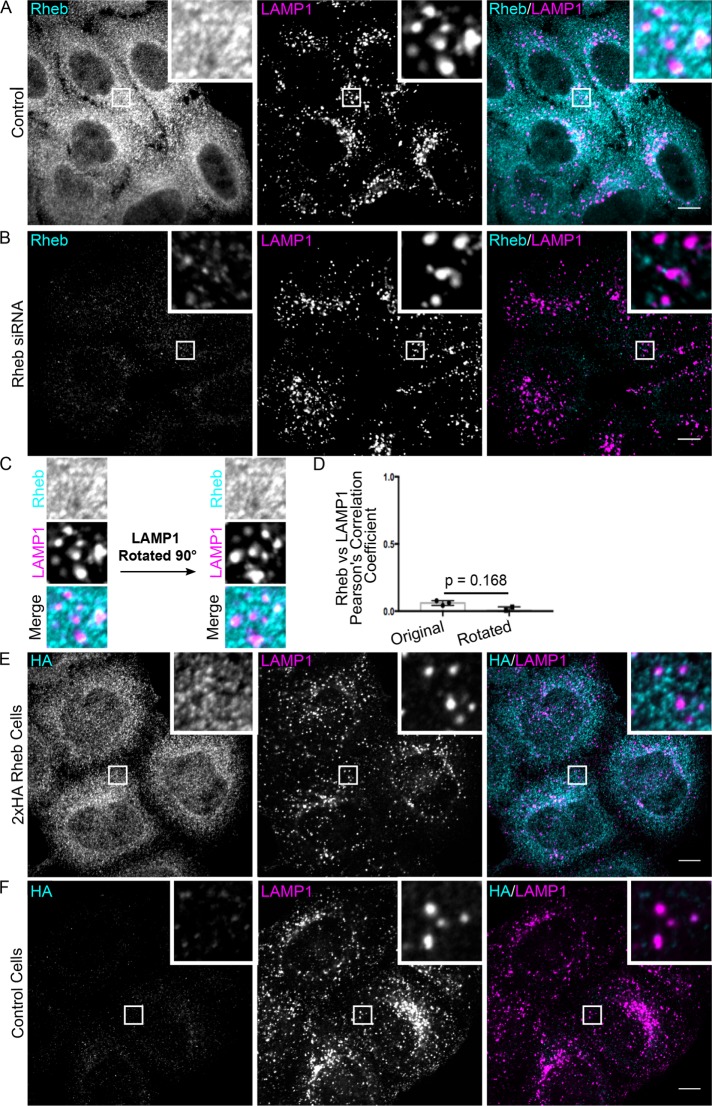
In contrast to the previously reported localization of Rheb to LAMP1-positive lysosomes in HeLa cells (Menon et al., 2014), we found that Rheb showed no major enrichment on such organelles (Figure 1A), even though we used the same combination of antibody for Rheb detection and Rheb small interfering RNA (siRNA) for verifying signal specificity (Figure 1B). This difference in conclusions arises mainly from improved resolution of individual lysosomes which allowed for a more stringent evaluation of colocalization. As Rheb is broadly found throughout cells, some Rheb inevitably overlaps with the LAMP1 signal. However, our analysis of the colocalization between Rheb and LAMP1 revealed that the degree of overlap between these two proteins was no greater than chance (Figure 1, C and D). As a control for the efficacy of the Rheb siRNA, immunoblotting experiments confirmed Rheb depletion following Rheb siRNA transfection and demonstrated that mTORC1 signaling was suppressed in these cells (Supplemental Figure S1, B–D).
FIGURE 1:
Rheb is not enriched on lysosomes. (A, B) Rheb and LAMP1 (late endosomes/lysosomes) immunofluorescence in HeLa cells that were transfected with control and Rheb siRNA, respectively. (C) Comparison of Rheb and LAMP1 colocalization in the insets from A before and after rotating the LAMP1 image by 90° to estimate the extent to which colocalization occurs by chance. (D) Quantification of the Pearson’s R value when comparing Rheb vs. LAMP1 in the original and rotated configurations as described in C (n = 3 biological replicates, 15 images quantified per replicate, t test). (E, F) Representative immunofluorescence images of anti-HA and anti-LAMP1 staining in genome edited 2xHA-Rheb HeLa cells and control HeLa cells, respectively. Scale bars, 10 μm.
To generate an alternative tool for detecting the endogenous Rheb protein, we used CRISPR-Cas9 gene editing to insert a 2xHA epitope tag immediately downstream of the start codon in the endogenous Rheb locus in HeLa cells (Supplemental Figure S1, E and F). However, the anti-HA immunofluorescence still did not show enrichment on lysosomes in these cells (Figure 1, E and F). Thus, two independent detection methods were successful in the specific immunofluorescent detection of the Rheb protein without generating a signal that exhibited any distinct lysosome enrichment.
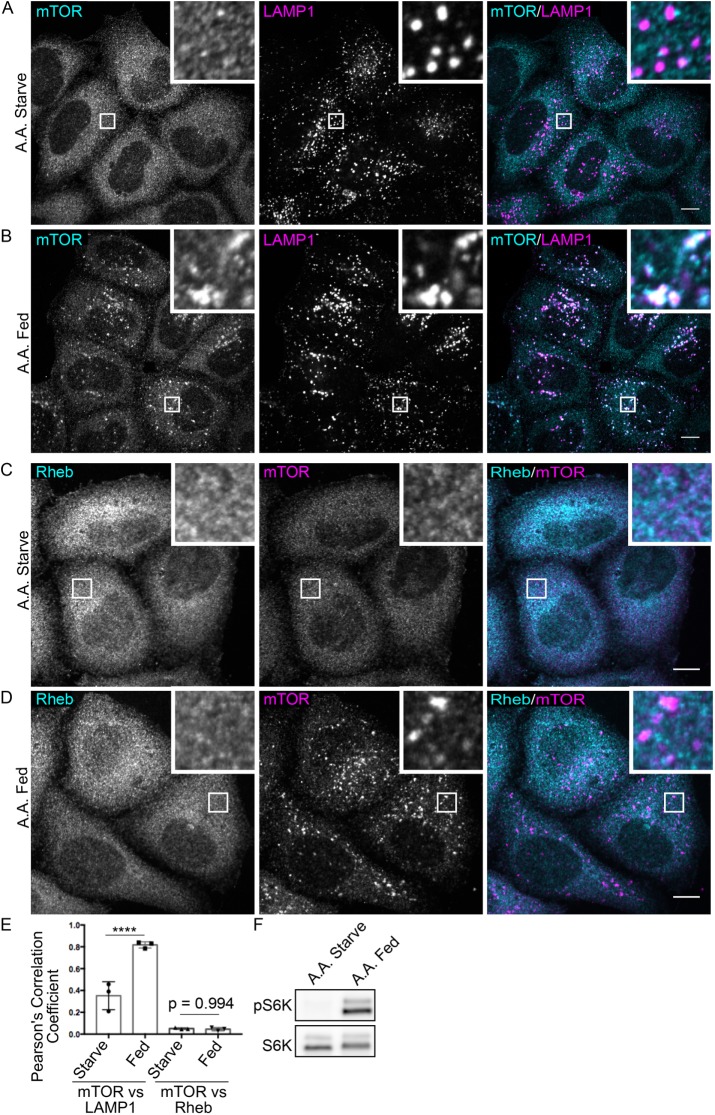
As mTOR and many mTORC1 regulatory proteins exhibit dynamic changes in their levels at lysosomes in response to acute changes in amino acid availability (Supplemental Figure S1A; Saxton and Sabatini, 2017), we next examined the effect of amino acid starvation and refeeding on Rheb localization. In contrast to mTOR, which showed enhanced recruitment to LAMP1-positive late endosomes and lysosomes in response to the refeeding of starved cells with amino acids (Figure 2, A and B, and Supplemental Figure S2, A and B), double labeling for mTOR and Rheb revealed that Rheb localization was not responsive to amino acid feeding and failed to coenrich with lysosomal mTOR puncta (Figure 2, C–E). Nonetheless, we still observed that mTORC1 signaling was activated in response to this amino acid refeeding protocol (Figure 2F). These results show a dramatic difference in the ability of mTOR and Rheb to localize to lysosomes and are surprising given the expectation that mTORC1 gets recruited to lysosomes in order to be activated by Rheb.
FIGURE 2:
Regulated recruitment of mTOR to lysosomes is not accompanied by significant colocalization with Rheb. (A, B) Immunofluorescence analysis of mTOR and LAMP1 localization in starved and amino acid refed cells, respectively. (C, D) Immunofluorescence analysis of mTOR and Rheb localization in starved and amino acid refed cells, respectively. (E) Quantification of the colocalization observed in experiments related to A–D (n = 3 biological replicates, 15 images quantified per replicate, one-way analysis of variance (ANOVA) with a Sidak’s multiple comparisons test). (F) Immunoblot analysis of phospho-S6K and S6K levels in starved and amino acid refed cells. Scale bars, 10 μm.
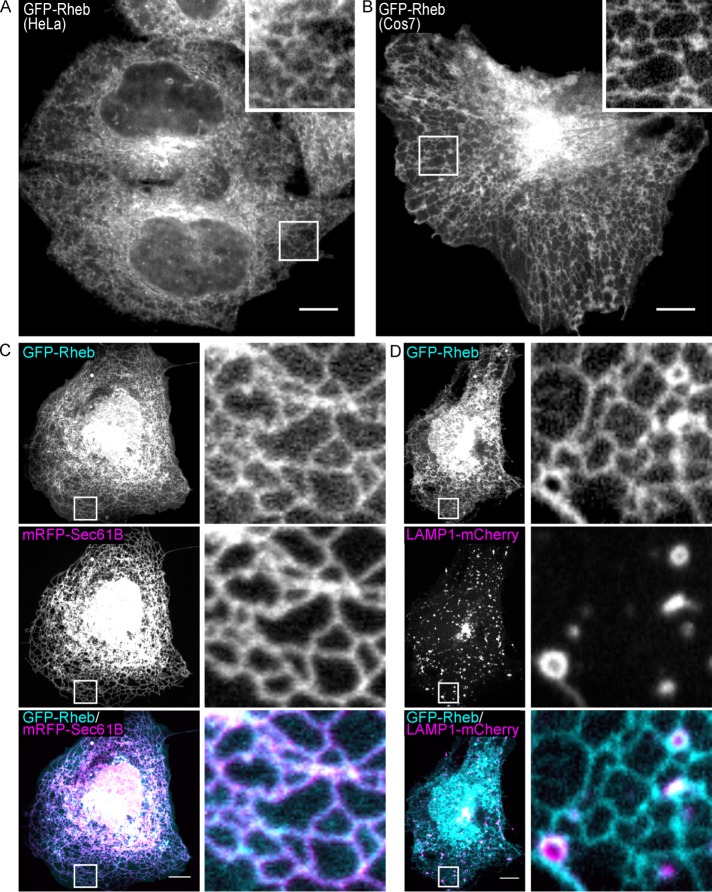
Live-cell imaging reveals enrichment of GFP-Rheb at the ER
Preserving Rheb on lysosomes could require specialized fixation, permeabilization, and/or antibody incubation conditions. Furthermore, our immunofluorescence experiments could have missed detecting a subpopulation of Rheb at lysosomes due to epitope masking by interacting proteins. To circumvent such issues, we next examined the localization of GFP-tagged Rheb expressed at moderate levels in live HeLa cells and observed a combination of cytosolic and ER-like localization patterns (Figure 3A). Rheb localization was further investigated in COS-7 cells as they contain a well-defined peripheral ER network that is highly suitable for live-cell imaging studies (Figure 3B) (Rowland et al., 2014). In addition to colocalizing extensively with mRFP-Sec61 (an ER protein), there was also a diffuse pool of Rheb in the cytosol (Figure 3C). In contrast, GFP-Rheb and LAMP1-mCherry (lysosome marker) had distinct, nonoverlapping, patterns of subcellular distribution. Interestingly, even though Rheb was not enriched on lysosomes, lysosomes were frequently adjacent to Rheb-positive ER tubules (Figure 3D; Supplemental Movie S1).
FIGURE 3:
GFP-Rheb localizes to the ER and cytosol. (A, B) Spinning disk confocal live-cell imaging of GFP-Rheb in HeLa cells and COS-7 cells, respectively. (C) GFP-Rheb and mRFP-Sec61B (ER marker) localization in COS-7 cells. (D) GFP-Rheb and LAMP1-mCherry (late endosomes and lysosomes) localization in COS-7 cells. Scale bars, 10 μm.
Movie S1.
GFP‐Rheb localizes to dynamic ER tubules that contact lysosomes. Spinning disk confocal live‐cell imaging of GFP‐Rheb (cyan) and Dextran‐loaded lysosomes (magenta; cells were labeled overnight with Alexa647‐Dextran and washed for 1 hour before imaging). Images were acquired at a rate of 15 frames/minute over an interval of 5 minutes. Scale bar, 5 μm.
Knockout (KO) cells reveal redundant functions for Rheb and RhebL1
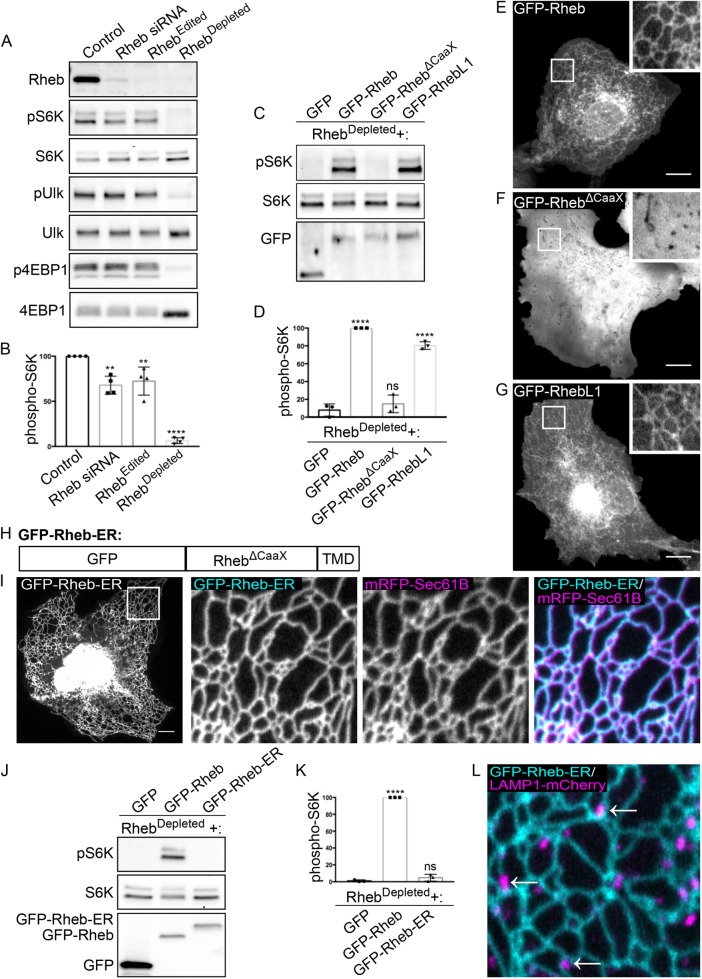
To further investigate the relationship between Rheb subcellular localization and function, we next generated Rheb KO HeLa cells to use as a platform for measuring the ability of Rheb targeted to distinct subcellular locations to rescue mTORC1 signaling defects. However, these Rheb KO cells maintained normal basal and serum-stimulated levels of mTORC1 signaling (Supplemental Figure S2C). This result indicated that Rheb is not absolutely essential for mTORC1 signaling and/or that cells can compensate for its long-term absence. Consistent with the possibility of Rheb-independent mechanisms for mTORC1 activation, Rheb KO mouse fibroblasts were previously reported to exhibit reduced, but not eliminated, phosphorylation of mTORC1 targets (Goorden et al., 2011; Groenewoud et al., 2013).
We next considered a potential role for the Rheb-like 1 (RhebL1, also known as Rheb2) protein as a way for cells to activate mTORC1 signaling in the absence of Rheb. Few studies have focused on the RhebL1 protein. However, in addition to sharing sequence similarity with Rheb, RhebL1 overexpression was previously shown to stimulate mTORC1 signaling (Tee et al., 2005). We therefore used CRISPR-Cas9 genome editing to mutate both the Rheb and RhebL1 genes in HeLa cells. After isolating clonal populations, we sought to identify cells harboring indels corresponding to frameshift mutations at the Cas9 target sites in both genes. The closest we came to achieving this goal was a cell line that contained one copy of Rheb with an in-frame 6-base-pair deletion and frameshift mutations in the remaining Rheb and RhebL1 alleles (Supplemental Figure S2D, henceforth referred to as RhebEdited cells). The absence of lines with frameshift mutations in all copies of both Rheb and RhebL1 likely reflects an essential role for Rheb/RhebL1 in supporting cell growth. Surprisingly, although Rheb protein levels were undetectable in the RhebEdited cells, basal levels of mTORC1 signaling were near normal (Figure 4, A and B). To test the possibility that traces of mutant Rheb protein containing the in frame mutation were responsible for the persistent mTORC1 signaling in the RhebEdited cells, we further treated them with Rheb siRNA. This resulted in near complete suppression of basal mTORC1 signaling as assessed by measuring the phosphorylation state of multiple downstream targets (Figure 4, A and B; Supplemental Figure S2, E and F). In light of these observations, we will subsequently refer to the RhebEdited cells that have been treated with Rheb siRNA as RhebDepleted. Although mTORC1 activity was lost in RhebDepleted cells, mTOR still localized to lysosomes in these cells (Supplemental Figure S2G). These results are consistent with the established model wherein mTORC1 localization to lysosomes is Rag-dependent. These results furthermore indicate that Rheb and RhebL1 have redundant functions that are essential for activation of mTORC1 signaling and provide an explanation for the previous reports of persistent mTORC1 signaling in Rheb KO mouse embryonic fibroblasts (Goorden et al., 2011; Groenewoud et al., 2013).
FIGURE 4:
Development of RhebDepleted cells as a tool for testing relationships between Rheb localization and function. (A) Immunoblot analysis of Rheb levels and phosphorylation status of mTORC1 substrates (S6K, ULK1, and 4EBP1) in Control, Rheb siRNA-treated, RhebEdited (Rheb hypomorph+RhebL1 KO), and RhebDepleted (Rheb hypomorph+RhebL1 KO+Rheb siRNA) HeLa cells. (B) Quantification of phospho-S6K levels under the indicated conditions where phospho-S6K levels were divided by S6K and normalized to Control (**P < 0.01; ****P < 0.0001; ANOVA with Dunnett’s multiple comparisons test, n = 4). (C) Immunoblot analysis of phospho-S6K levels in RhebDepleted cells transfected with the indicated plasmids. (D) Quantification of phospho-S6K levels from C. The phospho-S6K levels were divided by S6K and GFP values to control for loading and transfection efficiency. Values were normalized to GFP-Rheb. Statistics were calculated in comparison to the GFP transfection (****P < 0.0001; ANOVA with Dunnett’s multiple comparisons test; n = 3). (E–G) Live-cell images of GFP-Rheb, GFP-RhebΔCaaX, and GFP-RhebL1 in a COS-7 cells, respectively. (H) Schematic of GFP-Rheb-ER chimera that contains N-terminal GFP, RhebΔCaaX, and the transmembrane domain of cytochrome b5 (TMD). (I) Live-cell imaging of GFP-Rheb-ER localization. The leftmost image displays a low magnification view of GFP-Rheb-ER in a COS-7 cell. The three subsequent panels show higher magnifications of GFP-Rheb-ER and mRFP-Sec61B from the inset region. (J) Immunoblot analysis of phospho-S6K signaling in RhebDepleted cells transfected with the indicated plasmids. (K) Quantification of phospho-S6K levels in I. The phospho-S6K levels were divided by S6K and GFP values to control for loading and transfection. Values were normalized to GFP-Rheb. Statistics were calculated in comparison to GFP (****P < 0.0001; ANOVA with Dunnett’s multiple comparisons test; n = 3). (L) Image showing the spatial relationship between GFP-Rheb-ER on ER tubules and LAMP1-positive lysosomes. Scale bars, 10 μm.
Rheb C-terminal farnesylation is essential for mTORC1 signaling and Rheb localization to the ER
Having established that mTORC1 signaling is eliminated in the RhebDepleted cells, we next used this new model system to investigate the subcellular targeting mechanisms that support Rheb function. mTORC1 signaling (S6K-T389 phosphorylation) was restored following expression of either GFP-Rheb or GFP-RhebL1 in RhebDepleted cells (Figure 4, C and D). Similar to GFP-Rheb, GFP-RhebL1 exhibited an ER-like localization pattern (Figure 4, E and G; Supplemental Figure S3, A and B). Both Rheb and RhebL1 contain a C-terminal CaaX motif (cysteine followed by two aliphatic amino acids and X in the final position) wherein the cysteine is farnesylated (Clark et al., 1997). A Rheb mutant that lacked the CaaX motif was unable to rescue mTORC1 activity in RhebDepleted cells (Figure 4, C and D) and exhibited a diffuse subcellular distribution (Figure 4F). These results indicate that Rheb cannot efficiently activate mTORC1 signaling from the cytosol and that farnesylation-dependent membrane interactions are essential for its function.
Following the cytosolic farnesylation reaction, Rheb is further processed at the ER by Ras converting enzyme (RCE1) and isoprenylcysteine carboxymethyltransferase (ICMT) (Takahashi et al., 2005). This raised the possibility that the ER-localized pool of Rheb corresponds to newly synthesized Rheb protein that is undergoing these posttranslational modifications. However, treatment of cells with cycloheximide to block new protein synthesis and allow any recently synthesized Rheb to complete its posttranslational processing did not reduce Rheb abundance at the ER or result in the appearance of a lysosomal localization pattern (Supplemental Figure S4).
Constitutive ER localization of Rheb does not support mTORC1 signaling
The prominent membrane contact sites between ER and lysosomes suggested that ER-localized Rheb might reach across such regions of proximity to activate mTORC1 that has been recruited to lysosomes via interactions with the Rags. Indeed, such a model was proposed in another recent study (Walton et al., 2018). To test this model, we generated a chimeric protein composed of GFP-RhebΔCaaX fused to the transmembrane domain of cytochrome B5 (Figure 4H), a well-established ER targeting signal (Honsho et al., 1998). This chimeric protein localized to the ER (Figure 4I) but did not stimulate mTORC1 activity in RhebDepleted cells (Figure 4, J and K) even though we still observed frequent contact between ER and lysosomes in cells expressing GFP-Rheb-ER (Figure 4L).
Rheb C-terminal farnesylation supports mTORC1 signaling without any requirement for additional targeting motifs in the surrounding hypervariable region
As C-terminal farnesylation is essential for the function of Rheb, we next focused on dissecting the role played by Rheb farnesylation and the possibility of additional regulation conferred by adjacent sequences in the Rheb C-terminal hypervariable region. We focused in particular on the last 15 amino acids of Rheb as this region was previously reported to act as a lysosome targeting signal (Sancak et al., 2010) and might thus contain an additional determinant that cooperates with farnesylation to target Rheb to lysosomes. As both Rheb and RhebL1 are capable of supporting mTORC1 activation (Figure 4), we compared their C-terminal sequences for additional determinants that might help to target them to a common subcellular site of action but did not detect any obvious similarity beyond the C-terminal CaaX motifs (Figure 5A). In contrast to Rheb and RhebL1 whose ER and cytosolic localization are seemingly at odds with their ability to activate mTORC1 on lysosomes, other members of the Ras superfamily such as H-Ras localize robustly to their major site of action at the plasma membrane (Hancock et al., 1990; Choy et al., 1999). The plasma membrane localization of H-Ras depends on C-terminal farnesylation accompanied by two additional cysteine residues within the adjacent hypervariable region that are palmitoylated (Figure 5A) (Hancock et al., 1990; Choy et al., 1999).
FIGURE 5:
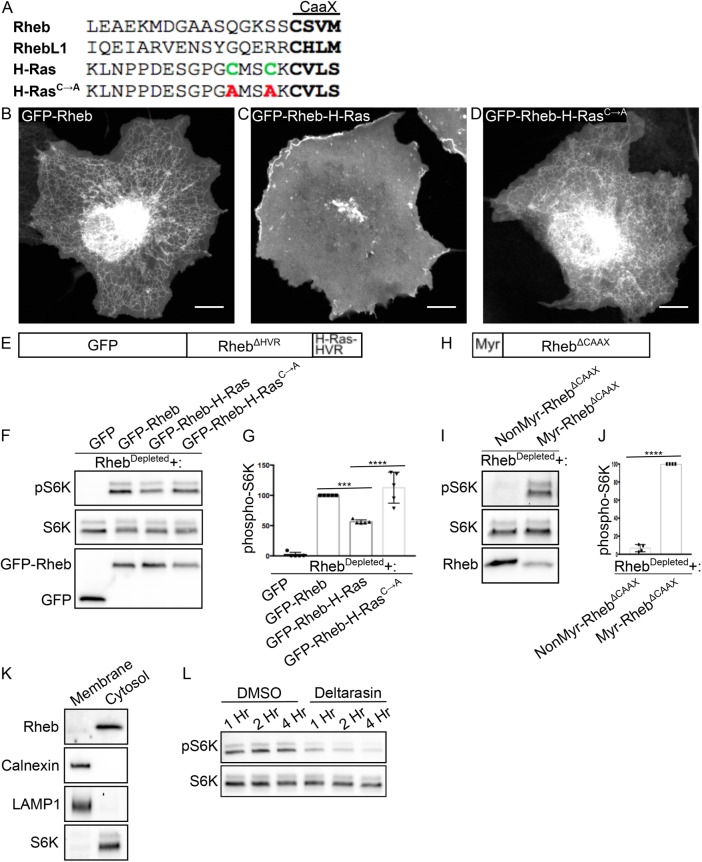
Weak membrane interactions are optimal for Rheb-dependent mTORC1 activation. (A) Alignment of the hypervariable regions of Rheb, RhebL1, and H-Ras proteins with CaaX motifs highlighted in bold and palmitoylated cysteines in green. These cysteines were mutated to alanines (red) in the GFP-Rheb-HRasC→A mutant. (B–D) Live-cell images of GFP-Rheb, GFP-Rheb-HRas, and GFP-Rheb-HRasC→A in COS-7 cells, respectively. Scale bars, 10 µm. (E) Schematic of GFP-Rheb-H-Ras chimera that contains N-terminal GFP, RhebΔHVR, and the HVR of H-Ras. (F) Immunoblot analysis of phospho-S6K levels in RhebDepleted cells transfected with the indicated plasmids. (G) Quantification of phospho-S6K levels from F. Phospho-S6K levels were divided by S6K and GFP values to control for loading and transfection (***P < 0.001; ****P < 0.0001; ANOVA with Tukey’s multiple comparisons test; n = 5). (H) Schematic of myr-Rheb chimera that contains RhebΔCaaX and a myristoylation consensus sequence. (I) Immunoblot analysis of phospho-S6K levels in RhebDepleted cells transfected with the indicated plasmids. (J) Quantification of phospho-S6K levels from I. Phospho-S6K levels were divided by S6K and Rheb values to control for loading and transfection (****P < 0.0001; unpaired t test). (K) Immunoblot analysis of proteins identified in membrane and cytosolic fractions of HeLa cells. (L) Immunoblot analysis of phospho-S6K levels in HeLa cells treated with DMSO vehicle and 5 μM deltarasin for the times indicated.
To investigate the role of the CaaX motif and its flanking sequences in determining the subcellular localization and function of Rheb, we generated a chimera wherein the Rheb C-terminus was replaced with the last 20 amino acids from H-Ras (Figure 5, A and E). This Rheb-H-Ras chimeric protein localized predominantly to the plasma membrane (Figure 5, B and C) and was less effective than full-length Rheb at activating mTORC1 signaling (Figure 5, F and G). Although the predominant localization of H-Ras to the plasma membrane is seemingly at odds with the ability of the Rheb-H-Ras chimera to moderately activate mTORC1 signaling, the H-Ras C-terminus is known to undergo cycles of depalmitoylation that allow it to transiently visit intracellular membranes (Rocks et al., 2005). To test the idea that farnesylation alone is the key determinant of Rheb localization and function, we generated a mutant Rheb-H-Ras chimera that lacks the palmitoylated cysteines in the hypervariable region (Figure 5A). This protein was no longer enriched at the plasma membrane and had a similar localization pattern to the wild-type (WT) Rheb (Figure 5, B and D). It also promoted mTORC1 activity just as well as the WT GFP-Rheb (Figure 5, E and F). As Rheb, RhebL1 and the palmitoylation-deficient H-Ras C-terminal regions lack sequence similarity beyond their CaaX motifs (Figure 5A), these results indicate that farnesylation, independent from any other major determinants within the Rheb C-terminus region, is both necessary and sufficient for the ability of Rheb to interact with membranes and support mTORC1 signaling.
N-terminal myristoylation can substitute for Rheb C-terminal farnesylation
Our results challenge expectations that Rheb stably resides at lysosomes via a specific targeting signal that resides within its C-terminus. To further test this conclusion, we sought an alternative method for dynamically targeting Rheb to membranes. For this purpose, we generated a Rheb mutant that lacks the C-terminal CaaX motif and therefore cannot be farnesylated but instead contains a myristoylation signal at its N-terminus (Figure 5H). This strategy was based on the following logic: 1) myristoylation, like farnesylation, supports only transient membrane interactions; 2) N-terminal myristoylation was previously shown to be an effective substitute for C-terminal farnesylation in H-Ras (Cadwallader et al., 1994); and 3) recent phylogenetic analysis identified Rheb genes in some species that lack C-terminal farnesylation but are predicted to be myristoylated at their N-termini (Zahonova et al., 2018). Interestingly, although a Rheb mutant that is neither farnesylated nor myrisotylated failed to promote mTORC1 signaling in RhebDepleted cells, myristoylated Rheb stimulated S6K phosphorylation (Figure 5, I and J). The functionality of myristoylated Rheb indicates that a weak, nonselective membrane interaction is the minimal targeting signal that is required for Rheb to function. This observation also argues against the possibility that farnesylation is essential for other aspects of Rheb function such as its ability to activate mTORC1. This interpretation is further corroborated by in vitro experiments that ruled out an essential requirement for Rheb farnesylation in mTORC1 activation (Sato et al., 2009). Consistent with a model wherein Rheb C-terminal farnesylation only supports transient membrane interactions, subcellular fractionation revealed that Rheb was predominantly found in the cytosolic fraction and only minimally present in the membrane fraction (Figure 5K).
Dynamic redistribution of Rheb between membranes is regulated by its interaction with PDEδ (Ismail et al., 2011; Kovacevic et al., 2018). The binding of farnesylated proteins to PDEδ can be blocked by drugs such as deltarasin that compete for binding to the hydrophobic cavity of PDEδ (Zimmermann et al., 2013). Consistent with previous reports (Zimmermann et al., 2013; Papke et al., 2016), we observed that mTORC1 signaling was suppressed in deltarasin-treated cells (Figure 5L). Although this could reflect effects on multiple farnesylated proteins beyond Rheb, this result is nonetheless consistent with a role for dynamic, PDEδ-dependent, redistribution of Rheb between subcellular membranes in supporting mTORC1 signaling.
Implications of weak Rheb membrane interactions for mTORC1 signaling
The minimal membrane targeting affinity and specificity of Rheb and RhebL1 contrasts with other members of the Ras superfamily that exhibit robust targeting to the membranes where they recruit their downstream effectors. However, Rheb and RhebL1 are distinct from other members of the Ras family in at least two important ways. First, Rheb and RhebL1 are not required for mTOR recruitment to the surface of lysosomes (Supplemental Figure S2G). mTORC1 localization to lysosomes is instead dependent on interactions between the Raptor subunit of mTORC1 and the Rag GTPases (Sancak et al., 2010). Second, recent structures of Rheb-mTORC1 complexes revealed that Rheb stimulates mTORC1 catalytic activity by binding to mTOR and inducing a conformational change that rearranges the mTOR kinase domain (Yang et al., 2017). Once this activation takes place, mTORC1 phosphorylates diverse substrates that reside throughout the cell (Ben-Sahra and Manning, 2017; Gonzalez and Hall, 2017; Saxton and Sabatini, 2017). Thus, the modest membrane affinity that is provided by Rheb farnesylation may represent an optimal solution for reducing dimensionality to facilitate interactions with lysosome-localized mTORC1 while also allowing the activated Rheb-mTORC1 complex to subsequently leave the lysosome to phosphorylate downstream targets that reside elsewhere in the cell.
Evidence in favor of dynamic interactions between Rheb and endo-lysosomal membranes as the key determinant mTORC1 activation
A predominantly ER and cytosolic steady-state localization of the Rheb protein challenges the idea that Rheb stably resides on lysosomes while waiting for the opportunity to activate mTORC1. Nonetheless, there are several arguments in support of a function of Rheb at lysosomes that could be mediated by transient interactions. First, considerable evidence indicates that Rag-dependent recruitment of mTORC1 to lysosomes is a prerequisite for mTORC1 activation and that such activation also depends on Rheb (Sancak et al., 2008, 2010; Bar-Peled et al., 2012, 2013; Betz and Hall, 2013; Petit et al., 2013; Tsun et al., 2013; Lim and Zoncu, 2016; Wolfson et al., 2017). Second, TSC, the Rheb GAP, is found at lysosomes, particularly in nutrient-starved cells (Demetriades et al., 2014; Menon et al., 2014; Carroll et al., 2016; Demetriades et al., 2016). Third, constitutively targeting TSC to lysosomes suppresses mTORC1 signaling (Menon et al., 2014). Fourth, although the Rheb protein in most commonly studied model organisms is targeted to membranes via farnesylation of a C-terminal CaaX motif, a recent phylogenetic analysis of Rheb protein sequences revealed that Rheb homologues in some species lack lipidation and are instead predicted to interact with membranes via an N-terminal FYVE domain (Zahonova et al., 2018). In others, the C-terminal CaaX motif is accompanied by an N-terminal PX domain (Zahonova et al., 2018). Although these bioinformatic predictions remain to be experimentally validated, PX and FYVE domains commonly recruit proteins to endosomal membranes via their ability to bind to phosphatidylinositol-3-phosphate (PI3P) with modest affinity (Burd and Emr, 1998; Gaullier et al., 1998). This evolutionary selection for Rheb targeting mechanisms that involve noncovalent interactions with endosomal lipids is consistent with a evolutionarily conserved function for Rheb in the endolysosomal pathway.
As an alternative to the possibility that Rheb dynamically visits the surface of lysosomes in order to activate mTORC1, it was recently reported that Rheb instead resides on the Golgi and activates mTORC1 via contact sites between the Golgi and the lysosomes (Hao et al., 2018). However, this study relied on assessing the ability of heavily overexpressed, Golgi-targeted Rheb to hyperactivate mTORC1 signaling in cells that also expressed endogenous WT Rheb and RhebL1. Furthermore, the Golgi-targeting strategy was based on engineering an artificial interaction between Rheb and Rab1a. Although this resulted in significant localization of Rheb to the Golgi and Rheb-dependent activation of mTORC1 signaling, Rab1a is a prenylated protein that cycles on and off of Golgi membranes (Smeland et al., 1994). Given our results with the predominantly plasma membrane-localized Rheb-H-Ras chimera (Figure 4) as well as our observations that only very low levels of Rheb are required for mTORC1 activation (Figure 3), caution must be applied with interpreting how the steady-state localization of an overexpressed Rheb-Rab chimeric protein that undergoes dynamic cycling on and off membranes relates to the actual site of mTORC1 activation.
Potential implications of Rheb localization to the ER
It was recently proposed that moving lysosomes toward the cell periphery suppresses mTORC1 activity by limiting their proximity with ER-localized Rheb (Walton et al., 2018). Although this idea is intriguing, we observed that even peripheral lysosomes still maintain contact with Rheb-positive ER tubules (Figure 3; Supplemental Movie S1). Furthermore, the inability of constitutively ER-localized Rheb to activate mTORC1 in spite of maintained ER-lysosome contact sites argues against a model wherein ER-localized Rheb activates mTORC1 by reaching across such contact sites (Figure 3). Although the presence of Rheb on the ER is suggestive of specific targeting mechanisms and functions, it has long been known that fusion to a farnesylated CaaX motif is sufficient to target proteins such as GFP to the ER (Choy et al., 1999). Therefore, Rheb localization to the ER appears to simply reflect the default localization for a farnesylated protein that lacks other strong subcellular localization signals. Finally, although we have focused on the relationship between Rheb membrane-targeting mechanisms and mTORC1 activation, it is also possible that the dynamic interactions of Rheb with the ER and other intracellular membranes might also be relevant for encounters with other effectors. Indeed, Rheb has been proposed to interact with and regulate several additional proteins in a GTP-dependent manner (Yee and Worley, 1997; Neuman and Henske, 2011; Sato et al., 2015).
Conclusions
Considerable evidence indicates that lysosomes are a major intracellular site where nutrient and growth factor signals are integrated in order to match mTORC1 signaling to ongoing environmental changes. Rheb-dependent activation of mTORC1 kinase activity is a central component of this model that has been accompanied by the idea that Rheb constitutively resides on the surface of lysosomes due to a selective targeting signal at its C-terminus. Nonetheless, our new observations argue against the presence of a large steady-state pool of Rheb at lysosomes. Nor did our results support the existence of a selective lysosome-targeting signal within the C-terminus of Rheb. Instead, we propose that transient, farnesylation-dependent, membrane interactions have been selected by evolution as optimal for the Rheb-mediated activation of mTORC1 at lysosomes. By overturning widely held beliefs concerning stable residence of Rheb at lysosomes, our new findings will guide future studies that focus on understanding how dynamic Rheb membrane interactions are coordinated with the Rag-dependent localization of mTORC1 to lysosomes to activate mTORC1 signaling in health and disease.
MATERIALS AND METHODS
Cell culture and transfection
HeLa M cells and COS-7 cells (both provided by P. De Camilli, Yale University, New Haven, CT) were grown in high-glucose DMEM with l-glutamine, 10% fetal bovine serum, and 1% penicillin/streptomycin supplement (ThermoFisher Scientific, Waltham, MA, and Corning, Corning, NY). To starve cells of amino acids, cells were incubated in amino acid-free RPMI media (US Biologicals, Swampscott, MA) for 2 h. Cells were refed with RPMI containing 1× MEM amino acid supplement (Invitrogen) for 20 min. To starve cells of growth factors, cells were incubated in serum-free DMEM overnight. Such cells were then refed for 30 min with complete media to measure the acute response to serum exposure.
To perform plasmid transfections, 500 ng of plasmid DNA, 1.5 µl of Fugene 6 transfection reagent (Promega, Madison, WI), and 100 µl of OptiMEM (ThermoFisher Scientific) were added to 80,000 cells per well in a six-well dish. For siRNA transfections, 5 µl of RNAiMAX (ThermoFisher Scientific), 500 µl of OptiMEM, and 5 µl of 20 µM siRNA stock were added to a subconfluent dish of cells (80,000 cells per well in a six-well dish). Cells were incubated for 48 h posttransfection prior to experiments. Control siRNA (5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′) was purchased from Integrated DNA Technologies (IDT, Coralville, IA) and a previously described Rheb siRNA (Menon et al., 2014) was purchased from Cell Signaling Technology (#14267; CST, Danvers, MA). Drugs utilized in this study: deltarasin (Cayman Chemical, Ann Arbor, MI, #9001536) and cycloheximide (Millipore Sigma, Burlington, MA, #239765).
Plasmids
pRK5 plasmid encoding HA-GST human Rheb was acquired from D. Sabatini (Massachusetts Institute of Technology, Cambridge, MA) via Addgene (Plasmid #14951; Sancak et al., 2007). Full-length Rheb was PCR amplified from this vector and cloned into SmaI-digested pEGFP-C1 by Gibson Assembly (New England Biolabs, Ipswich, MA). The Q5 Site-Directed Mutagenesis Kit (New England Biolabs) was used to generate GFP-RhebΔCAAX, GFP-Rheb-HRas, and GFP-Rheb-HRasC→A plasmids. To generate GFP-Rheb-ER, a cDNA (IDT, Coralville, IA) containing a fragment of cytochrome b5 (region from amino acids 95–134 (Itakura and Mizushima, 2010) with myc tag and glycosylation site (Honsho et al., 1998) was cloned into GFP-RhebΔCAAX (linearized at the C Terminus via PCR) by Gibson Assembly. Full-length RhebL1 cDNA was synthesized (IDT) and cloned into SmaI-digested pEGFPC1 by Gibson Assembly. The following plasmids were kind gifts: LAMP1-mCherry from J. Lippincott-Schwartz (Janelia Research Campus, Ashburn, VA) and mRFP-Sec61B from T. Rapoport (Harvard University, Cambridge, MA). Oligonucleotide primers and cDNA sequences used to generate these plasmids are listed in Supplemental Table S1. Our newly generated Rheb and RhebL1 plasmids will be made available via the Addgene plasmid repository.
Small guide RNAs (gRNAs) were designed using the CRISPR design tool (crispr.mit.edu) or selected from predesigned gRNA sequences (Sanjana et al., 2014). The gRNA-encoding DNA oligonucleotides (IDT) sequences were annealed and ligated into Bbs1-digested pX459 V2.0 plasmid that was provided by F. Zhang (Massachusetts Institute of Technology, Cambridge, MA) via Addgene (#62988; Ran et al., 2013) and transformed into Stabl3-competent Escherichia coli cells. The gRNA sequences are listed in Supplemental Table S2.
Immunoblotting
Cells were lysed in Tris-buffered saline + 1% Triton X-100 (TBST) with protease and phosphatase inhibitor cocktails (Roche Diagnostics, Florham Park, NJ). To remove insoluble materials, lysates were centrifuged for 6 min at 14,000 rpm. Lysate protein concentrations were measured via Bradford assay prior to denaturation with Laemmli buffer and 5 min at 95°C. Immunoblotting was performed with 4–15% gradient Mini-PROTEAN TGX precast polyacrylamide gels and nitrocellulose membranes (Bio-Rad, Hercules, CA). Membranes were blocked with 5% milk in TBS with 0.1% Tween 20 (TBST) buffer and then incubated with antibodies in 5% milk or bovine serum albumin (BSA) in TBST. Antibodies used in this study are summarized in the Supplemental Table S4. Horseradish peroxidase signal detection was performed using chemiluminescent detection reagents (ThermoFisher Scientific) and the Versadoc imaging station (Bio-Rad). ImageJ (National Institutes of Health [NIH]) was used to analyze the results and measure band intensities.
Cell fractionation
To perform cell fractionation, 2 million HeLa cells were plated in 10-cm dishes. Cells were rinsed, scraped into chilled phosphate-buffered saline (PBS), and spun at 1000 rpm for 10 min. PBS was aspirated and cells were resuspended in homogenization buffer (5 mM Tris-HCl, 250 mM sucrose, 1 mM egtazic acid, pH 7.4) with protease and phosphatase inhibitor cocktails. Cells were homogenized using a ball-bearing cell cracker (20 passages at 10 µm clearance; Isobiotec, Germany). Lysates were centrifuged at 1000 × g for 10 min to remove unlysed cells. Supernatant was spun at 100,000 × g for 1 h to pellet membrane. Supernatant containing cytosolic proteins and the resuspended membrane pellet were analyzed via immunoblotting.
Immunofluorescence and microscopy
Cells were grown on 12-mm No. 1 glass coverslips (Carolina Biological Supply) and fixed with 4% paraformaldehyde (PFA; Electron Microscopy Sciences, Hatfield, PA) in 0.1 M sodium phosphate buffer (pH 7.2) for 30 min. To best preserve cell structure, this was achieved by adding 1 vol of 8% PFA dropwise to cells growing on coverslips in growth medium. Coverslips were washed in PBS before permeabilization in PBS + 0.2% Triton X-100 for 10 min. Coverslips were blocked for 30 min in blocking buffer (5% NDS/PBS/0.2% Triton X-100). Cells were incubated in primary antibody overnight at 4°C in blocking buffer. Cells were subsequently washed 3× with 0.2% Triton X-100 and incubated in secondary antibody in blocking buffer for 30 min at room temperature. Cells were washed 3× with 0.2% Triton X-100 before slides were prepared with Prolong Gold Mounting media (Invitrogen, Carlsbad, CA). Antibodies used in this study are summarized in Supplemental Table S4.
For live-cell imaging, cells were grown on glass-bottom dishes (MatTek, Ashland, MA) prior to imaging. Subconfluent dishes were imaged via spinning disk confocal microscopy at room temperature in a buffer that contained 136 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.3 mM MgCl2, 10 mM HEPES, 0.2% glucose, 0.2% BSA, pH 7.4 (Brown et al., 2000). Dextran Alexa Fluor 647 (Invitrogen, #22914) was applied overnight and washed out 1 h before imaging. Our microscope consisted of the UltraVIEW VOX system (PerkinElmer, Waltham, MA) including the Ti-R Eclipse, Nikon inverted microscope (equipped with a 60× CFI Plan Apo VC, NA 1.4, oil immersion), a spinning disk confocal scan head (CSU-X1, Yokogawa), and Volocity (PerkinElmer) software. Images were acquired without binning with a 14-bit (1000 × 1000) EMCCD (Hamamatsu Photonics) and processed with ImageJ. Colocalization analysis was performed with the Fiji Coloc 2 plug-in available on GitHub (https://imagej.net/Coloc_2). To perform this analysis, an 8-bit, 54 × 54 pixel region of interest was selected (based on the presence of well-resolved lysosomes) split into separate channels and the Coloc 2 plug-in was used to calculate the Pearson’s R value (no threshold). The lysosome channel was then rotated 90° to the right and the Pearson’s R value (no threshold) was recalculated. This analysis was performed for three biological replicates per condition with 15 images analyzed per replicate.
CRISPR/Cas9 genome editing
Similar to a previously described protocol (Amick et al., 2016), to generate KO cells, HeLa cells were cotransfected with Rheb and RhebL1 sgRNAs encoded within the pX459 plasmid. On the next day, puromycin (2 µg/ml) was added and cells were selected for 2 d. Surviving cells were plated at single-cell density and allowed to recover. Putative KO colonies were initially identified via immunoblotting and subsequently confirmed by sequencing of the genomic loci. To obtain genomic DNA sequence for the target site in the Rheb gene, DNA was extracted (QuickExtract DNA extraction solution; Epicentre Biotechnologies), the region of interest was amplified by PCR (primers summarized in Supplemental Table S3), and PCR products were cloned into the pCR-Blunt TOPO vector (Zero Blunt TOPO PCR cloning kit; ThermoFisher Scientific) and transformed into XL-1 Blue Supercompetent Cells (Agilent Technologies, Santa Clara, CA). Plasmid DNA was isolated from multiple colonies and sequenced.
To insert a 2xHA epitope tag at the endogenous Rheb locus, we utilized a CRISPR/Cas9 genome-editing strategy as previously described (Petit et al., 2013; Leonetti et al., 2016; Richardson et al., 2016; Amick et al., 2018). The CRISPR RNA (crRNA), tracrRNA, and single-stranded DNA (ssDNA) oligo were resuspended in nuclease-free duplex buffer (IDT) to 100 µM. To form the RNA duplex, the crRNA and tracrRNA were added at a 1:1 ratio and heated for 5 min at 95°C and then cooled to room temperature. The RNA duplex (150 pmol), Cas9 (150 pmol), and OptiMem were mixed in a sterile tube and incubated for 10 min at room temperature to form RNP complex. HeLa cells (750,000 cells) were electroporated (Cell Line Kit V, electroporation program Q01; Lonza, Basel, Switzerland) with 10 µl RNP complex crRNA and ssDNA repair template (IDT Ultramer format). Sequences are provided in Supplemental Table S2. Clonal cell lines were generated and validated using Western blotting and immunofluorescence.
Statistical analysis
Data were analyzed using Prism (Graphpad software), and tests are denoted in the figure legends. All error bars represent SD. Data distribution was assumed to be normal, but this was not formally tested.
Supplementary Material
Acknowledgments
This research was supported in part by grants from the NIH (GM105718) and the Ellison Medical Foundation to S.M.F. B.A. was supported by a National Science Foundation Graduate Research Fellowship (DGE1752134). Pamela Torola contributed to early phases of this project. Both Agnes Ferguson and Arun Tharkeshwar provided helpful advice with respect to methods and experimental design. Microscopy studies were supported by the Yale University Program in Cellular Neuroscience, Neurodegeneration and Repair imaging facility.
Abbreviations used:
- BSA
bovine serum albumin
- crRNA
CRISPR RNA
- ER
endoplasmic reticulum
- gRNA
guide RNA
- KO
knockout
- mTORC1
mTOR complex 1
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- RhebL1
Rheb-like 1
- ssDNA
single-stranded DNA
- TBST
Tris-buffered saline + Tween 20.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E19-03-0146) on September 18, 2019.
REFERENCES
- Amick J, Roczniak-Ferguson A, Ferguson SM. (2016). C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell , 3040–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amick J, Tharkeshwar AK, Amaya C, Ferguson SM. (2018). WDR41 supports lysosomal response to changes in amino acid availability. Mol Biol Cell , 2213–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. (2013). A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science , 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. (2012). Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell , 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Manning BD. (2017). mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol , 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN. (2013). Where is mTOR and what is it doing there? J Cell Biol , 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PS, Wang E, Aroeti B, Chapin SJ, Mostov KE, Dunn KW. (2000). Definition of distinct compartments in polarized Madin-Darby canine kidney (MDCK) cells for membrane-volume sorting, polarized sorting and apical recycling. Traffic , 124–140. [DOI] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. (2006). Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun , 869–880. [DOI] [PubMed] [Google Scholar]
- Burd CG, Emr SD. (1998). Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell , 157–162. [DOI] [PubMed] [Google Scholar]
- Cadwallader KA, Paterson H, Macdonald SG, Hancock JF. (1994). N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol , 4722–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B, Maetzel D, Maddocks OD, Otten G, Ratcliff M, Smith GR, Dunlop EA, Passos JF, Davies OR, Jaenisch R, et al (2016). Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. Elife , e11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. (1999). Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell , 69–80. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ. (1997). The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J Biol Chem , 10608–10615. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA. (2014). Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell , 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C, Plescher M, Teleman AA. (2016). Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat Commun , 10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM. (2015). Beyond indigestion: emerging roles for lysosome-based signaling in human disease. Curr Opin Cell Biol , 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. (1998). FYVE fingers bind PtdIns(3)P. Nature , 432–433. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Hall MN. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J , 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, Hoogeveen-Westerveld M, Cheng C, van Woerden GM, Mozaffari M, Post L, Duckers HJ, Nellist M, Elgersma Y. (2011). Rheb is essential for murine development. Mol Cell Biol , 1672–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewoud MJ, Goorden SM, Kassies J, Pellis-van Berkel W, Lamb RF, Elgersma Y, Zwartkruis FJ. (2013). Mammalian target of rapamycin complex I (mTORC1) activity in ras homologue enriched in brain (Rheb)-deficient mouse embryonic fibroblasts. PLoS One , e81649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ. (1990). A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell , 133–139. [DOI] [PubMed] [Google Scholar]
- Hanker AB, Mitin N, Wilder RS, Henske EP, Tamanoi F, Cox AD, Der CJ. (2010). Differential requirement of CAAX-mediated posttranslational processing for Rheb localization and signaling. Oncogene , 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao F, Kondo K, Itoh T, Ikari S, Nada S, Okada M, Noda T. (2018). Rheb localized on the Golgi membrane activates lysosome-localized mTORC1 at the Golgi-lysosome contact site. J Cell Sci , jcs208017. [DOI] [PubMed] [Google Scholar]
- Honsho M, Mitoma JY, Ito A. (1998). Retention of cytochrome b5 in the endoplasmic reticulum is transmembrane and luminal domain-dependent. J Biol Chem , 20860–20866. [DOI] [PubMed] [Google Scholar]
- Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. (2011). Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol , 942–949. [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. (2010). Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy , 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic M, Klein CH, Rossmannek L, Konitsiotis AD, Stanoev A, Kraemer AU, Bastiaens PI. (2018). A spatially regulated GTPase cycle of Rheb 1 controls growth factor signaling to mTORC1. bioRxiv 472241; 10.1101/472241. [DOI]
- Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B. (2016). A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proc Natl Acad Sci USA , E3501–E3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Zoncu R. (2016). The lysosome as a command-and-control center for cellular metabolism. J Cell Biol , 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. (2014). Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell , 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman NA, Henske EP. (2011). Non-canonical functions of the tuberous sclerosis complex-Rheb signalling axis. EMBO Mol Med , 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke B, Murarka S, Vogel HA, Martin-Gago P, Kovacevic M, Truxius DC, Fansa EK, Ismail S, Zimmermann G, Heinelt K, et al (2016). Identification of pyrazolopyridazinones as PDEdelta inhibitors. Nat Commun , 11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. (2013). Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol , 1107–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc , 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. (2016). Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol , 339–344. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. (2005). An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science , 1746–1752. [DOI] [PubMed] [Google Scholar]
- Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK. (2014). ER contact sites define the position and timing of endosome fission. Cell , 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell , 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science , 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. (2007). PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell , 903–915. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods , 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Akasu H, Shimono W, Matsu C, Fujiwara Y, Shibagaki Y, Heard JJ, Tamanoi F, Hattori S. (2015). Rheb protein binds CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase) protein in a GTP- and effector domain-dependent manner and influences its cellular localization and carbamoyl-phosphate synthetase (CPSase) activity. J Biol Chem , 1096–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Nakashima A, Guo L, Tamanoi F. (2009). Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem , 12783–12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. (2017). mTOR signaling in growth, metabolism, and disease. Cell , 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvius JR, Bhagatji P, Leventis R, Terrone D. (2006). K-ras4B and prenylated proteins lacking “second signals” associate dynamically with cellular membranes. Mol Biol Cell , 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvius JR, l’Heureux F. (1994). Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry , 3014–3022. [DOI] [PubMed] [Google Scholar]
- Smeland TE, Seabra MC, Goldstein JL, Brown MS. (1994). Geranylgeranylated Rab proteins terminating in Cys-Ala-Cys, but not Cys-Cys, are carboxyl-methylated by bovine brain membranes in vitro. Proc Natl Acad Sci USA , 10712–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Nakagawa M, Young SG, Yamanaka S. (2005). Differential membrane localization of ERas and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. J Biol Chem , 32768–32774. [DOI] [PubMed] [Google Scholar]
- Tee AR, Blenis J, Proud CG. (2005). Analysis of mTOR signaling by the small G-proteins, Rheb and RhebL1. FEBS Lett , 4763–4768. [DOI] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. (2013). The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell , 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton ZE, Patel CH, Brooks RC, Yu Y, Ibrahim-Hashim A, Riddle M, Porcu A, Jiang T, Ecker BL, Tameire F, et al (2018). Acid suspends the circadian clock in hypoxia through inhibition of mTOR. Cell , 72–87.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Wyant GA, Gu X, Orozco JM, Shen K, Condon KJ, Petri S, Kedir J, Scaria SM, et al (2017). KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature , 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jiang X, Li B, Yang HJ, Miller M, Yang A, Dhar A, Pavletich NP. (2017). Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature , 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee WM, Worley PF. (1997). Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol Cell Biol , 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahonova K, Petrzelkova R, Valach M, Yazaki E, Tikhonenkov DV, Butenko A, Janouskovec J, Hrda S, Klimes V, Burger G, et al (2018). Extensive molecular tinkering in the evolution of the membrane attachment mode of the Rheb GTPase. Sci Rep , 5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, Waldmann H. (2013). Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature , 638–642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.