Abstract
Successful separation of two daughter cells (i.e., cytokinesis) is essential for life. Many eukaryotic cells divide using a contractile apparatus called the cytokinetic ring (CR) that associates dynamically with the plasma membrane (PM) and generates force that contributes to PM ingression between daughter cells. In Schizosaccharomyces pombe, important membrane–CR scaffolds include the paralogous F-BAR proteins Cdc15 and Imp2. Their conserved protein structure consists of the archetypal F-BAR domain linked to an SH3 domain by an intrinsically disordered region (IDR). Functions have been assigned to the F-BAR and SH3 domains. In this study we probed the function of the central IDR. We found that the IDR of Cdc15 is essential for viability and cannot be replaced by that of Imp2, whereas the F-BAR domain of Cdc15 can be swapped with several different F-BAR domains, including that of Imp2. Deleting part of the IDR results in CR defects and abolishes calcineurin phosphatase localization to the CR. Together these results indicate that Cdc15’s IDR has a nonredundant essential function that coordinates regulation of CR architecture.
INTRODUCTION
Cytokinesis is the final step in the cell division cycle when daughter cells separate. In Amoebozoa and Opisthokonta, cytokinesis employs an actin- and myosin-based proteinaceous apparatus termed the cytokinetic ring (CR; Mangione and Gould, 2019). The CR assembles at the future division plane and eventually constricts coincident with cleavage furrow ingression. Of the many proteins that compose the CR, membrane-bound scaffolds are crucial for transmitting CR-generated tension to the plasma membrane (PM) and for maintaining CR placement at the division site (Glotzer, 2017). F-BAR proteins are conserved membrane–CR scaffolds that function during cytokinesis as well as other processes involving coordination between the dynamic cytoskeleton and membranes (Lippincott and Li, 2000; Ahmed et al., 2010; Fricke et al., 2010; Roberts-Galbraith and Gould, 2010). The namesake F-BAR domain binds phospholipids and oligomerizes into a multivalent platform that can recruit proteins to membranes through interactions with the F-BAR domain itself or using additional functional domains (McDonald and Gould, 2016).
In the fission yeast Schizosaccharomyces pombe, the F-BAR protein Cdc15 is an essential protein that promotes CR stabilization and cell wall deposition (i.e., septation) that occurs coincident with CR constriction (Fankhauser et al., 1995; Wachtler et al., 2006; Huang et al., 2007; Pinar et al., 2008; Roberts-Galbraith et al., 2009; Arasada and Pollard, 2014; Cortes et al., 2015; Ren et al., 2015; Willet et al., 2015b; Sethi et al., 2016; Onwubiko et al., 2019). Cdc15 has an N-terminal F-BAR domain linked by a central intrinsically disordered region (IDR) to a C-terminal SH3 domain. The F-BAR domain directly binds the PM (McDonald et al., 2015) and the essential CR actin nucleator/elongator formin Cdc12 (Carnahan and Gould, 2003; Willet et al., 2015a). Additionally, the Cdc15 F-BAR domain oligomerizes into a multivalent scaffold with high avidity for the PM at the division site (McDonald et al., 2015). Disrupting the Cdc15-Cdc12 interaction delays CR formation (Willet et al., 2015a), while disrupting Cdc15 F-BAR membrane-binding and oligomerization destabilizes the CR and increases cytokinesis failures (McDonald et al., 2015). Indeed, the F-BAR domain is essential for Cdc15 function and cell viability (McDonald et al., 2015). In contrast, domain-swapping experiments showed that the Cdc15 SH3 domain is redundant with that of its paralogue Imp2, which is also a component of the CR (Demeter and Sazer, 1998; Roberts-Galbraith et al., 2009; Ren et al., 2015).
Imp2 shares Cdc15’s domain structure: an N-terminal F-BAR domain is linked by an IDR to a C-terminal SH3 domain. However, imp2 null cells are viable, though they have severe cytokinetic defects (McDonald et al., 2016) and Imp2 dynamics at the CR differ from those of Cdc15 (Demeter and Sazer, 1998; Ren et al., 2015). The defects of imp2 null cells are recapitulated by an F-BAR deletion mutant, imp2(C) (McDonald et al., 2016); however, replacing the F-BAR of Imp2 with that of Cdc15 or the Saccharomyces cerevisiae homologue Hof1 rescues the mutant phenotype (McDonald et al., 2016). These results not only emphasized the importance of membrane binding for F-BAR protein function but also suggested an unexpected plasticity among F-BAR domains in performing this critical function, and pointed to the IDRs of Imp2 and Cdc15 as the key determinants of their functional differences.
IDRs are flexible, unfolded structures for which predicting protein function is difficult. Their flexibility allows them to sample many conformations, thus enabling interaction with multiple binding partners (Tompa, 2012). Here, we have extended our comparative analyses of the relative importance of the F-BAR and IDR in the context of Cdc15’s essential function in cytokinesis. We found that a variety of F-BAR domains from human and S. pombe F-BAR proteins can substitute for the Cdc15 F-BAR. In contrast, the Cdc15 IDR is essential and apparently uniquely so, because it cannot be replaced by the Imp2 IDR. Further structure–function analysis of the Cdc15 IDR revealed that cells tolerate removal or exchange of parts of the IDR. However, such mutations are accompanied by cytokinesis defects. One section of the IDR is specifically needed to maintain the circularity of the CR during constriction and to recruit the calcineurin phosphatase to the division site. Taken together, our results reveal a remarkable functional plasticity among F-BAR domains and add to the growing appreciation that IDRs make critical contributions in a variety of proteins and biological contexts.
RESULTS
The Cdc15 IDR is essential for viability
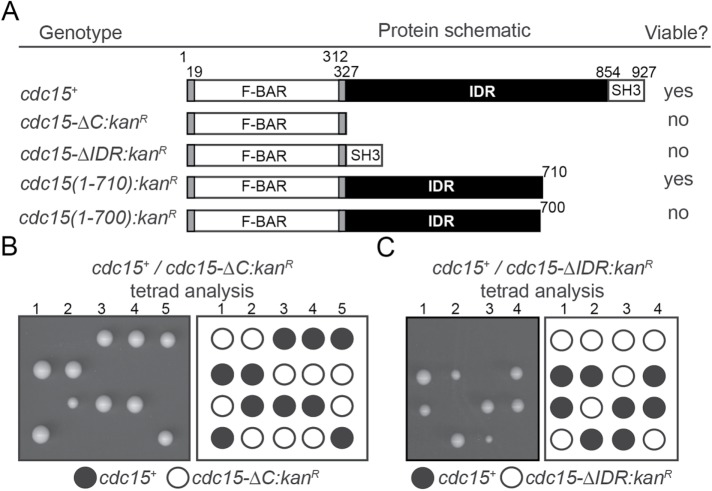
We tested the ability of various cdc15 mutants to support viability by integrating them at one cdc15 locus of diploid cells to make cdc15+/cdc15 mutant heterozygotes. Next, the diploids were sporulated and tetrads dissected to determine whether the cdc15 mutants could support viability when present as the sole cdc15 allele.
We found that the Cdc15 F-BAR domain alone was not sufficient to support cell viability (Figure 1, A and B). An allele encoding the Cdc15 F-BAR domain juxtaposed to the SH3 domain (cdc15-∆IDR) was also unable to support viability, verifying the necessity of Cdc15’s IDR (Figure 1, A and C). Deletion of the SH3 domain was previously determined to be viable (Roberts-Galbraith et al., 2009) and further C-terminal truncation was tolerated to aa 710 but not to aa 700 (Figure 1A). These results, and those from previous studies (McDonald et al., 2015), indicate that the F-BAR domain and part of the IDR are required to perform Cdc15’s essential function.
FIGURE 1:
The IDR of Cdc15 is essential. (A) Table of cdc15 alleles, schematic of gene product (drawn to scale), and ability to rescue cdc15 null. Numbers indicate aa position. (B, C) Representative tetrads from diploids of the indicated genotype that were induced to sporulate.
F-BAR domains can be exchanged
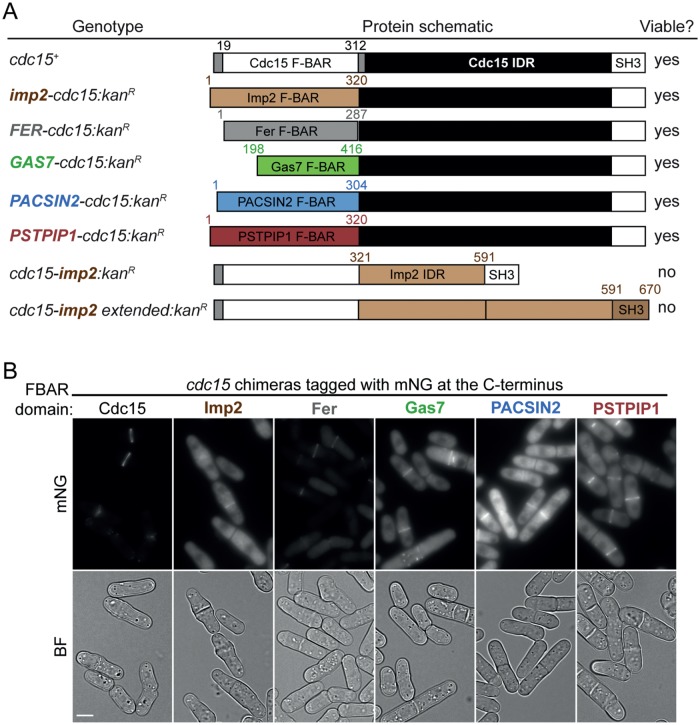
We next asked whether these features could be replaced by the comparable regions of other F-BAR proteins. We first generated a series of chimeras by fusing various F-BAR domains from human and yeast proteins to the Cdc15 C-terminus and then tested them for viability as previously described. Five of the 15 chimeras were viable: the fusions with F-BAR domains of the human proteins Fer, Gas7, PACSIN2, and PSTPIP1 and S. pombe Imp2 (Figure 2A; Supplemental Table S1). We tagged the viable chimeras with the fluorophore mNeonGreen (mNG) and they all localized to the CR (Figure 2B). The chimeras have septation defects (Figure 2B and Supplemental Figure S1, A and B) and are cold-sensitive (Supplemental Figure S1C). These defects are comparable among chimeras, suggesting a similar level of rescue.
FIGURE 2:
Cdc15 F-BAR, but not IDR, can be replaced by domains of homologous proteins. (A) Table of cdc15 alleles, schematic of gene product (drawn to scale), and ability to rescue cdc15 null. Numbers indicate aa position. (B) Live-cell images of the indicated haploid strains. A single BF z-slice and sum projections of mNG images are shown. Images of the Fer and PSTPIP1 chimeras were acquired on a separate day. Bar, 5 µm.
In contrast, replacement of only the Cdc15 IDR with that of Imp2 (cdc15-imp2 chimera) did not support viability (Figure 2A). Additionally, a mutant in which the Cdc15 F-BAR was fused to the Imp2 IDR duplicated (to match the length of Cdc15’s IDR) plus Imp2’s SH3 domain (cdc15-imp2 extended chimera) did not support viability (Figure 2A). These results indicate that the Cdc15 IDR is essential in a sequence-specific manner.
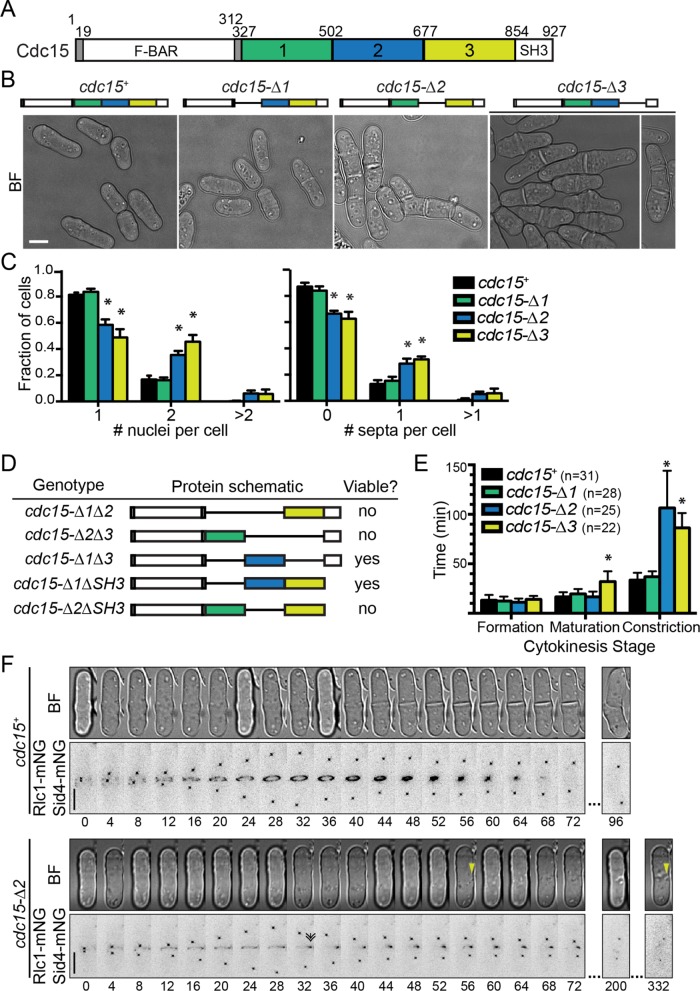
Partial loss of the IDR results in cytokinesis defects
To separate the function of the IDR from that of the SH3 domain, which coordinates CR constriction and septum deposition through multiple binding interactions (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Pinar et al., 2008; Roberts-Galbraith et al., 2009; Cortes et al., 2015; Ren et al., 2015; Sethi et al., 2016), we divided the IDR into three segments of 175 aa (Figure 3A) and deleted each segment individually. Each of the three deletion mutants was viable (Figure 3B). However, the three mutants had different phenotypes: cdc15-∆1 cells were morphologically indistinguishable from WT cells, while cdc15-∆2 and cdc15-∆3 cells had defects in morphology and cell division, as evidenced by an increased number of nuclei and septa per cell (Figure 3, B and C). We considered the possibility that the cytokinetic defects observed in cdc15-∆2 and cdc15-∆3 resulted from simultaneously removing key aa residues and shortening the separation between the F-BAR and SH3 domains. Therefore, we tested whether replacing the IDR with duplicated or triplicated segment 2 (aa 503–677) would rescue the morphological defects of cdc15-∆3 (Supplemental Figure S1D). However, both of these strains resembled the phenotype of cdc15-∆3 cells: a high percentage of cells were septated and length at septation was increased, indicating growth initiation before separation was accomplished (Supplemental Figure S1E). This points to a specific function of aa 503–854.
FIGURE 3:
Cdc15 IDR deletion mutants have cytokinesis defects. (A) Schematic of Cdc15 indicating the aa boundaries of IDR regions. (B) BF images of the indicated strains. Bar, 5 µm. (C) Quantification of the number of nuclei (left) and septa (right) per cell of the indicated strains, determined from cells stained with DAPI and methyl blue to visualize DNA and septum, respectively. Results are the means from three biological replicates, n > 198 cells per replicate. Error bars are SD. *, p < 0.0001. Ordinary two-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test. (D) Table of cdc15 alleles, schematic of gene product, and ability to rescue cdc15 null. Schematics are drawn to scale. Black lines indicate deletion. (E) Duration of cytokinesis stages determined from imaging of strains expressing the indicated cdc15 allele plus rlc1-mNG and sid4-mNG to label the CR and SPB, respectively. Results are the means from three biological replicates, n > 5 cells per replicate, and the total number of cells analyzed for each strain is indicated on the graph. Error bars are SD. *, p < 0.0001. Ordinary one-way ANOVA with Tukey’s multiple comparisons test. (F) Representative time-lapse series of the indicated strains. Images were acquired every 2 min. A single BF slice and max intensity z-projections of deconvolved mNG images are shown. Numbers indicate min from SPB separation. Bar, 5 µm. Double arrow indicates first frame when CR loses ringlike structure. Yellow arrowheads point out first and last frame with visible asymmetric septum deposition.
As described above, the shortest viable C-terminal truncation of Cdc15 was at aa 710 (Figure 1A). Therefore, although segment 3 (aa 678–854) could be deleted when the SH3 domain was present (Figure 3B), cells required part of segment 3 in the absence of the SH3 domain. This result suggested that there are likely multiple functional motifs along the length of Cdc15 and that loss of some combinations, but not others, might be tolerated. To expand on this idea, we combined deletions of IDR segments and/or the SH3 domain. While cdc15-∆1∆3 and cdc15-∆1∆SH3 are viable, cdc15-∆1∆2, cdc15-∆2∆3, and cdc15-∆2∆SH3 are not (Figure 3D). These results indicate that region 2 mediates a particularly important function such that its loss is barely tolerated.
Segments 2 and 3 of Cdc15’s IDR contain multiple short stretches that have similar sequence identity among Schizosaccharomyces species (Supplemental Figure S2B). Many of these conserved motifs also have high prediction for protein binding (ANCHOR; Dosztanyi et al., 2009; Meszaros et al., 2009; Supplemental Figure S2A). Therefore, deletions of these short conserved motifs were tested to better define important functional motifs within the IDR. However, none of the smaller deletions resulted in the cytokinetic phenotypes observed for cdc15-∆2 or cdc15-∆3 (Supplemental Figure S2C).
Cdc15 IDR mutants localize to the division site and are phosphoregulated
We confirmed by SDS–PAGE and immunoblotting that Cdc15-∆1, Cdc15-∆2, and Cdc15-∆3 were produced at levels comparable to WT Cdc15 (Supplemental Figure S3A). Additionally, the three single segment–deletion mutants localized to nodes and CRs (Supplemental Figure S3B).
Next, we examined whether the Cdc15 segment deletions were still subject to cell cycle–regulated phosphoregulation, as the IDR is the primary site of Cdc15 phosphorylation (Wilson-Grady et al., 2008; Roberts-Galbraith et al., 2010; Koch et al., 2011; Chen et al., 2013; Carpy et al., 2014; Kettenbach et al., 2015; Swaffer et al., 2016, 2018; Lee et al., 2018). During interphase, Cdc15 is highly phosphorylated, which is evidenced by slower mobility on SDS–PAGE (Fankhauser et al., 1995; Roberts-Galbraith et al., 2010). During mitosis and cytokinesis, Cdc15 is dephosphorylated (Fankhauser et al., 1995; Roberts-Galbraith et al., 2010). Mutants that abolish phosphorylation exhibit defects in cytokinesis and localize abnormally to the cell cortex during interphase (Roberts-Galbraith et al., 2010). Lambda phosphatase collapse revealed that Cdc15-∆1, Cdc15-∆2, and Cdc15-∆3 were still phosphorylated (Supplemental Figure S3C). Additionally, we determined that Cdc15-∆1, Cdc15-∆2, and Cdc15-∆3 maintained cell cycle changes in phosphorylation (Supplemental Figure S3D). Furthermore, mEGFP-Cdc15-∆2 and mEGFP-Cdc15-∆3 showed diffuse localization during interphase (Supplemental Figure S3E), suggesting that the division defects of cdc15-∆2 and cdc15-∆3 are not mimicking Cdc15 phospho-ablating mutants. In contrast, mEGFP-Cdc15-∆1 accumulated in cortical puncta during interphase despite the fact that cdc15-∆1 have normal morphology (Supplemental Figure S3E). These findings suggest that phosphorylation events throughout the IDR may have differential effects on Cdc15 function.
The Cdc15 IDR is important for normal cytokinesis dynamics and CR integrity
To better understand the defects observed in cdc15 IDR segment deletions, we measured the cytokinesis dynamics of mutants expressing spindle pole body (SPB) protein Sid4-mNG and CR protein Rlc1-mNG, which mark progression through mitosis and cytokinesis, respectively. As expected from the morphological analysis, the cytokinesis dynamics of cdc15-∆1 were like that of WT. In contrast, the CR of cdc15-∆2 cells took longer to constrict, while cdc15-∆3 cells showed increased duration of both CR maturation (defined as the time between CR assembly and constriction) and CR constriction (Figure 3E).
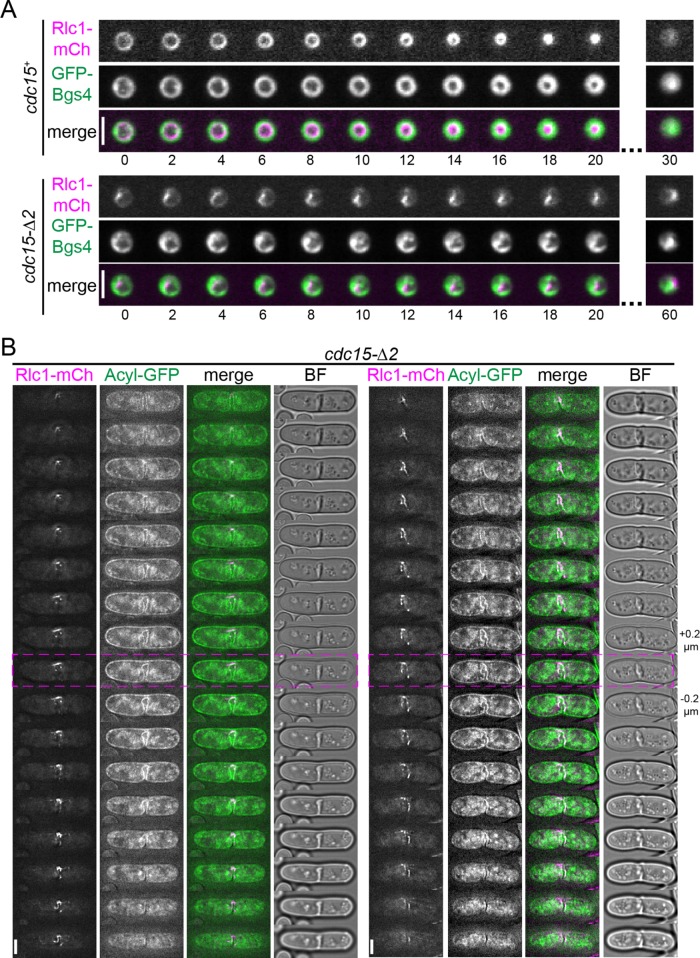
We were particularly interested in the cytokinesis defects of cdc15-∆2 cells, since segment 2 was the minimal IDR that supported viability (i.e., cdc15-∆1∆3 was viable; Figure 3D). Interestingly, the increased length of CR constriction in cdc15-∆2 mutants correlated with a loss of CR integrity upon constriction initiation and subsequent asymmetric deposition of cell wall material (Figure 3F). More precisely, the Rlc1-mNG signal in z-projections became asymmetric and accumulated at one point of the cell, appearing to collapse and lose circularity (Figure 3F, double arrowhead). Cdc15-∆2 and actin colocalized with Rlc1, indicating that the entire CR collapsed to this single point (Supplemental Figure S4, A and B). After CR collapse, septum formation became visible at the point(s) where the CR proteins remained (Figure 3F, yellow arrowheads). This was confirmed by imaging Rlc1-mCherry (mCh) to mark the CR and glucan synthase GFP-Bgs4 to mark the position of cell wall deposition in cells oriented vertically (Wang and Tran, 2014) (Figure 4A) and in the standard xy plane (Supplemental Figure S4C). A Z-series of images of Rlc1-mCh and the membrane marker Acyl-GFP also provided a clear view of asymmetric furrowing in cdc15-∆2 cells (Figure 4B). The observation that the position of septum formation corresponded to the asymmetric localization of Rlc1 is consistent with a previous report that the CR or actin remnants of the CR locally stimulate cell wall synthesis (Zhou et al., 2015). Asymmetric septation has not been observed in any previously characterized Cdc15 mutant with defective membrane association or oligomerization or reduced abundance (Huang et al., 2007; Arasada and Pollard, 2014; McDonald et al., 2015). Vice versa, the CR unraveling observed in the absence of both Cdc15’s and Imp2’s SH3 domains (Roberts-Galbraith et al., 2009) and the CR sliding observed in F-BAR oligomerization and membrane-binding mutants were not observed in cdc15-∆2 cells during time-lapse live-cell imaging (e.g., Figure 3F), nor did cdc15-∆2 cells have off-center septa (as measured by long-to-short cell ratio; Supplemental Figure S4D). These results suggest that the function of segment 2 is distinct from that of the F-BAR and SH3 domains.
FIGURE 4:
Characterizing the CR defect in cdc15-∆2. (A) End-on imaging of the indicated strains. Numbers indicate minutes elapsed since the start of the series. Bar, 5 µm. (B) Z-slices of two representative cells of the indicated genotype. The pink dashed lines mark the middle slice. Z-slices were taken every 0.2 µm through the volume of the cell. Fluorescence images are deconvolved. Bar, 5 µm.
Cell wall enzymes are trafficked to the division site and active in cdc15-∆2
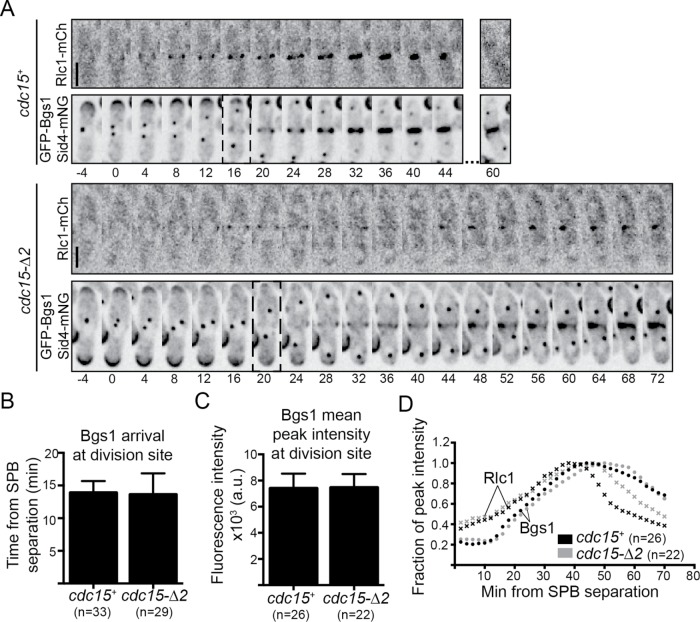
The septum is a trilaminar structure. The Bgs1-dependent primary septum is deposited first and sandwiched by secondary septa deposited by Bgs4 and Ags1; enzymatic digestion of the primary septum leads to daughter cell separation (reviewed in Willet et al., 2015b; García Cortés et al., 2016; Perez et al., 2016). We have already demonstrated that Bgs4 is recruited to the division site (Figure 4A and Supplemental Figure S4C). However, the Cdc15 IDR has been implicated in the recruitment of Bgs1 (Arasada and Pollard, 2014). Therefore, to test whether cdc15-∆2 cells failed to recruit Bgs1 adequately, we monitored GFP-Bgs1 localization in cdc15-∆2 cells. GFP-Bgs1 was recruited to the division site with similar temporal kinetics and abundance in WT and cdc15-∆2 (Figure 5). However, consistent with the asymmetric localization of GFP-Bgs4 (Figure 4A) and the deposition of septum material (Figures 3E and 4), GFP-Bgs1 distribution was not always circumferential in cdc15-∆2 cells (Figure 5A). Taken together, these data indicate that cell wall synthesis is activated at the proper time but not necessarily in the proper location in cdc15-∆2. In fact, cdc15-∆2 was synthetic lethal with bgs1-ts/cps1-191 (Supplemental Figure S5B), indicating that robust cell wall deposition is crucial for division and viability of cdc15-∆2.
FIGURE 5:
Cell wall synthases are properly trafficked in cdc15-∆2. (A) Representative time-lapse series of the indicated strains. Sum z-projections are shown. Images were acquired every 2 min. Numbers indicate minutes from SPB separation. Bar, 5 µm. (B) Timing of GFP-Bgs1 arrival to the division site in the indicated strains also expressing Sid4-mNG and Rlc1-mCh. Error bars are SD. p = 0.67. Student’s t test. (C) Average peak fluorescence of GFP-Bgs1 in the indicated strains also expressing Sid4-mNG and Rlc1-mCh. Error bars are SD. p = 0.85. Student’s t test. (D) Timing of Rlc1 (×) and Bgs1 (•) peak intensity relative to SPB separation for the indicated strains determined from strains imaged as in (A). Results in A–D are the means from two biological replicates, n > 9 cells per replicate, and total number of cells analyzed for each strain is indicated on the graph.
CR composition is altered in cdc15-∆2 cells
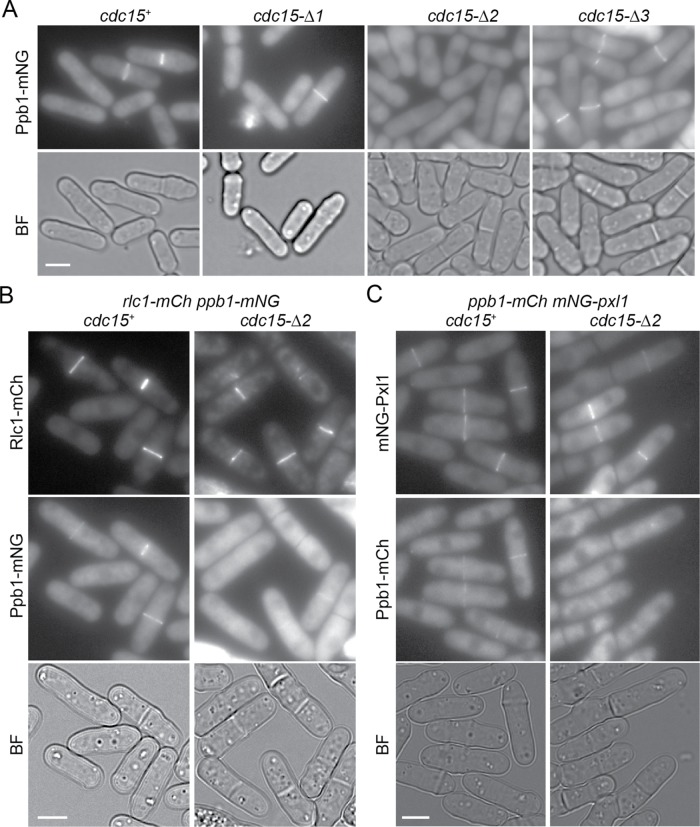
The results described above indicate that the asymmetry of CR constriction in cdc15-∆2 is not due to a defect in septation but rather results from a CR-intrinsic defect that leads to loss of a ring structure upon constriction onset and instead CR proteins remain at the division site as arcs or strands. Indeed, cdc15-∆2 mutants were sensitive to a low dose of latrunculin A (Supplemental Figure S5A) and were synthetically sick or lethal with numerous regulators of the actin cytoskeleton (Supplemental Figure S5B), indicating lower tolerance to further perturbations of the CR. Therefore, we used fluorescence microscopy to screen actin-binding proteins, actin regulators, Cdc15-binding partners, and other CR proteins for changes in their localization to the division site in cdc15-∆2 (Supplemental Figure S5C). The calcineurin phosphatase, which comprises catalytic subunit Ppb1 and regulatory subunit Cnb1, has been found to regulate Cdc15 (Martin-Garcia et al., 2018). Also, the morphologies of cdc15-∆2 and ppb1∆ (Yoshida et al., 1994) are similar. This prompted us to examine Ppb1’s localization in cdc15-∆2. We found that Ppb1 was greatly reduced at the CR of cdc15-∆2 cells, but it localized normally in other cdc15 segment deletions (Figure 6A). This was further confirmed by two-color imaging of Ppb1 and the CR marker Rlc1 (Figure 6B). The localization of Ppb1 to the CR was previously found to depend on the CR protein Pxl1 (Martin-Garcia et al., 2018), which associates with Cdc15 by Co-IP (Pinar et al., 2008; Roberts-Galbraith et al., 2009). Given that Ppb1 was essentially absent from the CR, it was surprising to find that Pxl1 localized normally to the division site in cdc15-∆2 (Figure 6C). This result suggests that Pxl1 is not sufficient for normal Ppb1 localization to the CR.
FIGURE 6:
Calcineurin is not recruited to CR in cdc15-∆2. (A–C) Representative sum projections of the indicated strains. Bar, 5 µm. (A) At least 100 cells from 3 d of imaging were examined. (B, C) At least 50 cells were examined.
DISCUSSION
It was unknown what protein feature(s) distinguish the essential Cdc15 from its nonessential paralogue Imp2. Using domain-swapping experiments, we determined that the essential function of the F-BAR domain can be replaced by Imp2’s F-BAR domain as well as other human F-BAR domains, whereas Cdc15’s IDR is uniquely essential and cannot be replaced by that of Imp2. Deletions of segments of the IDR exhibit either lengthened periods of CR maturation and constriction or loss of CR integrity upon constriction onset, resulting in asymmetric septum deposition (cdc15-∆3 and cdc15-∆2, respectively). Further, the calcineurin subunit Ppb1 is largely absent from the CR of cdc15-∆2 cells. This identifies a new function for the Cdc15 IDR in recruitment and/or maintenance of calcineurin at the CR, and we predict that the IDR has additional functions.
The exchangeability of F-BAR domains
Five of the 15 F-BAR domains tested replaced the essential function of Cdc15’s F-BAR domain: the F-BAR domains of S. pombe Imp2 and human Gas7, PSTPIP1, PACSIN2/syndapin2, and Fer (Supplemental Table S1). Given that the proteins in which these domains normally reside are involved in a variety of apparently unlinked functions and the F-BAR domains themselves have distinct biochemical characteristics and structures, we conclude that there is significant plasticity among F-BAR domains. This is perhaps not surprising, since all F-BAR domains share the ability to interact electrostatically with negatively charged phospholipids and to oligomerize (Supplemental Table S2; Tsujita et al., 2006; Shimada et al., 2010; Bai et al., 2012; Bai and Zheng, 2013; Goh et al., 2012; McDonald et al., 2015, 2016; McDonald and Gould, 2016). Although there are reported differences among F-BAR proteins in phospholipid or curvature specificity, F-BAR domains themselves interact relatively nonspecifically with negatively charged phospholipids, and it is only protein features adjacent to the F-BAR domain, such as the FX(C) domain (Itoh et al., 2009; Yamamoto et al., 2018), that convey phospholipid or curvature specificity to the core F-BAR domain (reviewed in Itoh and De Camilli, 2006). Thus, in the context of the full-length protein, an F-BAR domain acts as a module that concentrates the protein on membranes where other linked modules (e.g., SH3 domains, RhoGAP domains) provide the bulk of specific functions.
In terms of the F-BAR-Cdc15 chimeras’ defects or lethality, multiple explanations can be imagined. In addition to the strong possibility that some fusion proteins were not structurally sound, some F-BAR domains bind directly to other proteins (Shoham et al., 2003; Hansen et al., 2011; Senju et al., 2011; Begonja et al., 2015; Garabedian et al., 2018; Liu et al., 2019), including Cdc15’s F-BAR domain that binds the formin Cdc12 (Willet et al., 2015a). However, a mutant that disrupts the Cdc15-Cdc12 interaction is viable (Willet et al., 2015a), so this is unlikely to be the only explanation for defects/lethality. Loss or gain of other protein partners or variations in oligomerization strategies might also be factors contributing to the lack of function of some chimeras (reviewed in McDonald and Gould, 2016).
CR strands or fragments direct primary septum deposition
In cdc15-∆2, the CR appears to form normally and the glucan synthases are normally recruited, but upon constriction, the CR loses its structure and Bgs1 then colocalizes with the CR-protein strands/arclike structures that remain. As a result, the septum is deposited—albeit inefficiently—from that point. This behavior of the CR and Bgs1 in cdc15-∆2 cells is consistent with the idea that the primary purpose of the S. pombe CR may be to direct efficient cell wall deposition, which in turn provides the force for division (Proctor et al., 2012; Stachowiak et al., 2014; Thiyagarajan et al., 2015; Willet et al., 2015b; Zhou et al., 2015). The CR has been previously proposed to direct vesicular traffic (Vjestica et al., 2008), and the colocalization of glucan synthases with CR remnants throughout constriction in cdc15-∆2 indicates that the CR is indeed a landmark for glucan synthase delivery. CR contraction force has also been proposed to mechanically stimulate glucan synthase activity (Zhou et al., 2015), and it is possible that the CR strands observed in cdc15-∆2 are contraction-competent. However, in the absence of all myosin-II activity (N-degron-myo2 myp2∆), primary septum is still deposited (Okada et al., 2019), suggesting that mechanostimulation is not required, although it could provide positive feedback.
cdc15-∆2 is not the only mutant to display this type of CR behavior. A temperature-sensitive mutant of the essential myosin II heavy chain myo2-E1 displays similar CR dynamics at its nonpermissive temperature (Palani et al., 2017; Okada et al., 2019). Additionally, a phosphomutant of the formin Cdc12 that prevents its phosphorylation by the SIN kinase Sid2 also has CRs that lose their ring structure during a cps1-191 arrest when otherwise WT CRs do not (Bohnert et al., 2013). Single time-point images of the CR and/or septum suggest similar CR behavior in other mutants, including temperature-sensitive cofilin adf1-1 (Nakano and Mabuchi, 2006b) and deletions of both acp1 that encodes actin-capping protein (Kovar et al., 2005; Nakano and Mabuchi, 2006a) and fim1 that encodes the actin bundler fimbrin (Wu et al., 2001).
IDRs in F-BAR proteins
Our study revealed that like the F-BAR domain, Cdc15’s IDR is essential, but unlike the F-BAR domain, it cannot be replaced by that of its paralogue Imp2. In other contexts as well, the functional specificity of F-BAR proteins must be considered as the combined effect of their F-BAR domains and adjacent protein regions, including globular domains and IDRs. The IDR of S. cerevisiae Hof1 mediates its localization to the CR and is important for CR contraction; it also binds septins (Meitinger et al., 2011, 2013; Oh et al., 2013). The IDR in FCHO2 binds and allosterically activates AP-2 (Hollopeter et al., 2014; Umasankar et al., 2014). The IDR in Fer undergoes a disorder-to-order transition to convey a curvature sensing function (Yamamoto et al., 2018). In the case of Cdc15’s IDR, we found that it plays a key role in recruiting the phosphatase calcineurin. Calcineurin localization at the CR depends on the second region of Cdc15’s IDR (aa 503–677) and Pxl1 (Martin-Garcia et al., 2018), and determining how these two proteins collaborate for proper calcineurin localization will be an important next step. Cdc15-∆2 is dephosphorylated with timings similar to WT, which indicates that Cdc15’s role scaffolding calcineurin at the CR would be separate from its identity as a calcineurin substrate (Martin-Garcia et al., 2018), and furthermore that either other phosphatases are redundant with calcineurin or cytoplasmic calcineurin dephosphorylates Cdc15. Finally, because cdc15-∆3 also has cytokinesis defects, but is not required for calcineurin recruitment to the CR, it will be interesting to determine the other functions performed by Cdc15’s IDR.
MATERIALS AND METHODS
Yeast methods
S. pombe strains (Supplemental Table S3) were grown in yeast extract with supplements (YES), YES without additional adenine, or Edinburgh minimal media (EMM) plus selective supplements (Moreno et al., 1991; Forsburg and Rhind, 2006). Transformation of yeast with plasmid or linear DNA was accomplished using electroporation (Forsburg and Rhind, 2006) and lithium acetate methods (Keeney and Boeke, 1994; Forsburg and Rhind, 2006), respectively.
For integration of cdc15 alleles at the endogenous locus, cdc15+/cdc15::ura4+ was transformed with pIRT2 vector with the desired cdc15 allele plus cdc15 5′ and 3′ noncoding regions (Roberts-Galbraith et al., 2009). Spores were germinated on selective medium to select for cdc15::ura4+ haploids that have the vector. Haploid integrants were recovered based on resistance to 5-fluorourotic acid (FOA) and integration plus loss of vector was verified by growth on selective media, PCR and/or microscopy, and finally sequencing.
To confirm the lethality of cdc15 mutants, the ura4+ cassette in a cdc15+/cdc15::ura4+ diploid strain was replaced with a cassette consisting of a 500-base-pair 5′ cdc15+ flanking region, the mutant cdc15 coding sequence, kanR, and a 500-base-pair 3′ cdc15+ flanking region. Integrants resistant to 5-FOA and G418 were selected and confirmed by whole-cell PCR before sporulation and tetrad dissection on YES agar. Plates were grown at 32°C. At least 10 tetrads were dissected for each integrant. Production of the gene product was confirmed using lysates from the diploid and/or by expressing from the plasmid. In some cases, the experiment was repeated and tetrad plates were grown at alternative temperatures of 25 and/or 29°C; in all cases, the results were the same.
Genes were tagged at the 3′ ends of their ORFs with mNG:kanR, mNG:hygR, or mCh:natR using pFA6 cassettes as previously described (Wach et al., 1994; Bahler et al., 1998; Willet et al., 2015a). Integration of tags was verified using whole-cell PCR and/or microscopy.
The mNG-pxl1:kanR strain was generated by transforming KGY16851 with a cassette consisting of a 500-base-pair 5′ pxl1+ flanking region, the mNG-pxl1 coding sequence, kanR, and a 500-base-pair 3′ pxl1+ flanking region. Integrants resistant to G418 were selected and confirmed by whole-cell PCR.
Introduction of tagged loci or cdc15 mutants into other genetic backgrounds was accomplished using standard S. pombe mating, sporulation, and tetrad dissection techniques.
For growth assays, cells were grown to log phase in YES medium at 32°C for Supplemental Figure S2C and 25°C for Supplemental Figure S5, A and B. A suspension of 40 × 106 cells/ml YES was serially diluted 10-fold and 3 µl of each dilution was spotted on YES plates for growth at indicated temperatures. All spot assays were performed in duplicate.
Molecular biology
cdc15 plasmids were created by amplifying cdc15 fragments by PCR from pKG4456 (Roberts-Galbraith et al., 2009) and subsequent cloning into pIRT2 for integration at the endogenous locus (Roberts-Galbraith et al., 2009). All constructs were sequenced for verification.
Microscopy methods
Except for Figures 4A and 5, images of S. pombe cells were acquired using a Personal DeltaVision (GE Healthcare, Issaquah, WA) that includes an Olympus IX71 microscope, 60 × NA 1.42 Plan Apochromat and 100 × NA 1.40 U Plan S Apochromat objectives, fixed and live-cell filter wheels, a Photometrics CoolSnap HQ2 camera, and softWoRx imaging software (Applied Precision).
For live-cell fluorescence imaging, strains were grown overnight to log phase in YES media at 25°C in a shaking water bath, except for the chimeras in Figure 2B, which were grown at 32°C. For the brightfield (BF) images in Figure 3B and Supplemental Figure S1E, cells were grown at 32°C.
For Figures 2B, 3B, 4B, and 6 and Supplemental Figures S1E, S3B, S4C, and S5C, cells were imaged in YES media at 23–29°C.
For Supplemental Figure S4A, time-lapse imaging was performed on a 2% MAUL agarose pad. Images were acquired every 2 min.
For Figure 3, E and F, time-lapse imaging was performed using an ONIX microfluidics perfusion system (CellASIC ONIX; EMD Millipore). A suspension of 50 µl of 40 × 106 cells/ml YES was loaded into Y04C plates for 5 s at 8 psi. YES medium was flowed into the chamber at 5 psi throughout imaging. Time-lapse images were obtained at 2-min intervals. Representative fluorescence images are maximum-intensity or sum projections of z sections spaced at 0.2–0.5 µm that have been deconvolved with 10 iterations.
For Figure 4A, end-on images were acquired with a spinning-disk system, which includes a Zeiss Axiovert200m microscope, Yokogawa CSU-22, 63X NA 1.46 planApochromat and 100X numerical aperture (NA) 1.40 PlanApo oil immersion objectives, and a Hamamatsu ImageEM-X2 camera. Cells were grown to log phase in EMM plus adenine, uracil, and leucine, loaded into vertical chambers in a 4% MAUL agarose pad, and imaged every 2 min. Six z-slices at 0.5-µm intervals around the middle of the cell were acquired at each time point.
For Figure 5, time-lapse images were acquired with a Nikon spinning-disk system, which incorporates a Yokogawa CSU-X1 spinning disk head, Andor DU-897 EMCCD, a high-speed piezo [z] stage, a four-line high-powered solid state laser launch, and a Plan Apo Lambda (oil) 60 × 1.40 NA WD 0.13 mm. Cells were mounted on 2% MAUL agarose pads. Images were acquired every 2 min. Fifteen z-slices at 0.4-µm intervals were acquired at each time point. Sum projections were analyzed and used for representative images. Sum projections were corrected for background fluorescence before quantitation.
For Figure 3C and Supplemental Figures S1, A and B, and S4D, to visualize nuclei and septa, cells were grown to log phase at 32°C and fixed with ice-cold 70% ethanol at 4°C for at least 30 min. Approximately 0.5 OD of fixed cells were washed once with PBS, pH 7.5, and then incubated in 50 µl 1-mg/ml methyl blue (MB; Sigma; M6900) for 30 min at RT. MB-stained cells were concentrated by centrifugation and mixed with 5 µg/ml DAPI (4′,6-diamidino-2-phenylindole) (Sigma; D9542) at a 1:1 ratio immediately before imaging.
For Supplemental Figure S4B, cells were fixed in formaldehyde, permeabilized for 2 min in PBS with 0.1% NP-40, washed with PBS, and then stained with Alexa-Fluor-488-phalloidin for 60 min before imaging.
Image analysis was performed using ImageJ software (Schindelin et al., 2012).
Protein methods
Cell pellets were snap-frozen in dry ice–ethanol baths. Denatured lysates were prepared by bead disruption (Fastprep cell homogenizer; MP Biomedicals) in NP-40 buffer containing SDS as previously described (Gould et al., 1991), except with the addition of 0.5 mM diisopropyl fluorophosphates (Sigma-Aldrich).
For immunoprecipitation and lambda phosphatase collapse, lysates were incubated with anti-Cdc15 (Roberts-Galbraith et al., 2009) and Protein A magnetic beads or sepharose (GE Healthcare; 17-5280-04) for 2 h at 4°C. Beads were washed three times with NP-40 buffer and twice with phosphatase buffer (150 mM NaCl, 50 mM HEPES, pH 7.4) before being split in two and added to either lambda phosphatase reaction or control. Lambda phosphatase collapse was performed according to the manufacturer’s protocol (New England Biolabs; P0753). Reactions were stopped by the addition of sample buffer.
Protein samples were resolved by SDS–PAGE and transferred to a polyvinylidene fluoride membrane (Immobilon P, EMD Millipore). Anti–α-tubulin (B512) mouse monoclonal antibody (Sigma-Aldrich), anti-Cdc2 (PSTAIRE) mouse monoclonal antibody (Sigma-Aldrich; P7962), or anti-Cdc15(1-405) rabbit polyclonal antibody (Roberts-Galbraith et al., 2009) was used in immunoprecipitations and/or as a primary antibody in immunoblotting. Secondary antibodies were conjugated to IRDye800 or IRDye680 (LI-COR Biosciences). Blotted proteins were detected via an Odyssey machine (LI-COR Biosciences).
Statistical analysis
Statistical analysis was performed using Prism (GraphPad).
Supplementary Material
Acknowledgments
We are grateful to Anna Feoktistova, who cloned and integrated the F-BAR chimeras and performed the initial viability analysis, and we thank Nathan A. McDonald for designing the large deletions and making the initial cdc15 constructs, Jian-Qiu Wu for strains, Alaina H. Willet for critical comments on the manuscript, and all current and past members of the Gould lab for thoughtful discussion and encouragement. This work was supported by grants from the National Institute of General Medical Studies of the National Institutes of Health (NIH) (R01GM101035 to K.L.G., F31GM119252 to M.C.M., T32GM007347 to the Vanderbilt Medical-Scientist Training Program, and T32GM00855421 to C.E.S.) and from the American Heart Association (17PRE33410245 to C.E.S.). Experiments to acquire data for Figure 5 were performed in part through the use of the Vanderbilt Cell Imaging Shared Resource (supported by NIH Grants CA68485, DK20593, DK58404, DK59637, and EY08126).
Abbreviations used:
- BF
brightfield
- CR
cytokinetic ring
- IDR
intrinsically disordered region
- mCh
mCherry
- mNG
mNeonGreen
- PM
plasma membrane
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www .molbiolcell.org/cgi/doi/10.1091/mbc.E19-06-0314) on September 11, 2019.
REFERENCES
- Ahmed S, Bu W, Lee RT, Maurer-Stroh S, Goh WI. (2010). F-BAR domain proteins: families and function. Commun Integr Biol , 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. (2014). Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell Rep , 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast , 943–951. [DOI] [PubMed] [Google Scholar]
- Bai X, Meng G, Luo M, Zheng X. (2012). Rigidity of wedge loop in PACSIN 3 protein is a key factor in dictating diameters of tubules. J Biol Chem , 22387–22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Zheng X. (2013). Tip-to-tip interaction in the crystal packing of PACSIN 2 is important in regulating tubulation activity. Protein Cell , 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begonja AJ, Pluthero FG, Suphamungmee W, Giannini S, Christensen H, Leung R, Lo RW, Nakamura F, Lehman W, Plomann M, et al. (2015). FlnA binding to PACSIN2 F-BAR domain regulates membrane tubulation in megakaryocytes and platelets. Blood , 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Grzegorzewska AP, Willet AH, Vander Kooi CW, Kovar DR, Gould KL. (2013). SIN-dependent phosphoinhibition of formin multimerization controls fission yeast cytokinesis. Genes Dev , 2164–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. (2003). The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe . J Cell Biol , 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B. (2014). Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (fission yeast). Mol Cell Proteomics , 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Broadus MR, McLean JR, Feoktistova A, Ren L, Gould KL. (2013). Comprehensive proteomics analysis reveals new substrates and regulators of the fission yeast clp1/cdc14 phosphatase. Mol Cell Proteomics , 1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes JC, Pujol N, Sato M, Pinar M, Ramos M, Moreno B, Osumi M, Ribas JC, Perez P. (2015). Cooperation between Paxillin-like protein Pxl1 and glucan synthase Bgs1 is essential for actomyosin ring stability and septum formation in fission yeast. PLoS Genet , e1005358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, Sazer S. (1998). imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J Cell Biol , 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztanyi Z, Meszaros B, Simon I. (2009). ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics , 2745–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. (1995). The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell , 435–444. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N. (2006). Basic methods for fission yeast. Yeast , 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke R, Gohl C, Bogdan S. (2010). The F-BAR protein family actin’ on the membrane. Commun Integr Biol , 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabedian MV, Stanishneva-Konovalova T, Lou C, Rands TJ, Pollard LW, Sokolova OS, Goode BL. (2018). Integrated control of formin-mediated actin assembly by a stationary inhibitor and a mobile activator. J Cell Biol , 3512–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Cortés JC, Ramos M, Osumi M, Pérez P, Ribas JC. (2016). The cell biology of fission yeast septation. Microbiol Mol Biol Rev , 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. (2017). Cytokinesis in metazoa and fungi. Cold Spring Harb Perspect Biol , a022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh SL, Wang Q, Byrnes LJ, Sondermann H. (2012). Versatile membrane deformation potential of activated pacsin. PLoS one , e51628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Moreno S, Owen DJ, Sazer S, Nurse P. (1991). Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J , 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CG, Howard G, Nichols BJ. (2011). Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J Cell Sci , 2777–2785. [DOI] [PubMed] [Google Scholar]
- Hollopeter G, Lange JJ, Zhang Y, Vu TN, Gu M, Ailion M, Lambie EJ, Slaughter BD, Unruh JR, Florens L, Jorgensen EM. (2014). The membrane-associated proteins FCHo and SGIP are allosteric activators of the AP2 clathrin adaptor complex. eLife , e03648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chew TG, Ge W, Balasubramanian MK. (2007). Polarity determinants Tea1p, Tea4p, and Pom1p inhibit division-septum assembly at cell ends in fission yeast. Dev Cell , 987–996. [DOI] [PubMed] [Google Scholar]
- Itoh T, De Camilli P. (2006). BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta , 897–912. [DOI] [PubMed] [Google Scholar]
- Itoh T, Hasegawa J, Tsujita K, Kanaho Y, Takenawa T. (2009). The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci Signal , ra52. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. (1994). Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics , 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Deng L, Wu Y, Baldissard S, Adamo ME, Gerber SA, Moseley JB. (2015). Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the conserved fission yeast kinase pom1. Mol Cell Proteomics , 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Krug K, Pengelley S, Macek B, Hauf S. (2011). Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal , rs6. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. (2005). Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell , 2313–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ME, Rusin SF, Jenkins N, Kettenbach AN, Moseley JB. (2018). Mechanisms connecting the conserved protein kinases Ssp1, Kin1, and Pom1 in fission yeast cell polarity and division. Curr Biol , 84–92.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. (2000). Involvement of PCH family proteins in cytokinesis and actin distribution. Microsc Res Tech , 168–172. [DOI] [PubMed] [Google Scholar]
- Liu Y, McDonald NA, Naegele SM, Gould KL, Wu JQ. (2019). The F-BAR Domain of Rga7 relies on a cooperative mechanism of membrane binding with a partner protein during fission yeast cytokinesis. Cell Rep , 2540–2548.e2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangione MC, Gould KL. (2019). Molecular form and function of the cytokinetic ring. J Cell Sci , jcs226928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia R, Arribas V, Coll PM, Pinar M, Viana RA, Rincon SA, Correa-Bordes J, Ribas JC, Perez P. (2018). Paxillin-mediated recruitment of calcineurin to the contractile ring is required for the correct progression of cytokinesis in fission yeast. Cell Rep , 772–783.e774. [DOI] [PubMed] [Google Scholar]
- McDonald NA, Gould KL. (2016). Linking up at the BAR: oligomerization and F-BAR protein function. Cell Cycle , 1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Takizawa Y, Feoktistova A, Xu P, Ohi MD, Vander Kooi CW, Gould KL. (2016). The tubulation activity of a fission yeast F-BAR protein is dispensable for its function in cytokinesis. Cell Rep , 534–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. (2015). Oligomerization but not membrane bending underlies the function of certain F-BAR proteins in cell motility and cytokinesis. Dev Cell , 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. (2011). Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev , 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Palani S, Hub B, Pereira G. (2013). Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol Biol Cell , 1290–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B, Simon I, Dosztanyi Z. (2009). Prediction of protein binding regions in disordered proteins. PLoS Comput Biol , e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol , 795–823. [DOI] [PubMed] [Google Scholar]
- Morrell-Falvey JL, Ren L, Feoktistova A, Haese GD, Gould KL. (2005). Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J Cell Sci , 5563–5573. [DOI] [PubMed] [Google Scholar]
- Nakano K, Mabuchi I. (2006a). Actin-capping protein is involved in controlling organization of actin cytoskeleton together with ADF/cofilin, profilin and F-actin crosslinking proteins in fission yeast. Genes Cells , 893–905. [DOI] [PubMed] [Google Scholar]
- Nakano K, Mabuchi I. (2006b). Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol Biol Cell , 1933–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. (2013). Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol Biol Cell , 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Wloka C, Wu JQ, Bi E. (2019). Distinct roles of Myosin-II isoforms in cytokinesis under normal and stressed conditions. iScience , 69–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwubiko UN, Mlynarczyk PJ, Wei B, Habiyaremye J, Clack A, Abel SM, Das ME. (2019). A Cdc42 GEF, Gef1, through endocytosis organizes F-BAR Cdc15 along the actomyosin ring and promotes concentric furrowing. J Cell Sci . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani S, Chew TG, Ramanujam S, Kamnev A, Harne S, Chapa YLB, Hogg R, Sevugan M, Mishra M, Gayathri P, Balasubramanian MK. (2017). Motor activity dependent and independent functions of Myosin II contribute to actomyosin ring assembly and contraction in schizosaccharomyces pombe. Curr Biol , 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez P, Cortes JC, Martin-Garcia R, Ribas JC. (2016). Overview of fission yeast septation. Cell Microbiol , 1201–1207. [DOI] [PubMed] [Google Scholar]
- Pinar M, Coll PM, Rincon SA, Perez P. (2008). Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis. Mol Biol Cell , 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor SA, Minc N, Boudaoud A, Chang F. (2012). Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr Biol , 1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen JS, Huang H, Guillen R, Boone C, Sidhu SS, et al. (2015). The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell , 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. (2009). The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol , 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. (2010). Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle , 4091–4097. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. (2010). Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell , 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senju Y, Itoh Y, Takano K, Hamada S, Suetsugu S. (2011). Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J Cell Sci , 2032–2040. [DOI] [PubMed] [Google Scholar]
- Sethi K, Palani S, Cortes JC, Sato M, Sevugan M, Ramos M, Vijaykumar S, Osumi M, Naqvi NI, Ribas JC, Balasubramanian M. (2016). A new membrane protein Sbg1 links the contractile ring apparatus and septum synthesis machinery in fission yeast. PLoS Genet , e1006383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Takano K, Shirouzu M, Hanawa-Suetsugu K, Terada T, Toyooka K, Umehara T, Yamamoto M, Yokoyama S, Suetsugu S. (2010). Mapping of the basic amino-acid residues responsible for tubulation and cellular protrusion by the EFC/F-BAR domain of pacsin2/Syndapin II. FEBS Lett , 1111–1118. [DOI] [PubMed] [Google Scholar]
- Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, Kastner DL. (2003). Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci USA , 13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MR, Laplante C, Chin HF, Guirao B, Karatekin E, Pollard TD, O’Shaughnessy B. (2014). Mechanism of cytokinetic contractile ring constriction in fission yeast. Dev Cell , 547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. (2016). CDK substrate phosphorylation and ordering the cell cycle. Cell , 1750–1761.e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P. (2018). Quantitative phosphoproteomics reveals the signaling dynamics of cell-cycle kinases in the fission yeast Schizosaccharomyces pombe. Cell Rep , 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadura V, Garcia B, Garcia I, Garcia P, Sanchez Y. (2004). Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J Cell Sci , 6163–6174. [DOI] [PubMed] [Google Scholar]
- Thiyagarajan S, Munteanu EL, Arasada R, Pollard TD, O’Shaughnessy B. (2015). The fission yeast cytokinetic contractile ring regulates septum shape and closure. J Cell Sci , 3672–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. (2012). Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci , 509–516. [DOI] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. (2006). Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol , 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umasankar PK, Ma L, Thieman JR, Jha A, Doray B, Watkins SC, Traub LM. (2014). A clathrin coat assembly role for the muniscin protein central linker revealed by TALEN-mediated gene editing. eLife , e04137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjestica A, Tang XZ, Oliferenko S. (2008). The actomyosin ring recruits early secretory compartments to the division site in fission yeast. Mol Biol Cell , 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast , 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. (2006). Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell , 3254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tran PT. (2014). Visualizing single rod-shaped fission yeast vertically in micro-sized holes on agarose pad made by soft lithography. Methods Cell Biol , 227–234. [DOI] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. (2015a). The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol , 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Gould KL. (2015b). Regulation of contractile ring formation and septation in Schizosaccharomyces pombe. Curr Opin Microbiol , 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Grady JT, Villen J, Gygi SP. (2008). Phosphoproteome analysis of fission yeast. J Proteome Res , 1088–1097. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Bahler J, Pringle JR. (2001). Roles of a fimbrin and an alpha-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell , 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kondo A, Itoh T. (2018). A curvature-dependent membrane binding by tyrosine kinase Fer involves an intrinsically disordered region. Biochem Biophys Res Commun , 1522–1527. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Toda T, Yanagida M. (1994). A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J Cell Sci , 1725–1735. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Munteanu EL, He J, Ursell T, Bathe M, Huang KC, Chang F. (2015). The contractile ring coordinates curvature-dependent septum assembly during fission yeast cytokinesis. Mol Biol Cell , 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.