Abstract
BACKGROUND
To solve the problem of liver transplantation donor insufficiency, an alternative cell transplantation therapy was investigated. We focused on amniotic epithelial cells (AECs) as a cell source because, unlike induced pluripotent stem cells, they are cost-effective and non-tumorigenic. The utilization of AECs in regenerative medicine, however, is in its infancy. A general profile for AECs has not been comprehensively analyzed. Moreover, no hepatic differentiation protocol for AECs has yet been established. To this end, we independently compiled human AEC libraries, purified amniotic stem cells (ASCs), and co-cultured them with mesenchymal stem cells (MSCs) and human umbilical vein endothelial cell (HUVECs) in a 3D system which induces functional hepatic organoids.
AIM
To characterize AECs and generate functional hepatic organoids from ASCs and other somatic stem cells
METHODS
AECs, MSCs, and HUVECs were isolated from the placentae and umbilical cords of cesarean section patients. Amnion and primary AEC stemness characteristics and heterogeneity were analyzed by immunocytochemistry, Alkaline phosphatase (AP) staining, and flow cytometry. An adherent AEC subpopulation was selected and evaluated for ASC purification quality by a colony formation assay. AEC transcriptomes were compared with those for other hepatocytes cell sources by bioinformatics. The 2D and 3D culture were compared by relative gene expression using several differentiation protocols. ASCs, MSCs, and HUVECs were combined in a 3D co-culture system to generate hepatic organoids whose structure was compared with a 3D AEC sphere and whose function was elucidated by immunofluorescence imaging, periodic acid Schiff, and an indocyanine green (ICG) test.
RESULTS
AECs have certain stemness markers such as EPCAM, SSEA4, and E-cadherin. One AEC subpopulation was also either positive for AP staining or expressed the TRA-1-60 and TRA-1-81 stemness markers. Moreover, it could form colonies and its frequency was enhanced ten-fold in the adherent subpopulation after selective primary passage. Bioinformatics analysis of ribose nucleic acid sequencing revealed that the total AEC gene expression was distant from those of pluripotent stem cells and hepatocytes but some gene expression overlapped among these cells. TJP1, associated with epidermal growth factor receptor, and MET, associated with hepatocyte growth factor receptor, were upregulated and may be important for hepatic differentiation. In conventional flat culture, the cells turned unviable and did not readily differentiate into hepatocytes. In 3D culture, however, hepatic gene expression of the AEC sphere was elevated even under a two-step differentiation protocol. Furthermore, the organoids derived from the MSC and HUVEC co-culture showed 3D structure with polarity, hepatic-like glycogen storage, and ICG absorption/elimination.
CONCLUSION
Human amniotic epithelial cells are heterogeneous and certain subpopulations have high stemness. Under a 3D co-culture system, functional hepatic organoids were generated in a multicellular microenvironment.
Keywords: 3D micropattern, Amniotic epithelial cells, Amniotic stem cells, Hepatic differentiation, Heterogeneity, Human placental tissue, Human umbilical vein endothelial cells, Mesenchymal stem cells, Multicellular microenvironment, Organoid
Core tip: Amniotic stem cells were exploited as a cell source alternative to liver transplantation therapy instead of induced pluripotent stem cells. However, they presented with low hepatic function efficiency. We used 3D co-culture and a combination of supportive somatic stem cells to simulate an in vivo microenvironment. Our selected subpopulation of adherent amniotic stem cells self-organized ex vivo and generated functional organoids. Cell selection methods and bioinformatics may help refine the differentiation protocol.
INTRODUCTION
Liver cirrhosis and liver failure are global problems. They are caused by viral infections, alcoholic- or non-alcoholic steatohepatitis, autoimmune hepatitis, metabolic and hereditary diseases, and others[1]. The only curative treatment is liver transplantation. However, there is a worldwide shortage of liver donors. Moreover, liver transplantation is associated with high mortality and morbidity and high-risk patients with comorbidity do not meet the indication criteria[2,3]. Cell transplantation has been proposed as an alternative therapy to whole organ transplantation. Several cells have been investigated as hepatic cell sources. Human donor-derived hepatocyte transplantation was attempted to cure cirrhosis and it did have some therapeutic benefit[4,5]. However, it required many hepatocytes and failed to solve the problem of donor insufficiency.
Embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) are known to differentiate into hepatocytes[6]. Although they have a high potential for hepatic differentiation, there are ethical, tumorigenicity, and cost issues associated with them. Previous reports indicated that somatic cells such as fibroblasts were induced to differentiate into hepatocyte-like cells by direct reprogramming[7,8]. In this case, virus-mediated overexpression of lineage-specific transcription factors was needed. Other cell types include mesenchymal stem cells (MSCs), hematopoietic stem cells (HSCs), menstrual blood-derived stem cells, and amniotic stem cells (ASCs)[5,9]. Somatic stem cells require only differentiation factors but no gene editing. The latter may cause undesirable and unexpected side effects. Here, we attempted to induce ASCs to differentiate into hepatocytes because they have several beneficial characteristics.
Amniotic epithelial cells (AECs) are easily isolated from amniotic membranes after delivery. This process causes no harm to the donor. Embryologically, the amnion is derived from the epiblast which can differentiate into three germ layers. Even at full term pregnancy, this differentiation potential persists in ASCs which are an immature subpopulation of AECs[10]. AECs also have immune tolerance and are therefore suitable for allogenic transplantation[11,12]. Furthermore, they have certain features, in common with hepatocytes such as the expression of ALB, AAT, or CYP3A4[13].
Previous studies attempted to differentiate AECs into hepatocyte-like cells in 2D culture[14,15]. However, no differentiation protocol was established. Without systemic and comprehensive analyses in vivo or in vitro, AEC quality could not be verified and it may have been unsatisfactory and inferior to ESCs or iPSCs[5]. Therefore, its functional activity would be insufficient for clinical use when a higher level of hepatic function was required. Moreover, as the general AEC profile had not yet been elucidated, it was difficult to determine which differentiation protocol was appropriate for AECs. Therefore, we endeavored to clarify AEC stemness characteristics, heterogeneity, and general profile by using transcriptomes. We also tried to select an adherent subpopulation to purify ASCs.
A 3D culture system may promote and enhance stem cell differentiation potential in co-culture with endothelial, mesenchymal, and other cells[16,17]. Here, we combined 3D culture conditions and co-culture with other somatic stem cells and supportive stroma cells. We attempted to simulate an in vivo-like microenvironment using a selected subpopulation of adherent ASCs for ex-vivo self-organization and ultimately obtained functional organoids.
MATERIALS AND METHODS
Isolation of somatic stem cells
AECs: Human placenta was acquired from the University of Tsukuba Hospital with approval from the institutional review board (IRV code: H27-58). All samples were collected from patients who had provided informed consent. Emergent operation cases were excluded. The amniotic membrane was peeled off the placenta in the operating room immediately after birth. After washing in pre-digestion buffer (Hank’s balanced salt solution, HBSS; Wako Pure Chemical Industries Ltd., Osaka, Japan) with 0.02% egtazic acid (EGTA; Wako Pure Chemical Industries Ltd., Osaka, Japan), the membrane was incubated in 0.05% trypsin-EDTA (Thermo Fisher Scientific, Waltham, MA, United States) at 37 °C for 20-30 min. HBSS with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, United States) was added and gentle agitation followed to detach the epithelial cells from the membrane. The cells were incubated with 0.05% trypsin-EDTA and washed again to collect the AECs. A 100-µm strainer (Merck EMD, Darmstadt, Germany) was used to remove a clot from the cell suspension and isolate primary AECs. Viability was checked with 0.4% trypan blue dye (Dojindo Laboratories, Kumamoto, Japan) and found to be > 90%.
Human umbilical vein endothelial cells (HUVECs): After the umbilical cords were harvested, the umbilical veins were filled with collagenase buffer (HBSS with 0.24% 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; Merck EMD, Darmstadt, Germany), 0.005% trypsin inhibitor (Wako Pure Chemical Industries Ltd., Osaka, Japan), and 0.05% collagenase (Worthington Biochemical Corporation, Lakewood, NJ, United States). The veins were incubated at 37 °C for 15 min. The buffer was centrifuged at 450 × g and 4 °C for 5 min to obtain a cell pellet containing HUVECs. This pellet was seeded to a plate using an EGM Bullet Kit (Lonza, Basel, Switzerland). In most cases, the HUVECs grew to confluence within 4 d.
MSC: After HUVEC collection, Wharton's jelly was minced and seeded to a plate with Dulbecco’s modified Eagle’s medium and 10% FBS. Attached MSCs were isolated and in most cases, they grew to confluence within 2 wk.
Cell culture
2D culture: For the AECs, a custom culture medium was blended and named AEC basal medium (AECBM). The media and protocols are listed in Supplementary Figure 1. Primary AECs were used in most analyses. In the culture experiments, however, AECs were precultured with AECBM for 5-7 d then used for differentiation. In the present study, the multistep hepatic differentiation protocol of Nie et al[17] was applied. In that study, iPSCs were differentiated into hepatocytes. The AEC culture medium was changed step by step within 3 wk.
Another simple differentiation protocol from AECs to hepatocytes was reported by Maymó et al[14]. In brief, the cells were cultured for 1 wk in Iscove’s modified Dulbecco’s medium containing epidermal growth factor (EGF) then cultured with EGF and dexamethasone. This method was named the two-step protocol.
3D culture: A primary 2D culture was prepared to select a subpopulation of adherent AECs. Unattached cells were removed after 72 h. A 24-well 3D micropattern culture plate (Kuraray, Tokyo, Japan) was used for the next step. After coating the plate with lipidure (NOF Corporation, Tokyo, Japan), the precultured AECs were seeded at a density of 106/well. The AECs formed a sphere within 3-7 d. AECs, HUVECs, and MSCs were used for organoid propagation and they formed spheres within 1 d.
qRT-PCR
The primers used are listed in Table 1. The TaqMan PCR protocol from Integrated DNA Technologies, Inc. (Coralville, IA, United States) was used. In brief, the RNA was extracted from the cultured cells with Isogen (Nippon Gene, Tokyo, Japan). Then the isolated RNA was converted to cDNA with a Revert Aid RT kit (Thermo Fisher Scientific, Waltham, MA, United States). The cDNA, primers, probes, reference dye, and PrimeTime Gene Expression Master Mix (Integrated DNA Technologies, San Jose, CA, United States) containing hot-start DNA polymerase, dNTPs, enhancers, and stabilizers were combined. A 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, United States) ran the PCR.
Table 1.
List of primers
| Name | Primer sequence | |
| SOX2 | F | TGCACCGCTACGACGTGA |
| R | GGAGCCAAGAGCCATGCC | |
| NANOG | F | TGCTGAGATGCCTCACACG |
| R | TGCAGAAGTGGGTTGTTTGC | |
| OCT4 | F | GAAACCCACACTGCAGCAG |
| R | GACCCAGCAGCCTCAAAATC | |
| LIN28 | F | GGATGTCTTTGTGCACCAGAGTA |
| R | TGGATTCCAGACCCTTGGCT | |
| ALB | F | TGCCTGTTGCCAAAGCTCG |
| R | GCTACTGCCCATGCTTTGAAAG | |
| AFP | F | GCAAACGATGAAGCAAGAGTTTC |
| R | GCAGCATTTCTCCAACAGGC | |
| AAT | F | ACTGGAACCTATGATCTGAAGAGC |
| R | GCCTTATGCACGGCCTTGG | |
| CYP3A4 | F | GGATGAAAGAAAGTCGCCTCGA |
| R | TCCAGATCGGACAGAGCTTTG | |
| ACTB | F | CCTCGCCTTTGCCGATCC |
| R | CATGCCGGAGCCGTTGT |
Immunofluorescence analysis
Amnion and organoids were stained with following antibodies: Primary; SSEA4, TRA-1-60, TRA-1-81 (Cell Signaling Technology, Danvers, MA, United States), EPCAM (BD, Franklin Lakes, NJ, United States), E-cadherin (BD, Franklin Lakes, NJ, United States). Frozen sample sections were fixed with 50% acetone (Wako Pure Chemical Industries Ltd., Osaka, Japan) and 50% methanol (Wako Pure Chemical Industries Ltd., Osaka, Japan) for 30 min.
The samples were incubated with primary antibody at 4 °C overnight, washed 3 ×, and incubated with a fluorescence-conjugated secondary antibody for 1 h at 20-25 °C. Before each antibody incubation, blocking was performed with 10% normal goat serum (NGS; Merck EMD, Darmstadt, Germany). In the last step, the samples were mounted with 4’,6-diamidino-2-phenylindole (DAPI; Dojindo Laboratories, Kumamoto, Japan) and photographed under an All-in-One Fluorescence Microscope (BZ-X710; Keyence, Osaka, Japan) or a confocal microscope (Leica TCS SP6; Leica Microsystems, Wetzlar, Germany).
For live amnion staining, the amnion was incubated with anti-TRA-1-60 antibody for 30 min at 37 °C and then with Hoechst (Dojindo Laboratories, Kumamoto, Japan) for 15 min at 37 °C. Pictures were obtained with an Inverted System Microscope IX71 (Olympus, Tokyo, Japan).
Flow cytometry
The following antibodies and DAPI were used: SSEA4-FITC (BioLegend, San Diego, CA, United States), TRA-1-81-PE (Thermo Fisher Scientific, Waltham, MA, United States), TRA-1-60-PE (Thermo Fisher Scientific, Waltham, MA, United States), E-cadherin-Alexa Flour 647 (BD, Franklin Lakes, NJ, United States), and EPCAM-APC (BD, Franklin Lakes, NJ, United States). The cells were incubated with antibody on ice for 30 min then washed twice. The cells were analyzed in FACS Jazz (BD, Franklin Lakes, NJ, United States) and FlowJo v. 7.6.1 (FlowJo LCC, Ashland, CO, United States).
Colony formation assay
Primary and adherent AEC subpopulations were compared in a colony formation assay. To collect the adherent AEC subpopulation, a selective primary passage was used. This process was the same as the preculture step for the 3D culture, namely, nonattached cells were removed after 72 h culture. Primary and adherent AECs were seeded in a six-well plate at a density of 3000-20000 cells/well with AECBM. After 6 d of incubation, the numbers of colonies containing > 20 cells were counted and traced by scanning under an All-in-One Fluorescence Microscope (BZ-X710). The number of colonies was divided by the seeding number to calculate the colony formation frequency.
Whole-transcriptome shotgun sequencing and bioinformatics analysis
Total RNA was extracted from primary AECs and RNA sequencing was performed by Eurofins Genomics K.K. (Tokyo, Japan) who used the Illumina HiSeq 2500 (Illumina, San Diego, CA, United States). Gene expression data for the other cell sources were obtained from the National Center for Biotechnology Information Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra). Data sources are listed in Table 2.
Table 2.
RNAseq data sources used in the bioinformatics analysis
| Sample name | SRA number | Instrument | Memo | Ref. |
| AEC-1 | SRR9643783 | Illumina HiSeq 2500 | Primary AEC | |
| AEC-2 | SRR9643784 | Illumina HiSeq 2500 | Primary AEC | |
| MSC-1 | SRR6431450 | Illumina HiSeq 2000 | [44] | |
| MSC-2 | SRR6431451 | Illumina HiSeq 2000 | [44] | |
| hESC-1 | SRR4241924 | Illumina HiSeq 4000 | H9 | [45] |
| hESC-2 | SRR4241926 | Illumina HiSeq 4000 | H9 | [45] |
| NiPS-1 | SRR7592168 | Illumina HiSeq 2500 | Normal human iPSC | [46] |
| NiPS-2 | SRR7592169 | Illumina HiSeq 2500 | Normal human iPSC | [46] |
| DE-1 | SRR771468 | Illumina HiSeq 2000 | Definitive endoderm induced from H9 | [47] |
| DE-2 | SRR771469 | Illumina HiSeq 2000 | Definitive endoderm induced from H9 | [47] |
| hiHep-1 | SRR5974291 | Illumina HiSeq 2000 | Umbilical cord fibroblast derived hepatocyte-like like cell | [8] |
| hiHep-2 | SRR5974292 | Illumina HiSeq 2000 | Umbilical cord fibroblast derived hepatocyte-like cell | [8] |
| iPSCHLC-1 | SRR5974295 | Illumina HiSeq 2500 | iPSC-derived Hepatocyte-like cell | [8] |
| iPSCHLC-2 | SRR5974296 | Illumina HiSeq 2500 | iPSC-derived hepatocyte-like cell | [8] |
| Hepa-1 | SRR6176953 | Illumina HiSeq 2500 | Hepatocyte from clinical sample of adult | [48] |
| Hepa-2 | SRR6176948 | Illumina HiSeq 2500 | Hepatocyte from clinical sample of adult | [48] |
| Hepa-3 | SRR5974298 | Illumina HiSeq 2000 | Primary human hepatocyte 2 d | [8] |
| Hepa-4 | SRR5974299 | Illumina HiSeq 2000 | Primary human hepatocyte 4 d | [8] |
RNA Sequencing. All paired-end reads were trimmed and quality-filtered via CLC Genomics Workbench v. 10.1.1 (Qiagen, Hilden, Germany) which was also used to map the filtered reads against the human reference sequence (hg 19). Mapped reads were counted and transcript abundance was measured in TPM (transcripts per kilobase million) units.
Bioinformatics for RNA Sequencing Data. An original read count matrix was normalized with DESeq2 v. 3.5.2 for unified expression level. All normalized log-transformed expression values were ordered according to the median absolute deviation. The top 3500 genes were selected for a heatmap which was generated with the pheatmap package in R.
ICG test
AEC spheres or multiple stem cell-derived organoids were incubated with AECGM and 1 mg/mL indocyanine green (ICG) (Daiichi Sankyo, Tokyo, Japan) for 30 min at 37 °C. They were then observed under bright field with an Inverted System Microscope IX71 to confirm ICG absorption. They were then incubated with ICG-free AECGM for 6 h and checked for ICG elimination.
Statistical analysis
GraphPad Prism v. 8.0.1 (GraphPad Software, La Jolla, CA, United States) was used to process all data. A Wilcoxon signed-rank test compared means between pairs of groups. P < 0.05 was considered statistically significant.
RESULTS
Characterization of amnion-derived stem cells
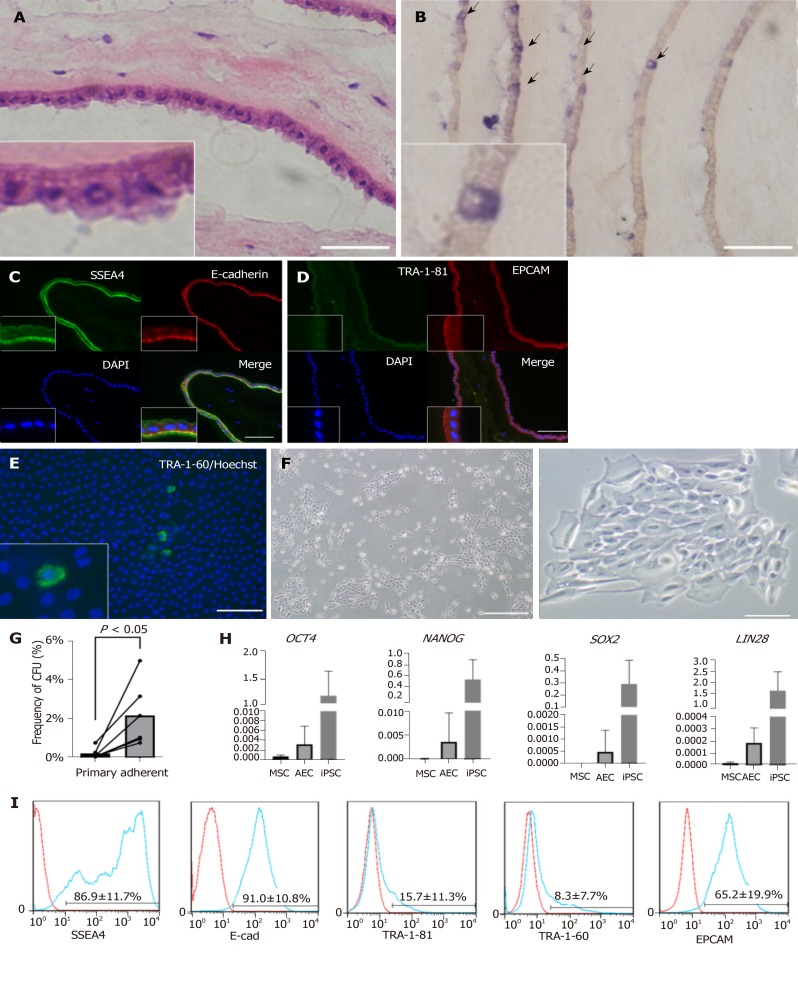
The amnion consisted of monolayer epithelium and mesenchyme (Figure 1A). We first examined the stemness characteristics of the amnion. Using an AP staining kit (Muto Pure Chemicals, Tokyo, Japan), we found that a small proportion of the epithelial cells were blue (Figure 1B). AP is often used as a multipotency marker. Immunofluorescence analysis of frozen amnion sections (Figures 1C and 1D) indicated that its epithelium was positive for the stemness markers SSEA4, E-cadherin, and EPCAM. However, other stemness markers such as TRA-1-60 (not shown) and TRA-1-81 were not detected. Previous studies[18,19] reported that a small subpopulation of AEC expresses TRA-1-60 or TRA-1-81. We attempted live staining of fresh amnion and found a low frequency of TRA-1-60-positive cells (Figure 1E). We concluded that amniotic epithelium has certain stemness or multipotency markers and only a subpopulation of its cells expresses stemness.
Figure 1.
Characteristics of human amniotic membrane and amniotic epithelial cells. A: H and E staining of amniotic membrane. Bar, 50 µm; B: AP staining of amniotic membrane. Positive cells are indicated with arrows. Bar, 100 µm. In A and B, the amniotic membrane was rolled before embedding. Therefore, many layers can be seen in one picture; C: Immunofluorescent staining of frozen section of amniotic membrane. Anti-SSEA4 antibody (green), E-cadherin antibody (red), and DAPI were used; D: Same as C. Anti-TRA-1-81 antibody (green), anti-EPCAM antibody (red), and DAPI were used; E: Direct tissue staining of amniotic membrane. Anti-TRA-1-60 antibody (green) and DAPI were used. Bars in C, D, and E represent 100 µm; F: Colonies formed from cultured amniotic epithelial cells (AECs) and observed by phase-contrast microscopy. Bar to left of F represents 200 µm. Bar to right of F represents 500 µm; G: Frequency of colony formation from primary AECs and adherent AECs. Cells which did not attach to the well surface were removed to purify the amniotic stem cells; H: Gene expression of primary AECs, mesenchymal stem cells(MSCs), and induced pluripotent stem cells(iPSCs) detected by qRT-PCR; I: surface markers of primary AECs verified by flow cytometry.
The AEC isolation method yielded 1-6 × 107 AECs with > 90% viability. We then examined AEC morphology, colony formation capability, and stemness markers. We seeded AECs into a plate and observed them by phase contrast microscopy (Figure 1F). Although only a few primary AECs attached to the plate as short, spindle-like cells within 1 d, they proliferated well and formed colonies. After the primary selective passage, the colony formation capacity increased ten-fold relative to that for the heterogenous primary AECs (Figure 1G). We also verified the gene expression of stemness markers by qRT-PCR. OCT4, NANOG, SOX2, and LIN28, which are important iPSC markers, were expressed at very low levels in AECs compared with those in iPSCs but at higher levels than those in MSCs (Figure 1H). We also analyzed the stemness surface markers by flow cytometry (Figure 1I). E-cadherin and EPCAM expression was highly positive. Certain SSEA4s were highly positive while others were only weakly positive. Only 10% of the TRA-1-81 and TRA-1-60 were positive. Based on the foregoing information, we concluded that amnion and AECs have stemness and amniotic stem cells widely express stemness markers.
Hepatic potentials and profiling by whole-transcriptome shotgun sequencing
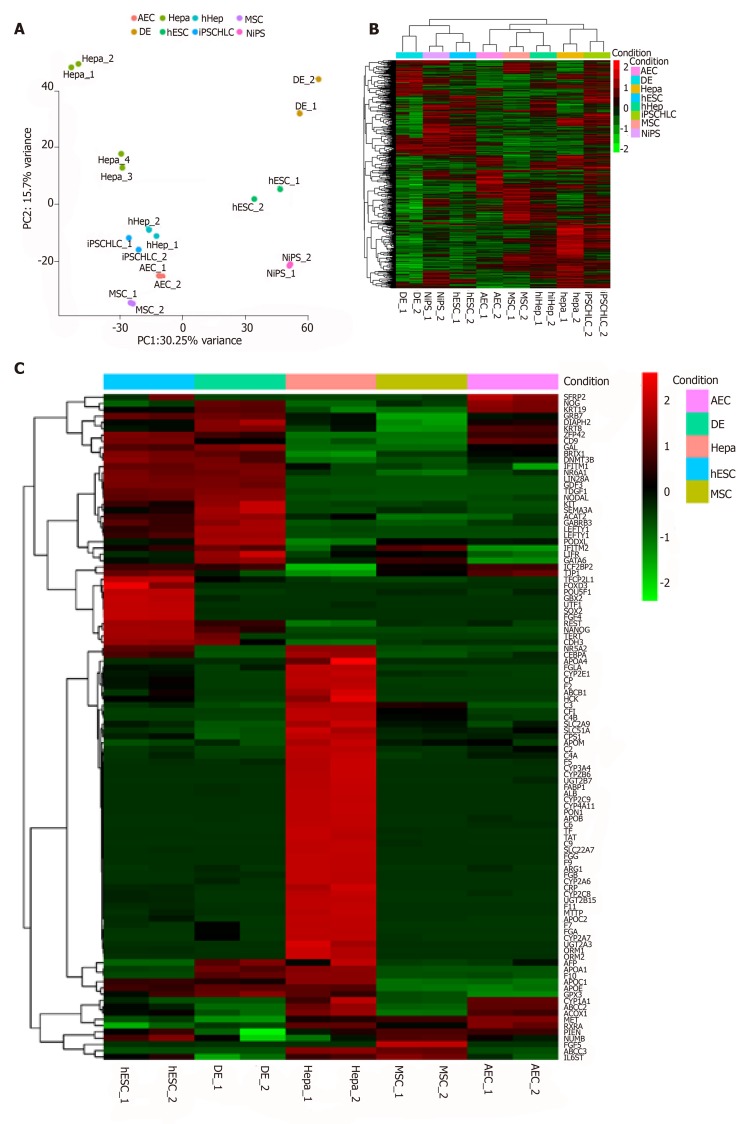
According to previous reports, AECs can differentiate into hepatocytes[20,21]. Here, we performed comprehensive profiling by whole-transcriptome shotgun sequencing to verify this potential in AECs. We also compared the AEC RNA sequences with those of pluripotent stem cells, hepatocytes or induced hepatocytes, and MSCs. MSCs are also often used as hepatocyte cell sources[9]. First, we normalized the data and checked its repeatability using Pearson’s correlation coefficient (Supplementary Figures 2A and 2B). A principal component analysis (PCA) and a general heatmap (Figures 2A and 2B) indicated that AECs and MSCs differ across the spectrum ranging from pluripotent stem cells to hepatocytes. Even human fibroblast-derived hepatocyte-like cells (hiHep) and iPSC-derived hepatocyte-like cells (iPSCHLCs), which had immature hepatic characteristics[8], did not closely resemble hepatocytes. An unexpected finding was that AECs were like hiHep and iPSCHLCs. We gathered 40 stem cell- and 66 hepatic cell markers to make a heatmap (Figure 2C). CD9 and TJP1 were highly expressed in both AECs and ESCs. IGF2BP2 and KRT19 were highly expressed in both AECs and the definitive endoderm. The hepatic markers MET, ABCC2, CYP1A1, etc. were highly expressed in AECs. The markers expressed in AECs are listed in Table 3. The features common to AEC, pluripotent stem cells, and hepatocytes may help elucidate the differentiation mechanism.
Figure 2.
Characteristics of primary amniotic epithelial cells and other cell sources verified by bioinformatics. A: Principal component analysis of all cell sources used in this assay; B: General heatmap for each cell source; C: Specific heatmap of stemness and hepatic markers. AEC: Amniotic epithelial cell; Hepa: Hepatocyte; hiHep: Human fibroblast-derived hepatocyte-like cell; MSC: Mesenchymal stem cell; DE: Definitive endoderm; hESC: Human embryonic stem cell; iPSCHLC, iPSC-derived hepatocyte-like cell; NiPS: Normal human iPSC.
Table 3.
Stemness (upper)- and hepatic (lower) genes expressed in amniotic epithelial cells.
| Gene symbol | Explanation | Ref. |
| CD9 | CD9 belongs to the transmembrane 4 superfamily. It is associated with cell proliferation, motility, and adhesion and regulates hematopoietic differentiation | [49] |
| TJP1 | Tight junction protein 1 is a determinant of plasma cell proteasome and is associated with EGFR, JAK1, and STAT3. It is regulated by TGF-b and involved with cell motility | [30,50] |
| IGF2BP2 | This member of the insulin–like growth factor 2 mRNA-binding protein family participates in normal embryonic growth and development. It is expressed in the pancreas and associated with type 2 diabetes mellitus | [51] |
| KRT19 | Keratin 19 is a cytoplasmic intermediate filament protein and belongs to the type 1 keratin family. It is used as a cholangiocyte marker and is not expressed in hepatocytes. It is associated with the progression of several cancers | [31,32] |
| GRB7 | Growth factor receptor-bound protein 7 mediates signal transduction and cell migration. It is associated with the metastasis of several cancers | [52] |
| KRT8 | Keratin 8 is a cytoplasmic intermediate filament protein of the type 2 keratin family. It is expressed in single layered epithelial cells. In cancer cells, it is associated with progression in the form of migration and adhesion | [33] |
| Gene symbol | Explanation | Ref. |
| ApoM | Apolipoprotein M (apoM) belongs to the lipocalin family. It is expressed in the liver and kidney. Hepatic apoM controls HDL metabolism | [53,54] |
| CPS1 | Carbamoyl phosphate synthase 1 is a mitochondrial enzyme and participates in the first step of the urea cycle in the liver. | [55] |
| SLC51A | Organic solute transporter subunit alpha is a bile acid transporter in the liver, small intestine, and kidney. It prevents the bile acid reflux | [56] |
| IL6ST | Interleukin 6 signal transducer controls IL-6 and other cytokines such as IL-11, IL-27, oncostatin M, and leukemia inhibitory factor | [57] |
| ACOX1 | Acyl-CoA oxidase 1 is a rate-limiting enzyme in fatty acid β-oxidation | [58] |
| RXRa | The nuclear hormone receptor retinoid X receptor belongs to the steroid hormone receptor family. It is a key factor of cholesterol synthesis | [59] |
| MET | MET encodes the hepatocyte growth factor (HGF) receptor and the key factor of hepatic regeneration. It activates epithelial migration and 3D morphogenesis | [35,36] |
| ABCC2 | ATP binding cassette subfamily C member 2 is expressed in the hepatocytes and is a biliary transporter. It is also related to drug elimination and multidrug resistance in several cancers | [60] |
| CYP1A1 | This member of the cytochrome P450 enzyme superfamily participates in fatty acid and steroid metabolism. It is associated with the detoxification of anthropogenic chemicals such as polycyclic aromatic hydrocarbons | [61] |
Hepatic differential induction with a 3D culture system
To induce hepatic differentiation, we raised AECs under a hepatic differentiation protocol and a 3D culture system (Figure 3A). First, we tested the hepatic differentiation protocol in 2D culture. We attempted a two-step differentiation protocol using AECBM as a negative control. In the 2D culture, the AECs were short-lived. Nearly all the cells were detached from the culture plate within 3 wk. Moreover, gene expression did not markedly increase during the culture period (Supplementary Figure 3).
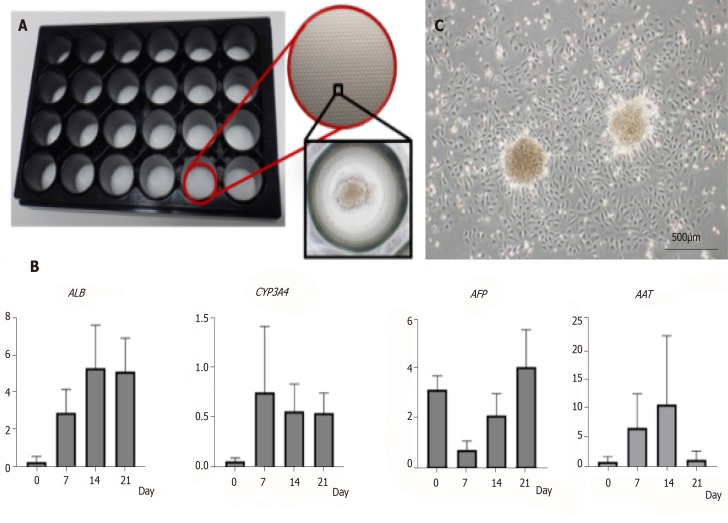
Figure 3.
Characteristics of amniotic epithelial cell spheres formed on 3D-micropattern plate. A: 3D-micropattern plate used in the present study. Round pits 500 µm in diameter are clustered on the surface. After culture, the amniotic epithelial cells (AECs) formed a sphere; B: Gene expression in the AEC sphere verified by qRT-PCR; C: After reseeding AEC sphere onto 2D culture dish, AEC proliferation was verified by phase-contrast microscopy. Bar, 500 µm.
We then tried 3D culture with three protocols including the two-step, multistep, and AECBM. With a 3D culture system in a micropattern plate, the seeded cells readily aggregated into a sphere and remained viable for 3 wk. We then checked hepatic gene expression by qRT-PCR. Relative to the multistep and AECBM protocols, hepatic gene expression in the two-step group protocol was higher (Figure 3B and Supplementary Figure 4). ALB and CYP3A4 were upregulated and sustained for 3 wk. Although AFP was upregulated in the 2D preculture then downregulated after 7 d, it gradually increased again within the 3-wk differentiation period. AAT was also upregulated over 14 d but was downregulated by 21 d. When we seeded the AEC sphere in a culture plate, we observed active AEC proliferation (Figure 3C). Based on the preceding data, we concluded that our 3D cultural system maintains AEC viability for > 3 wk and accelerates hepatic differentiation especially when a two-step protocol is used.
Hepatic function enhancement with multicellular organoids
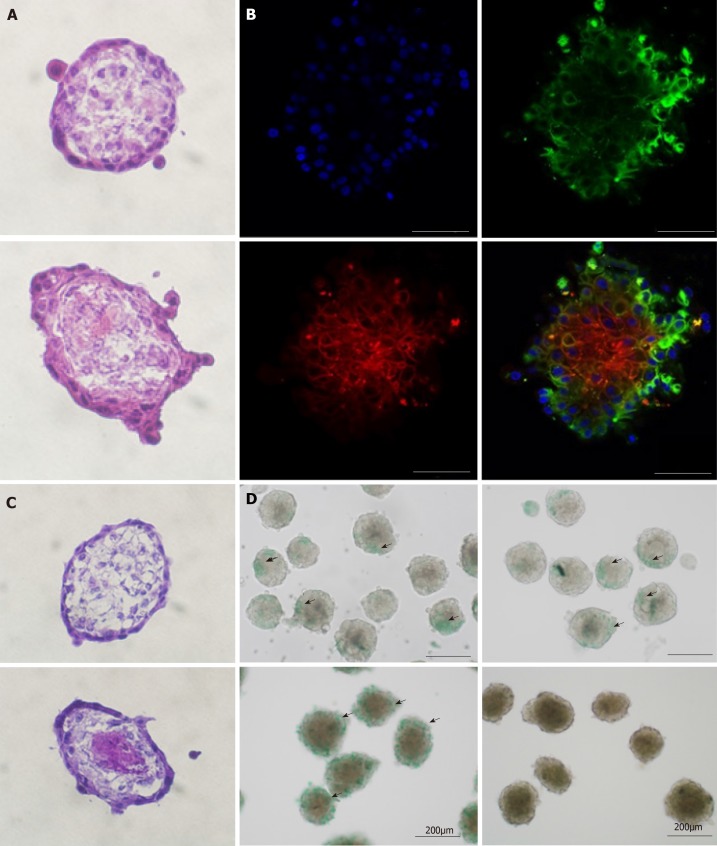
We then attempted co-culture by seeding AEC with MSC and HUVEC to form a multicellular sphere organoid. We examined its morphology, cellular distribution, and functional activity. Hematoxylin and eosin (H and E) staining disclosed that the AEC sphere and the organoid were morphologically similar (Figure 4A). However, immunofluorescence with CD31, CD90, SSEA4, and DAPI revealed inner-layer CD31-positive HUVECs (data not shown) and CD90-positive MSCs as well as outer-layer SSEA4-positive AECs (Figure 4B). This architecture resembles normal tissue disposition. Periodic acid Schiff staining showed glycogen accumulation in the center of the organoid (Figure 4C). Both the AEC sphere and the organoid absorbed ICG but only the organoid could excrete it (Figure 4D). From the aforementioned results, we concluded that AECs, MSCs, and HUVECs underwent 3D construction in our 3D co-culture system. Moreover, the organoid resembled a liver bud with complex liver function.
Figure 4.
Organoid morphology and hepatic function. A and C: Frozen sections of amniotic epithelial cell (AEC) sphere and organoid. The AEC sphere was placed in the upper layer and the organoid was placed in the lower layer. H&E staining is used in A, and Periodic acid Schiff staining is used in C; B: Immunofluorescent organoid staining observed under confocal microscopy. Anti-SSEA4 antibody (green) representing AECs; anti-CD90 antibody (red) representing mesenchymal stem cells and DAPI. Bars in A, B, and C, 50 µm; D: ICG tests on AEC sphere and organoid. The AEC sphere was placed in the upper layer and the organoid was placed in the lower layer.
DISCUSSION
AEC characteristics and stemness
AECs proliferate well and express stemness markers[20]. They also readily differentiate into hepatocyte-like cells in the presence of certain growth factors[14]. Miki et al[20] reported that AECs are derived from epiblasts and some of them remain immature then differentiate into three germ layers[18,20,22]. Amnion immunofluorescence disclosed that only a small proportion of AECs are TRA-1-81- and/or TRA-1-60-positive[19]. Here, it was confirmed by flow cytometry that a small primary AEC subpopulation is TRA-1-60- and/or TRA-1-81-positive and the proportions vary among samples. Izumi et al[18] reported that this subpopulation frequency is higher in early-stage than full-term pregnancies. However, all AECs used in the present study were derived from term pregnancies. Moreover, certain AEC lots with comparatively higher proportions of these stem markers had high colony formation frequencies and grew well. For this reason, it is important to select good AEC lots for differentiation.
Previous studies reported that primary AECs could be classified into three different culture layers, namely, floating, attached, or intermediate[20]. In the present study, we found that adherent cells derived from selective passage of primary AECs had a comparatively high colony formation capability. Based on the foregoing information, adherent subpopulations of passaged AECs were used in the 3D culture. Differentiation efficiency could be enhanced by purifying this high-stemness subpopulation of primary AECs.
Hepatic differentiation protocol
The iPSC-derived organoids have often been considered as models representing organs with highly complex structures and/or functions such as the intestine, lung, neural tissue, etc[23-25]. Cell-to-cell interactions[26] and hypoxic reactions were key factors in differentiation[27]. In the case of hepatic differentiation from iPSCs, mesenchymal and endothelial cells were reported as important factors[16]. However, only 2D culture was used in the attempt to differentiate AECs into hepatocytes[14,28] and the protocol used was not comparable to that applied in the present study.
Therefore, we compared 2D and 3D culture with several differentiation protocols. The 2D culture method was unsuitable for AECs because the cells did not remain viable in it. In 3D culture, AECs retain their viability and morphology for 21 d. A previous report stated for an iPSCs’ hepatic differentiation model in a 3D culture system that hypoxia suppresses cell proliferation and induces differentiation[26,27]. In the present study, AEC sphere proliferation was temporarily arrested during 3D culture then resumed after plate seeding. Therefore, a similar mechanism was assumed to occur under our AEC 3D culture conditions.
To date, no hepatic differentiation protocol has been established for AECs. In certain studies, an iPSC-to-hepatocyte differentiation protocol was used[15,29]. However, this stepwise protocol is complex and requires numerous growth factors. Since AECs are not pluripotent stem cells, it is uncertain whether every protocol step would be absolutely necessary. One report mentioned that activin A is not required for hepatic differentiation from AECs[15]. Activin A is required for the transformation of pluripotent stem cells to the definitive endodermal state. In contrast, the AEC-to-hepatocyte differentiation protocol is comparatively simple[14]. Our results indicated that this straightforward differentiation protocol upregulates AEC hepatic genes. Nevertheless, it may still be inadequate and must, therefore, be modified. Despite this deficiency, the co-culture method can nonetheless generate hepatic function from AECs. We concluded that a 3D co-culture system or a hepatic microenvironment is needed for hepatic differentiation from AECs.
Bioinformatics approach
Bioinformatics is a strong tool for the evaluation of cellular differentiation. It is often applied to iPSCs but not to AECs. Our PCA and general heatmap revealed that hiHep[8] and iPSCHLCs[8] remotely resemble mature hepatocytes. In fact, they are regarded as immature hepatocytes. AECs and MSCs are distant from iPSCs, ESCs, and DE but more closely resemble hiHep and iPSCHLCs. Therefore, AECs may be able to differentiate into hepatocytes in only a few steps.
The stemness and hepatic marker heatmaps disclosed certain genes common to all AECs. TJP1 is associated with EGFR/JAK1/STAT3 signaling[30]. Every two-step, multistep, and AECBM protocol included EGF and seemed reasonable. KRT8 and KRT19 are epidermal markers[31-33] and are also upregulated in AECs. In the hepatic transdifferentiation from fibroblasts, the first step is to transform mesenchymal cells into epidermal cells[34]. In the differentiation of ESC to DE, KRT8 and KRT19 are upregulated. Therefore, differentiation of AECs to hepatocytes may require fewer steps than pluripotent stem cell-to-hepatocyte differentiation.
The hepatic marker MET was upregulated in AECs. MET encodes the human growth factor (HGF) receptor which is important for hepatocyte proliferation or differentiation[35,36]. HGF may already be sensitive in AECs. In addition, IL6ST is upregulated in AECs. It transduces the signals of several chemicals containing oncostatin M. HGF and oncostatin M are normally added to iPSCs in the hepatocyte-like cell protocol but not in our two-step protocol[6]. Therefore, we may be able to improve differentiation by adding HGF or oncostatin M to our two-step protocol. Moreover, other markers of hepatic metabolism may also enhance liver function. A detailed list of the stemness and hepatic genes is presented in Table 3 A bioinformatics approach may help elucidate the differentiation mechanism and improve the protocol used for it.
Future prospects
An iPSC-derived hepatic organoid has already been created and used in basic research ex vivo[17]. However, iPSCs are costly, unstable, and tumorigenic[37,38]. In contrast, our organoid is cost-effective, stable, and non-tumorigenic. AEC organoids with hepatic character could be used both in basic ex vivo research and in vivo experiments.
The next step of our research is to develop and test a human-to-animal transplantation model with AEC-derived hepatic organoids. According to a previous report, terminal differentiation to a functional hepatocyte would be strongly induced in the recipient liver microenvironment[15]. Even in our organoids simulated the microenvironment, the process would not be as perfect as it would be in vivo. Therefore, in vitro tests of hepatic organoid function may be imperfect. Nevertheless, we must confirm hepatic function after transplantation. The present study demonstrated that the 3D culture condition temporarily suppressed proliferation but did not entirely cause a loss of this capability. Therefore, proliferation could still occur in vivo with the appropriate signal. Human organoids and animal transplantation models are required for pharmaceutical research and investigations into human-specific infectious disease such as hepatitis B and C[17,39].
A future intended application of our hepatic organoid is as an alternative to organ transplantation as a therapy for hepatic failure. More than one induction step from somatic cells to iPSCs is required and quality and safety must be verified[40,41]. On the other hand, AECs are directly isolated from tissues so they are comparatively safe and reliable.
Safety is of paramount importance in the clinical application of organoids. In this respect, AECs are superior to iPSCs. The safest transplantation model is a private placental and cord blood cell bank. Preserved blood cells can be used as a replacement therapy for hematopoietic disease and the regenerative treatment of congenital heart diseases and other developmental disorders[42,43]. Amnion and umbilical cords also have the same genetic profile as the baby. Therefore, private AEC, MSC, and HUVEC cell banks could be ideal sources of therapeutic materials for infants with developmental conditions.
In conclusion, we selected adherent AECs in which ASCs were purified as a cell source for differentiation into hepatocytes. The 3D- and co-culture systems with MSCs and HUVECs generated multicellular organoids with hepatic function. A bioinformatics approach and cell selection strategy would be useful for further improvement of the protocol and its hepatic function.
ARTICLE HIGHLIGHTS
Research background
Cell transplantation is a promising method to solve the problem of organ donor deficiency. Recently, amniotic epithelial cells (AECs) have been studied as a somatic stem cell source for regenerative medicine. It has been reported that AECs possess hepatic differentiation capability and they are more cost-effective and non-tumorigenic compared to other cell types. Therefore, AECs could provide a new cell source for cell transplantation in the future, particularly for liver diseases.
Research motivation
However, the general profile for AECs has not been comprehensively analyzed and hepatic differentiation protocol for AECs has also not been established. Therefore, by clarifying the comprehensive characteristics of AECs and refining the hepatic differentiation protocol, we could use it effectively as a transplantation cell source in the future.
Research objectives
This study aimed to elucidate the comprehensive characteristics of human AECs, and to establish a novel hepatic differentiation protocol. Additionally, 3D multicellular culture condition and purification of stem cells obtained from AECs were studied, with an intention to improve their differentiation capability.
Research methods
All examined AECs, mesenchymal stem cells (MSCs), and human umbilical vein endothelial cells (HUVECs) were isolated from the placentae and umbilical cords after cesarean section. Stemness characteristics and heterogeneity of amnion and primary AECs were analyzed by immunofluorescence and AP staining, and flow cytometry. In addition, AEC transcriptomes were analyzed and compared with those for other cell sources based on bioinformatics. An adherent AEC subpopulation was selected from primary isolated AECs and their amniotic stem cell (ASC) purification quality was evaluated based on a colony formation assay. Hepatic differentiation capacities of AECs which were cultured in varying 2D or 3D conditions were compared according to their relative gene expression. Finally, ASCs, MSCs and HUVECs were co-cultured in a 3D system to generate hepatic organoids, and the organoid structure and hepatic function were compared with those of 3D AEC spheres using immunofluorescence imaging, Periodic acid Schiff staining, and an indocyanine green (ICG) test.
Research results
AECs expressed stemness markers such as EPCAM, SSEA4, and E-cadherin, whereas only limited cells in the AEC subpopulation were AP-positive, or expressed TRA-1-60 and TRA-1-81 stemness marker in the flow cytometry. The colony formation assay revealed that primary AECs could form colonies and the frequency was enhanced ten-fold in the adherent subpopulation. According to bioinformatics analysis of RNA sequencing, the primary AEC gene expression was different from those of pluripotent stem cells and hepatocytes; however, some overlapped genes were detected. Compared with the 2D system, AECs could retain their viability for a longer time in 3D culture conditions and the hepatic gene expressions were comparatively elevated using a two-step differentiation protocol. Furthermore, organoids derived from 3D multicellular culture condition using ASCs, MSCs and HUVECs, showed a 3D hepatic structure with polarity, hepatic-like glycogen storage, and ICG absorption/elimination.
Research conclusions
Human AECs are heterogeneous and certain subpopulations exhibit high stemness. AEC transcriptome analysis suggests some advantages and factors related to hepatic differentiation. 3D multicellular culture conditions improve the differentiation of ASCs into functional hepatic organoids.
Research perspectives
The results of this comprehensive analysis indicate that AECs have high hepatic differentiation capability which should be optimized. By optimizing the selected high stemness subpopulations of AECs and using a 3D co-culture system, AEC-derived hepatic organoid can be used for performing transplantation experiments to evaluate its in vivo function.
ACKNOWLEDGEMENTS
We thank Chen Liang, Yoshio Shimizu, Roxanne Sabrina Ramirez, Dan Song, Yoshiaki Uchida and Rui Zhao for their kind support in isolating the human placental tissue specimens.
Footnotes
Institutional review board statement: All specimens and cells from the patients were obtained after their informed consent and ethical permission was obtained for participation in the study.
Conflict-of-interest statement: The authors report no relevant conflicts of interest.
Data sharing statement: Transcriptome datasets of primary amniotic epithelial cells from the Sequence Read Archive (SRA) of the NCBI are available in the website. SRA number is listed in Table 2. Participants gave informed consent for publication.
Manuscript source: Invited manuscript
Peer-review started: February 27, 2019
Article in press: August 27, 2019
Specialty type: Cell and tissue engineering
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Li W, Ku SY, Vagholkar KR S-Editor: Dou Y L-Editor: A E-Editor: Zhou BX
Contributor Information
Kinji Furuya, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan.
Yun-Wen Zheng, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan; Institute of Regenerative Medicine and Affiliated Hospital, Jiangsu University, Zhenjiang 212001, Jiangsu Province, China; Department of Regenerative Medicine, School of Medicine, Yokohama City University, Yokohama 236-0004, Japan. ywzheng@md.tsukuba.ac.jp.
Daisuke Sako, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan; Department of Medicinal and Life Sciences, Faculty of Pharmaceutical Sciences, Tokyo University of Science, Noda 278-8510, Japan.
Kenichi Iwasaki, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan.
Dong-Xu Zheng, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan.
Jian-Yun Ge, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan.
Li-Ping Liu, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan; Institute of Regenerative Medicine and Affiliated Hospital, Jiangsu University, Zhenjiang 212001, Jiangsu Province, China.
Tomoaki Furuta, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan.
Kazunori Akimoto, Department of Medicinal and Life Sciences, Faculty of Pharmaceutical Sciences, Tokyo University of Science, Noda 278-8510, Japan.
Hiroya Yagi, Department of Obstetrics and Gynecology, Faculty of Medicine, University of Tsukuba, Tsukuba 305-8575, Japan.
Hiromi Hamada, Department of Obstetrics and Gynecology, Faculty of Medicine, University of Tsukuba, Tsukuba 305-8575, Japan.
Hiroko Isoda, Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba 305-8572, Japan.
Tatsuya Oda, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan.
Nobuhiro Ohkohchi, Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Faculty of Medicine, University of Tsukuba, Tsukuba, Ibaraki 305-8575, Japan.
References
- 1.Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20:7312–7324. doi: 10.3748/wjg.v20.i23.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abad CL, Lahr BD, Razonable RR. Epidemiology and risk factors for infection after living donor liver transplantation. Liver Transpl. 2017;23:465–477. doi: 10.1002/lt.24739. [DOI] [PubMed] [Google Scholar]
- 3.Farkas S, Hackl C, Schlitt HJ. Overview of the indications and contraindications for liver transplantation. Cold Spring Harb Perspect Med. 2014:4. doi: 10.1101/cshperspect.a015602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietrosi G, Vizzini GB, Gruttadauria S, Gridelli B. Clinical applications of hepatocyte transplantation. World J Gastroenterol. 2009;15:2074–2077. doi: 10.3748/wjg.15.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tolosa L, Pareja E, Gómez-Lechón MJ. Clinical Application of Pluripotent Stem Cells: An Alternative Cell-Based Therapy for Treating Liver Diseases? Transplantation. 2016;100:2548–2557. doi: 10.1097/TP.0000000000001426. [DOI] [PubMed] [Google Scholar]
- 6.Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 8.Gao Y, Zhang X, Zhang L, Cen J, Ni X, Liao X, Yang C, Li Y, Chen X, Zhang Z, Shu Y, Cheng X, Hay DC, Lai D, Pan G, Wei G, Hui L. Distinct Gene Expression and Epigenetic Signatures in Hepatocyte-like Cells Produced by Different Strategies from the Same Donor. Stem Cell Reports. 2017;9:1813–1824. doi: 10.1016/j.stemcr.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar A, Kumar V, Rattan V, Jha V, Pal A, Bhattacharyya S. Molecular spectrum of secretome regulates the relative hepatogenic potential of mesenchymal stem cells from bone marrow and dental tissue. Sci Rep. 2017;7:15015. doi: 10.1038/s41598-017-14358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miki T. Amnion-derived stem cells: in quest of clinical applications. Stem Cell Res Ther. 2011;2:25. doi: 10.1186/scrt66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strom SC, Gramignoli R. Human amnion epithelial cells expressing HLA-G as novel cell-based treatment for liver disease. Hum Immunol. 2016;77:734–739. doi: 10.1016/j.humimm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Strom SC, Skvorak K, Gramignoli R, Marongiu F, Miki T. Translation of amnion stem cells to the clinic. Stem Cells Dev. 2013;22 Suppl 1:96–102. doi: 10.1089/scd.2013.0391. [DOI] [PubMed] [Google Scholar]
- 13.Takashima S, Ise H, Zhao P, Akaike T, Nikaido T. Human amniotic epithelial cells possess hepatocyte-like characteristics and functions. Cell Struct Funct. 2004;29:73–84. doi: 10.1247/csf.29.73. [DOI] [PubMed] [Google Scholar]
- 14.Maymó JL, Riedel R, Pérez-Pérez A, Magatti M, Maskin B, Dueñas JL, Parolini O, Sánchez-Margalet V, Varone CL. Proliferation and survival of human amniotic epithelial cells during their hepatic differentiation. PLoS One. 2018;13:e0191489. doi: 10.1371/journal.pone.0191489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marongiu F, Gramignoli R, Dorko K, Miki T, Ranade AR, Paola Serra M, Doratiotto S, Sini M, Sharma S, Mitamura K, Sellaro TL, Tahan V, Skvorak KJ, Ellis EC, Badylak SF, Davila JC, Hines R, Laconi E, Strom SC. Hepatic differentiation of amniotic epithelial cells. Hepatology. 2011;53:1719–1729. doi: 10.1002/hep.24255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 17.Nie YZ, Zheng YW, Miyakawa K, Murata S, Zhang RR, Sekine K, Ueno Y, Takebe T, Wakita T, Ryo A, Taniguchi H. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine. 2018;35:114–123. doi: 10.1016/j.ebiom.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izumi M, Pazin BJ, Minervini CF, Gerlach J, Ross MA, Stolz DB, Turner ME, Thompson RL, Miki T. Quantitative comparison of stem cell marker-positive cells in fetal and term human amnion. J Reprod Immunol. 2009;81:39–43. doi: 10.1016/j.jri.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Miki T, Mitamura K, Ross MA, Stolz DB, Strom SC. Identification of stem cell marker-positive cells by immunofluorescence in term human amnion. J Reprod Immunol. 2007;75:91–96. doi: 10.1016/j.jri.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Miki T, Lehmann T, Cai H, Stolz DB, Strom SC. Stem cell characteristics of amniotic epithelial cells. Stem Cells. 2005;23:1549–1559. doi: 10.1634/stemcells.2004-0357. [DOI] [PubMed] [Google Scholar]
- 21.Fanti M, Gramignoli R, Serra M, Cadoni E, Strom SC, Marongiu F. Differentiation of amniotic epithelial cells into various liver cell types and potential therapeutic applications. Placenta. 2017;59:139–145. doi: 10.1016/j.placenta.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Zavatti M, Beretti F, Casciaro F, Comitini G, Franchi F, Barbieri V, Bertoni L, De Pol A, La Sala GB, Maraldi T. Development of a novel method for amniotic fluid stem cell storage. Cytotherapy. 2017;19:1002–1012. doi: 10.1016/j.jcyt.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Nikolić MZ, Rawlins EL. Lung Organoids and Their Use To Study Cell-Cell Interaction. Curr Pathobiol Rep. 2017;5:223–231. doi: 10.1007/s40139-017-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krencik R, Seo K, van Asperen JV, Basu N, Cvetkovic C, Barlas S, Chen R, Ludwig C, Wang C, Ward ME, Gan L, Horner PJ, Rowitch DH, Ullian EM. Systematic Three-Dimensional Coculture Rapidly Recapitulates Interactions between Human Neurons and Astrocytes. Stem Cell Reports. 2017;9:1745–1753. doi: 10.1016/j.stemcr.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karve SS, Pradhan S, Ward DV, Weiss AA. Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS One. 2017;12:e0178966. doi: 10.1371/journal.pone.0178966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camp JG, Sekine K, Gerber T, Loeffler-Wirth H, Binder H, Gac M, Kanton S, Kageyama J, Damm G, Seehofer D, Belicova L, Bickle M, Barsacchi R, Okuda R, Yoshizawa E, Kimura M, Ayabe H, Taniguchi H, Takebe T, Treutlein B. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- 27.Ayabe H, Anada T, Kamoya T, Sato T, Kimura M, Yoshizawa E, Kikuchi S, Ueno Y, Sekine K, Camp JG, Treutlein B, Ferguson A, Suzuki O, Takebe T, Taniguchi H. Optimal Hypoxia Regulates Human iPSC-Derived Liver Bud Differentiation through Intercellular TGFB Signaling. Stem Cell Reports. 2018;11:306–316. doi: 10.1016/j.stemcr.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marongiu M, Serra MP, Contini A, Sini M, Strom SC, Laconi E, Marongiu F. Rat-derived amniotic epithelial cells differentiate into mature hepatocytes in vivo with no evidence of cell fusion. Stem Cells Dev. 2015;24:1429–1435. doi: 10.1089/scd.2014.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin JS, Zhou L, Sagayaraj A, Jumat NH, Choolani M, Chan JK, Biswas A, Wong PC, Lim SG, Dan YY. Hepatic differentiation of human amniotic epithelial cells and in vivo therapeutic effect on animal model of cirrhosis. J Gastroenterol Hepatol. 2015;30:1673–1682. doi: 10.1111/jgh.12991. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XD, Baladandayuthapani V, Lin H, Mulligan G, Li B, Esseltine DW, Qi L, Xu J, Hunziker W, Barlogie B, Usmani SZ, Zhang Q, Crowley J, Hoering A, Shah JJ, Weber DM, Manasanch EE, Thomas SK, Li BZ, Wang HH, Zhang J, Kuiatse I, Tang JL, Wang H, He J, Yang J, Milan E, Cenci S, Ma WC, Wang ZQ, Davis RE, Yang L, Orlowski RZ. Tight Junction Protein 1 Modulates Proteasome Capacity and Proteasome Inhibitor Sensitivity in Multiple Myeloma via EGFR/JAK1/STAT3 Signaling. Cancer Cell. 2016;29:639–652. doi: 10.1016/j.ccell.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saha SK, Kim K, Yang GM, Choi HY, Cho SG. Cytokeratin 19 <i>(KRT19)</i> has a Role in the Reprogramming of Cancer Stem Cell-Like Cells to Less Aggressive and More Drug-Sensitive Cells. Int J Mol Sci. 2018:19. doi: 10.3390/ijms19051423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govaere O, Komuta M, Berkers J, Spee B, Janssen C, de Luca F, Katoonizadeh A, Wouters J, van Kempen LC, Durnez A, Verslype C, De Kock J, Rogiers V, van Grunsven LA, Topal B, Pirenne J, Vankelecom H, Nevens F, van den Oord J, Pinzani M, Roskams T. Keratin 19: a key role player in the invasion of human hepatocellular carcinomas. Gut. 2014;63:674–685. doi: 10.1136/gutjnl-2012-304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang J, Wang H, Liu Y, Ding F, Ni Y, Shao S. High KRT8 expression promotes tumor progression and metastasis of gastric cancer. Cancer Sci. 2017;108:178–186. doi: 10.1111/cas.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim KT, Lee SC, Gao Y, Kim KP, Song G, An SY, Adachi K, Jang YJ, Kim J, Oh KJ, Kwak TH, Hwang SI, You JS, Ko K, Koo SH, Sharma AD, Kim JH, Hui L, Cantz T, Schöler HR, Han DW. Small Molecules Facilitate Single Factor-Mediated Hepatic Reprogramming. Cell Rep. 2016;15:814–829. doi: 10.1016/j.celrep.2016.03.071. [DOI] [PubMed] [Google Scholar]
- 35.Huh CG, Factor VM, Sánchez A, Uchida K, Conner EA, Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci U S A. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto K, Umitsu M, De Silva DM, Roy A, Bottaro DP. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci. 2017;108:296–307. doi: 10.1111/cas.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshihara M, Hayashizaki Y, Murakawa Y. Genomic Instability of iPSCs: Challenges Towards Their Clinical Applications. Stem Cell Rev. 2017;13:7–16. doi: 10.1007/s12015-016-9680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa T, Furue MK, Mizuguchi H. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzaki T, Matsumoto S, Kasai T, Yoshizawa E, Okamoto S, Yoshikawa HY, Taniguchi H, Takebe T. Defining Lineage-Specific Membrane Fluidity Signatures that Regulate Adhesion Kinetics. Stem Cell Reports. 2018;11:852–860. doi: 10.1016/j.stemcr.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Tang Y, Lü S, Zhou J, Du Z, Duan C, Li Z, Wang C. The tumourigenicity of iPS cells and their differentiated derivates. J Cell Mol Med. 2013;17:782–791. doi: 10.1111/jcmm.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazonson P, Kane M, Colberg K, Harris H, Brown H, Mohr A, Ziman A, Santas C. Prevalence of Medical Conditions Potentially Amenable to Cellular Therapy among Families Privately Storing Umbilical Cord Blood. Matern Child Health J. 2017;21:208–214. doi: 10.1007/s10995-016-2110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thornley I, Eapen M, Sung L, Lee SJ, Davies SM, Joffe S. Private cord blood banking: experiences and views of pediatric hematopoietic cell transplantation physicians. Pediatrics. 2009;123:1011–1017. doi: 10.1542/peds.2008-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrow JK, Cross LM, Reese RW, Jaiswal MK, Gregory CA, Kaunas R, Singh I, Gaharwar AK. Widespread changes in transcriptome profile of human mesenchymal stem cells induced by two-dimensional nanosilicates. Proc Natl Acad Sci U S A. 2018;115:E3905–E3913. doi: 10.1073/pnas.1716164115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Meyenn F, Berrens RV, Andrews S, Santos F, Collier AJ, Krueger F, Osorno R, Dean W, Rugg-Gunn PJ, Reik W. Comparative Principles of DNA Methylation Reprogramming during Human and Mouse In Vitro Primordial Germ Cell Specification. Dev Cell. 2016;39:104–115. doi: 10.1016/j.devcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yun W, Hong W, Son D, Liu HW, Kim SS, Park M, Kim IY, Kim DS, Song G, You S. Generation of Anterior Hindbrain-Specific, Glial-Restricted Progenitor-Like Cells from Human Pluripotent Stem Cells. Stem Cells Dev. 2019;28:633–648. doi: 10.1089/scd.2019.0033. [DOI] [PubMed] [Google Scholar]
- 47.Jiang W, Liu Y, Liu R, Zhang K, Zhang Y. The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep. 2015;11:137–148. doi: 10.1016/j.celrep.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu GB, Huang WJ, Zeng M, Zhou X, Wu HP, Liu CC, Wu H, Weng J, Zhang HD, Cai YC, Ashton C, Ding M, Tang D, Zhang BH, Gao Y, Yu WF, Zhai B, He ZY, Wang HY, Yan HX. Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell Res. 2019;29:8–22. doi: 10.1038/s41422-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoyama K, Oritani K, Yokota T, Ishikawa J, Nishiura T, Miyake K, Kanakura Y, Tomiyama Y, Kincade PW, Matsuzawa Y. Stromal cell CD9 regulates differentiation of hematopoietic stem/progenitor cells. Blood. 1999;93:2586–2594. [PubMed] [Google Scholar]
- 50.Lee SH, Paek AR, Yoon K, Kim SH, Lee SY, You HJ. Tight junction protein 1 is regulated by transforming growth factor-β and contributes to cell motility in NSCLC cells. BMB Rep. 2015;48:115–120. doi: 10.5483/BMBRep.2015.48.2.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao P, Wang H, Fang H, Gao Q, Zhang J, Song M, Zhou Y, Wang Y, Wang W. Association between IGF2BP2 Polymorphisms and Type 2 Diabetes Mellitus: A Case-Control Study and Meta-Analysis. Int J Environ Res Public Health. 2016:13. doi: 10.3390/ijerph13060574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paudyal P, Shrestha S, Madanayake T, Shuster CB, Rohrschneider LR, Rowland A, Lyons BA. Grb7 and Filamin-a associate and are colocalized to cell membrane ruffles upon EGF stimulation. J Mol Recognit. 2013;26:532–541. doi: 10.1002/jmr.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M, Seo J, Allegood J, Bi X, Zhu X, Boudyguina E, Gebre AK, Avni D, Shah D, Sorci-Thomas MG, Thomas MJ, Shelness GS, Spiegel S, Parks JS. Hepatic apolipoprotein M (apoM) overexpression stimulates formation of larger apoM/sphingosine 1-phosphate-enriched plasma high density lipoprotein. J Biol Chem. 2014;289:2801–2814. doi: 10.1074/jbc.M113.499913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu N, Dahlbäck B. A novel human apolipoprotein (apoM) J Biol Chem. 1999;274:31286–31290. doi: 10.1074/jbc.274.44.31286. [DOI] [PubMed] [Google Scholar]
- 55.Chen Z, Tang N, Wang X, Chen Y. The activity of the carbamoyl phosphate synthase 1 promoter in human liver-derived cells is dependent on hepatocyte nuclear factor 3-beta. J Cell Mol Med. 2017;21:2036–2045. doi: 10.1111/jcmm.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaffner CA, Mwinyi J, Gai Z, Thasler WE, Eloranta JJ, Kullak-Ublick GA. The organic solute transporters alpha and beta are induced by hypoxia in human hepatocytes. Liver Int. 2015;35:1152–1161. doi: 10.1111/liv.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwerd T, Twigg SRF, Aschenbrenner D, Manrique S, Miller KA, Taylor IB, Capitani M, McGowan SJ, Sweeney E, Weber A, Chen L, Bowness P, Riordan A, Cant A, Freeman AF, Milner JD, Holland SM, Frede N, Müller M, Schmidt-Arras D, Grimbacher B, Wall SA, Jones EY, Wilkie AOM, Uhlig HH. A biallelic mutation in <i>IL6ST</i> encoding the GP130 co-receptor causes immunodeficiency and craniosynostosis. J Exp Med. 2017;214:2547–2562. doi: 10.1084/jem.20161810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oaxaca-Castillo D, Andreoletti P, Vluggens A, Yu S, van Veldhoven PP, Reddy JK, Cherkaoui-Malki M. Biochemical characterization of two functional human liver acyl-CoA oxidase isoforms 1a and 1b encoded by a single gene. Biochem Biophys Res Commun. 2007;360:314–319. doi: 10.1016/j.bbrc.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kölsch H, Lütjohann D, Jessen F, Popp J, Hentschel F, Kelemen P, Friedrichs S, Maier TA, Heun R. RXRA gene variations influence Alzheimer's disease risk and cholesterol metabolism. J Cell Mol Med. 2009;13:589–598. doi: 10.1111/j.1582-4934.2009.00383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen X, Joy MS, Aleksunes LM. In Vitro Transport Activity and Trafficking of MRP2/ABCC2 Polymorphic Variants. Pharm Res. 2017;34:1637–1647. doi: 10.1007/s11095-017-2160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu HJ, Lee SH, Lee S, Choi YJ, Oh D, Nam KH, Yun Y, Ryu DY. Biochemical characterization of variants of canine CYP1A1 using heterologous expression. J Vet Med Sci. 2017;79:1327–1334. doi: 10.1292/jvms.17-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]