Abstract
N6-methyladenosine (m6A) acts as the most common mRNA modification in mammal cells. Fat Mass and Obesity-associated protein (FTO) is the firstly identified demethylase in the m6A modification. This research tries to discover the potential roles of FTO in the m6A modification in the hepatocellular carcinoma (HCC). FTO level was found to be up-regulated in the HCC tissue and cells. The over-expression of FTO was correlated to the poor prognosis of HCC individuals. Knockdown of FTO suppressed the proliferation and in vivo tumor growth, and induced the G0/G1 phase arrest. Mechanically, FTO triggered the demethylation of PKM2 mRNA and accelerated the translated production. Overall, this finding suggests the critical function of FTO in the HCC oncogenesis and its m6A modification.
Keywords: Hepatocellular carcinoma, N6-methyladenosine, FTO, PKM2
Introduction
Hepatocellular carcinoma (HCC) acts as one of the most common cause of cancer-related mortality, causing an extraordinarily prevalence rate worldwide [1,2]. The HCC patients are suffered from chronic hepatitis B virus infection or exposed to aflatoxin [3]. The clinical phenomenon that HCC patients are diagnosed at later or advanced stage make the therapeutic effect much worse [4]. Thus, the effective therapeutic strategy for the HCC requests us to discover the authentic molecular mechanism.
N6-methyladenosine (m6A) modification is regarded as the most prevalent chemical modifications in RNAs [5]. Outside besides the m6A there are still hundreds of chemical modifications for the eukaryocyte mRNA. High throughput sequencing technology could discover and identify the distribution and enrichment of RNA m6A modification [6]. Previous studies reveal that RNA m6A methylation occurs at the N-6 adenosine residue and the motif is specifically located in the RRACH. Among the motif, the R means the G or A purine and H means either A, C or U. The m6A methylation of mRNA was catalyzed by methyltransferase METTL3, METTL14 and WATP [7]. In breast cancer, the critical m6A demethylase FTO is up-regulated and promoted breast cancer cell proliferation, colony formation and metastasis in vitro and in vivo. Epigenetically, FTO mediated m6A demethylation on 3’-UTR of BNIP3 mRNA via an YTHDF2 independent manner to induce its degradation [8].
In the HCC tumorigenesis, the m6A modification is still unclear [9,10]. In this research, we found that Fat Mass and Obesity-associated protein (FTO) is up-regulated in the HCC tissue and indicated the poor prognosis of HCC patients. The demethylation of PKM2 was actuated by the FTO. This finding might provide interesting mechanism for the HCC tumorigenesis.
Materials and methods
Clinical HCC tissues collection
There were thirty pairs of HCC tissues as well as their adjacent normal tissues were collected for the study. The study had obtained the ethical approval from the ethics committee of No. 900 Hospital of the Joint Logistic Support Force. The obtained tissue was straightway frozen at -80°C for subsequent RNA isolation. Signed informed consents were gotten before this study.
Cell culture
HCC cell lines (HepG2, Hep3B, Huh7, SMMC-7721) and normal liver cell lines (Lo-2) were commercially provided by the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China). All cell lines were maintained in DMEM (Gibco, Gaithersburg, MD, USA) in a moist incubator (5% CO2 at 37°C) and added with 10% fetal bovine serum (FBS, Gibco).
Transfection
For FTO knockdown, shRNA targeting FTO and control (sh-NC) were synthesized by RiboBio (Guangzhou, China) and then transfected into the HepG2 cells. PKM2 overexpression plasmid was synthesized and constructed into the pcDNA3 vector by RiboBio, and then transfected into the cells. All the transfection was performed using Lipofectamine2000 reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The sequences were presented in the Table S1.
qRT-PCR analysis
Total RNAs for FTO and PKM2 were isolated from HCC tissues and cells by the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). For the complementary DNA (cDNA), the Transcriptor First-Strand cDNA Synthesis kit (Roche Diagnostics, Mannheim, Germany). qRT-PCR was performed by SYBR Green PCR Master Mix (Applied Biosystems) on an ABI7500 Real-time. Relative expression level was calculated using the 2-ΔΔCt methods normalized to GAPDH.
Colony formation assay
The cell proliferation of HCC cells were determined by performing colony formation assay. HCC cells (HepG2) were cultured in DMEM containing 10% FBS and placed to the 6-well plates (500 cells/well) and preserved in for about two weeks. After two weeks later, the colonies was fixed using methanol and stained by 0.1% crystal violet (SigmaAldrich, St. Louis, MO, USA). Finally, the colonies more than 50 cells were manually calculated under light microscope.
Cell cycle distribution assays
Cell cycle distribution assays were carried out using the flow cytometric analysis. After the cellular transfection with sh-RNA or the controls, cells were collected and stained with the propidium iodide (PI) using Annexin V/propidium iodide detection kit (KeyGen Biotech Co., Nanjing, China).
Western blot analysis
The protein lysate was isolated and then separated using SDS-PAGE onto the 10% gels. Proteins were electrophoretically transferred onto polyvinylidene difluoride membranes (PVDF, Roche). Anti-beta-actin monoclonal antibody and anti-PKM2 (ab137852, Abcam) monoclonal antibody were incubated with the PVDF membrane and then blocked with 10% skim milk. Extraction was probed with antibodies at 4°C overnight and with secondary antibody for 2 h at room temperature. Enhanced chemiluminescence system (ECL) was administrated to detect the proteins and X-ray film was used to expose it.
m6A quantification
The mRNA global m6A levels in the OS tissue and cells were detected using the EpiQuik m6A RNA Methylation Quantification Kit (Epigentek, Colorimetric) following the manufacturer’s protocol. 200 ng poly-A-purified RNA was used for each sample analysis.
m6A-RNA immunoprecipitation
m6A-RNA immunoprecipitation (Me-RIP) was performed as previously described [11]. Total RNA was extracted from the HepG2 cells. The extracted RNA was immunoprecipitated with m6A antibody using Magna methylated RNA immune-precipitation (MeRIP) m6A Kit (Merck Millipore) according to the manufacturer’s protocols. The immunoprecipitated RNA was analyzed using quantitative reverse-transcription polymerase chain reaction (RT-PCR).
Xenograft transplantation
The neonatal nude mice (5 weeks old) were provided by the Slac Laboratory Animal Center (Shanghai, China). The ethical approval for this animal assay was gained from the hospital and done in accordance with institutional guidelines. The transfected cells (2×106) were directly subcutaneously injected in to flank of mice. The width and length were measured every six days. After three weeks, the mice were killed and the necropsies were weighted.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism vision 7.0 and SPSS vision 19.0. The statistical difference was calculated by Student t-test or one-way ANOVA, Mann-Whitney U test. The correlation analysis was analyzed using the Pearson’s correlation and clinical prognosis was analyzed using Kaplan-Meier. P < 0.05 was statistical significance.
Results
The ectopic over-expression of FTO indicated the poor prognosis of HCC patients
The expression of FTO in the HCC tissue was detected in the recruited specimens, showing the high-expression of FTO in the HCC (Figure 1A). Similarly, the expression of FTO was also analysed in each paired HCC tissue and normal tissue (Figure 1B). The TCGA dataset showed that the level of FTO was highly expressed in the HCC tumor tissue compared to the normal control (Figure 1C), which was in accord with the clinical statistics. Moreover, the prognosis analysis of HCC patients who was divided into high FTO level and low FTO level indicated that the high expression of FTO predicted the lower survival rate for HCC individuals (Figure 1D). The percentage of m6A content in the total RNA was found to be reduced in the HCC tissue as compared to normal samples (Figure 1E). Overall, the ectopic over-expression of FTO indicated the poor prognosis of HCC patients, and correlated with the lower m6A content in HCC.
Figure 1.
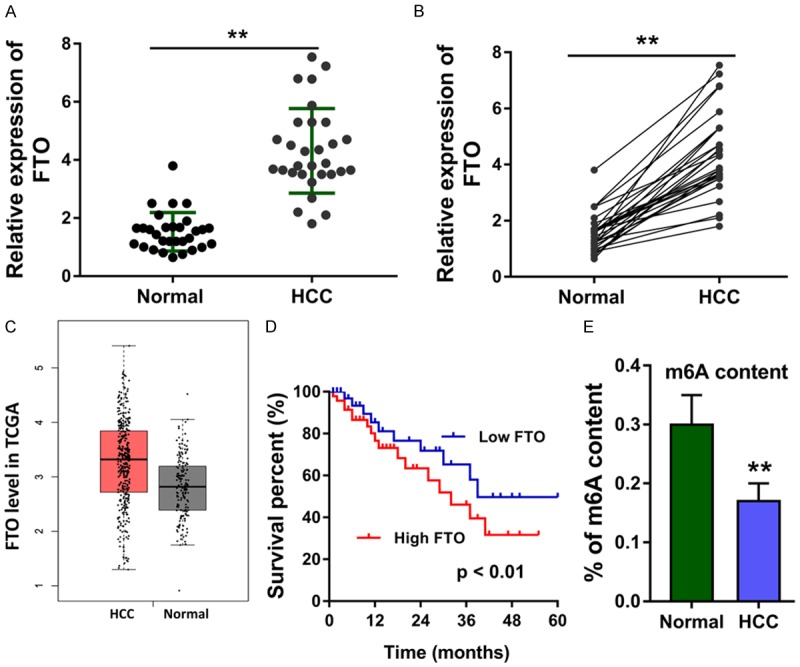
The ectopic over-expression of FTO indicated the poor prognosis of HCC patients. A. The expression of FTO in the HCC tissue was detected using RT-PCR. B. The expression of FTO was also analysed in each paired HCC tissue and normal tissue. C. The TCGA dataset showed the highly expressed level of FTO in the HCC tumor tissue compared to the normal control. D. The prognosis analysis of HCC patients who identified with high FTO level or low FTO level using Kaplan-Meier method. E. The percentage of m6A content in the total RNA was tested in the HCC tissue. Data are presented as means ± SD. **P < 0.01.
FTO knockdown represses the HCC progression and up-regulated the m6A level
The expression of FTO was measured using the RT-PCR, revealing the over-expression of FTO in the HCC cell lines (HepG2, Hep3B, Huh7, SMMC-7721) (Figure 2A). The relative m6A level in the HCC cells was measured by the m6A RNA methylation quantification, showing the low m6A level in the HCC cells (Figure 2B). The short hairpin RNA targeting FTO was transfected to construct the knockdown of FTO (Figure 2C). Moreover, the knockdown of FTO could facilitate the m6A level in HepG2 cells (Figure 2D). Colony formation assay illustrated that knockdown of FTO weaken the proliferative ability of HepG2 cells (Figure 2E). Cycle analysis conducted by the flow cytometry indicated that knockdown of FTO induced the G0/G1 phase arrest as compared to the controls (Figure 2F). In vivo heterotransplantation assay showed that knockdown of FTO alleviated the tumor growth of HCC cells (HepG2) (Figure 2G, 2H). Overall, our data found that FTO knockdown represses the HCC progression and up-regulated the m6A level.
Figure 2.
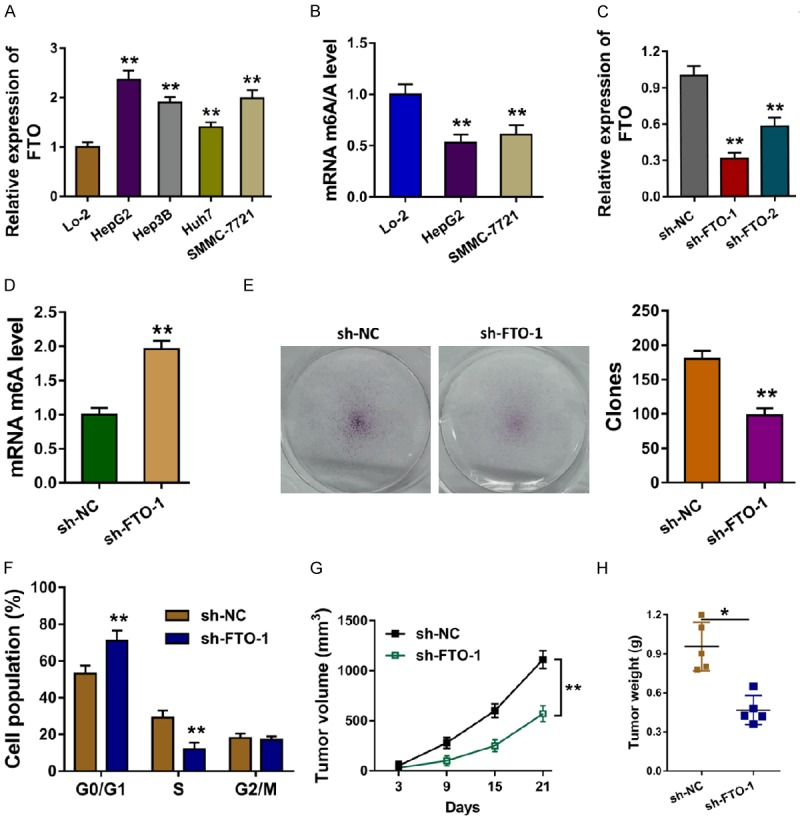
FTO knockdown represses the HCC progression and up-regulated the m6A level. A. RT-PCR revealed the over-expression of FTO in the HCC cell lines (HepG2, Hep3B, Huh7, SMMC-7721). B. The relative m6A level in the HCC cells was measured by the m6A RNA methylation quantification. C. The short hairpin RNA targeting FTO was transfected to construct the knockdown of FTO. D. The relative m6A level in the HepG2 cells transfected with the sh-FTO. E. Colony formation assay illustrated the proliferative ability of HepG2 cells. F. Cycle analysis conducted by the flow cytometry indicated the G0/G1 phase arrest. G, H. In vivo heterotransplantation assay showed the tumor growth of HCC cells (HepG2). Data are presented as means ± SD. **P < 0.01.
FTO triggers the demethylation of PKM2
Previous research indicated that there were series of proteins acted as the targets of FTO, including PKM2 [12]. The expression level of PKM2 was found to be up-regulated in the TCGA database (Figure 3A). In the HCC cells (HepG2), level of PKM2 was also found to be up-regulated (Figure 3B). The interaction of FTO and PKM2 was analyzed using the GEPIA database, suggesting the positive correlation within them (Figure 3C). The motif of FTO was analyzed by the RMBase V2.0 database (http://rna.sysu.edu.cn/rmbase/) (Figure 3D). Methylated RNA immune-precipitation qPCR (Me-RIP qPCR) showed that PKM2 mRNA was significantly enriched with m6A-specific antibody, and FTO knockdown decreased the precipitated m6A level (Figure 3E). The FTO knockdown could reduce the PKM2 mRNA (Figure 3F). Western blot analysis showed that FTO knockdown could reduce the PKM2 protein (Figure 3G). Therefore, this finding illustrates that FTO triggers the demethylation of PKM2.
Figure 3.
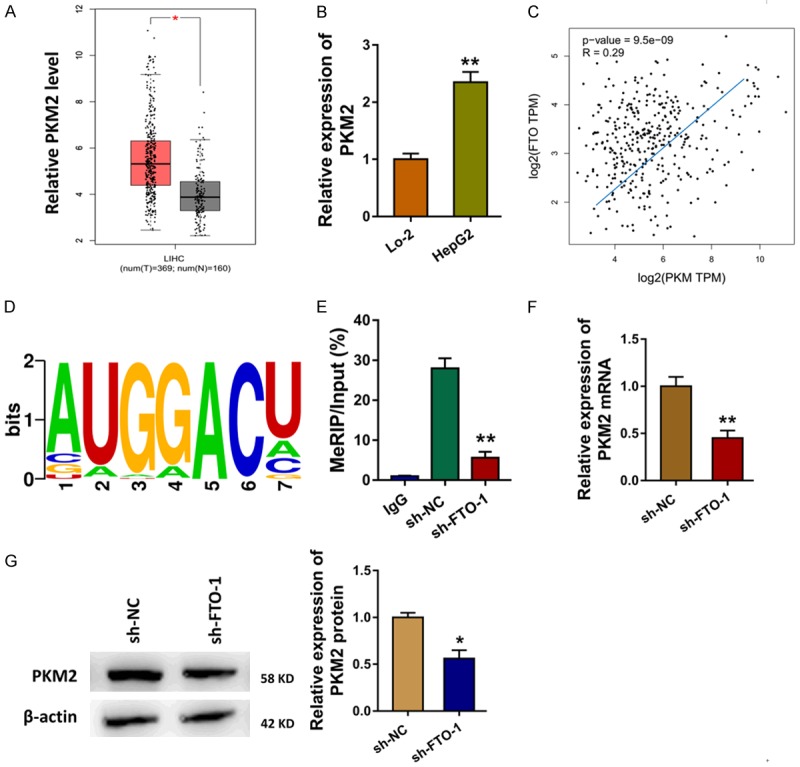
FTO triggers the demethylation of PKM2. A. The expression level of PKM2 was analyzed in the TCGA database. B. RT-PCR showed the level of PKM2 in the HCC cells (HepG2). C. The interaction of PKM and PKM2 was analyzed using the GEPIA database. D. The motif of FTO was analyzed by the RMBase V2.0 database (http://rna.sysu.edu.cn/rmbase/). E. Methylated RNA immune-precipitation qPCR (Me-RIP qPCR) showed the enrichment of PKM2 mRNA using the m6A-specific antibody. F. RT-PCR showed the PKM2 mRNA. G. Western blot analysis showed the PKM2 protein with or without FTO knockdown. Data are presented as means ± SD. **P < 0.01.
FTO coordinately targets PKM2 to regulate HCC progression
Given the demethylation of PKM2 was induced by the FTO, the expression of PKM2 was regulated by FTO. In further experiments, the PKM2 over-expression plasmid transfection could up-regulate the inhibition induced by FTO silencing (Figure 4A). Apoptosis by flow cytometry showed that PKM2 over-expression could reverse the G1/G2 phase arrest induced by the FTO silencing (Figure 4B). Colony formation assay showed that PKM2 over-expression could reverse the clones number induced by the FTO silencing (Figure 4C). The long term survival analysis calculated by the Kaplan Meier-plotter analysis showed that the higher PKM2 indicated the poorer survival of HCC patients (Figure 4D). Overall, this finding suggests that FTO coordinately targets PKM2 to regulate HCC progression.
Figure 4.
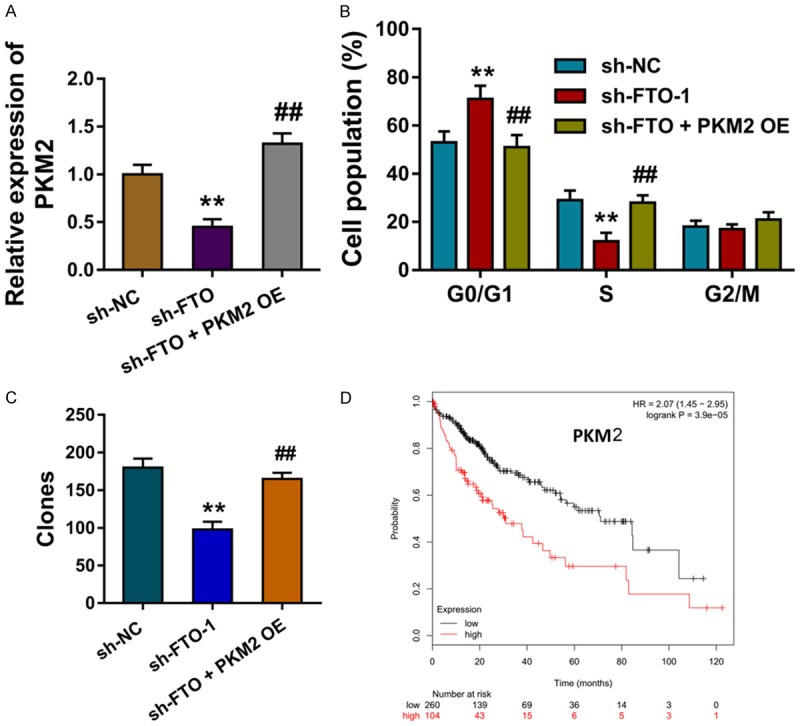
FTO coordinately targets PKM2 to regulate HCC progression. A. The level of PKM2 mRNA measured by the RT-PCR in HepG2 cells transfected with sh-FTO and PKM2 over-expression. B. Apoptosis by flow cytometry showed the cellular distribution of G0/G1, S and G2/M phase. C. Colony formation assay showed the clones number. D. The long term survival analysis calculated by the Kaplan Meier-plotter analysis for the survival of HCC patients. Data are presented as means ± SD. **P < 0.01.
Discussion
The critical roles of N6-methyladenosine (m6A) are increasingly important in recent years [13]. One thing researchers consistently agree on is that the m6A modification for mRNA is a reversible and dynamic modification in eukaryotes [14,15]. The reversible m6A modification regulates series of biological progression, such as RNA stability, alternative splicing, mRNA translation [16,17]. The sites of m6A modification always occurs at gene coding regions and 3’-UTRs with the consensus sequence RRACH region (R indicates G or A; H indicates A, C or U) [18,19]. In the HCC, multiple essential m6A regulatory enzymes have been reported, such as METTL3, METTL14.
Fat Mass and Obesity-associated protein (FTO) is a vital demethylase in the dynamic m6A modification. FTO protein is a subgroup of the Fe2+ and α-ketoglutarate (α-KG) dependent oxygenase family. The role of FTO in the m6A modification has been recognized. For example, Li J et al find that FTO level is overexpressed in human NSCLC and acts as an oncogene in the process. FTO could reduce the m6A content and increase the mRNA stability of ubiquitin-specific protease (USP7) [20]. In lung squamous cell carcinoma (LUSC), FTO is the major dysregulated protein that responsible for the aberrant m6A modification and FTO reduces the m6A level of MZF1 mRNA to enhance the mRNA stability and MZF1 protein expression [21].
In this research, we found that FTO was up-regulated in the HCC tissue and cells. Besides, the m6A level in the HCC cells was increased when the FTO was silenced, which was in accord with the demethylatal biological roles. The functional assay indicated that the knockdown of FTO could repress the HCC progression, e.g. proliferation and apoptosis. In vivo mice assay, we found that the knockdown of FTO could repress the tumor growth. Therefore, this finding suggests that the FTO functions as an oncogene in the HCC oncogenesis, and FTO might regulate the demethylation of mRNA thereby promoting the pathogenesis.
More than FTO, there are multiple regulators responsible for the methylation and demethylation [22]. Other m6A modification genes, including METTL3, METTL14 and WATP, could install the methylation of their target mRNA. Other m6A modification genes, including FTO and ALKBH5 could remove the methylation [23]. These factors could construct the dynamic and reversible process. Previous research have indicated that methyltransferase METLL3 was up-regulated in the HCC and METTL3 enhance the METTL3-mediated m6A modification for its target SOCS2 [24]. Based on this, METTL3 installs the methylation of HCC tissue. In our finding, we found that FTO could relieve the methylation of PKM2 mRNA, thereby up-regulating the expression of PKM2.
Pyruvate kinase (PKM2) is a group of glycolysis pyruvate kinase isoenzyme, encoded by the PKM gene [25,26]. PKM2 promotes the flux of glucose-derived carbons for ATP production via oxidative phosphorylation and accelerates the glucose metabolism towards anabolism through aerobic glycolysis [27]. Our results showed that the level of PKM2 was up-regulated in the HCC tissue, and the ectopic expression indicated the poor prognosis.
In conclusion, we found the overexpression of demethylase FTO in the HCC tissue and cells. FTO could regulate the demethylation of PKM2 in the HCC. This finding indicates the oncogenic role of FTO in the HCC, providing the potential target of HCC treatment.
Acknowledgements
This study was supported by Fujian Natural Science Foundation (2017J01218).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Gong X, Qin S. Study progression of anti-angiogenetic therapy and its combination with other agents for the treatment of advanced hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2018;7:466–474. doi: 10.21037/hbsn.2018.11.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y, Shi J, Liu X, Deng Q, Huang Y, Yang Z. Metabolic syndrome relates to high risk in hepatocellular carcinoma: a meta-analysis. Discov Med. 2018;26:185–196. [PubMed] [Google Scholar]
- 3.Rawla P, Thandra KC, Vellipuram A, Ali CDM. Efficacy and safety of megestrol in the management of hepatocellular carcinoma: a systematic review of the literature. Contemp Oncol (Pozn) 2018;22:209–214. doi: 10.5114/wo.2018.82641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcell Y, Copin P, Paulatto L, Pommier R, Vilgrain V, Ronot M. Hepatocellular carcinoma surveillance: eastern and western perspectives. Ultrasonography. 2019;38:191–199. doi: 10.14366/usg.18043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019;19:326. doi: 10.1186/s12885-019-5538-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F, Klemm L, Wu H, Zuo Z, Qin X, Dong Y, Zhou Y, Qin H, Tao S, Du J, Liu J, Lu Z, Yin H, Mesquita A, Yuan CL, Hu YC, Sun W, Su R, Dong L, Shen C, Li C, Qing Y, Jiang X, Wu X, Sun M, Guan JL, Qu L, Wei M, Muschen M, Huang G, He C, Yang J, Chen J. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567:414–419. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, Yu Z, Wang Y, Qi M, Zhu Y, Perry JM, Zhang K, Tao F, Zhou K, Hu D, Han Y, Zhao C, Alexander R, Xu H, Chen S, Peak A, Hall K, Peterson M, Perera A, Haug JS, Parmely T, Li H, Shen B, Zeitlinger J, He C, Li L. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28:904–917. doi: 10.1038/s41422-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, Wu X, Wan G. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, Liu Y, Ye L, Li Y, Zhang X. MicroRNA-145 modulates N(6)-methyladenosine levels by targeting the 3’-untranslated mRNA region of the N(6)-methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292:3614–3623. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, Xu W, Zhong L, Sun X. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21:859–868. doi: 10.3233/CBM-170791. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Peng C, Chen J, Chen D, Yang B, He B, Hu W, Zhang Y, Liu H, Dai L, Xie H, Zhou L, Wu J, Zheng S. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, Jia G, Chen J, He C. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973–985. e975. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin X, Chai G, Wu Y, Li J, Chen F, Liu J. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of snail. Nat Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Zhang C, Fu J, Zhou Y. A review in research progress concerning m6A methylation and immunoregulation. Front Immunol. 2019;10:922. doi: 10.3389/fimmu.2019.00922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Yin Z, Hou B, Yu M, Chen R, Jin H, Jian Z. Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manag Res. 2019;11:3921–3931. doi: 10.2147/CMAR.S191565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Qi X, Liu L, Hu X, Liu J, Yang J, Yang J, Lu L, Zhang Z, Ma S, Li H, Yun X, Sun T, Wang Y, Wang Z, Liu Z, Zhao W. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan Q, Liu PY, Haase J. The critical role of RNA m(6)A methylation in cancer. Cancer Res. 2019;79:1285–1292. doi: 10.1158/0008-5472.CAN-18-2965. [DOI] [PubMed] [Google Scholar]
- 18.Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuncel G, Kalkan R. Importance of m N(6)-methyladenosine (m(6)A) RNA modification in cancer. Med Oncol. 2019;36:36. doi: 10.1007/s12032-019-1260-6. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, Zheng H, Li B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512:479–485. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–464. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 22.Lence T, Paolantoni C, Worpenberg L, Roignant JY. Mechanistic insights into m(6)A RNA enzymes. Biochim Biophys Acta Gene Regul Mech. 2019;1862:222–229. doi: 10.1016/j.bbagrm.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Pinello N, Sun S, Wong JJ. Aberrant expression of enzymes regulating m(6)A mRNA methylation: implication in cancer. Cancer Biol Med. 2018;15:323–334. doi: 10.20892/j.issn.2095-3941.2018.0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, Tsang LH, Ho DW, Chiu DK, Lee JM, Wong CC, Ng IO, Wong CM. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 25.Lu DH, Lv WW, Li WX, Gao YD. High PKM2 expression is independently correlated with decreased overall survival in hepatocellular carcinoma. Oncol Lett. 2018;16:3603–3610. doi: 10.3892/ol.2018.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Q, Tu J, Dou C, Zhang J, Yang L, Liu X, Lei K, Liu Z, Wang Y, Li L, Bao H, Wang J, Tu K. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer. 2017;16:178. doi: 10.1186/s12943-017-0748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


