Abstract
Esophageal cancer is one of the most common malignant cancers worldwide. Long non-coding RNAs (lncRNAs) have been reported to be associated with different types of cancer; however, the precise function of lncRNAs in tumorigenesis remains largely unknown. Herein, we found that lncRNA NBAT-1 levels were lower in EC clinic tissues than in normal samples, and lncRNA NBAT-1 expression was negatively associated with prognosis and TNM stage in EC patients. More importantly, in EC cancer cells, lncRNA NBAT-1 overexpression strongly inhibited cell proliferation, cell growth and tumor glycolysis. Furthermore, a series of rescue experiments were performed to demonstrate that the role of lncRNA NBAT-1 in EC cell proliferation, cell growth and tumor glycolysis was partially dependent on the PKM2 protein, which is a key metabolic enzyme in tumor development. Taken together, our data illustrate a functional role for lncRNA NBAT-1 in metabolic reprogramming in EC cancer; thus, lncRNA NBAT-1 might be a potential clinical therapeutic target in EC patients.
Keywords: LncRNA-NBAT-1, proliferation, esophageal cancer, PKM2
Introduction
Esophageal cancer is one of the most common malignant cancers worldwide [1,2]. To date, surgical removal of early stage EC is the primary way to improve patient survival rates [3,4]. Current clinical strategies have been employed to diagnose early stage EC to improve the prognosis of EC patients. However, many patients are diagnosed with advanced tumors, which cause poor treatment outcomes for EC patients [5-7]. Thus, there is a great need to explore the molecular mechanisms underlying EC to develop effective therapies for treating advanced tumors.
Long noncoding RNAs (lncRNAs) are transcripts that are greater than 200 nucleotides in length and are reported to play important roles in many cellular processes [8,9]. Many studies have shown that lncRNAs commonly regulate gene expression via distinct regulator roles, such as transcriptional regulation and post-transcriptional modification [10,11]. However, the precise molecular mechanism by which lncRNAs regulate diverse cellular processes requires clarification. Many studies have shown that aberrant lncRNA expression is related to cancer initiation and progression [12,13]. Furthermore, recent studies have shown that lncRNAs are involved in multiple processes of tumorigenesis [14-16]. For example, Yang X. and his colleagues showed that long non-coding RNA HNF1A-AS1 mediates proliferation and metastasis through regulating lncRNA H19 in esophageal adenocarcinoma cells [17]. Regarding other aspects of tumor biology, lncRNA also plays important roles through different molecular mechanisms. Shahabi S. reported that LINC00261 was essential for activating the DNA damage response in lung carcinogenesis through epigenetic regulation [18]. There is much evidence that lncRNAs can be involved in the regulation of tumor metabolism, which is one of the hallmarks of human cancer development [19,20]. It has been demonstrated that lncRNA YIYA promotes breast growth by upregulating tumor glycolysis [21], indicating that lncRNAs are involved in tumor glycolysis. Recently, lncRNA NBAT-1 was shown to affect neuroblastoma development by regulating cell proliferation [22]. Another report also indicated that lncRNA NBAT-1 is involved in tumor progression in lung cancer [23]. However, the role of lncRNA NBAT-1 in EC progression is still unknown, and whether it can regulate tumor glycolysis metabolism remains unclear.
Here, our results demonstrate that lncRNA NBAT-1 expression correlated with the TNM stage of EC patients. More importantly, our data show that lncRNA NBAT-1 levels were higher in EC tumors than in adjacent normal tissues. In addition, silencing lncRNA NBAT-1 inhibited tumor growth and glycolysis. Mechanistically, we also revealed that lncRNA NBAT-1 enhances the expression of PKM2 to promote tumor proliferation. Thus, we discovered that lncRNA NBAT-1 could be a promising biomarker for EC patients and could be considered a novel target for tumor metabolism therapy.
Materials and methods
Cell culture, antibodies and transfection
Eca109 and KYSE150 cells were purchased from the American Type Culture Collection (ATCC) and were cultured with Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Stable cell lines expressing lncRNA NBAT-1 or the control vector were constructed using puromycin. Cells were transfected with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Antibodies against PKM2 and anti-β-actin were purchased from Santa Cruz Biotechnology. Antibodies against HK-II, anti-PFK, anti-PK and anti-PGK1 were obtained from Cell Signaling Technology.
RNA extraction and qRT-PCR
Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Then, total RNA was reverse transcribed into cDNA using a Prime-Script kit (TaKaRa, Dalian, China). The expression of related genes was examined by qRT-PCR using SYBR® Green Master Mix (TaKaRa, Dalian, China). The expression of related genes was normalized to that of GAPDH. The following primers were used in this study: lncRNA NBAT-1 5’-ATTTCTGCTCCTGGGTCTTAC-3’ and 5’-AGTGGCTTGTCTGTTAGAGTC-3’ and GAPDH 5’-CACCCACTCCTCCACCTTTG-3’ and 5’-CCACCACCCTGTTGCTGTAG-3’.
Western blot assay
Cells were lysed with NP40 lysis buffer (Thermo, USA) with protease and phosphatase inhibitor reagents (Thermo, USA) according to the manufacturer’s instructions. Then, an equal amount of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with the indicated primary antibody overnight at 4°C. A secondary antibody was used to identify the relevant bands. Finally, the bands were detected using chemiluminescence (Bio-Rad, USA).
Cell proliferation assay
For the CCK-8 assay, approximately 2000 cells from each group were seeded in a 96-well plate. Then, the cells were cultured for the indicated days. CCK-8 reagent (10 µl) was added to each well and incubated for 2 h. Finally, the optical density (OD) value of each well was examined at 450 nm.
For the cell count assay, approximately 1×104 cells from each group were plated into 6-well plates. After 24 h, the cells in each well were counted every 24 h, and the cell growth curve was plotted.
For the colony formation assay, approximately 100 cells from each group were seeded into 6-well plates. Then, fresh culture medium was added every three days. After 14 days of culture, cell colonies were fixed with 4% paraformaldehyde, and crystal violet staining was used to count the number of cell colonies.
Metabolite analysis of the culture media
Cells from each group were incubated with glucose-free DMEM supplemented with 10% dialyzed serum overnight. Then, glucose or [U13C] glucose (11 mM) was added to the media and incubated with the cells for 6 h. Finally, the cell culture media was harvested for metabolite analysis as previously described [19]. For glucose and lactate concentration analysis, cells from each group were seeded in 6-well plates for 24 h. Then, the cell culture media was harvested and analyzed using a Yellow Springs Instrument (YSI) 7100. The concentrations of glucose and lactate were detected according to the manufacturer’s instructions.
Statistical analysis
All data were analyzed using GraphPad Prism 5 software (San Diego, CA, USA). The results are shown as the mean ± standard error. An unpaired t test was used to examine lncRNA NBAT-1 expression between tumor tissues and paired non-tumor tissues, and a P value <0.05 was considered statistically significant.
Results
LncRNA NBAT-1 was downregulated in EC tissues
To identify the role of lncRNA NBAT-1 in EC development, we measured the expression of lncRNA NBAT-1 in paired EC samples and adjacent normal tissues. Interestingly, we found that lncRNA NBAT-1 expression levels were lower in EC tissues than in normal samples (Figure 1A). Furthermore, to evaluate the clinical significance of lncRNA NBAT-1 in EC patients, we analyzed multiple clinical parameters in 67 EC patients based on their level of lncRNA NBAT-1 expression. As shown in Table 1, we found that the expression of lncRNA NBAT-1 was negatively associated with TNM stage and tumor volume, indicating that lncRNA NBAT-1 might be involved in EC progression, particularly proliferation. To further examine the phenomenon observed in clinical EC tissues, we also determined the expression of lncRNA NBAT-1 in several normal and EC cell lines. Consistent with the above results, lower levels of lncRNA NBAT-1 were observed in EC cell lines compared to those in normal cell lines (Figure 1B). More importantly, a Kaplan-Meier analysis showed that the prognosis of EC patients with low lncRNA NBAT-1 expression was significantly poorer than that of patients with high lncRNA NBAT-1 expression (Figure 1C). In addition, we found that the expression of lncRNA NBAT-1 was negatively related to PKM2 (Figure 1D, 1E), which is an enzyme in glycolysis, but not other glycolytic enzymes (data not shown); these data suggest that lncRNA NBAT-1 might be involved in tumor glycolysis and thus regulate tumor proliferation.
Figure 1.
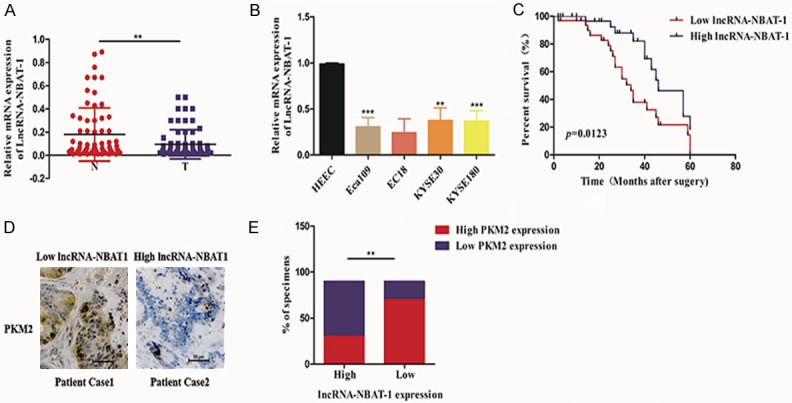
LncRNA NBAT-1 was downregulated in EC tissues. A. Relative mRNA expression levels of lncRNA NBAT-1 between EC tumor and adjacent normal tissues (n=67). **P<0.01. B. Relative mRNA expression levels of lncRNA NBAT-1 among different EC cell lines (Eca109, EC18, KYSE30 and KYSE180) and normal cell lines. C. Kaplan-Meier analysis showing the survival rate of EC patients stratified by lncRNA NBAT-1 expression. D. Relationship between the mRNA expression of lncRNA NBAT-1 and the protein expression of PKM2 in clinical EC cancer tissue samples. E. Spearman correlation analysis between lncRNA NBAT-1 and PKM2 in 67 EC cancer specimens. **P<0.01.
Table 1.
Association between clinicopathological characteristics and expression of lncRNA-NBAT-1 in the esophageal cancer patients (n=67)
| Characteristics | n | lncRNA-NBAT-1 | P-value | |
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | |||
| Sex | 0.548 | |||
| Male | 37 | 17 | 20 | |
| Female | 30 | 16 | 14 | |
| Age | 0.331 | |||
| <50 | 28 | 17 | 11 | |
| ≥50 | 39 | 19 | 20 | |
| Tumor size (cm) | 0.036 | |||
| <5 | 35 | 21 | 14 | |
| ≥5 | 32 | 11 | 21 | |
| Differential grade | 0.408 | |||
| Poor | 18 | 10 | 8 | |
| Middle | 23 | 14 | 9 | |
| Well | 26 | 11 | 15 | |
| LN metastasis | 0.193 | |||
| Positive | 25 | 16 | 9 | |
| Negative | 42 | 20 | 22 | |
| TNM stage | 0.026 | |||
| I | 28 | 20 | 8 | |
| II | 19 | 13 | 6 | |
| III | 20 | 7 | 13 | |
LN, lymph node; TNM, tumor, node, metastasis.
Overexpression of lncRNA NBAT-1 inhibited tumor proliferation
To examine whether lncRNA NBAT-1 affects tumor proliferation, we first constructed KYSE30 and Eca109 stable cell lines expressing lncRNA NBAT-1 using lentivirus. In addition, a negative vector lentivirus was used as a negative control. As shown in Figure 2A, the mRNA levels of lncRNA NBAT-1 were significantly increased in cells expressing the lncRNA NBAT-1 lentivirus. Next, we detected the ability of lncRNA NBAT-1 to affect proliferation in EC cell lines. As shown in Figure 2B, overexpression of lncRNA NBAT-1 strongly reduced the number of colonies formed. In addition, lncRNA NBAT-1 overexpression significantly decreased the growth rate of EC cell lines according to cell count assays. Furthermore, we used CCK-8 assays to analyze the effect of lncRNA NBAT-1 on the proliferative ability of EC cells (Figure 2C). Consistent with the above results, CCK-8 assays showed that overexpression of lncRNA NBAT-1 markedly abrogated EC cell viability (Figure 2D). These data suggest that lncRNA NBAT-1 is involved in tumor proliferation.
Figure 2.
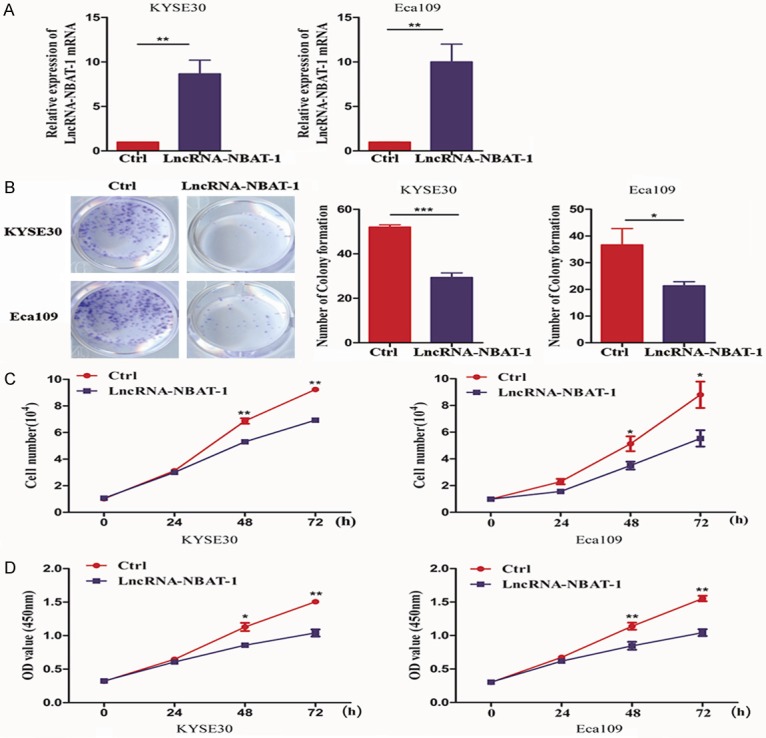
Overexpression of lncRNA NBAT-1 inhibited tumor proliferation. A. qPCR assay showing lncRNA NBAT-1 expression after lncRNA NBAT-1 overexpression plasmid transfection in EC cell lines. **P<0.01. B. Colony formation assays were used to evaluate the proliferative ability of Eca109 and KYSE150 cells expressing vector plasmid or lncRNA NBAT-1. *P<0.05, ***P<0.001. C. Cell count assays were performed to examine the growth ability of Eca109 and KYSE150 cells expressing vector plasmid or lncRNA NBAT-1. *P<0.05, **P<0.01. D. CCK-8 assays were used to determine the proliferative ability of Eca109 and KYSE150 cells expressing vector plasmid or lncRNA NBAT-1. *P<0.05, **P<0.01.
Overexpression of lncRNA NBAT-1 inhibited tumor glycolysis
Given that glycolysis is the most important hallmark of cancer, we further examined the role of lncRNA NBAT-1 in tumor glycolysis. Many metabolic enzymes have been reported to be involved in glucose metabolism. Thus, we examined the levels of related metabolic enzyme expression. As shown in Figure 3A, we surprisingly found that overexpression of lncRNA NBAT-1 reduced PKM2 protein expression, but not other metabolic enzymes such as HK-II, G-6-P, and PFK1. Next, we detected metabolic flux using a 13C-based strategy. As shown in Figure 3B, overexpression of lncRNA NBAT-1 decreased cellular 13C-glucose in EC cells. We also observed lower lactate production in lncRNA NBAT-1-overexpressing cells compared to that in control cells (Figure 3C). In addition, glucose consumption was also examined among the groups. As shown in Figure 3D, overexpression of lncRNA NBAT-1 suppressed glucose consumption, indicating that lncRNA NBAT-1 plays a key role in tumor glycolysis in EC.
Figure 3.
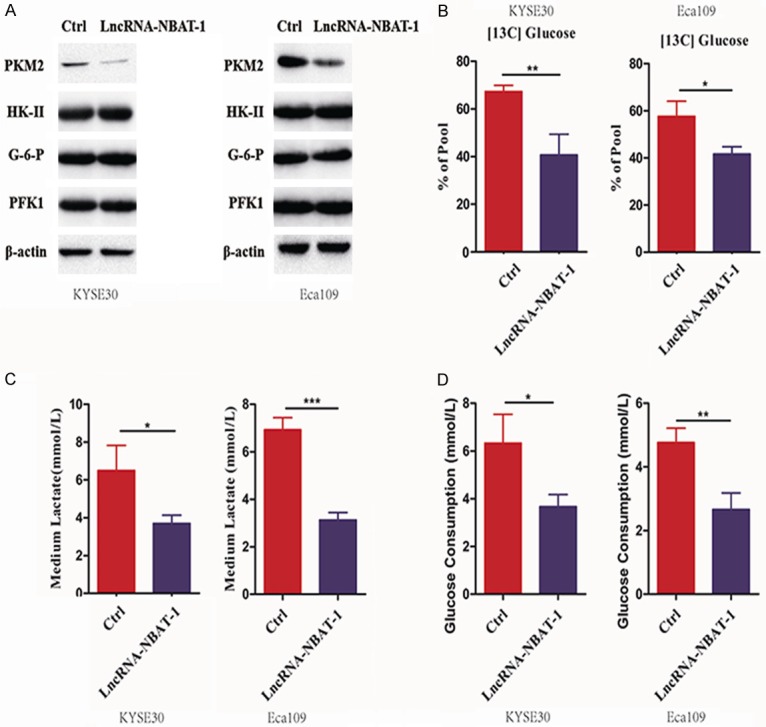
Overexpression of lncRNA NBAT-1 inhibited tumor glycolysis. A. The protein levels of related metabolic enzymes (PKM2, HK-II, G-6-P and PFK1) were examined by Western blot assay of control cells and lncRNA NBAT-1 overexpression cells. B. Percentage of [U-13C] glucose to unlabeled glucose in control cells and lncRNA NBAT-1 overexpression cells starved without glucose overnight, followed by [U-13C] glucose addition. *P<0.05, **P<0.01. C. Medium lactate levels were detected in control cells and lncRNA NBAT-1 overexpression cells. *P<0.05, ***P<0.001. D. Medium glucose consumption was detected in control cells and lncRNA NBAT-1 overexpression cells. *P<0.05, **P<0.01.
LncRNA NBAT-1 regulates tumor glycolysis through PKM2
The PKM2 protein is the only metabolic enzyme regulated by lncRNA NBAT-1. Thus, to determine the role of PKM2 in tumor glycolysis induced by lncRNA NBAT-1, we performed a series of rescue experiments. We overexpressed PKM2 in lncRNA NBAT-1-overexpressing cells to make the intracellular expression of PKM2 equal to that of the control group (Figure 4A). Interestingly, we found that overexpression of PKM2 could reverse the decreased lactate production induced by lncRNA NBAT-1 overexpression (Figure 4B). Furthermore, re-expression of PKM2 led to strongly increased glucose uptake inhibition by lncRNA NBAT-1 overexpression in EC cell lines (Figure 4C). Metabolic flux was also used to examine how the role of PKM2 in glycolysis was inhibited by lncRNA NBAT-1 overexpression. As shown in Figure 4D, expression of PKM2 significantly rescued the metabolic flux impaired by lncRNA NBAT-1 silencing. These data suggested that lncRNA NBAT-1 may modulate tumor glycolysis through regulating PKM2 expression.
Figure 4.
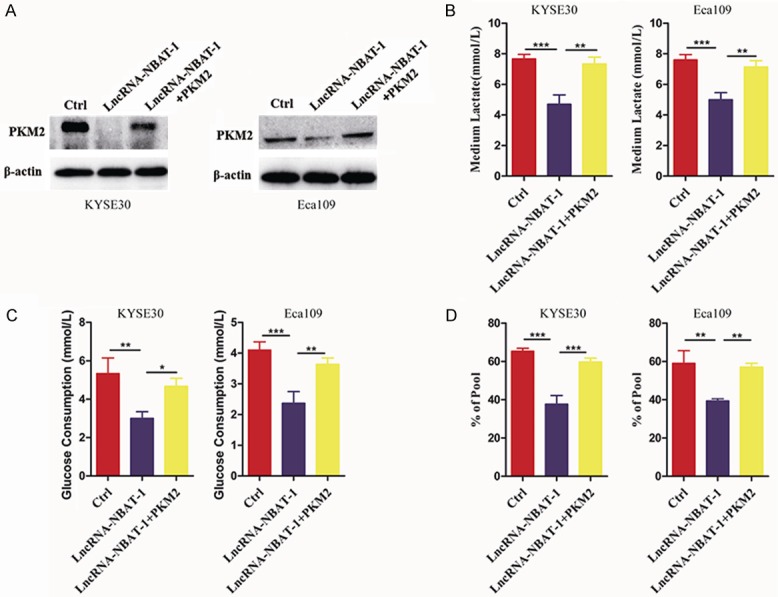
LncRNA NBAT-1 regulated tumor glycolysis through PKM2. A. The protein level of metabolic enzyme PKM2 were examined by Western blot assays of control cells and lncRNA NBAT-1 overexpression cells after transfected with PKM2 plasmids. B. Medium lactate levels in control cells and lncRNA NBAT-1 overexpression cells after transfected with PKM2 plasmids. **P<0.01, ***P<0.001. C. Medium glucose consumption examination in control cells and lncRNA NBAT-1 overexpression cells after transfected with PKM2 plasmids. **P<0.01, ***P<0.001. D. Percentage of [U-13C] glucose to unlabeled glucose in control cells and lncRNA NBAT-1 overexpression cells after transfected with PKM2 plasmids starved without glucose overnight, followed by [U-13C] glucose addition. **P<0.01, ***P<0.001.
PKM2 is necessary for tumor proliferation regulation by lncRNA NBAT-1
Given that glycolysis plays an important role in tumor progression, we next asked whether PKM2 is involved in tumor proliferation regulation by lncRNA NBAT-1. The colony formation assays results showed that re-expression of PKM2 strongly promoted colony formation (Figure 5A). Next, we further examined cell viability by CCK-8 assays. Unexpectedly, cell viability was reversed by the overexpression of PKM2 in lncRNA NBAT-1 overexpression cells (Figure 5B). Additionally, cell count analysis results also confirmed that overexpression of PKM2 could enhance cancer cell growth inhibited by lncRNA NBAT-1 (Figure 5C). These results imply that lncRNA NBAT-1 regulates EC proliferation via glycolysis in a manner dependent on PKM2 expression.
Figure 5.
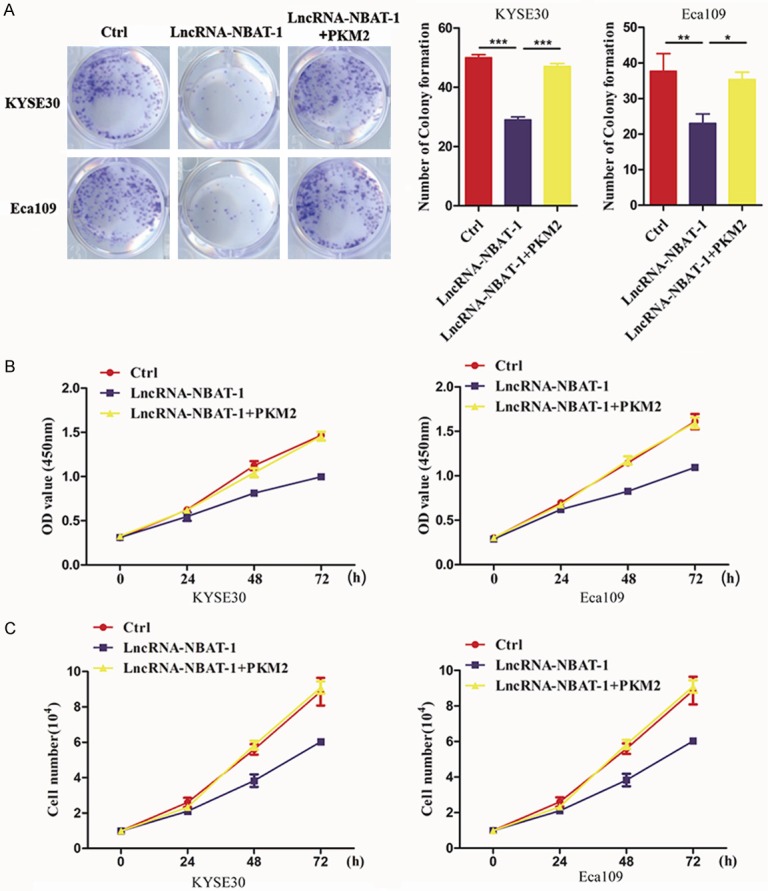
PKM2 is necessary for tumor proliferation regulation by lncRNA NBAT-1. A. A colony formation assay was used to evaluate the proliferative ability of EC109 and KYSE150 cells control cells and lncRNA NBAT-1 overexpression cells after transfected with PKM2 plasmids. *P<0.05, **P<0.01, ***P<0.001. B. Cell viability was assessed in control cells and lncRNA NBAT-1 overexpression cells after transfected with PKM2 plasmids by CCK-8 assay. C. Cell count assays were performed to examine the growth ability of EC109 and KYSE150 cells control cells and lncRNA NBAT-1 overexpression cells after transfected with PKM2 plasmids.
Discussion
Previous reports have revealed that lncRNA dysregulation is involved in diverse cellular processes [11,24]. In human cancer, there is increasing evidence that lncRNA dysregulation plays important roles in the initial progression and advanced development of tumors [25,26]. However, the precise molecular mechanisms underlying lncRNA-mediated tumor development remain obscure. In our study, we observed lower lncRNA NBAT-1 levels in EC tissues that in normal tissues, and the levels of lncRNA NBAT-1 expression were related to TNM stage and tumor size, indicating that lncRNA NBAT-1 may be involved in EC proliferation and might act as a potential suppressor biomarker in EC.
In fact, the suppressor roles of lncRNA NBAT-1 have been reported in lung cancer and neuroblastoma. These studies showed decreased lncRNA NBAT-1 levels in tumor samples compared with those in normal tissues. In support of this finding, our results also showed that low expression of lncRNA NBAT-1 was observed in EC tissues compared with that in paired normal tissues. Furthermore, we surprisingly found that the lncRNA NBAT-1 expression level was negatively related to the PKM2 protein level in clinical EC samples. Most studies on the role of lncRNAs in tumor development have shown that lncRNA dysregulation is involved in tumor proliferation and metastasis [27-29]. For example, a study demonstrated that lncRNA AB074169 inhibits papillary thyroid carcinoma cell proliferation via regulation of KHSRP-mediated CDKN1a expression [30]. Seiler J. and his colleague showed that lncRNA VELUCT significantly regulates lung cancer cell viability, indicating that the dysregulation of some types of lncRNAs contributes to tumor proliferation [31]. According to our data, we found that lncRNA NBAT-1 overexpression markedly inhibited EC proliferation through a series of experiments. Additionally, we found that overexpression of lncRNA NBAT-1 resulted in inhibited EC cell viability. These phenotypic experiments indicated that lncRNA NBAT-1 plays a crucial role in EC proliferation.
Glycolysis is the most important hallmark of cancer and has been shown to affect tumor progression [32,33]. More importantly, some documents have shown that dysregulation of lncRNAs is involved in tumor glycolysis. Linjie Zhao demonstrated that lncRNA LINC00092 could promote glycolysis to enhance the progression of ovarian cancer [34]. Zhen Xing and his colleagues also illustrated that lncRNA YIYA increased tumor glycolysis to promote breast cancer cell proliferation. In the present study, we identified that lncRNA NBAT-1 acts as a mediator of glycolysis in EC cells. Herein, we found that overexpression of lncRNA NBAT-1 strongly decreased the metabolic flux of EC cells and lactate in the medium of EC cells. In addition, our clinical data showed that the lncRNA NBAT-1 expression level was related to PKM2 expression, which is a key metabolic enzyme. Indeed, overexpression of lncRNA NBAT-1 markedly decreased the expression of PKM2, but not that of other metabolic enzymes. Furthermore, we found that the decreased proliferative ability mediated by lncRNA NBAT-1 was strongly reversed by PKM2 overexpression, suggesting that PKM2 may be a potential targeted effector of lncRNA NBAT-1. Moreover, the current research showed that the suppressive properties of lncRNA NBAT-1 were dependent on tumor glycolysis.
In conclusion, our results have shown decreased expression of lncRNA NBAT-1 in EC samples. Furthermore, the lncRNA NBAT-1 expression levels were associated with TNM in EC patients. The effects of lncRNA NBAT-1 on EC cell proliferation were dependent on tumor glycolysis and mediated by PKM2. Our study provides novel insight into understanding EC development and evidence that lncRNA NBAT-1 may act as a potential target for EC therapies.
Acknowledgements
This study was supported by funds from 1) the National Natural Science Foundation of China (grant number 81700174) and 2) the National Natural Science Foundation of China (grant number 81802160).
Disclosure of conflict of interest
None.
References
- 1.Short MW, Burgers KG, Fry VT. Esophageal cancer. Am Fam Physician. 2017;95:22–28. [PubMed] [Google Scholar]
- 2.Seton-Rogers S. Oesophageal cancer: model refinement. Nat Rev Cancer. 2015;15:511. doi: 10.1038/nrc4002. [DOI] [PubMed] [Google Scholar]
- 3.Bollschweiler E, Plum P, Monig SP, Holscher AH. Current and future treatment options for esophageal cancer in the elderly. Expert Opin Pharmacother. 2017;18:1001–1010. doi: 10.1080/14656566.2017.1334764. [DOI] [PubMed] [Google Scholar]
- 4.Kidane B, Korst RJ, Weksler B, Farrell A, Darling GE, Martin LW, Reddy R, Sarkaria IS. Neoadjuvant therapy versus upfront surgery for clinical T2N0 esophageal cancer: a systematic review. Ann Thorac Surg. 2019 doi: 10.1016/j.athoracsur.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. 2018;41:210–215. doi: 10.1016/j.asjsur.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Lagergren J, Lagergren P. Recent developments in esophageal adenocarcinoma. CA Cancer J Clin. 2013;63:232–248. doi: 10.3322/caac.21185. [DOI] [PubMed] [Google Scholar]
- 7.Moaven O, Wang TN. Combined modality therapy for management of esophageal cancer: current approach based on experiences from east and west. Surg Clin North Am. 2019;99:479–499. doi: 10.1016/j.suc.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol. 2017;1008:283–323. doi: 10.1007/978-981-10-5203-3_10. [DOI] [PubMed] [Google Scholar]
- 9.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majidinia M, Yousefi B. Long non-coding RNAs in cancer drug resistance development. DNA Repair (Amst) 2016;45:25–33. doi: 10.1016/j.dnarep.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrest ME, Khalil AM. Review: regulation of the cancer epigenome by long non-coding RNAs. Cancer Lett. 2017;407:106–112. doi: 10.1016/j.canlet.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 16.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108:1927–1933. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, Khashab M, Singh VK, Shin EJ, Yang X, Verma AK, Meltzer SJ, Mori Y. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahabi S, Kumaran V, Castillo J, Cong Z, Nandagopal G, Mullen DJ, Alvarado A, Correa MR, Saizan A, Goel R, Bhat A, Lynch SK, Zhou B, Borok Z, Marconett CN. LINC00261 is an epigenetically regulated tumor suppressor essential for activation of the DNA damage response. Cancer Res. 2019;79:3050–3062. doi: 10.1158/0008-5472.CAN-18-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan C, Tang Y, Wang J, Xiong F, Guo C, Wang Y, Zhang S, Gong Z, Wei F, Yang L, He Y, Zhou M, Li X, Li G, Xiong W, Zeng Z. Role of long non-coding RNAs in glucose metabolism in cancer. Mol Cancer. 2017;16:130. doi: 10.1186/s12943-017-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao ZD, Zhuang L, Gan B. Long non-coding RNAs in cancer metabolism. Bioessays. 2016;38:991–996. doi: 10.1002/bies.201600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xing Z, Zhang Y, Liang K, Yan L, Xiang Y, Li C, Hu Q, Jin F, Putluri V, Putluri N, Coarfa C. Expression of long noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer Res. 2018;78:4524–4532. doi: 10.1158/0008-5472.CAN-17-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyurek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Lei T, Lv ZY, Fu JF, Wang Z, Fan Z, Wang Y. LncRNA NBAT-1 is down-regulated in lung cancer and influences cell proliferation, apoptosis and cell cycle. Eur Rev Med Pharmacol Sci. 2018;22:1958–1962. doi: 10.26355/eurrev_201804_14721. [DOI] [PubMed] [Google Scholar]
- 24.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y, Zhao X. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/beta-catenin signaling. Nat Med. 2017;23:1331–1341. doi: 10.1038/nm.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan DSW, Chong FT, Leong HS, Toh SY, Lau DP, Kwang XL, Zhang X, Sundaram GM. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med. 2017;23:1167–1175. doi: 10.1038/nm.4401. [DOI] [PubMed] [Google Scholar]
- 27.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gou Q, Gao L, Nie X, Pu W, Zhu J, Wang Y, Liu X, Tan S, Zhou JK, Gong Y, He J, Wu K, Xie Y, Zhao W, Dai L, Liu L, Xiang R, Wei YQ, Zhang L, Peng Y. Long noncoding RNA AB074169 inhibits cell proliferation via modulation of KHSRP-mediated CDKN1a expression in papillary thyroid carcinoma. Cancer Res. 2018;78:4163–4174. doi: 10.1158/0008-5472.CAN-17-3766. [DOI] [PubMed] [Google Scholar]
- 31.Seiler J, Breinig M, Caudron-Herger M, Polycarpou-Schwarz M, Boutros M, Diederichs S. The lncRNA VELUCT strongly regulates viability of lung cancer cells despite its extremely low abundance. Nucleic Acids Res. 2017;45:5458–5469. doi: 10.1093/nar/gkx076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Hsu PP, Sabatini DM. Cancer cell metabolism: warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 34.Zhao L, Ji G, Le X, Wang C, Xu L, Feng M, Zhang Y, Yang H, Xuan Y, Yang Y, Lei L, Yang Q, Lau WB, Lau B, Chen Y, Deng X, Yao S, Yi T, Zhao X, Wei Y, Zhou S. Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res. 2017;77:1369–1382. doi: 10.1158/0008-5472.CAN-16-1615. [DOI] [PubMed] [Google Scholar]


