Abstract
Background: Cholangiocarcinoma (CCA) is a biliary malignancy, which is notoriously difficult to diagnose and associated with poor survival. Accumulating evidence indicates that long non-coding RNA Nicotinamide Nucleotide Transhydrogenase-antisense RNA1 (NNT-AS1) is overexpressed in several tumors and plays a crucial role in the development of neoplasm. However, the expression pattern and functional role of NNT-AS1 in CCA remain largely unknown. Methods: NNT-AS1 expression was assessed by RT-qPCR and In Situ Hybridization (ISH) assay. The clinical relevance of NNT-AS1 was analyzed using a CCA tissue microarray with follow-up data. The function role of NNT-AS1 and its underlying molecular mechanisms were evaluated using both in vitro/in vivo experiments and bioinformatics analysis. Luciferase reporter assay, western blot and RT-qPCR were conducted to identify the miRNA/target gene involved in the regulation of CCA progression. Results: LncRNA NNT-AS1 was found highly expressed in CCA. Upregulated NNT-AS1 expression was tightly associated with clinical malignancies and predicted poor prognosis of CCA patients. Functional studies showed that NNT-AS1 knockdown inhibited cell proliferation, migration and invasion of CCA cells in vitro. Conversely, NNT-AS1 overexpression showed the opposite biological effects. In a tumor xenograft model, we confirmed that NNT-AS1 knockdown could significantly inhibit the growth of CCA, while NNT-AS1 overexpression promoted CCA development. Mechanistically, we demonstrated that NNT-AS1 might function as a ceRNA in regulating HMGA2 (high mobility group AT-hook 2) through competitively binding to miR-142-5p in CCA. Moreover, we showed that NNT-AS1 regulated epithelial-mesenchymal transition in CCA. Conclusion: In summary, these findings suggest the potential prognostic and therapeutic value of NNT-AS1/miR-142-5p/HMGA2 axis in CCA patients.
Keywords: NNT-AS1, cholangiocarcinoma, proliferation, tumor progression, epithelial-mesenchymal transition
Introduction
Cholangiocarcinoma (CCA) is the second most common neoplasm arising in intrahepatic, perihilar, and extrahepatic biliary tree. Its incidence has significantly increased in recent decades worldwide [1]. Most patients are diagnosed with advanced stages which are not suitable to potentially curative surgical therapies [2]. To date, patients with CCA have poor survival despite treatment with chemotherapy. Dismal clinical outcome of CCA is due to, at least in part, aggressive biologic behaviors and high rate of tumor recurrence [3]. Thus, further elucidation of the underlying molecular mechanism of CCA is an important step toward developing more novel and effective biomarker and treatment strategies.
Long non-coding RNAs (lncRNAs) are a new class of noncoding RNA molecules over than 200 nt [4]. Over the past decade, mounting evidence has demonstrated that lncRNAs play critical roles in human diverse biological processes [5]. Additional, lncRNA dysregulation has been documented to contribute to neoplasm initiation, development and metastasis [6]. For example, AFAP1-AS1 was reported significantly overexpressed in CCA and promoted tumor progression through regulating the expression of MMP-2 and MMP-9 [7]. Jiang et al. reported that upregulated lncRNA CCAT1 was significantly associated with aggressive malignant behavior and was an independent prognostic biomarker for CCA [8]. In addition, lncRNA H19 and HULC could promote cell migration and invasion in CCA through functioning as competing endogenous RNAs (ceRNAs) [9]. However, the precise role and underlying mechanism of lncRNAs in the CCA development and progression remain unknown.
Long non-coding RNA NNT-AS1 locates at human chromosome 5p12 with 3 exons. Accumulating evidence indicated that NNT-AS1 was commonly overexpressed in several aggressive tumors, including osteosarcoma, cervical cancer and colorectal cancer [10-12]. High level of NNT-AS1 expression was associated with malignancies and poor prognosis, and could promote cancer cell proliferation and metastasis [10,13]. However, neither the biological role nor functional mechanism of NNT-AS1 has been reported in the progression of cholangiocarcinoma so far.
Herein, we investigated the expression profile of lncRNA NNT-AS1 and further explored its biological function in CCA. We found that NNT-AS1 was markedly overexpressed in CCA. High level of NNT-AS1 expression was associated with poor prognosis and clinical malignancies in CCA patients. NNT-AS1 knockdown impaired cell proliferation and invasion abilities. Meanwhile, silencing NNT-AS1 inhibited the tumor growth in a CCA xenograft tumors model. Mechanistically, we demonstrated that NNT-AS1 might function as a ceRNA in regulating HMGA2 through competitively binding to miR-142-5p in CCA. Moreover, we showed that NNT-AS1 regulated epithelial-mesenchymal transition in CCA. Taken together, these findings indicate that NNT-AS1/miR-142-5p/HMGA2 axis plays a crucial role in CCA progression and serve as potential biomarkers for diagnosis and therapeutic target in human CCA treatment.
Materials and methods
Patients and specimens
This study was approved by the research ethics committee of the First Affiliated Hospital of Zhengzhou University. A total of 98 primary CCA samples and matched normal tumor adjacent specimens used to analyze NNT-AS1 expression were obtained from patients who received surgical resection at the First Affiliated Hospital of Zhengzhou University, China (hereinafter referred to as the ZZU cohort). All patients were informed with consent and completed follow-up data records. Diagnoses and grading were confirmed by two experienced independent pathologists based on the 2010 CCA staging system of the AJCC. The clinicopathological features of CCA ZZU cohort were listed in Table 1.
Table 1.
Correlation of clinico-pathological features with NNT-AS1 expression in CCA TMA cohort
| Clinicopathological variables features | NNT-AS1 expression | P-value | ||
|---|---|---|---|---|
|
| ||||
| Low expression (n=42) | High expression (n=47) | |||
| Age (years) | ≤55 | 22 (52.4) | 21 (44.7) | 0.486 |
| >55 | 20 (47.6) | 26 (55.3) | ||
| Gender | Male | 25 (59.5) | 24 (51.1) | 0.423 |
| Female | 17 (40.5) | 23 (48.9) | ||
| Liver Cirrhosis | NO | 15 (35.7) | 16 (34.0) | 0.869 |
| YES | 27 (64.3) | 31 (66.0) | ||
| TNM stage | Stage I/II | 29 (69.0) | 20 (42.6) | 0.012 |
| Stage III | 13 (31.0) | 27 (57.4) | ||
| Tumor size (cm) | ≤5 | 26 (61.9) | 28 (59.6) | 0.822 |
| >5 | 16 (38.1) | 19 (40.4) | ||
| Intrahepatic metastasis | No | 28 (66.7) | 21 (44.7) | 0.037 |
| Yes | 14 (33.3) | 26 (55.3) | ||
| Vascular invasion | Absent | 26 (61.9) | 15 (31.9) | 0.004 |
| Present | 16 (38.1) | 32 (68.1) | ||
| Tumor multiplicity | Single | 27 (64.3) | 24 (51.1) | 0.208 |
| Multiple | 15 (35.7) | 23 (48.9) | ||
| Tumor envelope | No | 32 (76.2) | 30 (63.8) | 0.205 |
| Yes | 10 (23.8) | 17 (36.2) | ||
| HBsAg | No | 30 (71.4) | 29 (61.7) | 0.333 |
| Yes | 12 (28.6) | 18 (38.3) | ||
| Survival status | Live | 34 (81.0) | 23 (48.9) | 0.002 |
| Dead | 8 (19.0) | 24 (51.1) | ||
TCGA expression data sets analysis
The detailed procedures of TCGA analysis were described in our previous study [14,15]. Briefly, we downloaded and analyzed gene expression profiles and corresponding clinical data of cholangiocarcinoma (referred to as the TCGA-CHOL cohort) from The Cancer Genome Atlas Project (TCGA; http://tcga-data.nci.nih.gov/).
Reverse transcription and real-time quantitative PCR (RT-qPCR)
Total RNA was extracted by Trizol (Takara, Japan), and then reversely transcribed to cDNA using Reverse Transcription Kits (Applied Biosystems, USA). Quantitative PCR was performed by SYBR PCR kits (Takara) on Real Time PCR Detection System (Bio-Rad). RNA concentration was measured by Nanodrop (Invitrogen, USA). Relative lncRNA levels were normalized against β-actin, and calculated using the 2-ΔΔCt methods. Each sample was analyzed in triplicate.
Cell lines and cell culture
The human CCA cell lines (RBE, HuCCT1, QBC939 and TFK1) and nonmalignant cholangiocyte cell line HIBEpic was obtained from the Cell Bank of Shanghai Institute of Cell Biology. All Cells were grown in RPMI 1640 medium (Invitrogen, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA) with 100 U/ml penicillin/streptomycin at 37°C in a humidified 5% CO2 incubator. The cell lines used in present study were listed in Supplementary Figure 1.
Western blot
Total protein lysates was extracted and separated by SDS-PAGE, and then transferred to nitrocellulose membranes (Sigma, USA). Antibodies against N-cadherin, E-cadherin, MMP2 and MMP9 (Proteintech, China) were used, and actin was used as a loading control. After incubated with the secondary antibody (Proteintech, China), the membranes was detected and measured with the enhanced chemiluminescence system. The antibodies used in present study were listed in Supplementary Figure 1.
Knockdown or overexpression NNT-AS1 with shRNA or gene-encoded lentivirus
ShRNA used to silence NNT-AS1 and the lentiviral vector encoding the full NNT-AS1 sequence in present study were designed and provided by Hanbio company (Shanghai, China). Before transfection, cells (1 × 105) were cultured until 80% confluence and transfected with same virus MOI into CCA cell lines. Then cells were cultured with 1.5 μg/mL puromycin (Invitrogen, USA) for 36 hours for selection. The transfection efficacy was assessed by RT-qPCR analysis.
Cell growth and colony formation assay
CCK-8 kit (Beyotime, China) was applied to investigate cell proliferation. 1 × 104 cells were dispensed in a 96-well plate. At indicated time, 20 µL CCK-8 solution (5 mg/mL) was added into each well and dissolved in 150 µL dimethylsulphoxide (DMSO; USA). The 490 nm absorbance was measured by microplate reader (Molecular Devices, USA). For colony formation assay, treated cells (50 cells/well) were dispensed in a 24-well plate. Following 14 days culture, cells were fixed with 4% formaldehyde. Then colonies were stained with 0.5% crystal violet and calculated (defined as >50 cells).
5-ethynyl-20-deoxyuridine (EdU) assay
Detailed EdU assay methods were provided in the supplemental data of the material and methods (Supplementary Figure 1).
Migration and invasion assays
Detailed migration and invasion assays methods were provided in the supplemental data of the material and methods (Supplementary Figure 1).
Luciferase reporter assays
WT or mutated 3’-UTR of NNT-AS1 or HMGA2 sequences were cloned and constructed into the pGL3-Luc reporter vector (Promega, USA). HEK293 was transfected with the WT or mutated luciferase reporter vectors, together with miR-142-5p mimics or negative control. Relative luciferase activity was analyzed using a Dual-Luciferase Reporter Assay System (Promega, USA) 48 h later.
In situ hybridization (ISH) and immunohistochemistry (IHC) assay
ISH and IHC staining were performed according to our previous study [16,17]. In brief, TMA slices were washed and incubated with anti-NNT-AS1 oligodeoxy-nucleotide probes (RiboBio, China) at 37°C overnight. Next day, TMA were washed with 0.1% Tween-20 and re-stained by DAPI. For IHC analysis, paraffin-embedded mice tumor sections were deparaffinized and immunostained with antibodies against Ki67, N-cadherin, E-cadherin, MMP2 and MMP9 (Proteintech, China).
In vivo tumor growth
Xenograft tumors model was established according to our previous study [18]. BALB/c nude mice (Vital River Laboratory, China) were used and grouped into (1) NNT-AS1 silencing group (implanted with NNT-AS1 stable knockdown CCA cell lines) or NNT-AS1 overexpression group (implanted with NNT-AS1 stable overexpression CCA cell lines) (2) Control group (implanted with negative control cells). 1 × 107 cells were implanted into the right axillary fossa of nude mice. Tumors were photographed with an IVIS@ Lumina II system (Caliper Life Sciences, USA) every week, while tumor volumes and weights were recorded. Tumor volume was calculated by using the equation: length × width2 × 1/2. All procedures were approved by the Experimental Animal Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Statistical analysis
All statistical analyses were conducted using SPSS version 23.0 software. Results were presented as mean ± SD of at least three independent experiments. The differences between groups were analyzed by Student’s t-test (two-tailed) or Chi-square test. Student’s t-test (two-tailed) and Mann-Whitney test were performed to analyze the in vitro and in vivo data. The Kaplan-Meier and log-rank test method was performed to determine survival rate. P values less than 0.05 were considered to be statistically significant.
Results
NNT-AS1 is upregulated in human CCA tissues and cell lines
Through TCGA database analysis, we identified dysregulated expression of NNT-AS1 in a myriad of cancer types (Figure 1A). The results also showed that NNT-AS1 was markedly overexpressed in CCA tissues compared with normal tissues in TCGA CHOL cohort (Figure 1B). To further validate the expression pattern of NNT-AS1 in CCA, 30 pairs of CCA tissues and surrounding bile duct tissues were collected to determine the expression of NNT-AS1 by RT-qPCR and the results revealed the increased expression of NNT-AS1 in CCA tissues (Figure 1C and 1D). We next measured the expression levels of NNT-AS1 in human CCA cell lines. As shown in Figure 1E, NNT-AS1 expression was significantly higher in CCA cell lines (RBE, HuCCT1, QBC939 and TFK1) than that in control cell line HIBEpic.
Figure 1.
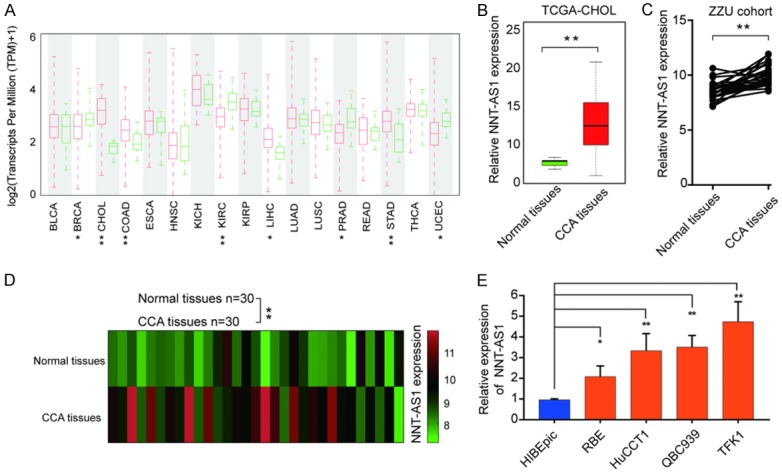
LncRNA NNT-AS1 is upregulated in human CCA tissues and cell lines. A. Analysis of NNT-AS1 expression in TCGA database. B. Analysis of NNT-AS1 expression level in normal tissues and CCA tissues in TCGA-CHOL. Bars represent median NNT-AS1 level. C, D. Analysis of NNT-AS1 expression in 20-paired normal tissues and CCA tissues. E. Expression level of NNT-AS1 in normal intrahepatic biliary epithelial and CCA cell lines. *P<0.05, **P<0.01, Mean ± SD, unpaired Student’s t-test.
High expression of NNT-AS1 is positively associated with clinical malignant and poor prognosis in CCA patients
We further performed in situ hybridization (ISH) analysis to explore the expression of NNT-AS1 in CCA tissues and non-tumor control tissues (Figure 2A and 2B). We found that NNT-AS1 expression was significantly higher in CCA tissues compared with that in paired normal bile duct tissues. Furthermore, high expression of NNT-AS1 was positively correlated with metastasis, vascular invasion and poor histological differentiation (Figure 2C-E). Kaplan-Meier analysis indicated CCA patients with higher NNT-AS1 level had significantly worse overall survival (OS) or disease-free survival (DFS) rate than those with a lower NNT-AS1 level (Figure 2F and 2G).
Figure 2.
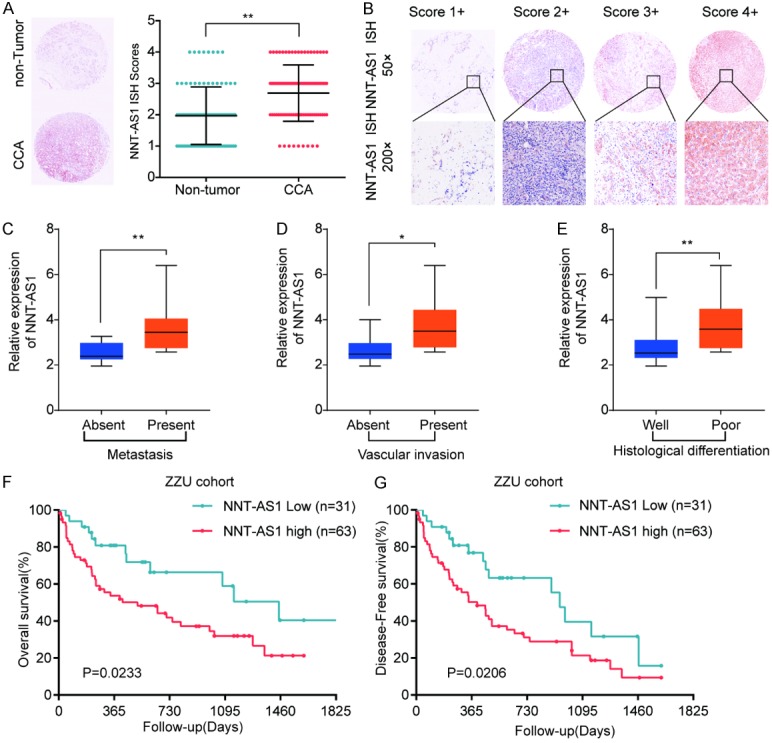
High expression of NNT-AS1 is positively associated with clinical malignant and poor prognosis in CCA patients. A. Representative ISH staining of NNT-AS1 (left panel) and the quantification of NNT-AS1 ISH staining scores in CCA tissues and corresponding non-tumor tissues. B. Representative pictures of NNT-AS1 expression with different staining scores in CCA tissues. C-E. Analysis the association between NNT-AS1 expression and metastasis, vascular invasion and histological differentiation. F, G. The overall survival and disease free survival were analyzed by Kaplan-Meier analysis, according to NNT-AS1 expression levels. *P<0.05, **P<0.01, Mean ± SD, unpaired Student’s t-test or Kaplan Meier analysis.
Furthermore, univariable and multivariable Cox proportional hazards analysis were performed to identify the prognostic risk factors of OS and RFS. Univariable analyses revealed that NNT-AS1 expression, TNM stage, intrahepatic metastasis and vascular invasion were significant prognostic factors for OS and PFS prediction (Table 2). Meanwhile, multivariable analysis showed that NNT-AS1 expression was an independent predictors of OS (P=0.002) and RFS (P=0.038) in patients with CCA in addition to TNM stage (Table 2). Taken together, these findings indicated that NNT-AS1 played a crucial role in the progression and may served as a potential target for prognosis prediction in CCA patients.
Table 2.
Univariate and multivariate analyses of overall survival of studied CCA patients
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Univariate and multivariate analysis of overall survival in CCA patients (n=95) | ||||||
| Age | 1.223 | 0.546-1.365 | 0.554 | |||
| Gender | 0.774 | 0.546-1.365 | 0.426 | |||
| Liver Cirrhosis | 1.156 | 0.876-1.464 | 0.279 | |||
| TNM stage | 2.551 | 1.837-2.905 | 0.014 | 2.622 | 1.960-4.119 | 0.032 |
| Tumor size (cm) | 1.321 | 1.108-1.543 | 0.065 | |||
| Intrahepatic metastasis | 1.900 | 1.030-2.966 | 0.037 | 2.036 | 1.754-3.245 | 0.036 |
| Vascular invasion | 2.006 | 1.199-3.378 | 0.022 | 1.621 | 0.915-2.814 | 0.044 |
| Tumor multiplicity | 1.352 | 1.238-1.659 | 0.455 | |||
| HBsAg | 1.307 | 1.102-1.683 | 0.200 | |||
| NNT-AS1 expression | 2.099 | 1.259-3.346 | 0.005 | 1.999 | 1.704-2.319 | 0.002 |
| Univariate and multivariate analysis of disease-free survival in CCA patients (n=95) | ||||||
| Age | 1.009 | 0.599-1.958 | 0.632 | |||
| Gender | 0.885 | 0.477-1.600 | 0.554 | |||
| Liver Cirrhosis | 0.674 | 0.396-1.303 | 0.222 | |||
| TNM stage | 2.663 | 1.701-3.339 | 2.0189 | 2.239 | 1.659-3.51 | 0.038 |
| Tumor size (cm) | 1.419 | 1.208-2.047 | 0.067 | |||
| Intrahepatic metastasis | 2.045 | 1.662-3.749 | 0.038 | 2.234 | 1.845-3.563 | 0.041 |
| Vascular invasion | 3.068 | 1.479-4.765 | 0.017 | 1.984 | 1.733-2.946 | 0.047 |
| Tumor multiplicity | 2.996 | 1.225-6.012 | 0.308 | |||
| HBsAg | 1.473 | 0.603-1.961 | 0.153 | |||
| NNT-AS1 expression | 2.108 | 1.561-2.594 | 0.025 | 2.201 | 1.695-2.602 | 0.032 |
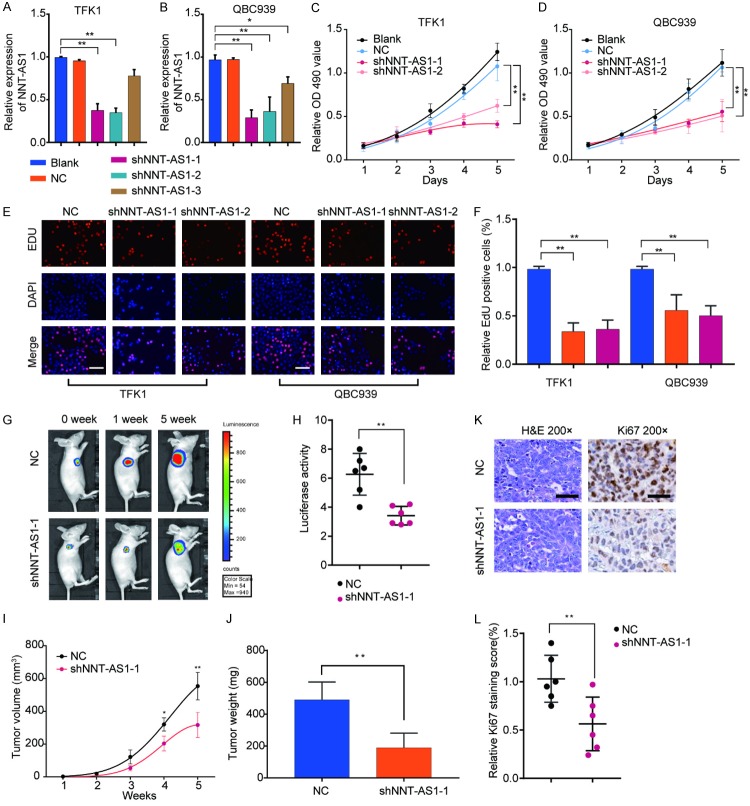
Knockdown of NNT-AS1 suppresses cell proliferation of CCA both in vitro and in vivo
To explore the biological function of NNT-AS1 in CCA, we firstly determined the effect of NNT-AS1 silencing on cell proliferation through shRNA knockdown approach. NNT-AS1 knockdown efficiency was validated in TFK1 and QBC939 cells (Figure 3A and 3B). CCK-8 assay showed that cell growth of the NNT-AS1-silencing TFK1 or QBC939 cells was significantly inhibited compared with that in cells transfected with negative controls (Figure 2C and 2D). In parallel, EdU assay also demonstrated that knockdown of NNT-AS1 effectively inhibited the cell proliferation (Figure 2E and 2F).
Figure 3.
Knockdown of NNT-AS1 suppresses cell proliferation of CCA both in vitro and in vivo. TFK1 or QBC939 cells were transfected with shRNA1/2/3 targeting NNT-AS1, respectively. A, B. The NNT-AS1 knockdown efficiency was determined by RT-qPCR. C, D. CCK-8 assay was performed to determine the viability of sh-NNT-AS1-1/2-transfected TFK1 or QBC939 cells at indicated time points. E, F. EdU staining assay were performed to determine the growth of sh- NNT-AS1-1/2-transfected TFK1 or QBC939 cells. TFK1 cells stably transfected with sh-NNT-AS1-1 or negative control (NC) were implanted into nude mice. Xenograft tumor was monitored for 5 weeks. G, H. Luciferase signal in the NC or sh-NNT-AS1-1 group was determined at indicated time points. I, J. Tumor growth and tumor volume were analyzed at indicated time points. Mice were euthanized and tumor tissues were extracted at week 5. Tumor weight was measured. Values are the mean ± SD (n=6/group). K, L. H&E and Ki67 IHC staining of xenograft tumors tissues from NC or sh-NNT-AS1-1 group. Mean ± SD, *P<0.05, **P<0.01, unpaired Student’s t-test or one-way ANNOVA followed by multiple t-test. Representative images and data are based on three independent experiments.
To further evaluate the function of NNT-AS1 on CCA cell proliferation in vivo, TFK1 cells stably transfected with NC or shNNT-AS1-1 were inoculated into nude mice. Consistent with the in vitro findings, results showed that xenograft tumors grown from NNT-AS1-silenced cells had markedly less mean luciferase signal than that of the tumors developed from NC cells (Figure 3G and 3H). Correspondingly, tumor volume and weight were markedly decreased in the sh-NNT-AS1 group compared with that in control group (Figure 3I and 3J). In addition, comparison with that in tumor tissues resected from control group, sh-NNT-AS1 derived tumors showed significantly weaker proliferation marker ki-67 staining (Figure 3K and 3L). These results confirmed the promo-oncogenic role of NNT-AS1 in CCA tumorigenesis.
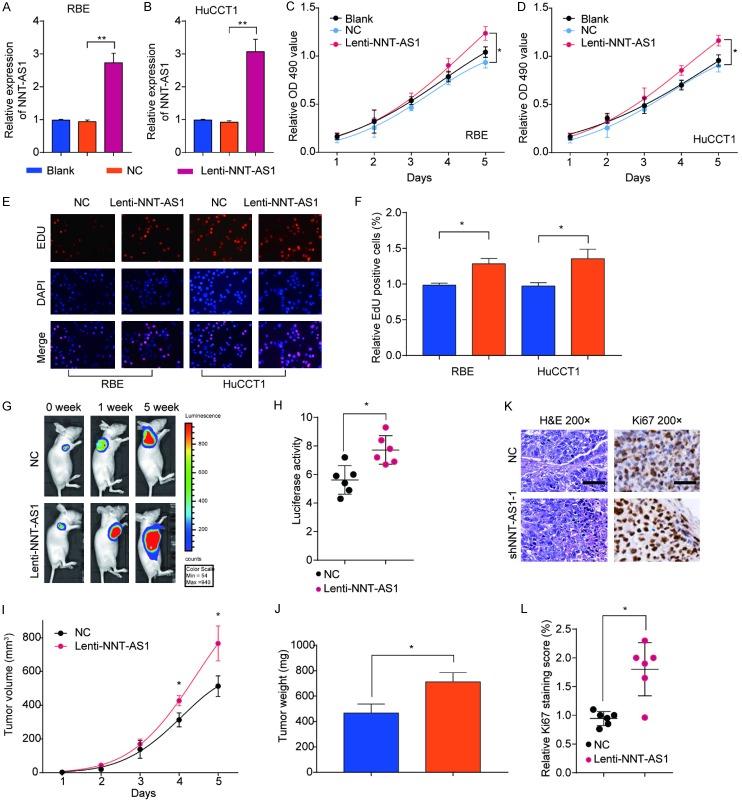
Overexpression of NNT-AS1 promotes cell proliferation of CCA both in vitro and in vivo
To further address the role of NNT-AS1 in CCA progression, we overexpressed NNT-AS1 in RBE and HuCCT1 cells with Lenti-NNT-AS1 transduction (Figure 4A and 4B). Functionally, we found that NNT-AS1 overexpression significantly enhanced the proliferative potential of RBE and HuCCT1 cells by CCK-8 assay and EdU assay (Figure 4C-F). Furthermore, we subcutaneously injected BALB/c nude mice with stably NNT-AS1 overexpression RBE cells. After 5 weeks, xenograft tumors grown from Lenti-NNT-AS1 cells showed dramatic increased mean luciferase signal than that in tumors developed from NC cells via ex vivo imaging analysis (Figure 4G and 4H). In addition, we observed that the tumor growth rate, tumor volume and tumor weight were markedly increased in Lenti-NNT-AS1 group compared to that in control group (Figure 4I and 4J). Moreover, tumors from Lenti-NNT-AS1 showed higher ki-67 expression than that in tumors from NC group (Figure 4K and 4L). These gain-of-function results combined with the loss-of-function data revealed that NNT-AS1 facilitated CCA growth.
Figure 4.
Overexpression of NNT-AS1 promotes cell proliferation of CCA both in vitro and in vivo. RBE or HuCCT1 cells were transfected with Lenti-NNT-AS1 or negative control (NC). A, B. NNT-AS1 expression in RBE or HuCCT1 cells was analyzed by RT-qPCR. C, D. CCK-8 assay was performed to assess the viability of RBE cells and HuCCT1 cells after NNT-AS1 overexpression. E, F. EdU staining assay was performed to determine the growth of RBE cells and HuCCT1 cells. RBE cells stably transfected with Lenti-NNT-AS1 or negative control (NC) were implanted into nude mice. Xenograft tumor was monitored for 5 weeks. G, H. Luciferase signal in the NC or Lenti-NNT-AS1 group was determined at indicated time points. I, J. Tumor growth and tumor volume were analyzed at indicated time points. Mice were euthanized and tumor tissues were extracted at week 5. Tumor weight was measured. Values are the mean ± SD (n=6/group). K, L. H&E and Ki67 IHC staining of xenograft tumors tissues from NC or Lenti-NNT-AS1 group. Mean ± SD, *P<0.05, **P<0.01, unpaired Student’s t-test or one-way ANNOVA followed by multiple t-test. Representative images and data are based on three independent experiments.
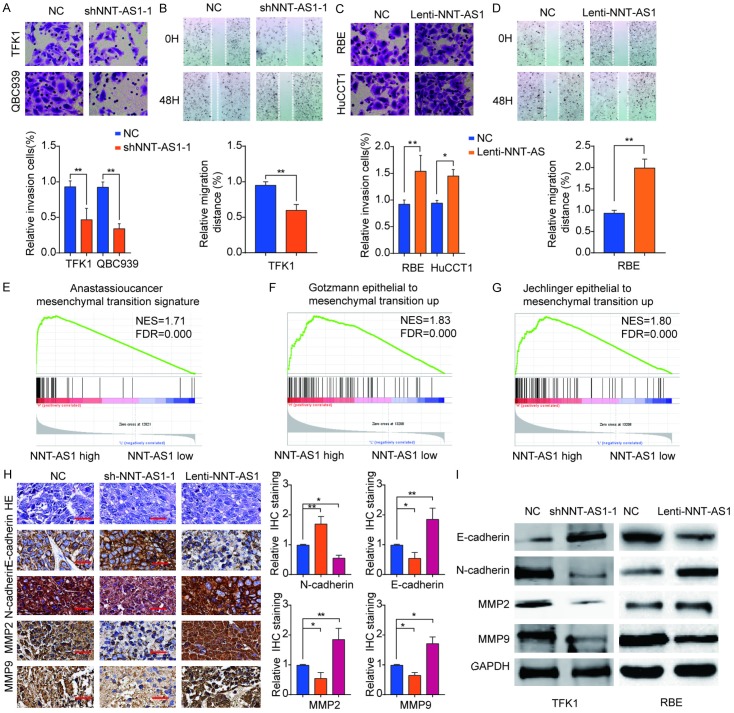
NNT-AS1 promotes CCA cell invasion and migration via promoting EMT
Given that high expression of NNT-AS1 was associated with vascular invasion and progression in patients with CCA, we further evaluated the effect of NNT-AS1 knockdown or overexpression on CCA cell migration and invasion. Transwell assay revealed that knockdown of NNT-AS1 impeded the invasive ability of CCA cells (Figure 5A). In parallel, a corresponding suppressive effect on migration was also observed via wound-healing assay (Figure 5B). In contrast, invasion and migration abilities of CCA cells were enhanced by NNT-AS1 overexpression (Figure 5C and 5D).
Figure 5.
NNT-AS1 promotes CCA cell invasion and migration via promoting EMT. A. TFK1 or QBC939 cells were transfected with shNNT-AS1-1 or negative control (NC). Transwell assay was performed to evaluate the cell invasion. B. TFK cells were transfected with shNNT-AS1-1 or NC. Wound-healing assay was conducted to evaluate the cell migration capability. C. RBE or HuCCT1 cells were transfected with Lenti-NNT-AS1-1 or NC. Transwell assay was performed to evaluate the cell invasion. D. RBE cells were transfected with Lenti-NNT-AS1-1 or NC. Wound-healing assay was conducted to evaluate the cell migration capability. E-G. Gene Set Enrichment Analysis indicated a significant correlation between NNT-AS1 expression and EMT. H. Western blot analysis of E-cadherin, N-cadherin, MMP2 and MMP9 in TFK1 cells transfected with shNNT-AS1-1 or NC, or in RBE cells transfected with Lenti-NNT-AS1 or NC. I. IHC staining analysis of E-cadherin, N-cadherin, MMP2 and MMP9 in tumor tissues from NNT-AS1 silencing or overexpression group. Scale bar, 100 μm. Mean ± SD, *P<0.05, **P<0.01, unpaired Student’s t-test. Representative images and data are based on three independent experiments.
To explore underlying mechanisms of NNT-AS1 involved in CCA metastasis. Gene set enrichment analysis (GSEA) of the CCA gene expression profile from TCGA database was conducted and results showed a significant correlation between NNT-AS1 expression and EMT (Figure 5E-G). We demonstrated that knockdown NNT-AS1 decreased the expression of N-cadherin, MMP2 and MMP9, and enhanced the expression of E-cadherin. Overexpression NNT-AS1 showed the opposite effect as shown by western blot results (Figure 5H). Furthermore, IHC analysis confirmed that tumor tissues from sh-NNT-AS1-1 group exhibited decreased expression of N-cadherin, MMP2 and MMP9, as well as increased expression of E-cadherin (Figure 5I). Collectively, these data suggested that repression of NNT-AS1 could suppress the migration and invasion of CCA cells, at least in part, by regulating the EMT.
NNT-AS1 directly interacts with miR-142-5p as miRNA sponge
To explore how NNT-AS1 regulates CCA progression and metastasis, we performed bioinformatics analysis to search for the potential targets of NNT-AS1. As shown in Figure 6A, miR-142-5p was predicted to have the putative binding sites with NNT-AS1. Pearson correlation analysis showed that NNT-AS1 expression was negatively associated with miR-142-5p expression (Figure 6B). We also found that the expression of miR-142-5p in CCA tissues was markedly lower than that in non-tumor control tissues in TCGA-CHOL cohort (Figure 6C). Consistently, the results showed that CCA cell lines (RBE, HuCCT1, QBC939 and TFK1) had significantly lower miR-142-5p expression than that in control cell line HIBEpic (Figure 6D). Previous study has shown that multiple lncRNAs are known to function as competing endogenous RNA (ceRNAs) for specific miRNAs [19]. While overexpression NNT-AS1 significantly inhibited miR-142-5p expression (Figure 6E), NNT-AS1 knockdown increased miR-142-5p expression (Figure 6F). Luciferase reporter assay showed that miR-142-5p mimics significantly inhibited the luciferase activity in HEK293 cells transfected with reporter vector containing wild type NNT-AS1, but not with mutant NNT-AS1 (Figure 6G).
Figure 6.
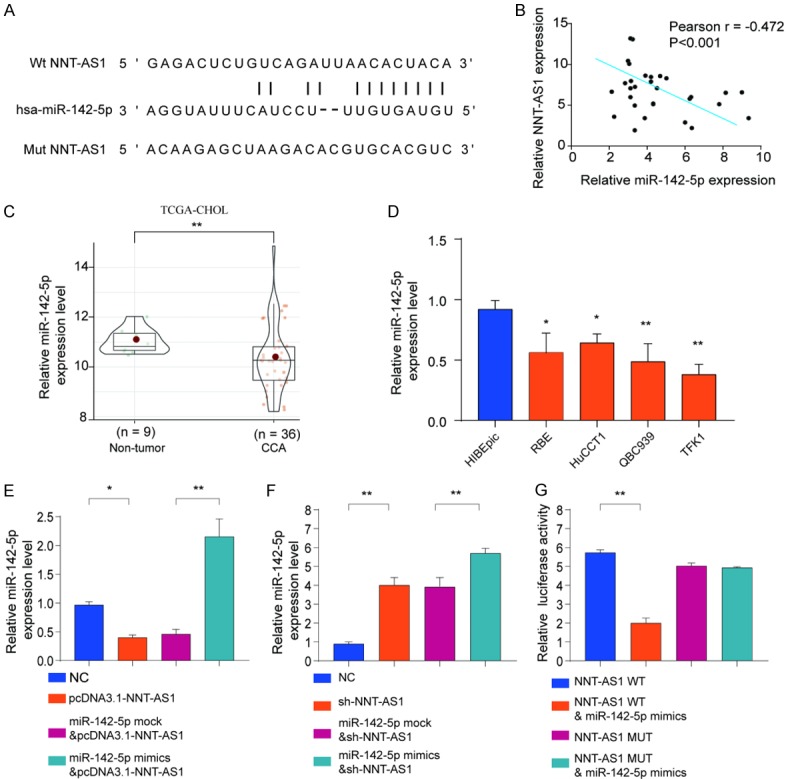
NNT-AS1 directly interacts with miR-142-5p as miRNA sponge. (A) Diagram of the putative binding sites of miR-142-5p on NNT-AS1 (WT) and the mutated sequences of NNT-AS1 (Mut). (B) Pearson correlation analysis of NNT-AS1 expression and miR-142-5p expression in CCA tissues. (C) The expression levels of miR-142-5p in CCA tissues and normal control tissues were analyzed based on TCGA. (D) The expression levels of miR-142-5p in CCA cell lines (RBE, HuCCT1, QBC939 and TFK1) and nonmalignant cell line HIBEpic were analyzed by qRT-PCR. (E, F) HEK293 cells were transfected with negative control (NC), pcDNA3.1-NNT-AS1, miR-142-5p mock, or miR-142-5p mimics and the expression of miR-142-5p (D) or NNT-AS1 (E) was analyzed by qRT-PCR. (G) The WT or mutated NNT-AS1 was fused to the luciferase-coding region and co-transfected into HEK293T cells with miR-142-5p mimics. Relative luciferase activity was determined 48 h after transfection. Data are presented as mean ± SD; Student’s t-test, *P<0.05 and **P<0.01. Pearson analysis was used to calculate the correlation between the expression of NNT-AS1 and miR-142-5p.
To further validate the interaction between miR-1425p and NNT-AS1, RBE or HuCCT1 cells were transfected with miR-142-5p mimics or mock control, with/without NNT-AS1 overexpression vector. Cell proliferation assessed by CCK-8, colony formation and transwell assays showed that while overexpression of miR-142-5p suppressed cell proliferation, colony formation and cell invasion of RBE or HuCCT1 cells, overexpression of NNT-AS1 partially reversed the suppressive effect of miR-142-5p in these cells (Figure 7A-C). These results confirmed the interaction between miR-142-5p and NNT-AS1 cooperatively regulated CCA cells proliferation and invasion.
Figure 7.
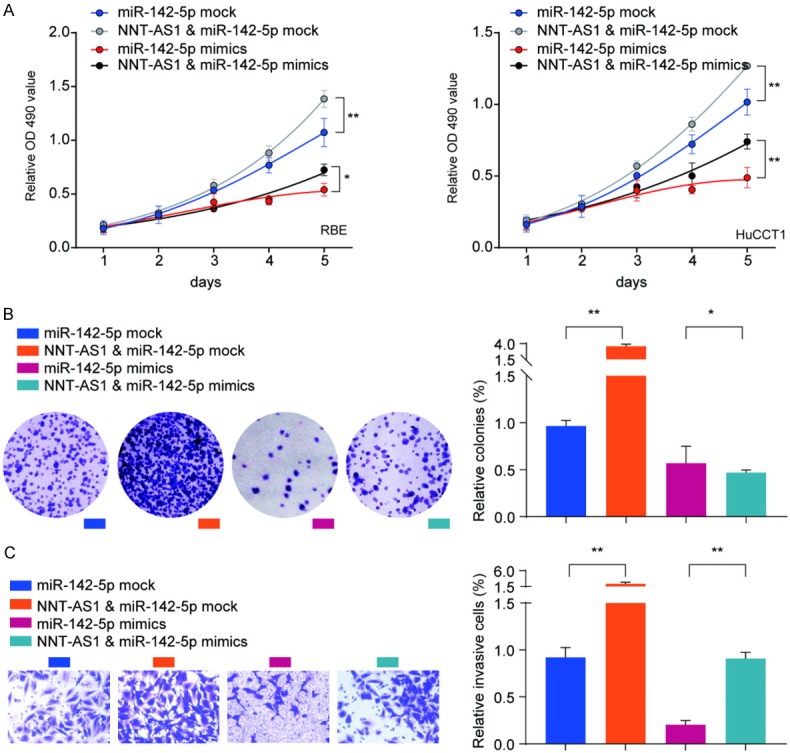
Overexpression of NNT-AS1 partially reverses the suppressive effect of miR-142-5p in CCA cells. RBE or HuCCT1 cells were transfected with miR-142-5p mock or miR-142-5p mimics, with or without NNT-AS1 overexpression vector. A. Cell proliferation of RBE or HuCCT1 was analyzed at indicated time points by CCK-8 kit. B, C. Colony formation and cell invasion of RBE cells were analyzed by colony formation assay and transwell assay, respectively. Data are presented as the mean ± SD; Using the Student’s t-test for statistical analysis. *P<0.05 and **P<0.01.
MiR-142-5p negatively regulates High Mobility Group AT-Hook 2 (HMGA2) expression by targeting 3’-UTR of HMGA2
To further investigate the potential target of miR-142-5p, we searched for miR-142-5p targets by online bioinformatics tool TargetScan. Intriguingly, HMGA2 was identified to have putative binding sites of miR-142-5p (Figure 8A). Consistently, the expression of HMGA2 was significantly negative-associated with the expression of miR-142-5p (Figure 8B). In addition, dual luciferase reporter assay showed that miR-142-5p mimics inhibited luciferase activity in HEK293 cells transfected with the reporter plasmid containing WT HMGA2 3’-UTR, while no significant suppression was found in HEK293 cells transfected with the reporter plasmid containing the mutant HMGA2 3’-UTR (Figure 8C). Moreover, HMGA2 mRNA and protein levels were dramatically decreased by ectopic miR-142-5p overexpression and increased when transfected with miR-142-5p inhibitor in RBE or HuCCT1 cells (Figure 8D, 8E). We also demonstrated that knockdown HMGA2 decreased the EMT-related N-cadherin, MMP2, and MMP9 expression while enhanced E-cadherin expression (Figure 8F). Together, these data validated that miR-142-5p directly regulated HMGA2 expression by targeting the 3’-UTR of HMGA2.
Figure 8.
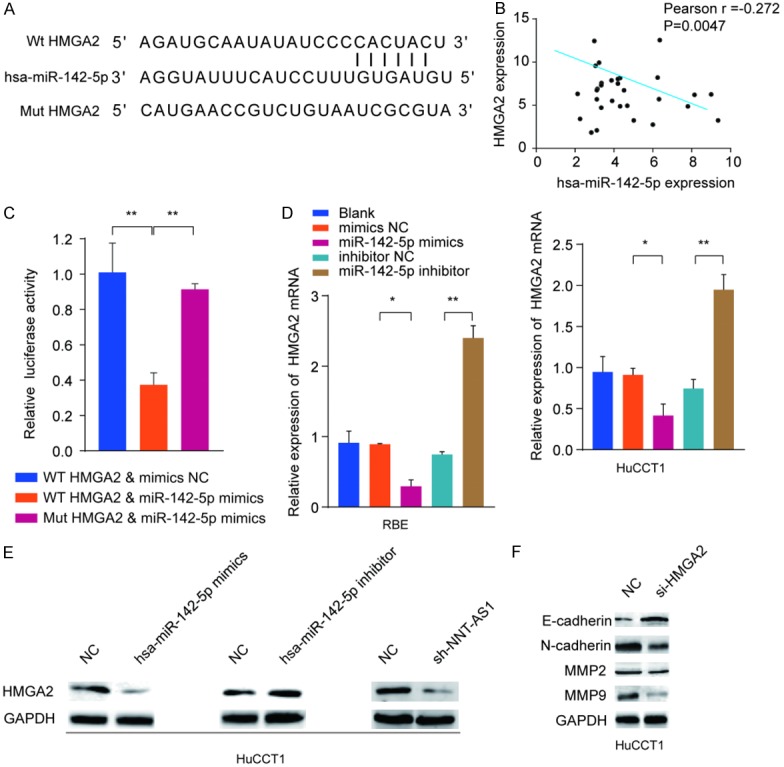
MiR-142-5p negatively regulates High Mobility Group AT-Hook 2 (HMGA2) expression by targeting 3’-UTR of HMGA2. A. Graphical representation of the potential binding sites between miR-142-5p and HMGA2. B. Correlation between HMGA2 and miR-142-5p was analyzed by Pearson’s correlation curve. C. Luciferase reporter assay was performed and the relative luciferase activity was tested after co-transfection with HMGA2 wide type, HMGA2 mutant type and miR-142-5p mimic. D, E. RBE or HuCCT1 cells were transfected with miR-142-5p mimics, inhibitor or negative controls. The mRNA and protein expression levels of HMGA2 were analyzed by RT-qPCR and western blot. F. Western blot analysis of E-cadherin, N-cadherin, MMP2 and MMP9 in HuCCT1 cells treated with siRNA targeting HMGA2 or negative group. Mean ± SD, *P<0.05 and **P<0.01, unpaired Student’s t-test. Representative images and data are based on three independent experiments.
Discussion
Cholangiocarcinoma is a highly malignant biliary tumor with poor outcome. It has been well proven that lncRNAs can influence various tumorigenesis processes by multiple mechanisms [6]. In the past decades, promising evidence has shown that lncRNAs are important factors in cancer and dysregulation of lncRNAs contributed to potential pathophysiological in various human cancer types [20]. Thus, the identification of lncRNAs associated with CCA progression may provide in-depth understanding on the progression of CCA.
In the current study, we revealed that lncRNA NNT-AS1 was dramatically upregulated in CCA and high NNT-AS1 expression was associated with differentiation, depth of invasion, TNM stage and worse prognosis. Multivariate analyses indicated that NNT-AS1 could be a novel and effective biomarker for risk prognostication. Consistent with our study, several studies reported that upregulated NNT-AS1 could be severed as a dependent negative prognostic factor for cancer. For example, NNT-AS1 was found with enhanced expression in human osteosarcoma, cervical, colorectal, breast, liver and ovarian cancer. Additional, up-regulated NNT-AS1 was correlated with clinical malignancies and poor prognosis of these malignancies [7,10-13,21-25]. Together, our findings suggest that NNT-AS1 overexpression represents an excellent biomarker of poor prognosis in CCA.
Next, we explored the biological function of NNT-AS1 in CCA. We found that NNT-AS1 knockdown led to significant inhibition of CCA cell proliferation, migration and invasion, while NNT-AS1 overexpression promoted cell invasion and cell viability in vitro and in vivo. These findings indicate that NNT-AS1 plays a crucial role in the CCA progression, stimulating novel therapeutic directions considering NNT-AS1 as a potential therapeutic target in CCA. In agreement with our results, Wang et al. reported that NNT-AS1 depletion inhibited cell proliferation and colony formation in colorectal cancer [13]. Hua et al. indicated that NNT-AS1 enhanced cell proliferation and invasion through activating Wnt/β-catenin pathway in cervical cancer [10]. Li et al. showed that NNT-AS1 accelerated cell migration and invasion in CCA and functioned as a sponge to bind to miR-142-3p [23]. Intriguingly, we also demonstrated that NNT-AS1 functioned as a ceRNA to bind to miR-142-5p. MiR-142-5p expression was negatively correlated with NNT-AS1 expression in CCA. These data indicate that NNT-AS1 might act as an oncogene in the development of CCA, also could be considered as a potential therapeutic target. However, the underlying mechanism of NNT-AS1-regulated CCA tumorigenicity remains to be elucidated.
It is widely accepted that EMT is involved in tumor metastasis and the characteristics of EMT include the inhibited E-cadherin, increased N-cadherin and Vimentin expression as well as enhanced MMP members protein [26,27]. A growing body of evidence revealed that LncRNA played a crucial role in the incidence of metastasis through regulating EMT [28,29]. Zhao et al. observed that lncRNA TUG1 promoted Pancreatic carcinoma progression through enhancing EMT phenotype [30]. Yan et al. demonstrated that upregulated SNHG6 was related with invasion and promoted gastric cancer cell EMT [31]. In addition, overexpressed lncRNA PlncRNA-1 could promote metastasis through inducing EMT in hepatocellular carcinoma [32]. In accordance with above previous reports, we found significant changed EMT markers after NNT-AS1 silencing or overexpression. Furthermore, we demonstrated that miR-142-5p directly targeting HMGA2, which was reported to induce EMT in human hepatocellular carcinoma cells [33]. Consistently, the GSEA showed high NNT-AS1 expression was markedly correlated with gene signatures of EMT. These results indicate that NNT-AS1/miR-142-5p/HMGA2 might promote the potential of CCA metastasis partly by enhancing EMT.
Although our study did not provide direct evidence whether the changes in EMT were required for the regulation of CCA cell proliferation and metastasis by NNT-AS1, numerous previous studies have implicated EMT process in the regulation of cell proliferation and metastasis. In this context, it is reasonable for us to conclude that NNT-AS1 promotes CCA cell metastasis via NNT-AS1 mediated regulation of EMT. Further investigation for NNT-AS1 relative to CCA metastasis is needed.
Conclusion
In summary, these findings suggest the potential prognostic and therapeutic value of NNT-AS1/miR-142-5p/HMGA2 axis in CCA patients. Although there is still a long way to go before the advent of lncRNA-based tumor therapy, our present study will undoubtedly enhance our knowledge of how NNT-AS1/miR-142-5p/HMGA2 functions in CCA, allowing us to better understand the pathogenesis and development of CCA.
Acknowledgements
This study was sponsored by grants from National S&T Major Project of China (2018ZX10301201), National Natural Science Foundation of China (81702757, 81500127), Youth innovation fund of First Affiliated Hospital of Zhengzhou University. The Medicine Science and Technology research project of Henan province (201702256). The funding sources had neither role in the design of this study nor any role during its execution, analyses, data interpretation, or decision to submit results.
The study was approved by the Institutional Review Board of the First Affiliated Hospital of Zhengzhou University and the First Affiliated Hospital. All participants provided a written informed consent upon enrolment.
Disclosure of conflict of interest
None.
Abbreviations
- CCA
Cholangiocarcinoma
- TCGA
The Cancer Genome Atlas Project
- EMT
epithelial-mesenchymal transition
- NNT-AS1
Nicotinamide Nucleotide Transhydrogenase-antisense RNA1
- lncRNAs
Long non-coding RNAs
- GSEA
gene set enrichment analysis
- EdU
5-ethynyl-20-deoxyuridine
- ISH
In situ hybridization
- IHC
immunohistochemistry
- OS
overall survival
- RFS
relapse-free survival
Supporting Information
References
- 1.Shin HR, Oh JK, Masuyer E, Curado MP, Bouvard V, Fang YY, Wiangnon S, Sripa B, Hong ST. Epidemiology of cholangiocarcinoma: an update focusing on risk factors. Cancer Sci. 2010;101:579–585. doi: 10.1111/j.1349-7006.2009.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberts SR, Gores GJ, Kim GP, Roberts LR, Kendrick ML, Rosen CB, Chari ST, Martenson JA. Treatment options for hepatobiliary and pancreatic cancer. Mayo Clin Proc. 2007;82:628–637. doi: 10.4065/82.5.628. [DOI] [PubMed] [Google Scholar]
- 4.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi AM, Tanzer A, Lagarde J, Lin W, Schlesinger F. Landscape of transcription in human cells. Nature. 2012;489:101. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–82. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nature Medicine. 2015;21:1253. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7.Shi X, Zhang H, Wang M, Xu X, Zhao Y, He R, Zhang M, Zhou M, Li X, Peng F, Shi C, Shen M, Wang X, Guo X, Qin R. LncRNA AFAP1-AS1 promotes growth and metastasis of cholangiocarcinoma cells. Oncotarget. 2017;8:58394–58404. doi: 10.18632/oncotarget.16880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang XM, Li ZL, Li JL, Zheng WY, Li XH, Cui YF, Sun DJ. LncRNA CCAT1 as the unfavorable prognostic biomarker for cholangiocarcinoma. Eur Rev Med Pharmacol Sci. 2017;21:1242–1247. [PubMed] [Google Scholar]
- 9.Wang WT, Ye H, Wei PP, Han BW, He B, Chen ZH, Chen YQ. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J Hematol Oncol. 2016;9:117. doi: 10.1186/s13045-016-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua F, Liu S, Zhu L, Ma N, Jiang S, Yang J. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/β-catenin signaling pathway in cervical cancer. Biomed Pharmacother. 2017;92:1128–1134. doi: 10.1016/j.biopha.2017.03.057. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Yang L, Hu X, Jiang Y, Hu Y, Liu Z, Liu J, Wen T, Ma Y, An G, Feng G. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8:3441–3453. doi: 10.18632/oncotarget.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye H, Lin J, Yao X, Li Y, Lin X, Lu H. Overexpression of long non-coding RNA NNT-AS1 correlates with tumor progression and poor prognosis in osteosarcoma. Cell Physiol Biochem. 2018;45:1904–1914. doi: 10.1159/000487966. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Ren M, Li Y, Hu J, Lu G, Ma W, Guo D, Lu X, He S. Long noncoding RNA NNT-AS1 promotes gastric cancer proliferation and invasion by regulating microRNA-363 expression. J Cell Biochem. 2019;120:5704–5712. doi: 10.1002/jcb.27855. [DOI] [PubMed] [Google Scholar]
- 14.Bao J, Yu Y, Chen J, He Y, Chen X, Ren Z, Xue C, Liu L, Hu Q, Li J, Cui G, Sun R. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018;9:1045. doi: 10.1038/s41419-018-1020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, He Y, Yu Y, Chen X, Cui G, Wang W, Zhang X, Luo Y, Li J, Ren F, Ren Z, Sun R. Upregulation of miR-374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med. 2018 doi: 10.1002/cam4.1576. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Y, Chen X, Yu Y, Li J, Hu Q, Xue C, Chen J, Shen S, Luo Y, Ren F, Li C, Bao J, Yan J, Qian G, Ren Z, Sun R, Cui G. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Mol Carcinog. 2018;57:772–783. doi: 10.1002/mc.22799. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Yu Y, Chen X, He Y, Hu Q, Li H, Han Q, Ren F, Li J, Li C, Bao J, Ren Z, Duan Z, Cui G, Sun R. MiR-139-5p is associated with poor prognosis and regulates glycolysis by repressing PKM2 in gallbladder carcinoma. Cell Prolif. 2018;51:e12510. doi: 10.1111/cpr.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Xue C, Yu Y, Chen J, Chen X, Ren F, Ren Z, Cui G, Sun R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag Res. 2018;10:3857–3865. doi: 10.2147/CMAR.S175681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su X, Xing J, Wang Z, Chen L, Cui M, Jiang B. microRNAs and ceRNAs: RNA networks in pathogenesis of cancer. Chin J Cancer Res. 2013;25:235–239. doi: 10.3978/j.issn.1000-9604.2013.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao MX, Jiang YP, Tang YL, Liang XH. The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity. Oncotarget. 2017;8:12472–12483. doi: 10.18632/oncotarget.13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Shi J, Xu Y. Long non-coding RNA NNT-AS1 contributes to cell proliferation, metastasis and apoptosis in human ovarian cancer. Oncol Lett. 2018;15:9264–9270. doi: 10.3892/ol.2018.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Zhang S, Qiu T, Wang Y, Ricketts DM, Qi C. Upregulation of long non-coding RNA NNT-AS1 promotes osteosarcoma progression by inhibiting the tumor suppressive miR-320a. Cancer Biol Ther. 2019;20:413–422. doi: 10.1080/15384047.2018.1538612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Lv M, Song Z, Lou Z, Wang R, Zhuang M. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed Pharmacother. 2018;103:939–946. doi: 10.1016/j.biopha.2018.04.087. [DOI] [PubMed] [Google Scholar]
- 24.Lu YB, Jiang Q, Yang MY, Zhou JX, Zhang Q. Long noncoding RNA NNT-AS1 promotes hepatocellular carcinoma progression and metastasis through miR-363/CDK6 axis. Oncotarget. 2017;8:88804–88814. doi: 10.18632/oncotarget.21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Q, Jiang Y. LncRNA NNT-AS1 promotes the proliferation, and invasion of lung cancer cells via regulating miR-129-5p expression. Biomed Pharmacother. 2018;105:176–181. doi: 10.1016/j.biopha.2018.05.123. [DOI] [PubMed] [Google Scholar]
- 26.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 28.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139:269–280. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 29.Liao JY, Wu J, Wang YJ, He JH, Deng WX, Hu K, Zhang YC, Zhang Y, Yan H, Wang DL, Liu Q, Zeng MS, Phillip Koeffler H, Song E, Yin D. Deep sequencing reveals a global reprogramming of lncRNA transcriptome during EMT. Biochim Biophys Acta Mol Cell Res. 2017;1864:1703–1713. doi: 10.1016/j.bbamcr.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Zhao L, Sun H, Kong H, Chen Z, Chen B, Zhou M. The lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMT phenotype formation through sponging Mir-382. Cell Physiol Biochem. 2017;42:2145–2158. doi: 10.1159/000479990. [DOI] [PubMed] [Google Scholar]
- 31.Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999–1012. doi: 10.1159/000478682. [DOI] [PubMed] [Google Scholar]
- 32.Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of long non-coding RNA PlncRNA-1 promotes metastasis and induces epithelial-mesenchymal transition in hepatocellular carcinoma. Cell Physiol Biochem. 2016;38:836–846. doi: 10.1159/000443038. [DOI] [PubMed] [Google Scholar]
- 33.Luo Y, Li W, Liao H. HMGA2 induces epithelial-to-mesenchymal transition in human hepatocellular carcinoma cells. Oncol Lett. 2013;5:1353–1356. doi: 10.3892/ol.2013.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.