Abstract
The etiology and pathogenesis of oral lichen planus have not achieved a consensus yet. This study aimed to explore the possible roles of exosomal miRNAs in the pathogenesis of oral lichen planus. Bioactive components from exosomes regulate intercellular communications that may be closely related to the occurrence and development of diseases, including oral lichen planus. Further, exosomes are expected to be a biomarker for the diagnosis and treatment of oral lichen planus. In this study, new advanced views about the biological characteristics, clinical significance, and involvement of exosomes in oral lichen planus were reviewed.
Keywords: Exosomes, etiology, microRNAs, oral lichen planus, pathogenesis
Introduction
The exosomes are cellular waste products in the metabolism of reticulocytes [1]. They were first discovered and named by Rose Johnstone from Canada in the 1980s [2,3]. In later decades, increasing evidence [4-7] showed the involvement of exosomes in cell proliferation, migration, inflammatory response, and tumor-associated immunomodulation in various diseases such as tumors [8,9], cardiovascular diseases [10], neurological diseases [11], and oral diseases [12]. Oral lichen planus (OLP) is a chronic immune-mediated inflammatory disease characterized by liquefaction degeneration in the basal keratinocytes and a band-shaped subepithelial infiltration of lymphocytes; it may also be accompanied by skin lesions in some cases [13,14]. The World Health Organization has classified OLP as “oral potentially malignant disorders (OPMDs)” [15] because of its malignant tendency, with undiscovered pathogenesis and no unified therapy. In recent years, exosomes have been found to be closely related to the occurrence and development of OLP [16,17].
Composition and biological properties of exosomes
Exosomes are nanosized vesicles between 40 and 100 nm in diameter [18,19]. They are generated from inwardly budding vesicles in multivesicular bodies (MVBs) released into the extracellular space after the fusion of MVBs with the plasma membrane [20]. Most cells of the human body, including epithelial cells [21], lymphocytes [22], mast cells [23], and dendritic cells [24], can secrete exosomes, which mediate cell-to-cell communications through blood plasma, breast milk, saliva, malignant ascites, amniotic fluid, urine, and many other body fluids during different biological processes [25,26].
Being rich in proteins, lipids, and nucleic acids [20,27], the conserved components of exosomal proteins are mostly located inside or on the surface of exosomes, contributing to the maintenance of the molecular structure and inner homeostasis of vesicles [4,28]. The cell type-specific components of exosomal proteins are related to the selected processes in target cells and the modulation of gene expression in recipient cells [29,30]. Moreover, the lipid bilayer membrane structure of exosomes contains a variety of lipid components similar to the plasma membrane [27,31], which are proved to be involved in the intercellular communication, thus supporting and stabilizing the structure and inner environment of exosomes [32]. Exosomes derived from different types of cells display different lipid compositions [33,34], which might help exosomes to target the recipient cells and then adapt to the intracellular space.
Nucleic acids, especially the microRNAs (miRNAs) [35-37] in exosomes, can be transferred to the target cells, affecting or even regulating the gene expression of recipient and target cells [38-40]. They protect circulating miRNAs from being degraded by RNA enzymes [41,42]. However, Chevillet et al. [37] proposed that most of the miRNAs in body fluids might exist independently of the exosomes, which meant that exosomes could not serve as vehicles for miRNAs to interfere in intercellular information transfer. Besides, previous studies [27,43] showed that the RNA components of exosomes also had cell specificity, which was similar to that of the protein components of exosomes. However, a few conclusions were reached regarding whether the RNA of exosomes was specifically sorted or randomly packaged [43]. Hence, further studies were needed to uncover the biogenesis of exosomes.
Clinical significance of exosomes
As the biological characteristics of exosomes and their roles in various diseases were deeply explored, their potential value in clinical diagnosis and therapy was gradually unveiled, implying that exosomes might serve as diagnostic and therapeutic tools [44-46] for many kinds of refractory diseases.
Role as diagnostic markers
Though derived from the same type of cells, the compositions and biological characteristics of exosomes are different under different physiological or pathological conditions [7,33]. The compositions of exosomes in patients with tumors [47-49], acute liver injury [50], or viral infection [34,51] reflect the abnormal states of their mother cells to some extent, suggesting that exosomes might become a biomarker for disease detection. Kim et al. [52] showed that the expression level of exosomal four-transmembrane proteins collected from the saliva of patients with oral cancer was different from that in healthy individuals, revealing that the four-transmembrane protein might be a potential biomarker of oral cancer detection. Furthermore, Mitchell et al. [53] found that miRNA-141 (a kind of specific miRNA expressed in prostate cancer) selectively aggregated in exosomes from mice with prostate cancer. They proposed that the serum level of miRNA-141 might assist in the examination and diagnosis of prostate cancer, implying that exosomes could be an important marker for the blood test of tumors. Recent studies further found a significant difference between exosomal miRNA profiles of patients with ovarian cancer and healthy individuals [54], indicating that the analysis of tumor-derived exosomes via blood test would provide the diagnostic evidence for cancer patients and even be beneficial to the disease therapy.
Regulation of tumor-related inflammation
Several studies [9,55-57] indicated that tumor-derived exosomes had anti-tumor effects by inducing the apoptosis of tumor cells or enhancing anti-tumor immunity. Kim et al. [52] discovered that the Fas ligand (FasL) binding mode was found to help tumor cells escape from host immune surveillance by inducing the apoptosis of CD8+ T cells, and the expression level of FasL from exosomes was affected by tumor growth and degree of lymphatic metastasis. Correspondingly, more studies explored the immunosuppressive functions of tumor-derived exosomes, which could stimulate the proliferation and migration of tumor cells to promote tumor growth through inhibiting the differentiation of dendritic cells or reducing the cytotoxicity of T cells and natural killer cells [58-60]. Comprehensively as Altevogt et al. reported [56], tumor-derived exosomes might be bifunctional to regulate alterations of immune cell functions by stimulating the production of inflammatory mediators or being directly delivered to target cells. The bidirectional effects of tumor exosomes on immune cells (whether stimulation or inhibition) were supposed to depend on the length of the exposure to inflammatory factors, namely between the exosomes and the immune cells.
Therefore, the exosomal regulation on tumor-associated immune responses could be applied to novel clinical anti-tumor therapy, which has attracted the attention of numerous researchers [17,61,62]. Notably, Viaud et al. [63] reported that the dendritic cell-derived exosomes loaded with tumor antigens could trigger CD4+ and CD8+ T-cell-mediated immune responses. This was because they conveyed exosomal major histocompatibility complexes to the antigen-presenting cells, promoting the anti-tumor immune responses and inhibiting tumor growth in mice. The dendritic cell-derived exosomes are now being widely studied as novel cell-free vaccines for cancer immunotherapy [64].
Role as drug delivery vehicles
Exosomes are expected to replace liposomes as an ideal drug carrier in the future, not only because of their good tolerance to the human body but also for their excellent homing ability toward targeted cells, which can be regulated by artificial membrane modification [46,65]. Meanwhile, exosomes can also protect drug proteins and nucleic acids from degradation in the circulation, help confirm the direction to specific targeted cells, and reduce adverse reactions caused by targeting deviation [57,66]. For instance, Alvarez-Erviti et al. [67] found that exosomal siRNA could cross the blood-brain barrier and knock out more than 60% of siRNA-targeting genes in neurons, microglia, oligodendrocytes, and their precursors, which can be applied to the treatment of nervous system diseases.
Exosomes and T-cell-specific immune response in OLP
The pathogenesis of OLP is related to the T-cell-specific immune response [14,68]. Corresponding to the pathological manifestations of OLP [13], the pathogenesis of OLP mainly comprises these processes: activation of T cells by antigen-presenting cells; T-cell proliferation, apoptosis, migration, and differentiation; and T-mediated apoptosis of keratinocytes. The depression of cell-mediated immunity, for example after taking cyclosporine, alleviates lymphocyte infiltration in lesions and clinical symptoms of OLP [69], with a limitation of an uncertainly recognized pathogen as a possible antigen. The roles of exosomes in immune responses have also attracted extensive attention in the field of oral diseases and hence researchers hope to uncover the exact pathogenesis and treatment of OLP.
Role as diagnostic markers for OLP
Due to easy operation, low cost, and noninvasiveness, saliva has become an ideal sample for diagnosing and treating various oral diseases [70,71]. However, the risk of contamination is the fatal weakness of saliva. Too many interfering impurities in saliva evidently lower the specificity and sensitivity of the test. This is well settled by the salivary exosome test for the diagnosis and antidiastole of oral diseases. Palanisamy et al. [72] clearly observed the intercellular signaling between fluorescence-labeled salivary exosomes and keratinocytes in vitro, indicating that salivary exosomes could be a potential biomarker for disease detection and diagnosis. OLP has been classified as an “OPMD” for its malignant tendency. Patients with OLP should better accept the lifelong follow-up to track the disease progression regularly [15,73]. In addition to higher acceptance of patients, the salivary exosome test is also more suitable for OLP compared with traditional invasive tests such as biopsy, which exhibits the advantage of salivary exosomes.
Several studies [70,74,75] demonstrated that the expression of specific miRNAs of the exosomes in the peripheral blood and saliva of patients with OLP was different from that in healthy individuals. Byun et al. [76] compared the exosomal miRNA profiles from the saliva of 16 patients with OLP and 8 healthy controls using gene-chip analysis and TaqMan quantitative polymerase chain reaction (PCR). They discovered that the miR-4484 was significantly upregulated, and might serve as a potential diagnostic marker and therapeutic target for OLP. The gene-expression profiles researched by Peng et al. [77] showed higher expression levels of miR-34a-5p and miR-130b-3p in circulating exosomes of patients with OLP than those of controls, while the levels of exosomal miR-301b-3p were lower. In addition, the positive association between the expression levels of miR-34a-5p and the severity of clinical symptoms of OLP was also discovered [77]. The higher the expression levels of circulating exosomal miR-34a-5p in patients with OLP, the more severe the clinical symptoms. This finding suggested that circulating exosomal miR-34a-5p might be a promising biomarker for assessing the clinical severity of OLP.
Moreover, exosomal miRNAs, as biomarkers, can not only be applied to the early screening and progress monitoring of OLP but also assist in the study of OLP pathogenesis. Studies of OLP-specific exosomal miRNAs [76,78] indicate that these nucleotide sequences might be involved in the development of OLP. Hopefully, a study on the function of OLP-specific exosomal miRNAs might promote the investigation of OLP pathogenesis. Besides the positive correlation between OLP severity and the expression levels of miR-34a-5p demonstrated by Peng et al., the target genes of miR-34a-5p are possibly associated with the modulation of cellular communication, signal transduction, metabolic processes, and gene expression, and hence the regulation of OLP progression, via phosphatidylinositol 3 kinase (PI3K)/protein kinase B (AKT) signaling pathways [77] (Figure 1).
Figure 1.
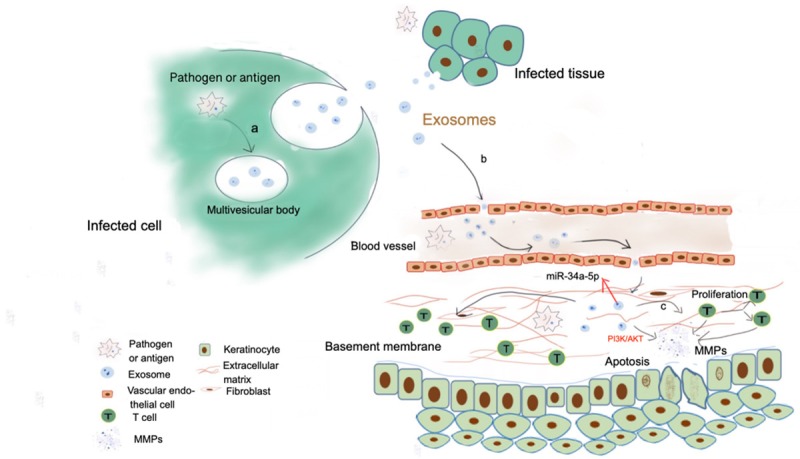
Possible relations among pathogen/antigen, exosomes and OLP via circulation: a. Pathogen miRNAs in infected cells are integrated and uptaked by multivesicular bodies, and finally released into the extracellular space as cargoes in exosomes; b. Exosomes with pathogen miRNAs target and march into oral mucosa lamina propria through circulation; c. Exosomes carrying infected messages could promote proliferation of CD4+ and CD8+ T cells, as well as up-regulate activation of matrix metalloproteinases (MMPs) to stimulate destruction of the basement membrane and liquefaction degeneration of basal cells.
Exosomes and immune disorders in OLP
The main pathological features of OLP include band-shaped lymphocytes of subepithelial infiltration and liquefaction degeneration of the basal keratinocytes [13]. Studies proved that the pathogenesis of OLP involves immune system disorders, according to which OLP is speculated to be a T-cell-mediated autoimmune disease [14]. Interestingly, besides the discovery of specific expression of some immune-related miRNAs in OLP-derived exosomes, the following in vitro experiments [16] also demonstrated that OLP-derived exosomes had certain regulatory effects on T cells and related cytokines, further confirming the role of T-cell-specific immune response in the pathogenesis of OLP in the perspective of exosomes. Peng et al. [16] investigated the biological characteristics of T cells co-cultured with plasma-derived exosomes from patients with OLP using confocal laser scanning microscopy. They found that OLP-derived exosomes could significantly enhance T-cell proliferation and inhibit T-cell apoptosis. Hence, it was speculated that exosomes might affect the progress of OLP by regulating the T-cell-mediated inflammatory response. Moreover, exosomes from different subtypes and severities of OLP might affect T-cell vital movements to varying degrees. Compared with reticular OLP, as Brant et al. [79] reported, less apoptosis but more lymphocytes were found in the inflammatory infiltrate of erosive OLP, indicating that the exosomes of erosive OLP might have a greater impact on T cells. The exosomes of erosive OLP can possibly enhance the T-cell-mediated immune response, leading to the aggravation of the clinical symptoms of OLP, which corresponds with relatively longer duration and higher recurrence rate of erosive OLP [80]. Besides, combining the different performances of T-cell subsets between erosive and nonerosive OLP with the hypothesis that circulating exosomes might help T cells differentiate into different subsets [81], Wang et al. [82] speculated that exosomes could also regulate the different directions of the differentiation of T cells through different immune mechanisms resulting in the clinical symptoms of OLP (Figure 2).
Figure 2.
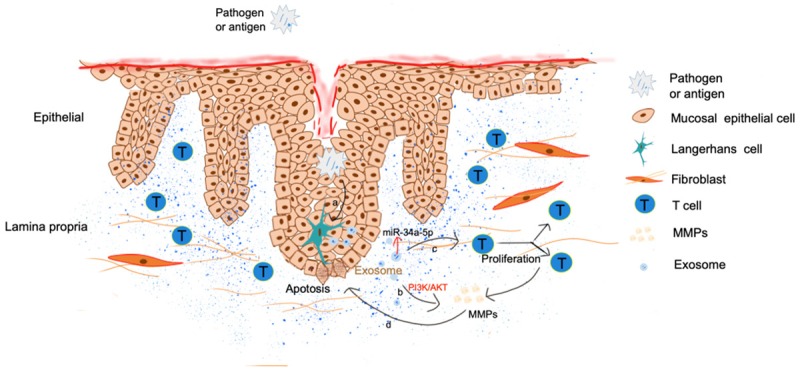
Possible relations among pathogen/antigen, exosomes and OLP via keratinocytes: a. Pathogens invading into mucosal epithelium stimulate Langerhans cells/dendritic cells to secrete exosomes; b. Exosomes carrying pathogenic RNA enter the lamina propria and secrete MMPs through PI3K/AKT signaling pathways; c. Exosomes carrying pathogenic RNA enter the lamina propria, and stimulate the proliferation of CD4+/CD8+ T cells and the secretion of MMPs; d. MMPs suppress the proliferation of epithelial cells and promote the apoptosis of keratinocytes.
Methodological sense of viruses and exosomes for OLP research
Another study showed that exosomes carried viral RNAs to contaminate noninfected cells [51], indicating the same mechanisms of viral infection in OLP pathogenesis [68,83]. Recent studies [84,85] demonstrated that the intracellular infection in keratinocytes might give rise to a vicious circle: T-cell-specific immune response induced by infection, followed by liquefaction degeneration, epithelial barrier dysfunction, aggravation of viral infection, and finally stimulation of local immune response. The long-term persistent infection induced by this vicious circle might be the cause for chronic and long-term features of OLP [83]. Furthermore, exosomes can deliver infectious proteins and viral RNAs among cells to contaminate noninfected cells [34,51], which might be a powerful force for the viral transmission and immune escape in infection mechanisms.
Hepatitis C virus-exosomes-OLP
Lichen planus is acknowledged as one of the extrahepatic manifestations of hepatitis C virus (HCV) infection [86]. Hence, HCV is also considered to be closely involved in OLP. An earlier epidemiological survey [87] showed that the positive detection rate of HCV in Japanese patients with OLP was much higher than that in healthy people, but other studies [88,89] at home and abroad revealed no significant correlation between HCV and OLP. Still, the meta-analyses [90,91] of the association between HCV and OLP consistently reported a significantly higher detection rate of HCV in the serum of patients with OLP than that in the control group, implying that a certain connection between HCV and OLP might be influenced by geographical differences. Meanwhile, Cosset et al. [92] detected infectious viral RNAs in exosomes collected from hepatocytes and serum of patients with HCV, suggesting that exosomes could carry HCV-related cargo and fuse with noninfected cells to help in viral transmission. In addition, similar to OLP lesions, the lymphocyte infiltration is also a typical feature in liver lesions of patients with HCV [93], indicating a similar immune background between these two diseases.
Human cytomegalovirus-exosomes-OLP
Human cytomegalovirus (HCMV) is a ubiquitous opportunistic pathogen with no obvious clinical effects on healthy people [94] and an infection rate as high as 70%-100% in the global population. Ding et al. [95] analyzed the plasma of patients with OLP through RT-qPCR. They found five significantly upregulated HCMV-encoded miRNAs (hcmv-miR-UL112-3p, hcmv-miR-UL22a-5p, hcmv-miR-UL148d, hcmv-miR-UL36-5p, and hcmv-miR-UL59). These miRNAs were found to indirectly or directly regulate the expression of immune-related active molecules [96-99], suggesting that specific HCMV-miRNAs might participate in OLP-specific immune response and play a certain role in the development of OLP. In addition, Ding et al. [95] also found that HCMV-DNA in peripheral blood leukocytes of patients with OLP was significantly higher than that in the control group. It was speculated that the higher the HCMV content in the body, the more likely the occurrence of OLP, further confirming that HCMV might be involved in the pathogenesis of OLP.
Ding et al. [95] also detected that most viral miRNAs were encapsulated in exosomes. They proposed that exosomes secreted by infectious cells and delivered into plasma might be the source of HCMV-encoded miRNAs in patients with OLP. Persistent intracellular viral replication may also allow the infectious cells to secrete exosomes loaded with specific viral miRNAs into body circulation [100,101]. As these viral-encoded miRNAs are nonimmunogenic molecules [85], the exosomes can protect the inner viral information from the attack of the immune system. The mechanism underlying the interaction between HCMV-encoded miRNAs and OLP is still unclear. However, HCMV-encoded miRNAs may have significance in patients with OLP [95,102], providing a hint for the study of the pathogenic role of HCMV in OLP, with an extraordinary significance for the etiology, pathogenesis, diagnosis, and treatment of OLP.
Other viruses-exosomes-OLP
Except for the aforementioned two viruses, several other viruses are reported to be connected with OLP [103,104], such as Epstein-Barr virus, human papillomavirus, and herpes virus. It is reasonable to speculate that these OLP-related viruses can use exosomes as vehicles to take part in the pathogenesis of OLP.
All these hypotheses about viruses-exosomes-OLP need further verification.
Conclusion
As vesicles mediating intercellular communication, exosomes are found to be involved in cell proliferation, migration, and various inflammatory responses. The lipid content of differently derived exosomes is different. The difference in lipid composition may help exosomes better adapt to the environment where the target cells are located. The different composition and biological characteristics of exosomes can possibly reflect the abnormal state of the mother cells under different physiological and pathological conditions. Whether the exosomal RNA is specifically sorted or randomly packaged in exosomes needs further exploration to uncover the biogenesis of exosomes. Researchers have found that many diseases, including OLP, are closely associated with exosomes. Exosomal microRNA-34a-5p in body fluids was found to be positively correlated with the severity of the clinical symptoms of OLP. The higher the level of exosomal microRNA-34a-5p, the more severe the clinical symptoms. Circulating exosomal microRNA-34a-5p may become a potential biomarker for diagnosing and evaluating the severity of OLP, whose target genes are mostly linked to the regulation of gene expression, signal transduction, and cellular metabolism. Further, PI3K/Akt signaling pathways might be involved in these progressions.
Exosomes may participate in OLP mainly by inducing cellular immunity and evading immune surveillance. Hence, the study of the relationships among exosomes, OLP-related immune responses, and microbial infections might provide a new perspective and direction for the study of the etiology of OLP and the mechanisms of the progression of the disease. Moreover, exosomes could be applied as the biomarkers for the evaluation and diagnosis of OLP and used as the drug delivery vehicles for the treatment of OLP, indicating great prospects for the clinical application of exosomes. However, the application of exosomes in the diagnosis and treatment of OLP still needs further investigation to design a correct therapeutic strategy for the disease.
Acknowledgements
This study was supported by the Co-construction Program WKJ-ZJ-1623 from the Provincial Bureau and National Commission of Hygiene & Health in China, the Nonprofit Specific Fund 201502018 from the National Hygiene and Health Commission of China, and the Education Reform Program (yxyb20172030) from the School of Medicine of Zhejiang University in China.
Disclosure of conflict of interest
None.
Abbreviations
- HCV
Hepatitis C virus
- HCMV
human cytomegalovirus
- miRNAs
microRNAs
- OLP
oral lichen planus
- OPMD
oral potentially malignant disorders
- PI3K/AKT
phosphatidylinositol 3 kinase/protein kinase B
- MMPs
metalloproteinases
References
- 1.Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 2.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 3.Johnstone RM. Revisiting the road to the discovery of exosomes. Blood Cells Mol Dis. 2005;34:214–219. doi: 10.1016/j.bcmd.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. Int J Biochem Cell Biol. 2012;44:11–15. doi: 10.1016/j.biocel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Kowal J, Tkach M, Thery C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Wiklander OP, Nordin JZ, O’Loughlin A, Gustafsson Y, Corso G, Mager I, Vader P, Lee Y, Sork H, Seow Y, Heldring N, Alvarez-Erviti L, Smith CI, Le Blanc K, Macchiarini P, Jungebluth P, Wood MJ, Andaloussi SE. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316. doi: 10.3402/jev.v4.26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Toro J, Herschlik L, Waldner C, Mongini C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol. 2015;6:203. doi: 10.3389/fimmu.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S, Cao H, Shen B, Feng J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget. 2015;6:37151–37168. doi: 10.18632/oncotarget.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo D, Chen Y, Wang S, Yu L, Shen Y, Zhong H, Yang Y. Exosomes from heat-stressed tumour cells inhibit tumour growth by converting regulatory T cells to Th17 cells via IL-6. Immunology. 2018;154:132–143. doi: 10.1111/imm.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loyer X, Vion AC, Tedgui A, Boulanger CM. Microvesicles as cell-cell messengers in cardiovascular diseases. Circ Res. 2014;114:345–353. doi: 10.1161/CIRCRESAHA.113.300858. [DOI] [PubMed] [Google Scholar]
- 11.Yuan M, Liu N, Wang X, Tian C, Ren X, Zhang H, Yang X, Li X, Zhu H, Zhu L, Shang H, Xing Y, Gao Y. The mechanism of exosomes function in neurological diseases: a progressive review. Curr Pharm Des. 2018;24:2855–2861. doi: 10.2174/1381612824666180903113136. [DOI] [PubMed] [Google Scholar]
- 12.Huang CC, Narayanan R, Alapati S, Ravindran S. Exosomes as biomimetic tools for stem cell differentiation: applications in dental pulp tissue regeneration. Biomaterials. 2016;111:103–115. doi: 10.1016/j.biomaterials.2016.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta S, Jawanda MK. Oral lichen planus: an update on etiology, pathogenesis, clinical presentation, diagnosis and management. Indian J Dermatol. 2015;60:222–229. doi: 10.4103/0019-5154.156315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurago ZB. Etiology and pathogenesis of oral lichen planus: an overview. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122:72–80. doi: 10.1016/j.oooo.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 15.van der Waal I. Potentially malignant disorders of the oral and oropharyngeal mucosa; present concepts of management. Oral Oncol. 2010;46:423–425. doi: 10.1016/j.oraloncology.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Peng Q, Zhang J, Zhou G. Circulating exosomes regulate T-cell-mediated inflammatory response in oral lichen planus. J Oral Pathol Med. 2019;48:143–150. doi: 10.1111/jop.12804. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Han Y, Zhao Z, Ji X, Wang X, Jin J, Wang Q, Guo X, Cheng Z, Lu M, Wang G, Wang Y, Liu H. Oral mucosal mesenchymal stem cellderived exosomes: a potential therapeutic target in oral premalignant lesions. Int J Oncol. 2019;54:1567–1578. doi: 10.3892/ijo.2019.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalra H, Drummen GP, Mathivanan S. Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci. 2016;17:170. doi: 10.3390/ijms17020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan JL, Lau SN, Leaw B, Nguyen HPT, Salamonsen LA, Saad MI, Chan ST, Zhu D, Krause M, Kim C, Sievert W, Wallace EM, Lim R. Amnion epithelial cell-derived exosomes restrict lung injury and enhance endogenous lung repair. Stem Cells Transl Med. 2018;7:180–196. doi: 10.1002/sctm.17-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown M, Johnson LA, Leone DA, Majek P, Vaahtomeri K, Senfter D, Bukosza N, Schachner H, Asfour G, Langer B, Hauschild R, Parapatics K, Hong YK, Bennett KL, Kain R, Detmar M, Sixt M, Jackson DG, Kerjaschki D. Lymphatic exosomes promote dendritic cell migration along guidance cues. J Cell Biol. 2018;217:2205–2221. doi: 10.1083/jcb.201612051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Inca F, Pucillo CE. Exosomes: tiny clues for mast cell communication. Front Immunol. 2015;6:73. doi: 10.3389/fimmu.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng L, Li Z, Ling W, Zhu D, Feng Z, Kong L. Exosomes derived from dendritic cells attenuate liver injury by modulating the balance of treg and Th17 cells after ischemia reperfusion. Cell Physiol Biochem. 2018;46:740–756. doi: 10.1159/000488733. [DOI] [PubMed] [Google Scholar]
- 25.Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36:301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soares Martins T, Catita J, Martins Rosa I, A B da Cruz E Silva O, Henriques AG. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS One. 2018;13:e0198820. doi: 10.1371/journal.pone.0198820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, Gangoda L, Mathivanan S. ExoCarta: a web-based compendium of exosomal cargo. J Mol Biol. 2016;428:688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014:3. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao H, Lasser C, Shelke GV, Wang J, Radinger M, Lunavat TR, Malmhall C, Lin LH, Li J, Li L, Lotvall J. Mast cell exosomes promote lung adenocarcinoma cell proliferation - role of KIT-stem cell factor signaling. Cell Commun Signal. 2014;12:64. doi: 10.1186/s12964-014-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glebov K, Lochner M, Jabs R, Lau T, Merkel O, Schloss P, Steinhauser C, Walter J. Serotonin stimulates secretion of exosomes from microglia cells. Glia. 2015;63:626–634. doi: 10.1002/glia.22772. [DOI] [PubMed] [Google Scholar]
- 31.Vallejo MC, Nakayasu ES, Longo LV, Ganiko L, Lopes FG, Matsuo AL, Almeida IC, Puccia R. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS One. 2012;7:e39463. doi: 10.1371/journal.pone.0039463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60:9–18. doi: 10.1194/jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841:108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Chapuy-Regaud S, Dubois M, Plisson-Chastang C, Bonnefois T, Lhomme S, Bertrand-Michel J, You B, Simoneau S, Gleizes PE, Flan B, Abravanel F, Izopet J. Characterization of the lipid envelope of exosome encapsulated HEV particles protected from the immune response. Biochimie. 2017;141:70–79. doi: 10.1016/j.biochi.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 35.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. miRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1568–1575. doi: 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M, De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 40.Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139:1443–1448. doi: 10.1002/ijc.30179. [DOI] [PubMed] [Google Scholar]
- 41.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014:3. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nedaeinia R, Manian M, Jazayeri MH, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia SH, Avan A, Ghayour-Mobarhan M. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017;24:48–56. doi: 10.1038/cgt.2016.77. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Salo T, Vered M. Morphological and molecular features of oral fluid-derived exosomes: oral cancer patients versus healthy individuals. J Cancer Res Clin Oncol. 2016;142:101–110. doi: 10.1007/s00432-015-2005-3. [DOI] [PubMed] [Google Scholar]
- 46.Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi: 10.1016/j.pharmthera.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 47.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 48.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wan Z, Gao X, Dong Y, Zhao Y, Chen X, Yang G, Liu L. Exosome-mediated cell-cell communication in tumor progression. Am J Cancer Res. 2018;8:1661–1673. [PMC free article] [PubMed] [Google Scholar]
- 50.Motawi TK, Mohamed MR, Shahin NN, Ali MAM, Azzam MA. Time-course expression profile and diagnostic potential of a miRNA panel in exosomes and total serum in acute liver injury. Int J Biochem Cell Biol. 2018;100:11–21. doi: 10.1016/j.biocel.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Alenquer M, Amorim MJ. Exosome biogenesis, regulation, and function in viral infection. Viruses. 2015;7:5066–5083. doi: 10.3390/v7092862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11:1010–1020. [PubMed] [Google Scholar]
- 53.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen J, Zhu X, Fei J, Shi P, Yu S, Zhou J. Advances of exosome in the development of ovarian cancer and its diagnostic and therapeutic prospect. Onco Targets Ther. 2018;11:2831–2841. doi: 10.2147/OTT.S159829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whiteside TL. Immune modulation of T-cell and NK (natural killer) cell activities by TEXs (tumour-derived exosomes) Biochem Soc Trans. 2013;41:245–251. doi: 10.1042/BST20120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altevogt P, Bretz NP, Ridinger J, Utikal J, Umansky V. Novel insights into exosome-induced, tumor-associated inflammation and immunomodulation. Semin Cancer Biol. 2014;28:51–57. doi: 10.1016/j.semcancer.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 57.Urbanelli L, Buratta S, Sagini K, Ferrara G, Lanni M, Emiliani C. Exosome-based strategies for diagnosis and therapy. Recent Pat CNS Drug Discov. 2015;10:10–27. doi: 10.2174/1574889810666150702124059. [DOI] [PubMed] [Google Scholar]
- 58.Saleem SN, Abdel-Mageed AB. Tumor-derived exosomes in oncogenic reprogramming and cancer progression. Cell Mol Life Sci. 2015;72:1–10. doi: 10.1007/s00018-014-1710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Y, Wang D, Jin F, Bian Z, Li L, Liang H, Li M, Shi L, Pan C, Zhu D, Chen X, Hu G, Liu Y, Zhang CY, Zen K. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat Commun. 2017;8:14041. doi: 10.1038/ncomms14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B, Tan Z, Guan F. Tumor-derived exosomes mediate the instability of cadherins and promote tumor progression. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20153652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goh WJ, Lee CK, Zou S, Woon EC, Czarny B, Pastorin G. Doxorubicin-loaded cell-derived nanovesicles: an alternative targeted approach for anti-tumor therapy. Int J Nanomedicine. 2017;12:2759–2767. doi: 10.2147/IJN.S131786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu L, Kalimuthu S, Gangadaran P, Oh JM, Lee HW, Baek SH, Jeong SY, Lee SW, Lee J, Ahn BC. Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics. 2017;7:2732–2745. doi: 10.7150/thno.18752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Viaud S, Ploix S, Lapierre V, Thery C, Commere PH, Tramalloni D, Gorrichon K, Virault-Rocroy P, Tursz T, Lantz O, Zitvogel L, Chaput N. Updated technology to produce highly immunogenic dendritic cell-derived exosomes of clinical grade: a critical role of interferon-gamma. J Immunother. 2011;34:65–75. doi: 10.1097/CJI.0b013e3181fe535b. [DOI] [PubMed] [Google Scholar]
- 64.Lu Z, Zuo B, Jing R, Gao X, Rao Q, Liu Z, Qi H, Guo H, Yin H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J Hepatol. 2017;67:739–748. doi: 10.1016/j.jhep.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 65.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31:543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 66.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 68.Mutafchieva MZ, Draganova-Filipova MN, Zagorchev PI, Tomov GT. Oral lichen planus - known and unknown: a review. Folia Med (Plovdiv) 2018;60:528–535. doi: 10.2478/folmed-2018-0017. [DOI] [PubMed] [Google Scholar]
- 69.Sun SL, Liu JJ, Zhong B, Wang JK, Jin X, Xu H, Yin FY, Liu TN, Chen QM, Zeng X. Topical calcineurin inhibitors in the treatment of oral lichen planus: a systematic review and meta-analysis. Br J Dermatol. 2019 doi: 10.1111/bjd.17898. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 70.Wu DT, Tao O, Trinh N, Javaid MA, Ahmed AS, Durand R, Tran SD. Saliva - a promising tool for diagnosing oral diseases. Current Oral Health Reports. 2018;5:242–249. [Google Scholar]
- 71.Zheng X, Chen F, Zhang J, Zhang Q, Lin J. Exosome analysis: a promising biomarker system with special attention to saliva. J Membr Biol. 2014;247:1129–1136. doi: 10.1007/s00232-014-9717-1. [DOI] [PubMed] [Google Scholar]
- 72.Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;5:e8577. doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alrashdan MS, Cirillo N, McCullough M. Oral lichen planus: a literature review and update. Arch Dermatol Res. 2016;308:539–551. doi: 10.1007/s00403-016-1667-2. [DOI] [PubMed] [Google Scholar]
- 74.Hung KF, Liu CJ, Chiu PC, Lin JS, Chang KW, Shih WY, Kao SY, Tu HF. MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol. 2016;53:42–47. doi: 10.1016/j.oraloncology.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 75.Wang J, Luo H, Xiao Y, Wang L. miR-125b inhibits keratinocyte proliferation and promotes keratinocyte apoptosis in oral lichen planus by targeting MMP-2 expression through PI3K/Akt/mTOR pathway. Biomed Pharmacother. 2016;80:373–380. doi: 10.1016/j.biopha.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 76.Byun JS, Hong SH, Choi JK, Jung JK, Lee HJ. Diagnostic profiling of salivary exosomal microRNAs in oral lichen planus patients. Oral Dis. 2015;21:987–993. doi: 10.1111/odi.12374. [DOI] [PubMed] [Google Scholar]
- 77.Peng Q, Zhang J, Zhou G. Differentially circulating exosomal microRNAs expression profiling in oral lichen planus. Am J Transl Res. 2018;10:2848–2858. [PMC free article] [PubMed] [Google Scholar]
- 78.El-Sakka H, Kujan O, Farah CS. Assessing miRNAs profile expression as a risk stratification biomarker in oral potentially malignant disorders: a systematic review. Oral Oncol. 2018;77:57–82. doi: 10.1016/j.oraloncology.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 79.Brant JM, Aguiar MC, Grandinetti HA, Rodrigues LV, Vasconcelos AC. A comparative study of apoptosis in reticular and erosive oral lichen planus. Braz Dent J. 2012;23:564–569. doi: 10.1590/s0103-64402012000500016. [DOI] [PubMed] [Google Scholar]
- 80.Chiang CP, Yu-Fong Chang J, Wang YP, Wu YH, Lu SY, Sun A. Oral lichen planus - differential diagnoses, serum autoantibodies, hematinic deficiencies, and management. J Formos Med Assoc. 2018;117:756–765. doi: 10.1016/j.jfma.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, Zhou J, Fu S, Wang C, Zhou B. A study of association between oral lichen planus and immune balance of Th1/Th2 cells. Inflammation. 2015;38:1874–1879. doi: 10.1007/s10753-015-0167-4. [DOI] [PubMed] [Google Scholar]
- 82.Wang H, Bai J, Luo Z, Fu J, Wang H, Sun Z. Overexpression and varied clinical significance of Th9 versus Th17 cells in distinct subtypes of oral lichen planus. Arch Oral Biol. 2017;80:110–116. doi: 10.1016/j.archoralbio.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 83.Baek K, Choi Y. The microbiology of oral lichen planus: is microbial infection the cause of oral lichen planus? Mol Oral Microbiol. 2018;33:22–28. doi: 10.1111/omi.12197. [DOI] [PubMed] [Google Scholar]
- 84.Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahsan NA, Sampey GC, Lepene B, Akpamagbo Y, Barclay RA, Iordanskiy S, Hakami RM, Kashanchi F. Presence of viral RNA and proteins in exosomes from cellular clones resistant to rift valley fever virus infection. Front Microbiol. 2016;7:139. doi: 10.3389/fmicb.2016.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramos-Casals M, Zignego AL, Ferri C, Brito-Zeron P, Retamozo S, Casato M, Lamprecht P, Mangia A, Saadoun D, Tzioufas AG, Younossi ZM, Cacoub P International Study Group of Extrahepatic Manifestations related to HCV (ISG-EHCV) Evidence-based recommendations on the management of extrahepatic manifestations of chronic hepatitis C virus infection. J Hepatol. 2017;66:1282–1299. doi: 10.1016/j.jhep.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 87.Nagao Y, Sata M, Fukuizumi K, Ryu F, Ueno T. High incidence of oral lichen planus in an HCV hyperendemic area. Gastroenterology. 2000;119:882–883. doi: 10.1053/gast.2000.17936. [DOI] [PubMed] [Google Scholar]
- 88.Giuliani M, Lajolo C, Miani MC, Lodi G, Minenna P, Mangia A. Hepatitis C virus chronic infection and oral lichen planus: an Italian case-control study. Eur J Gastroenterol Hepatol. 2007;19:647–652. doi: 10.1097/MEG.0b013e32821f6134. [DOI] [PubMed] [Google Scholar]
- 89.Song J, Zhang Z, Ji X, Su S, Liu X, Xu S, Han Y, Mu D, Liu H. Lack of evidence of hepatitis in patients with oral lichen planus in China: a case control study. Med Oral Patol Oral Cir Bucal. 2016;21:e161–168. doi: 10.4317/medoral.20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Petti S, Rabiei M, De Luca M, Scully C. The magnitude of the association between hepatitis C virus infection and oral lichen planus: meta-analysis and case control study. Odontology. 2011;99:168–178. doi: 10.1007/s10266-011-0008-3. [DOI] [PubMed] [Google Scholar]
- 91.Alaizari NA, Al-Maweri SA, Al-Shamiri HM, Tarakji B, Shugaa-Addin B. Hepatitis C virus infections in oral lichen planus: a systematic review and meta-analysis. Aust Dent J. 2016;61:282–287. doi: 10.1111/adj.12382. [DOI] [PubMed] [Google Scholar]
- 92.Cosset FL, Dreux M. HCV transmission by hepatic exosomes establishes a productive infection. J Hepatol. 2014;60:674–675. doi: 10.1016/j.jhep.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 93.Chigbu DI, Loonawat R, Sehgal M, Patel D, Jain P. Hepatitis C virus infection: host(-)virus interaction and mechanisms of viral persistence. Cells. 2019;8 doi: 10.3390/cells8040376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fielding CA, Aicheler R, Stanton RJ, Wang EC, Han S, Seirafian S, Davies J, McSharry BP, Weekes MP, Antrobus PR, Prod’homme V, Blanchet FP, Sugrue D, Cuff S, Roberts D, Davison AJ, Lehner PJ, Wilkinson GW, Tomasec P. Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog. 2014;10:e1004058. doi: 10.1371/journal.ppat.1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ding M, Wang X, Wang C, Liu X, Zen K, Wang W, Zhang CY, Zhang C. Distinct expression profile of HCMV encoded miRNAs in plasma from oral lichen planus patients. J Transl Med. 2017;15:133. doi: 10.1186/s12967-017-1222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim Y, Lee S, Kim S, Kim D, Ahn JH, Ahn K. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 2012;8:e1002577. doi: 10.1371/journal.ppat.1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lisboa LF, Egli A, O’Shea D, Asberg A, Hartmann A, Rollag H, Pang XL, Tyrrell DL, Kumar D, Humar A. Hcmv-miR-UL22A-5p: a biomarker in transplantation with broad impact on host gene expression and potential immunological implications. Am J Transplant. 2015;15:1893–1902. doi: 10.1111/ajt.13222. [DOI] [PubMed] [Google Scholar]
- 98.Guo X, Huang Y, Qi Y, Liu Z, Ma Y, Shao Y, Jiang S, Sun Z, Ruan Q. Human cytomegalovirus miR-UL36-5p inhibits apoptosis via downregulation of adenine nucleotide translocator 3 in cultured cells. Arch Virol. 2015;160:2483–2490. doi: 10.1007/s00705-015-2498-8. [DOI] [PubMed] [Google Scholar]
- 99.Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA. Human cytomegalovirus miR-UL112-3p targets TLR2 and modulates the TLR2/IRAK1/NFkappaB signaling pathway. PLoS Pathog. 2015;11:e1004881. doi: 10.1371/journal.ppat.1004881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anderson MR, Kashanchi F, Jacobson S. Exosomes in viral disease. Neurotherapeutics. 2016;13:535–546. doi: 10.1007/s13311-016-0450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kouwaki T, Okamoto M, Tsukamoto H, Fukushima Y, Oshiumi H. Extracellular vesicles deliver host and virus RNA and regulate innate immune response. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18030666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lucchese A. A potential peptide pathway from viruses to oral lichen planus. J Med Virol. 2015;87:1060–1065. doi: 10.1002/jmv.24131. [DOI] [PubMed] [Google Scholar]
- 103.Pol CA, Ghige SK, Gosavi SR. Role of human papilloma virus-16 in the pathogenesis of oral lichen planus--an immunohistochemical study. Int Dent J. 2015;65:11–14. doi: 10.1111/idj.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shariati M, Mokhtari M, Masoudifar A. Association between oral lichen planus and Epstein-Barr virus in Iranian patients. J Res Med Sci. 2018;23:24. doi: 10.4103/jrms.JRMS_438_17. [DOI] [PMC free article] [PubMed] [Google Scholar]


