Abstract
The combination of anti-angiogenesis and chemotherapy can significantly prolong the survival period of patients with non-squamous non-small cell lung cancer (NSCLC). But drug resistance will inevitably occur, thereby causing increased tumor invasion and metastasis. Claudin-5 (CLDN5) is a protein member of tight junction (TJ) structures expressed in endothelial and epithelial cells and confirmed to be involved in the proliferation and leakage of endothelial cells (ECs) and malignant metastases. This study aimed to investigate how bevacizumab, a vascular endothelial growth factor A (VEGFA) neutralizing antibody applied in clinic, affects the tight junction protein CLDN5 and subsequently influences tumor cell invasion and potential metastasis. Western-blot, quantitative real-time polymerase chain reaction (q-PCR), immunofluorescence and immunohistochemistry revealed that low-dose bevacizumab up-regulated CLDN5, whereas high-dose bevacizumab down-regulated CLDN5. Cell migration, invasion and permeation assay demonstrated that high-dose bevacizumab enhanced the migration, invasion and permeation abilities of human umbilical vein endothelial cells (HUVECs). The migration and permeation abilities of HUVECs were also enhanced by silencing the CLDN5 expression. CLDN5 was regulated by JNK, PI3K and transforming growth factor-β-1 (TGFβ1), and these findings were confirmed by the inhibitor or siRNA of JNK, PI3K and TGFβ1. Our data indicated that high-dose bevacizumab likely increased tumor invasion and potential metastatic abilities by down-regulating CLDN5, which was down regulated by TGFβ1. Low-dose bevacizumab increased CLDN5 expression by up-regulating PI3K and JNK expression.
Keywords: Bevacizumab, claudin5, tight junction, VEGF, TGFβ1, PI3K
Introduction
Lung cancer is the leading cause of cancer mortality worldwide. Although the American Cancer Society reported that the incidence of lung cancer in the United States decreased in 2017, the mortality rate of this disease still ranks first [1].
Bevacizumab was the first monoclonal antibody to vascular endothelial growth factor (VEGF-A) approved by US FDA in 2004 in association with the first-line platinum-based chemotherapy for metastatic non-squamous non-small cell lung cancer (NSCLC) [2]. Many clinical trials have confirmed that bevacizumab improves the prognosis of patients with advanced non-squamous NSCLC [3-9]. However, many tumors still metastasize, even when patients undergo such therapy. Therefore, underlying mechanisms should be explored.
Numerous studies have shown that tight junction (TJ) proteins are closely related to tumor cell invasion and metastasis [10-12]. Claudin5 (CLDN5) is an endothelial cell-specific component of TJ strands that was first reported by Morita [13]. The reduction of CLDN5 increased the progression of glioma cells [14] and the leakage of the brain-blood barrier (BBB) [15,16]. A study has revealed that CLDN5 deficient mice displayed a higher permeability of small molecules (< 800 Da) [17]. Therefore, our study focused on how bevacizumab affected CLDN5 and tumor cell invasion and potential metastasis during anti-angiogenic therapy.
Materials and methods
Cell culture and reagents
Human umbilical vein endothelial cells (HUVECs, Peking Union Medical College Cell Bank) and mouse retinal microvascular endothelial cells (MRMECs, Tianjin Medical University Eye Hospital) were cultured in DMEM containing 10% FBS (Gibco, Thermo Fisher Scientific, USA) with growth supplements and 1% penicillin and streptomycin (15140-122; Life Technologies, USA). A549 (American Type Culture Collection, USA), a human lung adenocarcinoma cell line, was cultured in RPMI medium 1640 (Gibco). The cultures were maintained in a humidified atmosphere (5% CO2/95% air) at 37°C. Bevacizumab was purchased from Roche Company (Batch No. H0180B01). The dose of bevacizumab in HUVECs was 100 μg/mL in the high-dose group and 10 μg/mL in the low-dose group. Anlotinib, a multi-targeted antiangiogenic agent, was a gift from the Nanjing Chia Tai-Tianqing Pharmaceutical Co., Ltd.
Anti-claudin5 antibody (ab15106), goat anti-rabbit IgG secondary antibody (ab96899) and anti-TGFβ1 antibody (ab66957) were obtained from Abcam Inc. (Abcam, USA). (Phospho-)SAPK/JNK rabbit antibody (#9252, #4668), PI3K p110α antibody (#4255), phosphor-PI3K antibody (#4228), anti-GAPDH (#5174) and PI3K inhibitor (LY294002) were purchased from CST Inc. (CST, USA). Goat anti-rabbit conjugated with HRP and anti-rabbit immunohistochemistry secondary antibody were procured from Santa Cruz Biotechnologies Inc. (Santa Cruz, USA). TGFβ1 and JNK inhibitor (SB431542, SP600125) were purchased from Selleck Inc. (Selleck, USA).
Animal model and treatment protocol
Six to eight-week-old female nude mice (BALB/c-nu), were purchased from the Model Animal Center of Nanjing University (Nanjing, China). A549 cells were resuspended in PBS and injected subcutaneously into the left abdomen of the mice (1 × 107 cells/mouse). The tumor-bearing mice were randomly and equally divided into three groups, intraperitoneally injected with 0, 5 and 50 mg/kg bevacizumab for 3 weeks, and anesthetized with 7% chloral hydrate to harvest tumor tissues for immunohistochemistry analyses.
Western blot analysis
HUVECs were homogenized, subjected to 12% SDS/PAGE and transferred to PVDF membranes (Millipore, USA), which were blocked with 5% nonfat milk powder in TBS-T buffer for 1 hour at room temperature (RT) and incubated with primary antibodies at 4°C overnight. On the next day, the blots were incubated with secondary antibody for 1 h, and developed with chemiluminescence (ECL, Millipore, USA). The gray values of protein bands were quantified by using ImageJ software (National Institute of Health, NY), and the data were subjected to unpaired Student’s t-test.
Quantitative real-time polymerase chain reaction (Q-PCR)
The total RNA of HUVECs was extracted with Trizol reagent and reverse transcribed to cDNA by using Mix (TransGen Biotech, Beijing, China) in a 20 μL reaction. Subsequently, q-PCR was conducted in accordance with the protocol. Gene expression was normalized to GAPDH mRNA. The specific primers used in this experiment were synthesized by Sangon Biotech, and the sequences are displayed in Table 1. The relative quantification of genes was calculated by using the 2-ΔΔCt method.
Table 1.
Primers used in this study
| Gene | Primer sequence (Forward) | Primer sequence (Reverse) |
|---|---|---|
| GAPDH | CCTTCATTGACCTCAACTA | GGAAGGCCATGCCAGTGAGC |
| Claudin5 | CACGGGAGGAGCGCTTTAT | GGCACCGTTGGATCATAGAACT |
| siCLDN5-1 | CCUUAACAGACGGAAUGAATT | UUCAUUCCGUCUGUUAAGGTT |
| siCLDN5-2 | CUGCUGGUUCGCCAACAUUTT | UUCAUUCCGUCUGUUAAGGTT |
| Negative control | UGACCUCAACUACAUGGUUTT | ACGUGACACGUUCGGAGAATT |
Immunofluorescence assay
HUVECs treated with bevacizumab were seeded on cell slides precoated in a 24-well plate and incubated for 24 hours. Then the slides were fixed with cold methanol for 30 min and blocked with 5% goat serum. Next, the cells were incubated with primary antibody at 4°C overnight and then with secondary antibody for 1 h at RT and stained with DAPI (Solarbio, Beijing, China). Images were captured with Axiovert200 microscope (Carl Zeiss, Thornwood, NY, USA). Fluorescence quantitative analysis was performed by ImageJ software, and the data were subjected to one-way ANOVA.
Immunohistochemistry
The tumors of mice were fixed with 4% paraformaldehyde, embedded with paraffin, and cut at 6 μm. Histological sections were dewaxed, antigen retrieved with citric acid buffer, blocked with 3% hydroperoxide for 20 minutes, incubated with primary antibody at 4°C overnight and incubated with secondary antibody for 1 h at RT. Diaminobenzidine was used as a chromogen to visualize the antibody/antigen complex. The sections were counterstained with hematoxylin for 1.5 min, dehydrated, cleared and mounted in neutral gum. Images were captured with Olympus Bx61 microscope (Olympus Corporation, Japan). Immunohistochemical scoring was performed by ImageJ software, and the data were subjected to one-way ANOVA.
Cell proliferation assay
HUVECs were pre-seeded into five 96-well plates (3000 cells/well). Then, 20 μL MTT (5 mg/mL, Solarbio, Beijing, China) was added after 6, 24, 48, 72 and 96 h of incubation. After 4 hours, the supernatants were removed and 150 μL DMSO was added and shaken for 10 min. The optical density (OD) was read using Microplate Reader (Bio-Rad Laboratories, Hercules, CA, USA) at 490 nm.
Cell migration assay
HUVECs were pre-seeded into a 6-well plate (3 × 105 cells/well). After the cells reached confluency, three horizontal straight lines simulation “wound” were made. The extent of wound closure was monitored under a microscope. After 24 h, imaging was conducted under a microscope at 10 × magnification. The distance cells migrated was analyzed with ImageJ software, and the data were subjected to one-way ANOVA.
HUVECs with 300 μL serum-free DMEM were seeded into the upper chamber and 600 μL DMEM containing 20% FBS was added to the wells of 24-well plate, incubated for 6 h, fixed with 4% paraformaldehyde for 20 min, and stained with crystal violet. Each group of the migrating cells was photographed with five fields under the microscope. The number of HUVECs migrated was analyzed with ImageJ software, and the data were subjected to one-way ANOVA.
In vitro permeation assay
HUVEC suspension with 1 × 105 cells/well was seeded into the upper chamber, and DMEM containing 20% FBS was added to the wells of 24-well plates. After 6 h incubation, A549 cells were labeled with CM-DiI (orange red; 40726ES10, Invitrogen, USA) and seeded onto the HUVEC monolayer. After 12 h, the invading cells were fixed with cold methanol for 20 min. The membranes were inverted on the slides and stained with DAPI for 15 min. The cells stained with orange-red membrane and blue cores represented A549 cells, while the cells with blue cores represented HUVECs. Images were captured with Axiovert200 microscope (Carl Zeiss, Thornwood, NY, USA). The number of A549 cells was analyzed by ImageJ software, and the data were subjected to one-way ANOVA.
In vivo permeation assay
The mice were randomly divided into three groups, and intraperitoneally injected with 0, 5 and 50 mg/kg bevacizumab for 14 days. Then, Evans blue solution (2% solution 4 mL/kg) was injected into the tail vein and circulated for 10 min. The mice were sacrificed by cervical dislocation to harvest the abdomen skin, and the weight of each of these abdominal skin specimens was measured. And the Evans blue of these specimens was extracted by formamide as described [18]. Evans blue was dissolved in formamide, and the solution was diluted to obtain a standard curve at a wavelength of 635 nm [18]. The amount of Evans blue (μg) in the abdominal skin (g) was calculated from the OD of these specimens by using the formula from the standard curve [18].
Gene knockdown and overexpression
CLDN5 siRNA and its control siRNA were synthesized by Sangon Biotech (Shanghai, China), and the siRNA of JNK or PI3K was purchased from CST Inc. (#6232, #6359, CST, USA). The target sequences are listed in Table 1. When 70% confluency was reached, HUVECs were transfected with 3 μL siRNA, 6 μL Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and 900 μL serum-free Opti-MEM medium (Invitrogen, USA) without antibiotics. After 24 h of incubation, the cells were used for the experiments described in the previous sections. For the JNK overexpression, lentiviruses were used. The lentiviruses of JNK were obtained from Genechem (Shanghai, China) and then transfected into HUVECs by using Lipofectamine 2000 Reagent in accordance with the manufacturer’s instructions.
Statistical analysis
All these experiments were repeated three times and the data were presented as mean ± SD. The data were subjected to unpaired Student’s t-test or one-way ANOVA. P-values less than 0.05 were considered statistically significant. Statistical analyses were performed using SPSS 20.0 software (SPSS Inc., IL, USA).
Results
CLDN5 is regulated by bevacizumab
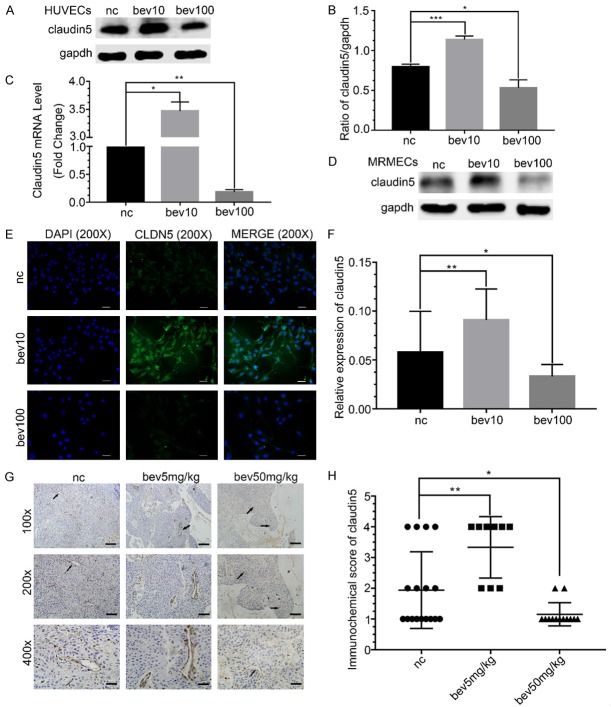
CLDN5 was significantly increased at protein and mRNA levels following low-dose bevacizumab treatment in HUVECs, reduced by high-dose bevacizumab (P < 0.05, Figure 1A-C). The regulation of CLDN5 by different concentrations of bevacizumab was verified in MRMECs (Figure 1D). The immunofluorescence staining of HUVECs and immunohistochemistry in the tumor tissues of mice further validated that CLDN5 was up-regulated by low-dose bevacizumab but was down-regulated by high-dose bevacizumab (P < 0.05, Figure 1E-H).
Figure 1.
Effect of different concentrations of bevacizumab on tight junction protein claudin5. A. Western blot showed the change in claudin5 under the different concentrations of bevacizumab (bevacizumab 10 μg/mL, 100 μg/mL) treatment in HUVECs, the results showed that low-dose bevacizumab up-regulated claudin5 but high-dose down-regulated it. B. Quantitative analyses of claudin5 protein levels following bevacizumab treatment. Data represented the mean ± SD, *P < 0.05, ***P < 0.0005; one-way ANOVA. C. Changes of claudin5 mRNA in response to different concentrations of bevacizumab, which were consistent with the western blot. Data represented the mean ± SD, *P < 0.05, **P < 0.005; one-way ANOVA. D. Western blot showed the change in claudin5 under bevacizumab treatment in MRMECs, which were consistent with HUVECs. E. Fluorescence microscopy images of HUVECs treated with different concentrations of bevacizumab. Blue, DAPI; green, claudin5; magnification, × 200; scale bar: 200 μm. F. Quantitative analyses of fluorescence intensities of claudin5 relative expression. Data represented the mean ± SD, *P < 0.05, **P < 0.005; one-way ANOVA. G. Typical images of claudin5 (brown) expression in different groups of adenocarcinoma xenograft tumor model. Black arrows: claudin5. Magnification, row 1: × 100; row 2: × 200; row 3: × 400. Scale bar: 300 μm. H. Scatter diagram showed the immunochemistry score of claudin5 in the adenocarcinoma xenograft tumor. Data represented the mean ± SD, *P < 0.05, **P < 0.005; non-parameter test.
Low-dose bevacizumab upregulates CLDN5 via PI3K
Low-dose bevacizumab up-regulated the phosphorylated PI3K (P-PI3K, Figure 2A). The results indicated that the inhibitor of PI3K (LY294002) effectively inhibited P-PI3K and CLDN5, which were reversed by low-dose bevacizumab (Figure 2B). The results of CLDN5 were also verified on mRNA levels (P < 0.05, Figure 2C).
Figure 2.
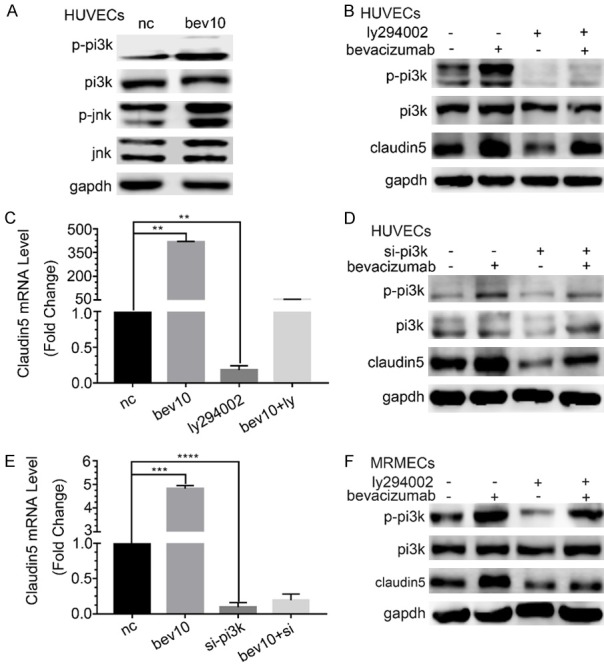
Regulation of low-dose bevacizumab on claudin5 through PI3K. A. Western blot showed that low-dose bevacizumab (bevacizumab 10 μg/mL) up-regulated P-JNK and P-PI3K in HUVECs. B. Western-blot showed the changes of claudin5 and P-PI3K protein levels after PI3K inhibitor (LY294002) and/or low-dose bevacizumab treatment in HUVECs. LY294002 inhibited both the expression of P-PI3K and claudin5. C. Changes of claudin5 mRNA levels after bevacizumab and/or LY294002 treatment, which were consistent with the western blot, **P < 0.005. D. Western blot showed the changes of claudin5 and P-PI3K protein levels under low-dose bevacizumab and/or PI3K siRNA treatment in HUVECs. E. mRNA levels of claudin5 following low-dose bevacizumab and/or siRNA of PI3K treatment in HUVECs, ***P < 0.0005, ****P < 0.0001. F. Changes of claudin5 and P-PI3K protein under LY294002 and/or low-dose bevacizumab treatment by western-blot in MRMECs, which were consistent with HUVECs. All data were expressed as mean ± SD, and all statistical analyses were performed by one-way ANOVA.
As an alternative approach, the siRNA of PI3K was introduced. The results revealed that P-PI3K, PI3K and CLDN5 were effectively down-regulated by PI3K siRNA at protein levels, which were reversed by low-dose bevacizumab (Figure 2D). The results of CLDN5 were also verified at mRNA levels (P < 0.05, Figure 2E). The results of P-PI3K emerging under the regulation of low-dose bevacizumab on CLDN5 were also validated in MRMECs (Figure 2F).
JNK participates in the regulation of low-dose bevacizumab on CLDN5
Low-dose bevacizumab up-regulated the phosphorylated JNK (P-JNK, Figure 3A). The results indicated that P-JNK and CLDN5 increased by low-dose bevacizumab were down-regulated by the inhibitor of JNK (SP600125, P < 0.05, Figure 3B), and the changes in the mRNA levels of CLDN5 were consistent with its protein levels (P < 0.05, Figure 3C). The results caused by the siRNA of JNK were the same as those caused by SP600125 (P < 0.05, Figure 3D and 3E). The overexpression and inhibition of JNK were also used to observe the regulation of JNK on CLDN5, and the results showed that CLDN5 was up-regulated by the JNK overexpression and down-regulated by SP600125 (Figure 3F). The results of JNK involved in the regulation of low-dose bevacizumab on CLDN5 were also validated in MRMECs (Figure 3G).
Figure 3.
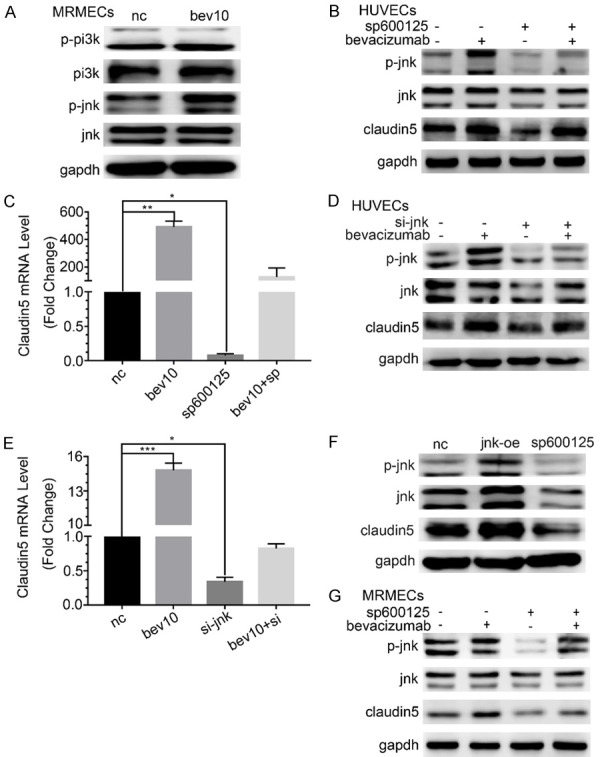
The regulation on claudin5 through JNK by low dose bevacizumab. A. The changes of P-PI3K and P-JNK protein under low-dose bevacizumab (bevacizumab 10 μg/mL) treatment by western-blot in MRMECs, which were consistent with HUVECs. B. Changes of claudin5 and P-JNK protein under JNK inhibitor (SP600125) and/or low-dose bevacizumab treatment by western-blot in HUVECs. SP600125 inhibited both the expression of P-JNK and claudin5. C. mRNA levels of claudin5 following bevacizumab and/or SP600125 treatment, *P < 0.05, **P < 0.005. D. Changes of claudin5 and P-JNK protein under low-dose bevacizumab and/or JNK siRNA treatment by western-blot in HUVECs. JNK siRNA inhibited the expression of JNK, P-JNK and claudin5. E. Claudin5 mRNA expression following bevacizumab and/or JNK siRNA treatment, *P < 0.05, ***P < 0.0005. F. Changes of claudin5 protein following JNK overexpression (jnk-oe) or SP600125. Claudin5 rose and fell together with JNK. G. Changes of claudin5 and P-NK protein under SP600125 and/or low-dose bevacizumab treatment by western-blot in MRMECs, which were consistent with HUVECs. All data were expressed as mean ± SD, and all statistical analyses were performed by one-way ANOVA.
To study the relationship between JNK and PI3K, the siRNA of JNK or PI3K was introduced. The results indicated that P-JNK decreased after si-PI3K stimulation, by contrast, P-PI3K increased after JNK silencing (P < 0.05, Figure 4A and 4B). Therefore, JNK should be located at the down-stream of PI3K.
Figure 4.
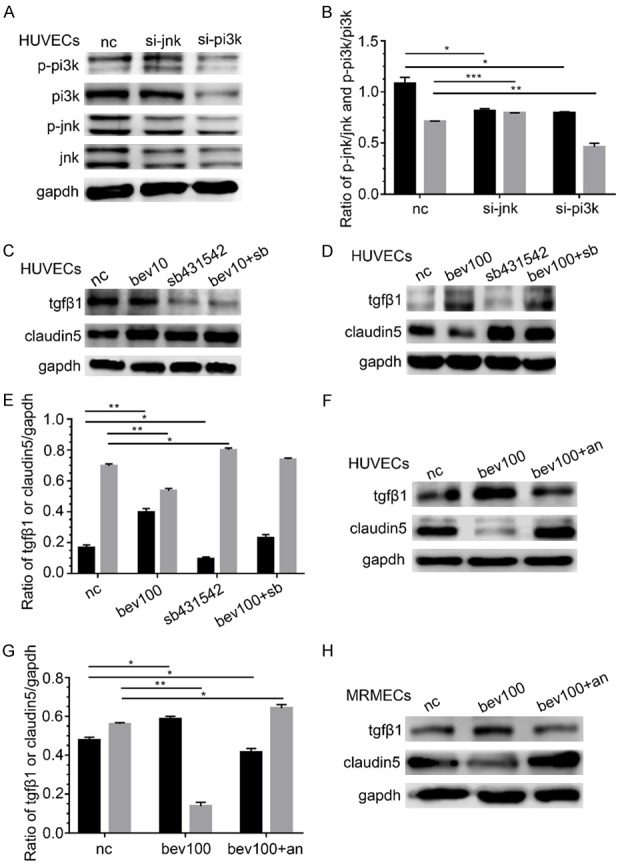
The up-regulation on clauidn5 by low-dose bevacizumab via up-regulation PI3K and JNK, the down regulation on claudin5 by high-dose bevacizumab through up-regulation TGFβ1, and the reversal by anlotinib on regulation on claudin5 induced by high-dose bevacizumab. (A) Changes of P-PI3K and P-JNK protein after siRNA of JNK or PI3K treatment. P-JNK decreased after PI3K siRNA stimulation and P-PI3K increased after JNK silencing. (B) Densitometry analyses of P-PI3K and P-JNK in (A), *P < 0.05, **P < 0.005, ***P < 0.0005; black, P-JNK/JNK; grey, P-PI3K/PI3K. (C) Changes in TGFβ1 and claudin5 protein by western-blot after low-dose bevacizumab (bevacizumab 10 μg/mL) and/or TGFβ1 inhibitor (SB431542) treatment. Low-dose bevacizumab had no significant influence on TGFβ1. (D) Changes in TGFβ1 and claudin5 protein after high-dose bevacizumab (bevacizumab 100 μg/mL) and/or SB431542 treatment. High-dose bevacizumab increased the expression of TGFβ1, and claudin5 increased after SB431542 treatment. (E) Densitometry analyses of TGFβ1 and claudin5 in (D), *P < 0.05, **P < 0.005; black, TGFβ1/GAPDH; grey, claudin5/GAPDH. (F) Changes in TGFβ1 and claudin5 protein after bevacizumab or bevacizumab and anlotinib (1 μM) sequentially (bev100+an) treatment on HUVECs. Anlotinib reversed the expression of TGFβ1 and claudin5 regulated by bevacizumab 100 μg/mL. (G) Densitometry analyses of TGFβ1 and claudin5 in (F), *P < 0.05, **P < 0.005; black, TGFβ1/GAPDH; grey, claudin5/GAPDH. (H) Changes in TGFβ1 and claudin5 protein after high dose-bevacizumab or bevacizumab and anlotinib sequentially treatment in MRMECs, which were consistent with HUVECs. All data were expressed as mean ± SD, and all statistical analyses were performed by one-way ANOVA.
TGFβ1 is involved in the down regulation of CLDN5 by high-dose bevacizumab
Low-dose bevacizumab had no significant influence on TGFβ1, and the expression of CLDN5 was slightly changed by the stimulation of TGFβ1 inhibitor (SB431542, Figure 4C). Conversely, high-dose bevacizumab enhanced the expression of TGFβ1, and CLDN5 increased after SB431542 treatment obviously (P < 0.05, Figure 4D and 4E).
Anlotinib reverses the regulation of high-dose bevacizumab on CLDN5
HUVECs were treated with bevacizumab for 12 h and then with anlotinib (1 μM) for 12 h. The results showed that CLDN5 down-regulated by high-dose bevacizumab was increased by anlotinib, and the expression of TGFβ1 regulated by high-dose bevacizumab was significantly reversed by anlotinib at the same time (P < 0.05, Figure 4F and 4G). The results were confirmed in MRMECs (Figure 4H).
High-dose bevacizumab promotes the abilities of migration, invasion and permeation of HUVECs
The effect of high-dose bevacizumab on HUVECs migration ability was examined using wound healing assay and Transwell migration assay. In the wound healing experiment, the distance of cell migrated was longer after 24 hours in the high-dose group than in the control group (P < 0.05, Figure 5A and 5B). In the Transwell migration experiment, more cells were perforated across the membrane in the high-dose group compared with those of the control (P < 0.05, Figure 5C and 5D). Transwell invasion assay showed that the number of invasive HUVECs increased in the high-dose group and decreased in the low-dose group, and difference was statistically significant (P < 0.05, Figure 5E and 5F).
Figure 5.
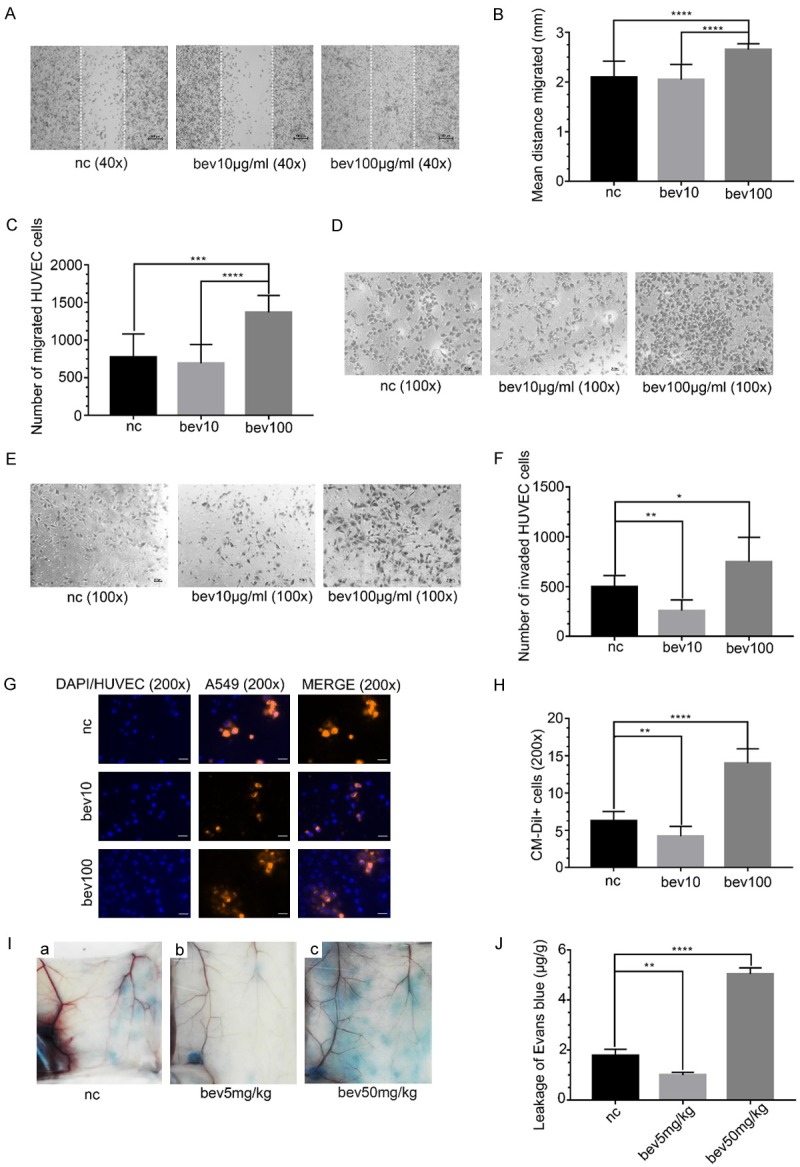
The abilities of HUVECs in migration, invasion and permeation promoted by high-dose bevacizumab. (A) Microscopic images of wound healing assay of HUVECs treated with bevacizumab (0, 10, 100 μg/mL) for 24 hours. The distance between the dashed lines was the scratch. Magnification: × 40. Scale bar: 100 μm. (B) Quantitative analyses of the distance HUVECs migrated in (A), ****P < 0.0001. The results showed that the migration distance was longer in the high-dose bevacizumab group than the control. (C) Quantitative analyses of the number of HUVECs migrated in (D), ***P < 0.0005, ****P < 0.0001. The results showed that the number of migrated HUVECs was greatest in the high-dose bevacizumab group. (D) Microscopic images of Transwell migration experiment of HUVECs treated with bevacizumab for 6 hours. Magnification: × 100. Scale bar: 100 μm. (E) Microscopic images of Transwell invasion assay of HUVECs treated with bevacizumab for 12 hours. Magnification: × 100. Scale bar: 100 μm. (F) Quantitative analyses of the number of invasive HUVECs in (E), *P < 0.05, **P < 0.0005. The results showed that the number of invasive HUVECs was greatest in the high-dose bevacizumab group. (G) Fluorescence microscopy images of A549 cells across HUVECs in in vitro permeation assay. HUVECs were pre-treated with different concentrations of bevacizumab. Blue, DAPI or HUVECs; orange red, A549 cells. Magnification, × 200. Scale bar: 200 μm. (H) Quantitative analyses of the number of CM-DiI+ cells, i.e., the number of A549 cells, **P < 0.005, ****P < 0.0001. The results showed that the number of A549 cells leaking out through HUVECs was the greatest in the high-dose bevacizumab group. (I) In vivo permeation assay showed the Evans blue leaked from blood vessels of mice. The mice were pretreated with different concentrations of bevacizumab (0, 5, 50 mg/kg) for 14 days. (J) The amount of Evans blue (μg) in the abdominal skin (g) for each group, **P < 0.005, ****P < 0.0001. The results indicated that the Evans blue exudation was highest in the high-dose group. All data were expressed as mean ± SD, and all statistical analyses were performed by one-way ANOVA.
In vitro permeation assay revealed that more A549 cells stained in orange-red invaded in HUVECs in the high-dose group, but fewer in the low-dose group compared with those in the control, and difference was statistically significant (P < 0.05, Figure 5G and 5H). The results were verified by the permeation assay in vivo, and the amount of Evans blue leaked from blood vessels in the high-dose group was much more than that in the control group, but was reduced in the low-dose group, and difference was statistically significant (P < 0.05, Figure 5I and 5J), i.e., the permeation of blood vessels was promoted by high-dose bevacizumab.
Knockdown of CLDN5 in HUVECs and the effects of CLDN5 on HUVECs’ bio-behaviors
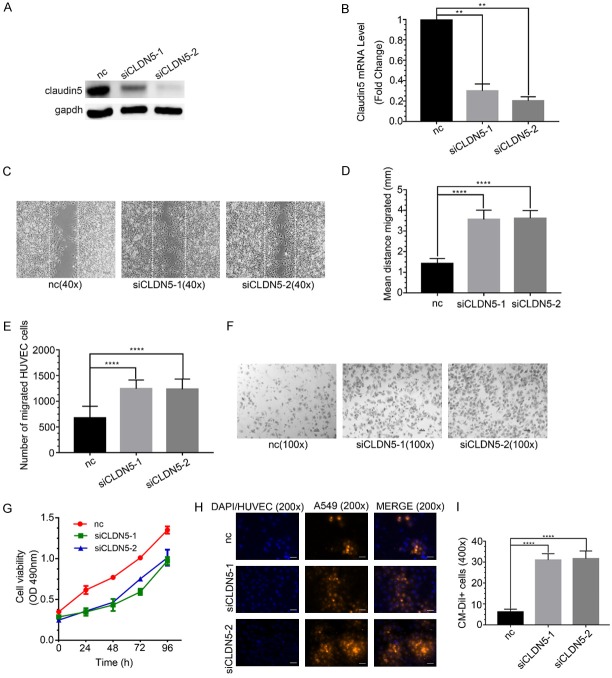
To explore the effect of CLDN5 on HUVECs’ bio-behaviors, we silenced CLDN5 by using two transcript-specific siRNAs (siCLDN5-1 and siCLDN5-2). The results indicated that the protein and mRNA levels of CLDN5 were reduced markedly in siCLDN5-1 and siCLDN5-2 groups (P < 0.05, Figure 6A and 6B).
Figure 6.
The abilities of migration and permeation enhanced and the ability of proliferation decreased by claudin5 siRNA in HUVECs. A. Claudin5 protein levels following claudin5 siRNAs treatment by western blot. B. mRNA levels of claudin5 after claudin5 siRNAs treatment by q-PCR, **P < 0.005. The results indicated that claudin5 siRNAs knocked down claudin5 expression obviously. C. Microscopic images of wound healing assay of HUVECs treated with siRNAs of claudin5 for 24 hours. The distance between the dashed lines was the scratch. Magnification: × 40. Scale bar: 50 μm. D. Quantitative analyses of the distance HUVECs migrated, ****P < 0.0001. The results showed that the migration distance was longer in siCLDN5 groups than in the control. E. Quantitative analyses of the number of migrated HUVECs, ****P < 0.0001. The results showed that the number of migrated HUVECs was greater in siCLDN5 groups than in the control. F. Microscopic images of Transwell migration experiment of HUVECs treated with claudin5 siRNAs for 6 hours. Magnification: × 100. Scale bar: 100 μm. G. The absorbance values at 490 nm was introduced to analyze the proliferation ability of HUVECs after treated with claudin5 siRNAs. The proliferation ability was decreased in the siCLDN5 groups. H. Fluorescence microscopy images of A549 cells across HUVECs in in vitro permeation assay. HUVECs were pre-treated with siclaudin5s. Blue, DAPI or HUVECs; orange red, A549 cells. Magnification, × 200. Scale bar: 200 μm. I. Quantitative analyses of the number of A549 cells, ****P < 0.0001. The number of A549 cells leaking out through HUVECs was greater in the siCLDN5 groups than in the control. All data were expressed as mean ± SD, and all statistical analyses were performed by one-way ANOVA.
The wound healing and Transwell migration experiments indicated that the migration ability of HUVECs was significantly promoted by silencing the CLDN5 expression than that by the control, and difference was statistically significant (P < 0.05, Figure 6C-F). The results indicated that CLDN5 inhibited the migration of ECs. The cell proliferation assay revealed that siclaudin5-1 and siclaudin5-2 decreased the proliferation ability of HUVECs compared with the control group (Figure 6G), indicating that CLDN5 promoted endothelial cell proliferation.
In vitro permeation assay indicated that the permeation ability of HUVECs were enhanced in the siclaudin5 groups compared with the control group, and difference was statistically significant (P < 0.05, Figure 6H, 6I). The results indicated that CLDN5 protected the conjunction of TJ so that reduce the permeation of ECs.
Discussion
Bevacizumab exhibits anti-VEGF efficacy, but probably induces the resistance of tumor [19]. As such, some tumors can re-grow even after sufficiently anti-angiogenic therapy [20-23]. Furthermore, several studies have shown that bevacizumab can increase the invasiveness of tumors at the later stage of treatment [21,24,25]. Therefore, the underlying mechanism should be elaborated to avoid clinical risks. Some studies have indicated that the main reason is hypoxia [21,26,27]. However, many other hypotheses, such as epithelial-to-mesenchymal transition programs, phenotypic progression of malignancy, and latent signaling circuits (e.g., c-Met), have been presented [21]. In our study, high-dose bevacizumab promoted the migration, invasion and leakage abilities of HUVECs under oxygen-rich conditions, indicating that high-dose bevacizumab is apt to directly promote the metastasis of NSCLC.
Experimental evidence has further suggested that TJ proteins are crucial players in the progression and metastasis of cancers [10,28,29]. CLDN5 is an important component of TJs, which exist in the paracellular space, and is highly expressed in ECs. We found that high-dose bevacizumab down-regulated CLDN5, whereas low-dose bevacizumab up-regulated CLDN5. We found that the proliferation ability of HUVECs decreased, whereas the migration and permeation abilities increased after si-CLDN5 stimulation, suggesting that CLDN5 might protect the ECs’ stability by increasing their proliferation ability and reducing their migration and permeation abilities.
In summary, the increased migration, invasion and permeation of HUVECs caused by high-dose bevacizumab could be explained by the decreased expression of CLDN5 by high-dose bevacizumab. However, the upregulation of CLDN5 by low-dose bevacizumab could maintain the integrity of TJs, which reduced the migration and permeation of ECs.
Previous studies described that CLDN5 is regulated via MAPK, JAK-STAT, PI3K-Akt, Wnt, Notch, CAMs, NFκB, N-WASP and ROCK signaling pathways [16,30-33]. In our study, PI3K and JNK were involved in the regulation of low-dose bevacizumab on CLDN5 and JNK was the down-stream molecule of PI3K, which was in accordance with those reported in the previous papers [34]. We also found that TGFβ1 was involved in the regulation of high-dose bevacizumab on CLDN5, which was consistent with previous finding that TGFβ1 down-regulates CLDN5 expression [35,36]. Considering that bevacizumab is a neutralizing antibody against human VEGF, and the standard way to measure the efficacy of bevacizumab is by determining the amount of VEGF neutralized by bevacizumab, we found that high-dose bevacizumab effectively blocked the VEGF expression (figure not shown). High-dose bevacizumab down-regulated CLDN5 expression possibly through the direct effect of bevacizumab on TGFβ1. Nevertheless, the specific mechanisms will be further explored in subsequent experiment. The schematic of the regulation of bevacizumab on CLDN5 is illustrated in Figure 7.
Figure 7.
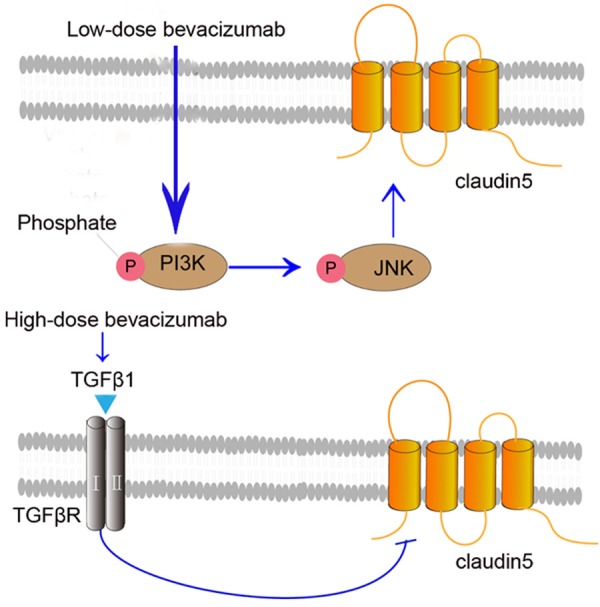
The theoretical regulation of bevacizumab on claudin5 signaling pathway. Low-dose bevacizumab up-regulates the expression of P-PI3K, which up-regulates P-JNK and claudin5, so that maintain the integrity of tight junction protein that reduces permeation of vascular ECs. While high-dose bevacizumab up-regulates TGFβ1, then down-regulates claudin5, which increases the permeation of vascular ECs.
Anlotinib could reverse the effect of high-dose bevacizumab on CLDN5 by reversing the TGFβ1 expression. Based on the results, our conclusion was that low-dose bevacizumab up-regulated CLDN5 by up-regulating PI3K and JNK, whereas high-dose bevacizumab down-regulated CLDN5 by up-regulating TGFβ1.
Conventional wisdom holds that hypoxia could induce tumor cell metastasis and vascular ECs activation, but the uniqueness of this study is to report the causes of tumor cell permeation through ECs in normoxic environment, which may make special sense since probably hypoxia not always existed inside of tumor during whole anti-angiogenic therapy in clinic. Previous studies have revealed different mechanisms of cancer progression at the later stage of bevacizumab treatment, but our study reported that high-dose bevacizumab could increase cancer cell invasion by down-regulating CLDN5 in vascular ECs. This phenomenon might show the risk of cancer metastasis even at normal dosage of bevacizumab treatment and provide potential markers to indicate cancer resistance to antiangiogenic therapy, because the clinically normal dosage of bevacizumab for major populations of patients could also become ‘high dose’ in some individuals and cause the malignant cells’ to easily migrate through unconsolidated connection of endothelium of micrangium.
The concentration of bevacizumab in our experiment was determined on the bases of the biological behaviors of ECs in previous studies. The clinical standard dose of bevacizumab is 7.5-15 mg/kg, and the cursory conversion is as follows: according to the standard weight and blood volume of a patient (60 kg and 4 L), regardless of the pharmacokinetics of bevacizumab, the plasma concentration for this patient is 60*(7.5 to 15)/4 mg/L, equal to 112.5-225.0 mg/L (i.e., 112.5-225.0 μg/mL). Furthermore, in a clinical study, the plasma concentration of bevacizumab has reached 136.3 μg/mL after patients are treated with 15 mg/kg of bevacizumab for 21 days [37]. Therefore, in our study, the concentration of bevacizumab was close to the clinical dosage and the maximum dose that the mice and cells could tolerate.
Amazingly and interestingly, the ‘vicious function’ by ‘improper dosage’ from the single-targeted anti-angiogenic drug bevacizumab could be reversed by anlotinib, a multi-targeted antiangiogenic agent [38]. This might indicate a new way to abrogate the potential risk in clinics and provide more possibilities to recover vascular normalization form disordered malignant micrangium, which may help chemotherapeutic drugs infiltrate into tumor tissues more easily [39].
On the bases of the data presented here, we could determine the link between bevacizumab and TJ proteins. High-dose bevacizumab likely increased tumor cell permeation through ECs by down-regulating the expression of TJ proteins.
Acknowledgements
This study was supported by grants from Tianjin Municipality Science and Technology Commission Projects (12ZCDZSY15600 to Kai Li), CSCO (Chinese Society of Clinical Oncology) Special Foundation for Tumor anti-angiogenesis Therapy (Y-S2014-011 to Kai Li) and Nature Science Foundation of Tianjin (18JCQNJC82500 to Tingting Qin), and The Science & Technology Development Fund of Tianjin Education Commission for Higher Education (2017KJ201 to Tingting Qin). We are grateful to Nanjing CHIA TAI TIANQING company for providing the anlotinib. We appreciated the Tianjin medical university eye hospital for providing the MRMEC cell lines. The experimental animals used in this study were performed based on the protocols approved by Tianjin Medical University Cancer Institute and Hospital Ethics Committee on Pre-Clinical Studies (SYXK2012-0005).
Disclosure of conflict of interest
None.
Abbreviations
- CLDN5
Claudin-5
- TJ
tight junction
- ECs
endothelial cells
- VEGFA
Vascular Endothelial Growth Factor A
- PI3K
phosphatidylinositol 3-kinase
- JNK
c-Jun N-terminal kinase
- TGFβ
transforming growth factor β
- PVDF
polyvinylidene fluoride
- DAPI
4’,6-diamidino-2-phenylindole
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- DMSO
Dimethyl sulfoxide
- siRNA
small interfering RNA
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Assoun S, Brosseau S, Steinmetz C, Gounant V, Zalcman G. Bevacizumab in advanced lung cancer: state of the art. Future Oncol. 2017;13:2515–2535. doi: 10.2217/fon-2017-0302. [DOI] [PubMed] [Google Scholar]
- 3.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 4.Reck M, von Pawel J, Zatloukal P, Ramlau R, Gorbounova V, Hirsh V, Leighl N, Mezger J, Archer V, Moore N, Manegold C. Phase III trial of cisplatin plus gemcitabine with either placebo or bevacizumab as first-line therapy for nonsquamous non-small-cell lung cancer: AVAil. J. Clin. Oncol. 2009;27:1227–1234. doi: 10.1200/JCO.2007.14.5466. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Wu YL, Chen G, Liu X, Zhu Y, Lu S, Feng J, He J, Han B, Wang J, Jiang G, Hu C, Zhang H, Cheng G, Song X, Lu Y, Pan H, Zheng W, Yin AY. BEYOND: a randomized, double-blind, placebo-controlled, multicenter, phase III study of first-line carboplatin/paclitaxel plus bevacizumab or placebo in Chinese patients with advanced or recurrent nonsquamous non-small-cell lung cancer. J. Clin. Oncol. 2015;33:2197–2204. doi: 10.1200/JCO.2014.59.4424. [DOI] [PubMed] [Google Scholar]
- 6.Barlesi F, Scherpereel A, Rittmeyer A, Pazzola A, Ferrer Tur N, Kim JH, Ahn MJ, Aerts JG, Gorbunova V, Vikstrom A, Wong EK, Perez-Moreno P, Mitchell L, Groen HJ. Randomized phase III trial of maintenance bevacizumab with or without pemetrexed after first-line induction with bevacizumab, cisplatin, and pemetrexed in advanced nonsquamous non-small-cell lung cancer: AVAPERL (MO22089) J. Clin. Oncol. 2013;31:3004–3011. doi: 10.1200/JCO.2012.42.3749. [DOI] [PubMed] [Google Scholar]
- 7.Barlesi F, Scherpereel A, Gorbunova V, Gervais R, Vikstrom A, Chouaid C, Chella A, Kim JH, Ahn MJ, Reck M, Pazzola A, Kim HT, Aerts JG, Morando C, Loundou A, Groen HJ, Rittmeyer A. Maintenance bevacizumab-pemetrexed after first-line cisplatin-pemetrexed-bevacizumab for advanced nonsquamous nonsmall-cell lung cancer: updated survival analysis of the AVAPERL (MO22089) randomized phase III trial. Ann Oncol. 2014;25:1044–1052. doi: 10.1093/annonc/mdu098. [DOI] [PubMed] [Google Scholar]
- 8.Reck M, von Pawel J, Zatloukal P, Ramlau R, Gorbounova V, Hirsh V, Leighl N, Mezger J, Archer V, Moore N, Manegold C. Overall survival with cisplatin-gemcitabine and bevacizumab or placebo as first-line therapy for nonsquamous non-small-cell lung cancer: results from a randomised phase III trial (AVAiL) Ann Oncol. 2010;21:1804–1809. doi: 10.1093/annonc/mdq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaked Y, Henke E, Roodhart JM, Mancuso P, Langenberg MH, Colleoni M, Daenen LG, Man S, Xu P, Emmenegger U, Tang T, Zhu Z, Witte L, Strieter RM, Bertolini F, Voest EE, Benezra R, Kerbel RS. Rapid chemotherapy-induced acute endothelial progenitor cell mobilization: implications for antiangiogenic drugs as chemosensitizing agents. Cancer Cell. 2008;14:263–273. doi: 10.1016/j.ccr.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Runkle EA, Mu D. Tight junction proteins: from barrier to tumorigenesis. Cancer Lett. 2013;337:41–48. doi: 10.1016/j.canlet.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González-Mariscal L, Lechuga S, Garay E. Role of tight junctions in cell proliferation and cancer. Prog Histochem Cytochem. 2007;42:1–57. doi: 10.1016/j.proghi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Martin TA. The role of tight junctions in cancer metastasis. Semin Cell Dev Biol. 2014;36:224–231. doi: 10.1016/j.semcdb.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karnati HK, Panigrahi M, Shaik NA, Greig NH, Bagadi SA, Kamal MA, Kapalavayi N. Down regulated expression of claudin-1 and claudin-5 and up-regulation of β-catenin: association with human glioma progression. CNS Neurol Disord Drug Targets. 2014;13:1413–1426. doi: 10.2174/1871527313666141023121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu WY, Wang ZB, Wang Y, Tong LC, Li Y, Wei X, Luan P, Li L. Increasing the permeability of the blood-brain barrier in three different models in vivo. CNS Neurosci Ther. 2015;21:568–574. doi: 10.1111/cns.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma SC, Li Q, Peng JY, Zhouwen JL, Diao JF, Niu JX, Wang X, Guan XD, Jia W, Jiang WG. Claudin-5 regulates blood-brain barrier permeability by modifying brain microvascular endothelial cell proliferation, migration, and adhesion to prevent lung cancer metastasis. CNS Neurosci Ther. 2017;23:947–960. doi: 10.1111/cns.12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luissint AC, Federici C, Guillonneau F, Chretien F, Camoin L, Glacial F, Ganeshamoorthy K, Couraud PO. Guanine nucleotide-binding protein Galphai2: a new partner of claudin-5 that regulates tight junction integrity in human brain endothelial cells. J Cereb Blood Flow Metab. 2012;32:860–873. doi: 10.1038/jcbfm.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shim KH, Jeong KH, Bae SO, Kang MO, Maeng EH, Choi CS, Kim YR, Hulme J, Lee EK, Kim MK, An SS. Assessment of ZnO and SiO2 nanoparticle permeability through and toxicity to the blood-brain barrier using Evans blue and TEM. Int J Nanomedicine. 2014;9(Suppl 2):225–233. doi: 10.2147/IJN.S58205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becherirat S, Valamanesh F, Karimi M, Faussat AM, Launay JM, Pimpie C, Therwath A, Pocard M. Discontinuous schedule of bevacizumab in colorectal cancer induces accelerated tumor growth and phenotypic changes. Transl Oncol. 2018;11:406–415. doi: 10.1016/j.tranon.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gampenrieder SP, Rinnerthaler G, Hackl H, Pulverer W, Weinhaeusel A, Ilic S, Hufnagl C, Hauser-Kronberger C, Egle A, Risch A, Greil R. DNA methylation signatures predicting bevacizumab efficacy in metastatic breast cancer. Theranostics. 2018;8:2278–2288. doi: 10.7150/thno.23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu XD, Hoang A, Zhou L, Kalra S, Yetil A, Sun M, Ding Z, Zhang X, Bai S, German P, Tamboli P, Rao P, Karam JA, Wood C, Matin S, Zurita A, Bex A, Griffioen AW, Gao J, Sharma P, Tannir N, Sircar K, Jonasch E. Resistance to antiangiogenic therapy is associated with an immunosuppressive tumor microenvironment in metastatic renal cell carcinoma. Cancer Immunol Res. 2015;3:1017–1029. doi: 10.1158/2326-6066.CIR-14-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunkel P, Ulbricht U, Bohlen P, Brockmann MA, Fillbrandt R, Stavrou D, Westphal M, Lamszus K. Inhibition of glioma angiogenesis and growth in vivo by systemic treatment with a monoclonal antibody against vascular endothelial growth factor receptor-2. Cancer Res. 2001;61:6624–6628. [PubMed] [Google Scholar]
- 25.Rubenstein JL, Kim J, Ozawa T, Zhang M, Westphal M, Deen DF, Shuman MA. Anti-VEGF antibody treatment of glioblastoma prolongs survival but results in increased vascular cooption. Neoplasia. 2000;2:306–314. doi: 10.1038/sj.neo.7900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns RA, Kalliomaki T, Hill RP. Acute (cyclic) hypoxia enhances spontaneous metastasis of KHT murine tumors. Cancer Res. 2001;61:8903–8908. [PubMed] [Google Scholar]
- 27.Escudero-Esparza A, Jiang WG, Martin TA. Claudin-5 is involved in breast cancer cell motility through the N-WASP and ROCK signalling pathways. J Exp Clin Cancer Res. 2012;31:43. doi: 10.1186/1756-9966-31-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latorre IJ, Roh MH, Frese KK, Weiss RS, Margolis B, Javier RT. Viral oncoprotein-induced mislocalization of select PDZ proteins disrupts tight junctions and causes polarity defects in epithelial cells. J Cell Sci. 2005;118:4283–4293. doi: 10.1242/jcs.02560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta. 2009;1788:872–891. doi: 10.1016/j.bbamem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Escudero-Esparza A, Jiang WG, Martin TA. Claudin-5 participates in the regulation of endothelial cell motility. Mol Cell Biochem. 2012;362:71–85. doi: 10.1007/s11010-011-1129-2. [DOI] [PubMed] [Google Scholar]
- 31.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Six I, Kureishi Y, Luo Z, Walsh K. Akt signaling mediates VEGF/VPF vascular permeability in vivo. FEBS Lett. 2002;532:67–69. doi: 10.1016/s0014-5793(02)03630-x. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Stahl A, Krah NM, Seaward MR, Dennison RJ, Sapieha P, Hua J, Hatton CJ, Juan AM, Aderman CM, Willett KL, Guerin KI, Mammoto A, Campbell M, Smith LE. Wnt signaling mediates pathological vascular growth in proliferative retinopathy. Circulation. 2011;124:1871–1881. doi: 10.1161/CIRCULATIONAHA.111.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Singh UK, Kini SG, Garg V, Agrawal S, Tomar PK, Pathak P, Chaudhary A, Gupta P, Malik A. JNK pathway signaling: a novel and smarter therapeutic targets for various biological diseases. Future Med Chem. 2015;7:2065–2086. doi: 10.4155/fmc.15.132. [DOI] [PubMed] [Google Scholar]
- 35.Argaw AT, Gurfein BT, Zhang Y, Zameer A, John GR. VEGF-mediated disruption of endothelial CLN-5 promotes blood-brain barrier breakdown. Proc Natl Acad Sci U S A. 2009;106:1977–1982. doi: 10.1073/pnas.0808698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunzel D, Yu AS. Claudins and the modulation of tight junction permeability. Physiol Rev. 2013;93:525–569. doi: 10.1152/physrev.00019.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiu HH, Liao HW, Shao YY, Lu YS, Lin CH, Tsai IL, Kuo CH. Development of a general method for quantifying IgG-based therapeutic monoclonal antibodies in human plasma using protein G purification coupled with a two internal standard calibration strategy using LC-MS/MS. Anal Chim Acta. 2018;1019:93–102. doi: 10.1016/j.aca.2018.02.040. [DOI] [PubMed] [Google Scholar]
- 38.Han B, Li K, Wang Q, Zhang L, Shi J, Wang Z, Cheng Y, He J, Shi Y, Zhao Y, Yu H, Zhao Y, Chen W, Luo Y, Wu L, Wang X, Pirker R, Nan K, Jin F, Dong J, Li B, Sun Y. Effect of Anlotinib as a third-line or further treatment on overall survival of patients with advanced non-small cell lung cancer: the ALTER 0303 phase 3 randomized clinical trial. JAMA Oncol. 2018;4:1569–1575. doi: 10.1001/jamaoncol.2018.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong RT, Boucher Y, Kozin SV, Winkler F, Hicklin DJ, Jain RK. Vascular normalization by vascular endothelial growth factor receptor 2 blockade induces a pressure gradient across the vasculature and improves drug penetration in tumors. Cancer Res. 2004;64:3731–3736. doi: 10.1158/0008-5472.CAN-04-0074. [DOI] [PubMed] [Google Scholar]