Abstract
Long non-coding RNA (lncRNA) was recognized as crucial regulator for cancer progression. The functions of small nucleolar RNA host gene 14 (SNHG14) in cancers have been appreciated in recent years. However, its role in hepatocellular carcinoma (HCC) remains to be elucidated. SNHG14 expression level in HCC cells and tissues was analyzed at first. Effects of SNHG14 on HCC cell proliferation, colony formation, and apoptosis were analyzed by Cell Counting Kit-8 (CCK-8), colony formation assay and flow cytometry, respectively. The binding relationship of microRNA-4673 (miR-4673) with SNHG14 or suppressor of cytokine signaling 1 (SOCS1) was examined by bioinformatic analysis tools and luciferase reporter gene assays. Recue experiments were performed to analyze whether SHHG14 affect HCC cell proliferation, colony formation, and apoptosis via regulating miR-4673/SOCS1 axis. SNHG14 was found highly expressed in HCC cells and tissues. In addition, we found SNHG14 overexpression could accelerate HCC cell proliferation and colony formation bur inhibit cell apoptosis. On the contrary, knockdown of SNHG14 could cause the exactly opposite effects on HCC cells. Dual-Luciferase reporter assays confirmed miR-4673 could bind with SNHG14 and SOCS1. In addition, we showed overexpression of miR-4673 or knockdown of SOCS1 could partially reverse the effects of SNHG14 overexpression on HCC cells. SNHG14 was revealed could promote HCC cell proliferation, colony formation but inhibit cell apoptosis by sponging miR-4673 to regulate SOCS1 expression.
Keywords: SNHG14, miR-4673, SOCS1, hepatocellular carcinoma, ceRNA
Introduction
Hepatocellular carcinoma (HCC) accounts for 75-85% of all liver cancer cases in a worldwide range [1]. It was estimated there were 466.1 thousand newly diagnosed liver cancer patients in China in 2015, which cover over 50% of all liver cancer cases worldwide [1,2]. Apparently, HCC is a heavy public health threat for people in China. Therefore, it is imperative to investigate mechanisms behind HCC progression with the purpose to provide more targets for HCC treatment.
Human genome project indicated approximately 90% of genome transcribed into non-coding RNAs [3]. Non-coding RNAs, including long-non coding RNAs (lncRNA, over 200 nucleotides in length) and microRNAs (miRNA, 18-25 nucleotides in length), have been reported to function as crucial regulators for cancer and other human diseases [4-7]. In addition, it was reported lncRNA and miRNA were able to regulate almost all processes related to cancer initiation and progression [4-7].
Small nucleolar RNA host gene 14 (SNHG14) is a lncRNA that reported could promote the initiation and progression of multiplies human cancers including colorectal cancer, non-small cell lung cancer, and ovarian cancer [8-10]. SNHG14 was found upregulated expression in colorectal cancer tissues and cell lines [8]. In addition, they reported SNHG14 could contribute to colorectal cancer progression in via regulating miR-186-5p/enhancer of zeste homolog 2 both in vitro and in vivo [8]. Another study on non-small cell lung cancer showed knockdown the expression SNHG14 could inhibit tumor progression and enhance the sensitivity of cancer cell response to cisplatin via regulating miR-34a and high mobility group box 1 axis [9]. In addition, SNHG14 was demonstrated to be an oncogene in ovarian cancer and promoted cancer metastasis through regulating miR-219a-5p expression [10]. However, it was unclear whether SNHG14 has a role in the development of HCC.
Here, we analyzed SNHG14 expression in HCC tissues at StarBase. Also, SNHG14 expression in HCC cell lines and normal cell line was analyzed using quantitative real-time PCR (RT-qPCR) method. Effects of SNHG14 on HCC cell growth and apoptosis were analyzed by cell counting kit-8 (CCK-8), colony formation assay, and flow cytometry assay. Moreover, the mechanisms related to SNHG14 regulated HCC progression were also investigated.
Materials and methods
StarBase tool to analyze SNHG14 in HCC tissues
StarBase v3.0 database, a platform contains gene expression levels in pan-cancer, was utilized to analyze SNHG14 expression level in HCC tumor tissues and normal tissues [11].
Cell line and cell culture
HCC cell lines (Hep3B and Huh-7) and normal liver cell line L02 were purchased from American Type Culture Collection (Manassas, VA, USA). DMEM containing 10% of fetal bovine serum (FBS) and 1% of penicillin/streptomycin (all purchased from Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to incubate these cells in a 37°C humidified atmosphere containing 95% air/5% CO2.
Cell transfection
pcDNA3.1 contains the sequence of SNHG14 (pSNHG14) or suppressor of cytokine signaling 1 (pSOCS1) was bought from GenScript (Nanjing, Jiangsu, China). Small interfering RNA targeting SNHG14 (si-SNHG14) and negative control (si-NC) were purchased from RiboBio (Guangzhou, Guangdong, China). miR-4673 mimic and the nontargeting control (mi-NC) were also bought from RiboBio. Lipofectamine 2000 (Invitrogen) was utilized for cell transfection according to the provided instructions.
RNA isolation and RT-qPCR
RNA from cultured cells was extracted using Trizol reagent (Invitrogen) and reverse transcribed into complementary DNA using PrimeScript RT kit (Takara, Dalian, Liaoning, China) on the basis of the standard protocols. Expression levels of SNHG14 or miR-4673 were analyzed with SYBR Premix ExTaq II or SYBR PrimeScript miRNA kit (Takara) at the ABI 7700 system (Applied Biosystems, Foster City, CA, USA) with GAPDH or U6 snRNA as internal controls. Primers were synthesized by GenScript and listed as follows: SNHG14: F-5’-GGGTGTTTACGTAGACCAGAACC-3’; R-5’-CTTCCAAAAGCCTTCTGCCTTAG-3’; GAPDH: F-5’-CGGAGTCAACGGATTTGGTCGTAT-3’; R-5’-AGCCTTCTCCATGGTGGTGAAGAC-3’; miR-4673: F-5’-ACACTCCAGCTGGGAGGUCAGGCCGAGGAC-3’; R-5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGTCCGT-3’; U6 snRNA: F-5’-TCCGATCGTGAAGCGTTC-3’; R-5’-GTGCAGGGTCCGAGGT-3’. Relative expression levels were calculated with 2-ΔΔCt method. RT-qPCR procedure was as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 12 s and 58°C for 40 s.
Protein isolation and western blot
Protein from cultured cells was isolated using RIPA lysis buffer contains protease inhibitors (Beyotime, Haimen, Jiangsu, China) and then quantified with bicinchoninic acid kit (Beyotime). Same amount of protein sample (50 μg) was separated at 10% SDS-PAGE and transferred to PVDF membrane. After blocked with fat-free milk at 37°C for 4 h, membranes were incubated with primary antibodies (rabbit anti-SOCS1: ab62584, anti-GAPDH: ab181602, Abcam, Cambridge, MA, USA) at 4°C for overnight. Subsequently, membranes were washed with Tris-Buffered Saline and Tween 20, and incubated with goat anti-rabbit secondary antibody (ab6721, Abcam) at 37°C for 4 h. Finally, BeyoECL kit obtained from Beyotime was utilized to visualize the band signals.
CCK-8 assay
Cells were seeded into 96-well plate with the density of 2 × 103 cells/well. CCK-8 reagent (Beyotime) was added to well at 0, 24, 48, and 72 h after seeding. Subsequently, cells were incubated at above-mentioned conditions for another 2 h. Finally, optical density was measured at 450 nm using microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation assay
800 cells were seeded into 6-well plate and incubated for 2 weeks at the described conditions. Colonies formed were fixed by methanol, stained with crystal violet, and counted manually under microscope.
Flow cytometry
Cells were seeded into 96-well plate and double stained with Annexin V-FITC (fluorescein isothiocyanate) and Propidium Iodide (PI) obtained from Beyotime at dark according to the provided protocol. Cell apoptosis rate was analyzed with FACS Calibur system (BD Biosciences, San Jose, CA, USA).
Bioinformatic analysis
Targets of SNHG14 were analyzed at LncBase Predicted v2 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php) and the results showed miR-4671 was a potential target for SNHG14. Targets for miR-4673 were analyzed at TargetScan (http://www.targetscan.org/vert_72/) and showed SOCS1 was a putative target for miR-4673.
Luciferase activity reporter assay
Sequences of SNHG14 or SOCS1 contained the putative binding sequence of miR-4673 were amplified from genome and then cloned into pmirGLO (Promega, Madison, WI, USA) to generate wild-type (wt)-SNHG14/SOCS1 luciferase construct. The mutant (mt)-SNHG14/SOCS1 luciferase constructs were built using site-direct mutagenesis kit (Takara) from wt-SNHG14/SOCS1. Cells were co-transfected with luciferase constructs or synthetic miRNAs using Lipofectamine 2000. 48 h after co-transfection, relative luciferase activity was measured using dual-luciferase activity kit (Promega).
Statistical analysis
SPSS V_17.0 (SPSS Inc., Chicago, IL, USA) was utilized for statistical analyses. Data were presented as mean ± standard deviation. Differences were analyzed with Student’s t-test and one-way ANOVA and Tukey post-hoc test. P value less than 0.05 was considered as statistically significance.
Results
Elevated SNHG14 expression in HCC
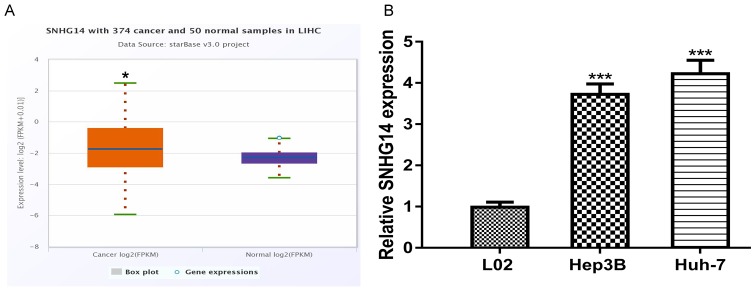
Using StarBase algorithm, we found SNHG14 expression level was significantly upregulated in HCC tumor tissues compared with normal tissues (Figure 1A). Consistent with this result, RT-qPCR analysis result showed SNHG14 expression level in HCC cell lines (Hep3B and Huh-7) was significantly upregulated compared with normal cell line (Figure 1B).
Figure 1.
SNHG14 expression was upregulated in HCC. Expression of SNHG14 in (A) HCC tissues and (B) cell lines. SNHG14: small nucleolar RNA host gene 14; HCC: hepatocellular carcinoma.
SNHG14 overexpression promotes HCC cell proliferation, colony formation, but inhibits apoptosis
pSNHG14 was used to elevate SNHG14 expression level in Hep3B cell line whose expression was relatively low in HCC cells investigated. It was found pSNHG14 transfection significantly increased the levels of SNHG14 in Hep3B (Figure 2A). CCK-8 assay showed SNHG14 overexpression significantly increased cell proliferation rate (Figure 2B). Colony formation assay showed introduction of pSNHG14 increased colony formation ability (Figure 2C). Cell apoptosis assay further showed overexpression of SNHG14 inhibited cell apoptosis rate (Figure 2D).
Figure 2.
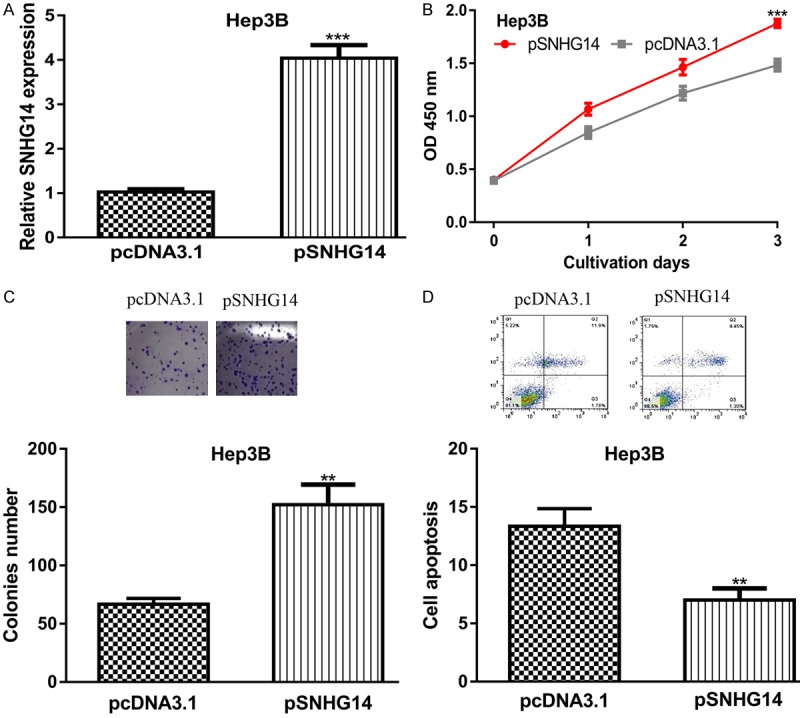
SNHG14 overexpression promotes HCC cell growth but inhibits cell apoptosis. (A) Relative SNHG14 expression, (B) Cell proliferation, (C) Colony formation, and (D) Cell apoptosis in HCC cell with pSNHG14 or pcDNA3.1 transfection. SNHG14: small nucleolar RNA host gene 14; HCC: hepatocellular carcinoma.
SNHG14 knockdown inhibits cell proliferation, colony formation, but promotes apoptosis
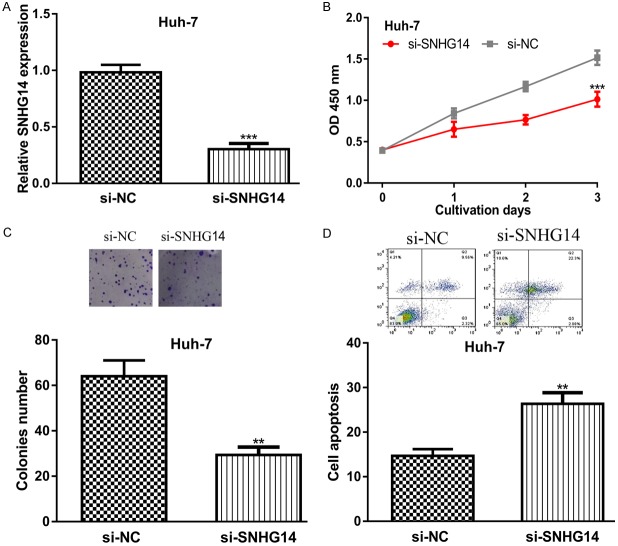
We further assessed the biological functions of SNHG14 by transferring si-SNHG14 into Huh-7 cell line. Not surprisingly, the introduction of si-SNHG14 significantly decreased SNHG14 expression in Huh-7 (Figure 3A). CCK-8 assay and colony formation assay showed that knockdown of SNHG14 inhibits HCC cell proliferation and colony formation (Figure 3B and 3C). Flow cytometry assay indicated SNHG14 downregulation could promote HCC cell apoptosis (Figure 3D).
Figure 3.
SNHG14 knockdown inhibits HCC cell growth but promotes cell apoptosis. (A) Relative SNHG14 expression, (B) Cell proliferation, (C) Colony formation, and (D) Cell apoptosis in HCC cell with si-SNHG14 or si-NC transfection. SNHG14: small nucleolar RNA host gene 14; HCC: hepatocellular carcinoma; si-SNHG14: small interfering RNA targeting SNHG14; si-NC: negative control siRNA.
SNHG14 regulates miR-4673 expression in HCC
Bioinformatic analysis showed SNHG14 could bind with miR-4673 (Figure 4A). Luciferase activity reporter assay showed miR-4673 mimic could decrease the luciferase activity of cells transfected with wt-SNHG14 (Figure 4B). In addition, we found miR-4673 expression level was decreased in HCC cell lines compared with normal cell line (Figure 4C). Importantly, we showed that miR-4673 mimic transfection decreased SNHG14 expression in HCC cell (Figure 4D).
Figure 4.
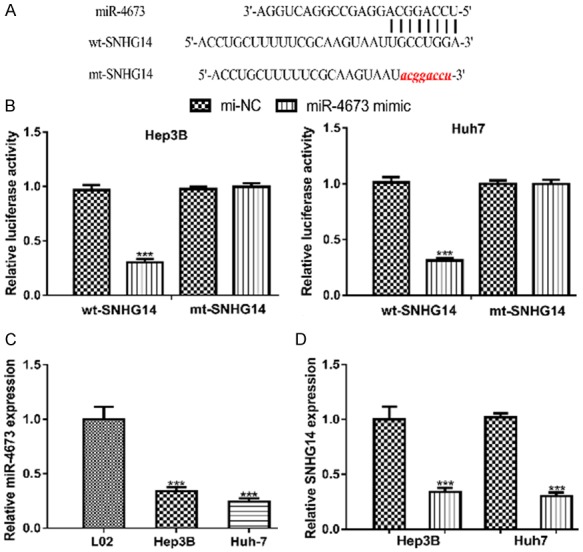
SNHG14 could directly bind with miR-4673 in HCC. A. Predicted binding model of SNHG14 and miR-4673. B. Luciferase activity in HCC cell with wt or mt-SNHG14 and miR-4673 mimic or mi-NC transfection. C. Relative expression of miR-4673 in HCC cells and normal cell line. D. Relative SNHG14 expression in HCC cell with miR-4673 mimic or mi-NC transfection. SNHG14: small nucleolar RNA host gene 14; HCC: hepatocellular carcinoma; miR-4673: microRNA-4673; wt: wild-type; mt: mutant; mi-NC: negative control miRNA.
miR-4673 regulates SOCS1 expression in HCC
TargetScan showed that miR-4673 could directly bind with the 3’-untranslated region of SOCS1 (Figure 5A). Luciferase activity showed miR-4673 mimic significantly decreased luciferase activity in cells transfected with wt-SOCS1 (Figure 5B). We also found SOCS1 expression level was significantly increased in HCC cells compared with normal cell line (Figure 5C). Furthermore, we showed the transfection of miR-4673 mimic decreased the expression level of SOCS1 in HCC cell (Figure 6A).
Figure 5.
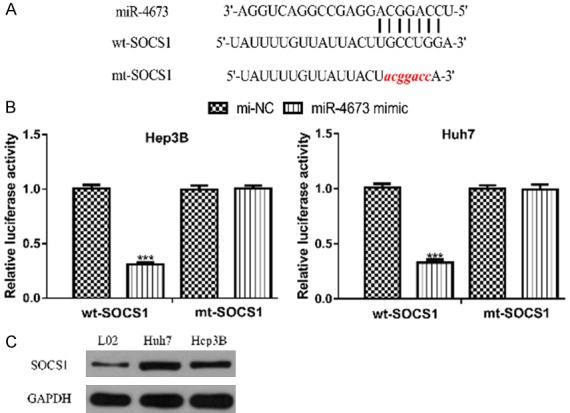
miR-4673 could directly bind with SOCS1 in HCC. A. Predicted binding model of SOCS1 and miR-4673. B. Luciferase activity in HCC cell with wt or mt-SOCS1 and miR-4673 mimic or mi-NC transfection. C. Expression of SOCS1 in HCC cells and normal cell line. SOCS1: suppressor of cytokine signaling 1; HCC: hepatocellular carcinoma; miR-4673: microRNA-4673; wt: wild-type; mt: mutant; mi-NC: negative control miRNA.
Figure 6.
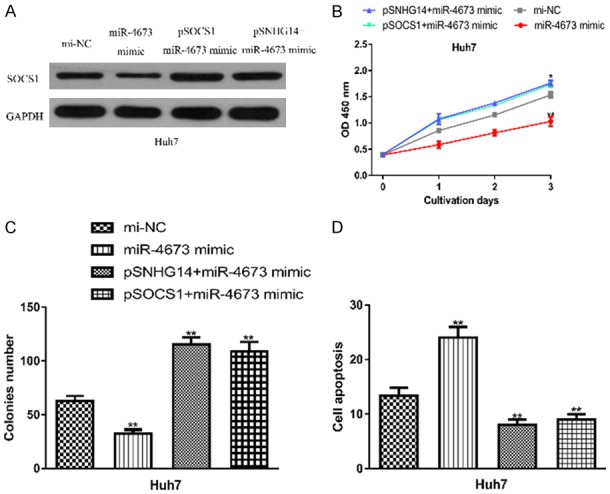
SNHG14 modulating HCC cell behaviors via the miR-4673/SOCS1 axis. (A) SOCS1 expression, (B) Cell proliferation, (C) Colony formation, and (D) Cell apoptosis in HCC cell transfected with miR-4673 mimic, mi-NC, pSNHG14+miR-4673 mimic, or pSOCS1+miR-4673 mimic. SNHG14: small nucleolar RNA host gene 14; SOCS1: suppressor of cytokine signaling 1; HCC: hepatocellular carcinoma; miR-4673: microRNA-4673; mi-NC: negative control miRNA.
SNHG14 regulates HCC cell behaviors via targeting miR-4673/SOCS1 axis
Rescue experiments were performed to investigate the mechanisms behind SNHG14 mediated HCC progression. It was found pSOCS1 increased the levels of SOCS1 in HCC cell (Figure 6A). Moreover, co-transfection of miR-4673 mimic and pSNHG14 or pSOCS1 partially reversed their effects on SOCS1 expression level (Figure 6A). CCK-8 assay, colony formation assay, and flow cytometry assay showed that miR-4673 mimic transfection decreased cell proliferation, colony formation but increased cell apoptosis (Figure 6B-D). In addition, these rescue experiments showed miR-4673 mimic transfection partially abolished the effects of pSNHG14 or pSOCS1 (Figure 6B-D).
Discussion
Risk factors for HCC progression including hepatitis C transfection, hepatitis B transfection, Non-Alcoholic Fatty Liver disease or Hereditary Hemochromatosis [12]. In recent years, multiple genes have been reported to have crucial roles in regulating HCC progression [6,13-15]. For example, SNHG7 was revealed to be increased expression in HCC tissues in comparison with normal adjacent tissues, and its knockdown could inhibit HCC cell migration and invasion via regulating RNA binding motif protein 5 [13]. SNHG16 was revealed to be upregulated expression in HCC and correlated with large tumor size, and late tumor stages [14]. Also, it was found SNHG16 could accelerate HCC progression in vitro and in vivo via regulating miR-186, indicating targeting SNHG16 is a high promising therapeutic target for HCC treatment [14]. Moreover, LINC00339 was found could interact with miR-1182 and spindle and kinetochore-associated protein 1 to promote HCC progression [15].
SNHG14 was reported to act as an oncogene in multiple human cancers [8-10]. Here, we showed SNHG14 expression level was significantly increased in both HCC tissues and cell lines in comparison with the normal tissues or cell line, respectively. Importantly, we showed overexpression of SNHG14 could promote HCC cell proliferation and colony formation but inhibit cell apoptosis. Interestingly, we showed the knockdown of SNHG14 could cause the exactly opposite effects on HCC cell behaviors as comparison with SNHG14 overexpression. These results collectively suggested that SNHG14 also has an oncogenic role in HCC.
The competing endogenous RNA (ceRNA) theory suggested that lncRNA could regulate miRNA expression at a competitive manner [16,17]. LncBase Predicted v2 algorithm showed that miR-4673 was a putative target of SNG14. Forced miR-4673 expression was found could inhibit non-small cell lung cancer cell growth and promote cell apoptosis through targeting 8-oxoguanine-DNA glycosylase-1 [18]. Luciferase activity reporter assay showed SNHG14 could directly bind with miR-4673, indicating miR-4673 might play a crucial role in HCC progression.
lncRNA was report could harbor miRNA and hence indirectly regulate the expression of target genes. TargetScan showed that miR-4673 has a putative binding site within the 3’-UTR of SOCS1. The luciferase activity reporter assay showed that miR-4673 mimic transfection decreased the luciferase activity in cells with wt-SOCS1 transfection. Importantly, it was found miR-4673 mimic transfection could decrease the expression of SOCS1. SOCS1 was reported to have dual roles in human cancers [19]. For instance, SOCS1 was found could be regulated by miR-4458 in triple-negative breast cancer to promote cancer progression, indicating an oncogenic role of SOCS1 [20]. Moreover, SOCS1 was also shown to be able to inhibit prostate cancer metastasis and attenuate tumor growth, suggesting the tumor suppressive role of SOCS1 [21].
Followingly these analyses results, we are interested to investigate whether SNHG14 was able to regulate HCC cell behaviors via the miR-4673/SOCS1 axis. Hence, recue experiments were performed in this work by co-transferring miR-4673 mimic with pSNHG14 or pSOCS1. It was found the introduction of miR-4673 mimic decreased HCC cell proliferation, and colony formation but increased cell apoptosis rate. More importantly, we found the overexpression of SNHG14 or SOCS1 could reversed the inhibitory effects of miR-4673 on HCC cells. Our work provide the first evidence that SNHG14/miR-4673/SOCS1 axis functions as crucial roles in HCC progression, indicating SNHG14 may be a potential target for HCC treatment. The limitation in this work was that we only explored these results at in vitro cell lines. Further studies are needed to validate the conclusions in this work using in vivo animal models.
Conclusions
Together, our results indicated that SNHG14 was elevated expression in both HCC tissues and cells. Molecular mechanisms exploration revealed SNHG14 plays an oncogenic role in HCC by acting as sponge for miR-4673 to regulate SOCS1 expression. Our results provided valuable insights into the mechanisms behind HCC progression and offer novel targets to develop treatment measures targeting HCC.
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu L, Tang Q, Li G, Chen K. Long non-coding RNAs as biomarkers and therapeutic targets: recent insights into hepatocellular carcinoma. Life Sci. 2017;191:273–282. doi: 10.1016/j.lfs.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Wu D, Xue W, Li X, Yu Z, Gu X, Tong J, Li M. Long noncoding RNA SNHG14 facilitates colorectal cancer metastasis through targeting EZH2-regulated EPHA7. Cell Death Dis. 2019;10:514. doi: 10.1038/s41419-019-1707-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao P, Hou J, Yao M, Wu J, Ren G. SNHG14 silencing suppresses the progression and promotes cisplatin sensitivity in non-small cell lung cancer. Biomed Pharmacother. 2019;117:109164. doi: 10.1016/j.biopha.2019.109164. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Zhang R, Li SJ. Long noncoding RNA SNHG14 promotes ovarian cancer cell proliferation and metastasis via sponging miR-219a-5p. Eur Rev Med Pharmacol Sci. 2019;23:4136–4142. doi: 10.26355/eurrev_201905_17915. [DOI] [PubMed] [Google Scholar]
- 11.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–159. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Sun BZ, Ji DG, Feng ZX, Wang Y. Long noncoding RNA SNHG7 represses the expression of RBM5 to strengthen metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:5699–5704. doi: 10.26355/eurrev_201907_18307. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Li M, Huang P. LncRNA SNHG16 promotes hepatocellular carcinoma proliferation, migration and invasion by regulating miR-186 expression. J Cancer. 2019;10:3571–3581. doi: 10.7150/jca.28428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao J, Yu H, Ma Z. LINC00339 promotes growth and invasiveness of hepatocellular carcinoma by the miR-1182/SKA1 pathway. Onco Targets Ther. 2019;12:4481–4488. doi: 10.2147/OTT.S207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C, Wu D, Gao L, Liu X, Jin Y, Wang D, Wang T, Li X. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479–13490. doi: 10.18632/oncotarget.7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang HL, Shi YP, He HJ, Wang YH, Chen T, Yang LW, Yang T, Chen J, Cao J, Yao WM, Liu G. MiR-4673 modulates paclitaxel-induced oxidative stress and loss of mitochondrial membrane potential by targeting 8-oxoguanine-DNA glycosylase-1. Cell Physiol Biochem. 2017;42:889–900. doi: 10.1159/000478644. [DOI] [PubMed] [Google Scholar]
- 19.Beaurivage C, Champagne A, Tobelaim WS, Pomerleau V, Menendez A, Saucier C. SOCS1 in cancer: an oncogene and a tumor suppressor. Cytokine. 2016;82:87–94. doi: 10.1016/j.cyto.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Wang J, Zhang G. miR-4458 regulates cell proliferation and apoptosis through targeting SOCS1 in triple-negative breast cancer. J Cell Biochem. 2019;120:12943–12948. doi: 10.1002/jcb.28565. [DOI] [PubMed] [Google Scholar]
- 21.Villalobos-Hernandez A, Bobbala D, Kandhi R, Khan MG, Mayhue M, Dubois CM, Ferbeyre G, Saucier C, Ramanathan S, Ilangumaran S. SOCS1 inhibits migration and invasion of prostate cancer cells, attenuates tumor growth and modulates the tumor stroma. Prostate Cancer Prostatic Dis. 2017;20:36–47. doi: 10.1038/pcan.2016.50. [DOI] [PubMed] [Google Scholar]