Abstract
The CDKN2B-AS1 gene, also called ANRIL, is located at the human CDKN2A/B locus at 9p21.3 and transcribed by RNA polymerase II into a long non-coding RNA of 3834 bp. The CDKN2B-AS1 gene overlaps a critical region of 125 kb covering the CDKN2B gene. The CDKN2A/B locus encompasses three major tumor suppressors juxtaposed and joined into a p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster. CDKN2A encodes splice variants p16-CDKN2A and p14-ARF, and CDKN2B encodes p15-CDKN2B. ANRIL shares a bidirectional promoter with the p14-ARF gene and is transcribed from the opposite strand to the cluster. We performed an analysis of the expression level of ANRIL and tumor suppressor p16-CDKN2A, p15-CDKN2B, and p14-ARF genes using quantitative RT-PCR in a multitumor panel. We observed the overexpression of the four genes ANRIL, p16-CDKN2A, p15-CDKN2B, and p14-ARF in the great majority of the 17 different cancer types. ANRIL was upregulated in 13/17 tumors compared to normal tissues, ranging from 5% (prostate cancer) to 91% (cervix cancer), with variable expression of p16-CDKN2A, p15-CDKN2B, and p14-ARF genes. A high positive correlation was identified between levels of expression of ANRIL and the three tumor suppressors. The strongest positive association was observed with p14-ARF (p < 0.001) in all but one (lung squamous cell carcinoma) of the examined tumor types. This correlation suggests coordinated deregulated mechanisms in all cancer types through aberrant activation of a bidirectional p14-ARF/ANRIL promoter. Furthermore, significant positive correlation was unexpectedly established in prostatic carcinomas, in contradiction with previous data.
Keywords: lncRNA, ANRIL overexpression, p16-CDKN2A/p15-CDKN2B/p14-ARF cluster
1. Introduction
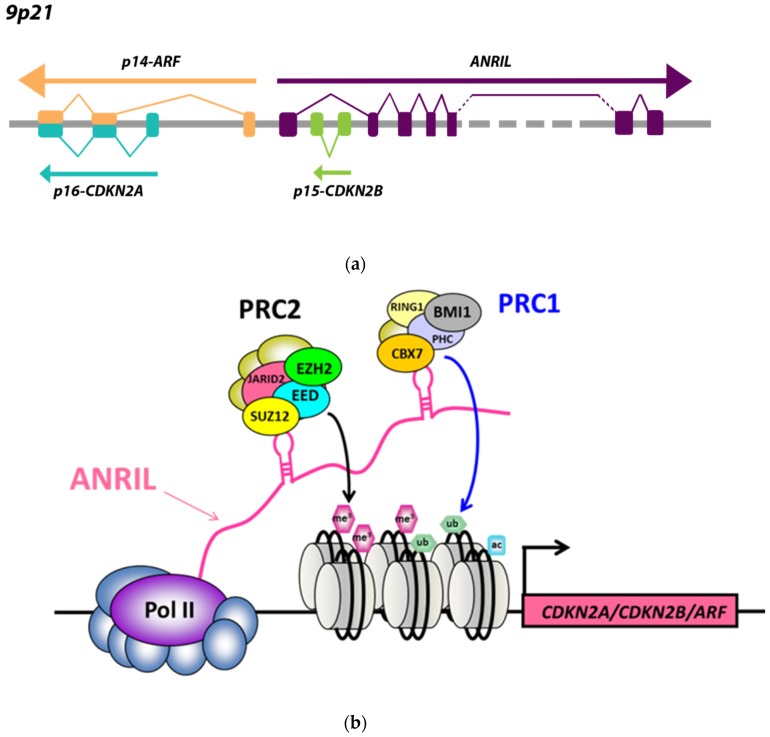
The CDKN2B-AS1 gene, also called ANRIL, is located at the human CDKN2A/B locus at 9p21.3, and is transcribed by RNA polymerase II into a long non-coding RNA of 3834 bp. The CDKN2B-AS1 gene contains 21 reported exons over a critical region of 125 kb covering the CDKN2B gene [1]. This long non-coding RNA (lncRNA) includes LINE, SINE, and Alu repetitive sequences, and comprises numerous linear and circular splice variants [2]. The CDKN2A/B locus encompasses three major tumor suppressors juxtaposed and joined into a p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster [1]. CDKN2A encodes splice variants p16-CDKN2A and p14-ARF, and CDKN2B encodes p15-CDKN2B (Figure 1a). ANRIL shares a bidirectional promoter with the p14-ARF gene, and is transcribed from the opposite strand to the cluster [1] (Figure 1a). Expression of these two genes is therefore coordinated, and activation of this divergent promoter is provided by the transcription factor E2F1 [3,4]. ANRIL intervenes mainly at the transcriptional level through epigenetic mechanisms. This lncRNA acts mainly in cis at the p16-CDKN2A/p15-CDKN2B/p14-ARF cluster, whose three tumor suppressor genes are involved in stem cell renewal, senescence, and apoptosis by promoting anti-proliferative and pro-apoptotic activities of Rb1 and p53 [5]. ANRIL functions as a docking platform for the polycomb complexes PRC1 and PRC2, which are epigenetic regulators acting in coordination to modify the histone code [6,7,8] (Figure 1b). The PRC2 complex mainly comprises subunits JARID2, EED, SUZ12, and EZH2, which maintain chromatin repression by catalyzing mono-, di-, and trimethylation of histone H3 lysine 27 (H3K27me1, H3K27me2, and H3K27me3) [9]. The PRC1 complex is composed of several subunits, such as PHC, CBX7, BMI1, and RING1a/1b, which catalyze the mono-ubiquitination of H2A on K119 (H2AK119ub1), so as to firmly maintain chromatin inactivation [10]. During this process, ANRIL promotes specific interactions with the two proteins SUZ12 (PRC2) and CBX7 (PRC1), in order to stabilize PRC1/PRC2 complexes, modify the histone code of the p16-CDKN2A/p15-CDKN2B/p14-ARF locus, and ensure a firm and perennial transcriptional repression [5]. ANRIL is overexpressed in many malignant tumors, including leukemias and stomach, prostate, kidney, esophagus, colon, breast, ovary, and lung carcinomas [6,11,12,13,14,15]. In these cancers, statistical studies and meta-analyses have revealed that the overexpression of ANRIL was positively correlated with advanced TNM stage and occurrence of lymph node metastases, and constituted an independent predictor of adverse overall survival [16]. Mounting data from the literature suggests that this lncRNA could function as an oncogenic driver, consistent with the association of multiple single nucleotide polymorphisms (SNPs) in 9p21.3 with numerous cancers [17]. Structural alterations of ANRIL (deletions and translocations) were described in neurofibromas and gliomas [18], and MTAP-ANRIL fusion transcripts were observed in melanomas [19]. During carcinogenesis, DNA alterations favor activation of the ATM-E2F1 pathway and induce ANRIL dysregulation, resulting in its aberrant chronic overexpression. Aberrant ANRIL overexpression causes inefficiency of DNA damage repair mechanisms, leading to genomic instability and tumor progression [20] via cell cycle progression, inhibition of apoptosis and senescence, tumor proliferation, and angiogenesis [21]. A study on human prostatic tumors suggested that ANRIL overexpression was mainly accompanied by transcriptional inactivation of the p16-CDKN2A/p15-CDKN2B/p14-ARF locus through cis direct interaction with the two repressive polycomb complexes PRC2 and PRC1 [6]. However, little is known about the mechanisms of ANRIL deregulation in most other types of cancers. In a recent article concerning a series of 456 breast carcinomas, we identified an unexpected strong positive link between ANRIL and the p16-CDKN2A/p15-CDKN2B/p14-ARF locus and a complex pattern of interactions between ANRIL, PRC2/PRC1, and several suppressive/oncogenic miRNAs [15]. We identified an altered repressive function of PRC2 and PRC1 at RNA and protein levels, resulting in the absence of p16-CDKN2A/p15-CDKN2B/p14-ARF cluster inactivation with frequent p16-CDKN2A, p15-CDKN2B, and p14-ARF overexpressions. This discrepancy between prostate and breast cancer suggests a large variability of the ANRIL/p16-CDKN2A/p15-CDKN2B/p14-ARF interaction network depending on the tumor type. Here, we analyzed expression levels of ANRIL and the p16-CDKN2A/p15-CDKN2B/p14-ARF locus in a multi-tumor panel of 702 malignant tumor samples among 17 different types of cancers.
Figure 1.
(a) ANRIL–ARF bidirectional promoter; (b) Interactions between the long non-coding RNA (lncRNA) ANRIL and polycomb PRC2/PRC1 repressive complexes at the p16-CDKN2A/p15-CDKN2B/p14-ARF locus (see Supplementary Table S2).
2. Materials and Methods
In this study, we investigated the expression of ANRIL and p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster by quantitative RT-PCR in a multi-tumor panel of different types of cancers.
2.1. Patients and Samples
Samples from 17 different types of primary tumors (total, n = 702; Supplementary Table S1) were obtained from multi-center Departments of Pathology from 1978 to 2010. All patients were informed that their tumor samples might be used for scientific purposes, and had the opportunity to decline. Since 2007, patients treated in our institutions have given their approval by signed informed consent. This study followed institutional guidelines as put forth by the French Ethical Committee and the Ethics Committee of Curie institute (Agreement number C75-05-18). Samples were immediately stored in liquid nitrogen until RNA extraction and then stored at −80 °C. A tumor sample was considered suitable for this study if the proportion of tumor cells exceeded 70%. Normal tissues belong to non-cancerous patients (e.g., mammoplasties for breast tissues) or adjacent normal tissues from cancer patients (distance >1 cm from tumor) for which a morphological analysis has eliminated a micrometastatic disease that was confirmed by combining an immunohistochemical study that has revealed a Ki67 index of epithelial cells of <2% and a RT-PCR analysis using MKI67 gene expression.
2.2. RNA Extraction
Total RNA was extracted by using acid-phenol guanidium, as previously described [22]. RNA quality was determined by electrophoresis through agarose gels, staining with ethidium bromide, and visualization of the 18S and 28S RNA bands under ultraviolet light. Determination of RNAs’ integrity is based on combination of three criteria: the 28S/18S ratio, the RIN (RNA integrity number) value, and the electrophoretic profile. A qualitative assessment of the type “Good, Medium, or Bad” was attributed for each of the three criteria: 28S/18S ratio (≥1.5, Good; 1.3 < ratio < 1.5, Medium; <1.3, Bad), RIN value (≥7, Good; 6 < RIN < 7, Medium; <6, Bad), and electrophoretic profile (flat baseline and RIN >7, Good; high baseline in the “fast region”, Medium; relatively high baseline, Bad). We selected RNAs with high integrity suitable for this study when a minimum of two out of three criteria were classed.
2.3. Real-Time RT-PCR
ANRIL, p16-CDKN2A, p15-CDKN2B, and p14-ARF gene mRNA expression levels were quantified using real-time RT-PCR. Quantitative values were obtained from the cycle number (Ct value) at which the increase in the fluorescence signal associated with exponential growth of PCR products started to be detected by the laser detector of the ABI Prism 7900 Sequence Detection System (Perkin-Elmer Applied Biosystems), using PE Biosystems analysis software according to the manufacturer’s manuals. The precise amount of total RNA added to each reaction mix (based on optical density) and its quality (i.e., lack of extensive degradation) are both difficult to assess. We therefore also quantified transcripts of the TBP gene (Genbank accession: NM_003194) encoding the TATA box-binding protein (a component of the DNA-binding protein complex TFIID) as an endogenous RNA control, and normalized each sample on the basis of its TBP content. We selected TBP as an endogenous control because the prevalence of its transcripts is moderate and because there are no known TBP retropseudogenes (retropseudogenes lead to coamplification of contaminating genomic DNA and thus interfere with RT-PCR, despite the use of primers in separate exons). We also selected RPLP0 because the prevalence of its transcripts is high as compared with TBP and because this gene is used widely as an endogenous control for Northern blot analysis (known better as 36B4). Results, expressed as N-fold differences in target gene expression relative to the TBP (or RPLP0) gene and termed “Ntarget,” were determined as Ntarget = 2ΔCt_sample, where the ΔCt value of the sample was determined by subtracting the average Ct value of the target gene from the average Ct value of the TBP (or RPLPO) gene. The smallest amount of mRNA that was detectable and quantifiable by RT-qPCR (ΔCt = 35) was used as a reference (basal mRNA level = 1) to normalize the data for normal and tumoral tissue samples. For each tumor type, the Ntarget values of the samples were also subsequently normalized such that the median of the Ntarget values for each normal tissue type was 1. Ntarget values of 0.33 or less were considered to represent under-expression, and values of 3 or more to represent overexpression of these genes in tumor samples. We previously used the same approach to determine cut-off points for the altered expression of tumor genes [15]. The primers for TBP, RPLPO, ANRIL, p16-CDKN2A, p15-CDKN2B, and p14-ARF genes were chosen with the assistance of the Oligo 6.0 program (National Biosciences). We scanned the dbEST and nr databases to confirm the total gene specificity of the nucleotide sequences chosen for the primers and the absence of SNPs. To avoid amplification of contaminating gDNA, one of the two primers was placed at the junction between two exons or on two different exons. The primer pairs for each CDKN2A/B cluster genes were selected to be unique when compared to the sequences of the two other CDKN2A/B cluster genes—in particular, comparison between p14ARF/CDKN2A and p16/CDKN2A. Moreover, to avoid amplification of contaminating genomic DNA, one of the two primers was placed in a different exon. For example, the upper primer of TBP was placed at the junction between exons 5 and 6, whereas the lower primer was placed in exon 6. The nucleotide sequences of the oligonucleotide hybridization primers are shown in Supplementary Table S2. Agarose gel electrophoresis was used to verify the specificity of PCR amplicons. The conditions of cDNA synthesis and PCR were as described [23]. QPCRs were performed with duplicates for each data point.
2.4. Database Sources
RNA Sequencing data and mRNA expression levels in the TCGA series used for statistical analysis between ANRIL, p16-CDKN2A, p14-ARF, and p15-CDKN2B were downloaded from the publicly available database cBioPortal for Cancer Genomics (https://www.cbioportal.org). Graphical view of the ANRIL (CDKN2B-AS1) gene was adapted from the University of California Santa Cruz (UCSC) Genome Browser on Human Dec. 2013 (GRCh38/hg38) Assembly. RNA and protein expressions overviews of p14-ARF, p15-CDKN2B, and p16-CDKN2A in various types of cancer and protein expression of p14-ARF, p15-CDKN2B, and p16-CDKN2A in prostate cancer TMA samples were adapted from Protein Atlas website—version 18.1 (https://www.proteinatlas.org).
2.5. Statistical Analysis
Relationships between mRNA levels of the different target genes were analyzed using the nonparametric Spearman rank correlation test (relation between two quantitative parameters). Differences were considered significant at confidence levels greater than 95% (p < 0.05).
3. Results
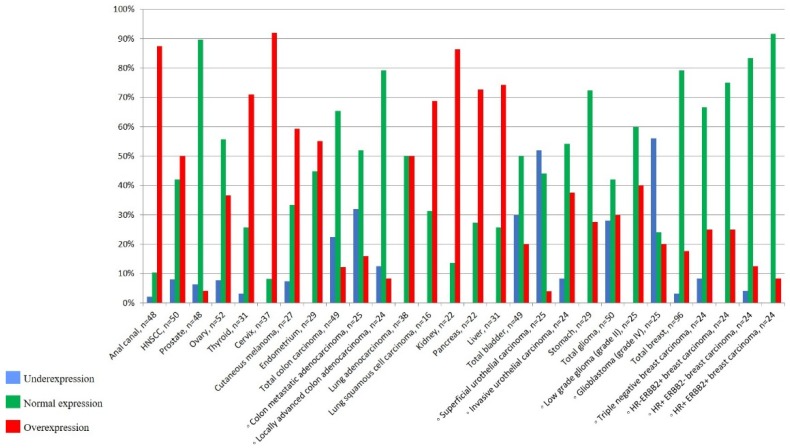
We identified the overexpression of ANRIL in 13 among the 17 cancer types compared to normal tissues. Interestingly, the expression status of ANRIL was different depending on histological subtype, tumor stage, and tumor grade. ANRIL overexpression varied according to histological subtype of malignant tumors from 5% (prostate) to 91% (cervix), and was predominant in anal canal (88%), kidney (87%), and liver (74%) carcinomas (Figure 2, Supplementary Figure S1). ANRIL upregulation was more frequent in organs that mainly develop carcinomas of squamous cell type, such as cervix (91%) and anal canal (88%). Accordingly, overexpression of ANRIL in lung cancer was more marked in squamous cell carcinomas (68%) than adenocarcinomas (50%). In colon carcinomas, ANRIL overexpression was observed in invasive tumors (12%) compared to normal tissue (0%), and was higher in carcinomas at metastatic stage (16%) compared to locally invasive primary colon carcinomas (8%). In bladder carcinomas, ANRIL upregulation was more pronounced in invasive carcinomas (38%) than in superficial non-invasive urothelial tumors (4%). In invasive breast carcinomas, ANRIL overexpression was higher in triple-negative (25%) and HER2+ (24%) subtypes than in luminal carcinomas (12%).
Figure 2.
Percentage of underexpression, normal expression, and overexpression of ANRIL in tumor tissues.
Different profiles of expression of the three tumor suppressor genes p16-CDKN2A, p15-CDKN2B, and p14-ARF were observed in the multi-tumor panel. P16-CDKN2A was mostly overexpressed in cervix (100%), anal canal (83%), and ovary (81%) (Supplementary Figure S2A,B). P15-CDKN2B was mainly upregulated in pancreas (91%), thyroid (78%), and kidney (77%) carcinomas, whereas p15-CDKN2B underexpression was identified in colon (90%) carcinomas, skin melanomas (59%), and head and neck squamous cell carcinoma (HNSCC; 38%) (Supplementary Figure S2C,D). P14-ARF was overexpressed in all tumors, mainly in kidney (100%), cervix (95%), and anal canal (94%) carcinomas (Supplementary Figure S2E,F). mRNA levels calculated as described in the Materials and Methods showed abundance of the target relative to the endogenous control (TBP), to normalize the starting amount and quality of total RNA. Similar results were obtained with the second endogenous control, RPLP0 (data non shown).
A very high positive correlation was identified between ANRIL and the p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster levels of expression in the vast majority of tumors as compared to normal tissues. Most normal tissues showed no or few significant associations between ANRIL and the p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster, and marked positive correlation values (p < 0.001) were only found between ANRIL and p14-ARF in head and neck (p = 0.000026) and colon (p = 0.00014); to a lesser degree between ANRIL and p15-CDKN2B in anal canal (p = 0.024), liver (p = 0.027), and lung (p = 0.0013); and between ANRIL and p16-CDKN2A in ovary (p = 0.019) (Table 1). Conversely, most tumors revealed a strong positive correlation between ANRIL and the p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster levels of expression, especially HNSCC, ovary, thyroid, cervix, colon, kidney, bladder, brain, and breast (p < 0.0000001) (Table 2). The strongest positive correlation was observed with the tumor suppressor p14-ARF. All examined tumors except lung squamous cell carcinoma revealed a very high positive association between ANRIL and p14 expression, as compared with p15-CDKN2B and p16-CDKN2A. No negative association was identified, particularly in prostatic carcinomas. We confirmed our experimental results by performing an in-silico analysis of mRNA expression levels in the TCGA series (RNA sequencing data from https://www.cbioportal.org), which showed positive correlation between ANRIL and p14-ARF and p16-CDKN2A (p < 0.0001), and between ANRIL and p15-CDKN2B (p < 0.0001) in both breast carcinomas and prostate carcinomas (Supplementary Table S3).
Table 1.
Correlations between ANRIL and P14, P15 and P16 mRNA expression levels in the series of normal tissues.
| Normal Tissues | Nbr | P14 | P16 | P15 | |||
|---|---|---|---|---|---|---|---|
| r | p-value a | r | p-value a | r | p-value a | ||
| Anal canal | 17 | +0.297 | 0.25 (NS) | +0.385 | 0.12 (NS) | +0.542 | 0.024 |
| Head and neck | 27 | +0.727 | 0.000026 | +0.137 | 0.50 (NS) | +0.159 | 0.43 (NS) |
| Prostate | 7 | +0.094 | 0.83 (NS) | −0.668 | 0.10 (NS) | +0.256 | 0.58 (NS) |
| Ovary | 27 | +0.423 | 0.026 | +0.447 | 0.019 | +0.010 | 0.96 (NS) |
| Thyroid | 9 | +0.852 | 0.0038 | −0.248 | 0.53 (NS) | −0.201 | 0.61 (NS) |
| Cervix | 14 | +0.534 | 0.047 | −0.295 | 0.31 (NS) | +0.116 | 0.69 (NS) |
| Skin | 9 | +0.438 | 0.24 (NS) | +0.479 | 0.19 (NS) | −0.114 | 0.77 (NS) |
| Endometrium | 8 | +0.286 | 0.50 (NS) | +0.218 | 0.61 (NS) | +0.571 | 0.14 (NS) |
| Colon | 30 | +0.647 | 0.00014 | +0.209 | 0.27 (NS) | −0.126 | 0.51 (NS) |
| Lung | 16 | +0.644 | 0.0069 | +0.433 | 0.091 (NS) | +0.732 | 0.0013 |
| Kidney | 18 | +0.058 | 0.81 (NS) | +0.073 | 0.77 (NS) | +0.124 | 0.63 (NS) |
| Pancreas | 11 | +0.609 | 0.045 | +0.202 | 0.56 (NS) | +0.212 | 0.54 (NS) |
| Liver | 10 | +0.547 | 0.099 (NS) | +0.291 | 0.42 (NS) | +0.688 | 0.027 |
| Bladder | 14 | +0.459 | 0.096 (NS) | +0.034 | 0.90 (NS) | +0.341 | 0.23 (NS) |
| Stomach | 11 | +0.361 | 0.28 (NS) | −0.391 | 0.23 (NS) | +0.267 | 0.43 (NS) |
| Brain | 21 | +0.619 | 0.0028 | +0.419 | 0.056 (NS) | +0.366 | 0.099 (NS) |
| Breast | 11 | −0.136 | 0.69 (NS) | −0.195 | 0.57 (NS) | −0.027 | 0.93 (NS) |
a Spearman’s rank correlation.
Table 2.
Statistical analysis and correlation between ANRIL and P14, P15 and P16 mRNA expression levels in the series of tumoral tissues.
| Tumoral Tissues | Nbr | P14 | P16 | P15 | |||
|---|---|---|---|---|---|---|---|
| r | p-value a | r | p-value a | r | p-value a | ||
| Anal canal | 48 | +0.571 | 0.000037 | +0.413 | 0.0036 | +0.247 | 0.087 (NS) |
| HNSCC | 50 | +0.709 | <0.0000001 | +0.609 | 0.000006 | +0.667 | 0.00000042 |
| Prostate | 48 | +0.467 | 0.00093 | +0.427 | 0.0026 | +0.059 | 0.69 (NS) |
| Ovary | 52 | +0.728 | <0.0000001 | +0.581 | 0.000013 | +0.502 | 0.0002 |
| Thyroid | 31 | +0.854 | <0.0000001 | +0.459 | 0.0092 | +0.690 | 0.000027 |
| Cervix | 37 | +0.843 | <0.0000001 | +0.563 | 0.00035 | +0.548 | 0.00053 |
| Cutaneous melanoma | 27 | +0.797 | 0.0000013 | +0.392 | 0.041 | +0.854 | <0.0000001 |
| Endometrium | 29 | +0.801 | 0.00000042 | +0.742 | 0.000007 | +0.634 | 0.00027 |
| Total colon carcinoma | 49 | +0.832 | <0.0000001 | +0.095 | 0.52 (NS) | +0.664 | 0.00000062 |
| Lung adenocarcinoma | 38 | +0.553 | 0.0003 | +0.325 | 0.047 | +0.493 | 0.0016 |
| Lung squamous cell carcinoma | 16 | +0.380 | 0.15 (NS) | +0.398 | 0.13 (NS) | +0.332 | 0.21 (NS) |
| Kidney | 22 | +0.947 | <0.0000001 | +0.700 | 0.00033 | +0.363 | 0.093 (NS) |
| Pancreas | 22 | +0.684 | 0.0005 | +0.571 | 0.0054 | +0.801 | 0.000011 |
| Liver | 31 | +0.642 | 0.00013 | -0.292 | 0.11 (NS) | +0.340 | 0.058 (NS) |
| Total bladder | 49 | +0.865 | <0.0000001 | +0.727 | <0.0000001 | +0.762 | <0.0000001 |
| Stomach | 29 | +0.733 | 0.00001 | +0.219 | 0.25 (NS) | +0.426 | 0.02 |
| Total glioma | 50 | +0.902 | <0.0000001 | +0.858 | <0.0000001 | +0.812 | <0.0000001 |
| Total breast | 96 | +0.758 | <0.0000001 | +0.576 | <0.0000001 | +0.576 | <0.0000001 |
a Spearman’s rank correlation.
4. Discussion
Concerning CDKN2A-B locus deregulation in cancers, literature data provided contradictory results. Against commonly accepted ANRIL- and PRC2/PRC1-induced epigenetic inactivation of the three oncosuppressors, positive correlations between ANRIL, p16-CDKN2A, and p15-CDKN2B genes were frequently identified in numerous tissues and non-cancerous diseases [24,25,26,27,28]. In accordance with these data, in our multitumor panel we observed overexpression of the four genes ANRIL, p15-CDKN2B, p16-CDKN2A, and p14-ARF in a great majority of the 17 tumor types. Although most normal tissues showed non-significant association between the expression of ANRIL and genes belonging to the p16-CDKN2A/p15-CDKN2B/p14-ARF gene cluster, high positive correlations were identified between ANRIL and p16-CDKN2A, p15-CDKN2B, and p14-ARF genes in the great majority of malignant tumors, suggesting coordinated alterations of their regulation in all types of cancers. Furthermore, in agreement with the presence of an E2F1-activated divergent promoter shared between ANRIL (CDKN2B-AS1) and p14-ARF genes, we observed the strongest positive correlation between ANRIL and the tumor suppressor p14-ARF, and to a lesser degree with p15-CDKN2B and p16-CDKN2A. This very significant positive association is in favor of a mechanism of deregulation in cancer involving the aberrant activation of a bidirectional ANRIL–p14-ARF promoter. Moreover, a positive correlation was unexpectedly observed in prostatic carcinomas of our series between ANRIL and p14-ARF, p15-CDKN2B, and p16-CDKN2A. These results are contradict previous data from the literature suggesting that in prostatic carcinomas ANRIL overexpression was mainly accompanied by transcriptional inactivation of the p16-CDKN2A/p15-CDKN2B/p14-ARF locus via repressive action of the two polycomb complexes PRC2 and PRC1 [6]. Our experimental and in-silico results are in accordance with recent data showing transcriptional co-activation of ANRIL, p14-ARF, p15-CDKN2B, and p16-CDKN2A in cancers induced by a bidirectional ANRIL–CDKN2A promoter in cell lines of colon cancer [29]. Despite important progress, ANRIL deregulation in cancer remains largely unknown, and only a few mechanisms have recently been evocated, such as ANRIL and p16-CDKN2A promoter methylation status regulating ANRIL/p16-CDKN2A and ANRIL/p14-ARF transcription [29,30] and transcription factors activating ANRIL transcription [31,32,33]. Splicing was recently implicated in ANRIL deregulation, and ANRIL transcript stability seems to be influenced by miRNAs and proteins [34,35]. ANRIL–miRNAs and PRC2/PRC1–miRNAs interacting networks’ deregulation has also been implicated in carcinogenesis [36,37,38].
In conclusion, the lncRNA ANRIL is an oncogenic driver that is deregulated in most cancers, and a better knowledge of its mechanisms of deregulation is a pivotal step in confirming its therapeutic potential.
Supplementary Materials
The supplementary materials are available online at https://www.mdpi.com/2311-553X/5/3/44/s1.
Author Contributions
Conceptualization: D.M., K.D.A. and I.B.; Methodology: D.M., K.D.A., E.P. and I.B.; Investigation: D.M., A.N., M.L., K.D.A. and I.B.; Resources: Y.A., E.L., J.C., M.L., D.M. and I.B.; Software and Data Curation: S.V., A.N., A.S., W.C., W.C., G.P. and C.C.; Visualization: S.V., K.D.A. and D.M.; Validation: E.P. and I.B.; Writing—Original Draft Preparation, K.D.A. and D.M.; Writing—Review & Editing: K.D.A., D.M. and I.B.; Supervision: I.B.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Kong Y., Hsieh C.H., Alonso L.C. ANRIL: A lncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018;9:405. doi: 10.3389/fendo.2018.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato K., Nakagawa H., Tajima A., Yoshida K., Inoue I. ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. Oncol. Rep. 2010;24:701–707. doi: 10.3892/or_00000910. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez C., Borgel J., Court F., Cathala G., Forné T., Piette J. CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochem. Biophys. Res. Commun. 2010;392:129–134. doi: 10.1016/j.bbrc.2009.12.159. [DOI] [PubMed] [Google Scholar]
- 5.Popov N., Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: In sickness and in health. Epigenetics. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap K.L., Li S., Muñoz-Cabello A.M., Raguz S., Zeng L., Mujtaba S., Gil J., Walsh M.J., Zhou M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. Long non-coding RNA anril is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achour C., Aguilo F. Long non-coding RNA and Polycomb: An intricate partnership in cancer biology. Front. Biosci. 2018;23:2106–2132. doi: 10.2741/4693. [DOI] [PubMed] [Google Scholar]
- 9.Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C.L., Dynlacht B.D., Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Leung F.C. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 11.Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A.P., Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunnington M.S., SantibanezKoref M., Mayosi B.M., Burn J., Keavney B. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D., Zhang Z., Mao C., Zhou Y., Yu L., Yin Y., Wu S., Mou X., Zhu Y. ANRIL inhibits p15 (INK4b) through the TGFb1 signaling pathway in human esophageal squamous cell carcinoma. Cell. Immunol. 2014;289:91–96. doi: 10.1016/j.cellimm.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Zhang E.B., Kong R., Yin D.D., You L.H., Sun M., Han L., Xu T.P., Xia R., Yang J.S., De W., et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meseure D., Vacher S., Drak Alsibai K., Nicolas A., Chemlali W., Caly M., Lidereau R., Pasmant E., Callens C., Bieche I. Expression of ANRIL-Polycomb Complexes-CDKN2A/B/ARF Genes in Breast Tumors: Identification of a Two-Gene (EZH2/CBX7) Signature with Independent Prognostic Value. Mol. Cancer Res. 2016;14:623–633. doi: 10.1158/1541-7786.MCR-15-0418. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Yang T., Xu Z., Cao X. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed. Pharmacother. 2016;82:406–412. doi: 10.1016/j.biopha.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 17.De los Campos G., Gianola D., Allison D.B. Predicting genetic predisposition in humans: The promise of whole-genome markers. Nat. Rev. Genet. 2010;11:880–886. doi: 10.1038/nrg2898. [DOI] [PubMed] [Google Scholar]
- 18.Pasmant E., Sabbagh A., Masliah-Planchon J., Ortonne N., Laurendeau I., Melin L., Ferkal S., Hernandez L., Leroy K., Valeyrie-Allanore L., et al. Role of noncoding RNA ANRIL in genesis of plexiform neurofibromas in neurofibromatosis type 1. J. Natl. Cancer Inst. 2011;103:1713–1722. doi: 10.1093/jnci/djr416. [DOI] [PubMed] [Google Scholar]
- 19.Xie H., Rachakonda P.S., Heidenreich B., Nagore E., Sucker A., Hemminki K., Schadendorf D., Kumar R. Mapping of deletion breakpoints at the CDKN2A locus in melanoma: Detection of MTAP-ANRIL fusion transcripts. Oncotarget. 2016;7:16490–16504. doi: 10.18632/oncotarget.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan G., Mathur R., Hu X., Liu Y., Zhang X., Peng G., Lu X. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013;25:1086–1095. doi: 10.1016/j.cellsig.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S., Zhang J.Q., Chen J.Z., Chen H.X., Qiu F.N., Yan M.L., Chen Y.L., Peng C.H., Tian Y.F., Wang Y.D. The over expression of long non-coding RNA ANRIL promotes epithelial-mesenchymal transition by activating the ATM-E2F1 signaling pathway in pancreatic cancer: An in vivo and in vitro study. Int. J. Biol. Macromol. 2017;102:718–728. doi: 10.1016/j.ijbiomac.2017.03.123. [DOI] [PubMed] [Google Scholar]
- 22.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidiniumthiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 23.Meseure D., Vacher S., Lallemand F., Alsibai K.D., Hatem R., Chemlali W., Nicolas A., De Koning L., Pasmant E., Callens C., et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer. 2016;114:1395–1404. doi: 10.1038/bjc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Congrains A., Kamide K., Katsuya T., Yasuda O., Oguro R., Yamamoto K., Ohishi M., Rakugi H. CVD-associated non-coding RNA, ANRIL, modulates expression of atherogenic pathways in VSMC. Biochem. Biophys. Res. Commun. 2012;419:612–616. doi: 10.1016/j.bbrc.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 26.Motterle A., Pu X., Wood H., Xiao Q., Gor S., Liang Ng F., Chan K., Cross F., Shohreh B., Poston R.N., et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum. Mol. Genet. 2012;21:4021–4029. doi: 10.1093/hmg/dds224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson A.D., Hwang S.J., Voorman A., Morrison A., Peloso G.M., Hsu Y.H., Thanassoulis G., Newton-Cheh C., Rogers I.S., Hoffmann U., et al. Resequencing and clinical associations of the 9p21.3 region: A comprehensive investigation in the Framingham heart study. Circulation. 2013;127:799–810. doi: 10.1161/CIRCULATIONAHA.112.111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong Y., Sharma R.B., Nwosu B.U., Alonso L.C. Islet biology, the CDKN2A/B locus and type 2 diabetes risk. Diabetologia. 2016;59:1579–1593. doi: 10.1007/s00125-016-3967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gan Y., Ma W., Wang X., Qiao J., Zhang B., Cui C., Liu Z., Deng D. Coordinated transcription of ANRIL and P16 genes is silenced by P16 DNA methylation. Chin. J. Cancer Res. 2018;30:93–103. doi: 10.21147/j.issn.1000-9604.2018.01.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lillycrop K., Murray R., Cheong C., Teh A.L., Clarke-Harris R., Barton S., Costello P., Garratt E., Cook E., Titcombe P., et al. ANRIL promoter DNA methylation: A perinatal marker for later adiposity. EBioMedicine. 2017;19:60–72. doi: 10.1016/j.ebiom.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang M., Chen W., Qi F., Xia R., Sun M., Xu T., Yin L., Zhang E.B., De W., Shu Y.Q. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J. Hematol. Oncol. 2015;8:57. doi: 10.1186/s13045-015-0153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y., Zhou X., Xu L., Rong C., Shen C., Bian W. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. OncoTargets Ther. 2016;9:3077–3084. doi: 10.2147/OTT.S102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J.-H., Tang J.-M., Li J., Li X.-W. Upregulation of SOX2-activated lncRNA ANRIL promotes nasopharyngeal carcinoma cell growth. Sci. Rep. 2018;8:3333. doi: 10.1038/s41598-018-21708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holdt L.M., Beutner F., Scholz M., Gielen S., Gäbel G., Bergert H., Schuler G., Thiery J., Teupser D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler. Thromb. Vasc. Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 35.Sethuraman S., Gay L.A., Jain V., Haecker I., Renne R. microRNA dependent and independent deregulation of long non-coding RNAs by an oncogenic herpesvirus. PLoS Pathog. 2017;13:e1006508. doi: 10.1371/journal.ppat.1006508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J.J., Wang D.D., Du C.X., Wang Y. Long noncoding RNA ANRIL promotes cervical cancer development by acting as a sponge of miR-186. Oncol. Res. 2017;26:345–352. doi: 10.3727/096504017X14953948675449. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ma J., Li T., Han X., Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2018;144:205–214. doi: 10.1007/s00432-017-2543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P., Zhang M., Niu Q., Zhang F., Yang Y., Jiang X. Knockdown of long non-coding RNA ANRIL inhibits tumorigenesis in human gastric cancer cells via microRNA-99a-mediated down-regulation of BMI1. Braz. J. Med. Biol. Res. 2018;51:e6839. doi: 10.1590/1414-431x20186839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.