Significance
Chromatin modifications are broadly believed to influence gene expression, and are thought to transmit epigenetic memory of gene expression across mitotic generations. The best available evidence about the fate of nucleosomes during DNA replication is at odds with this intuition. This work studies the biochemistry of chromatin replication in living cells, and demonstrates that nucleosomes can, in fact, transmit epigenetic memory. Furthermore, contrary to published models, nucleosomes were not observed to translocate locally during transcription. This work motivates a substantial reevaluation of published biochemical data about chromatin replication and transcription.
Keywords: chromatin, epigenetics, Saccharomyces, Dpb3, Mcm2
Abstract
Nucleosomes are the fundamental structural unit of chromatin. In addition to stabilizing the DNA polymer, nucleosomes are modified in ways that reflect and affect gene expression in their vicinity. It has long been assumed that nucleosomes can transmit memory of gene expression through their covalent posttranslational modifications. An unproven assumption of this model, which is essential to most models of epigenetic inheritance, is that a nucleosome present at a locus reoccupies the same locus after DNA replication. We tested this assumption by nucleating a synthetic chromatin domain in vivo, in which ∼4 nucleosomes at an arbitrary locus were covalently labeled with biotin. We tracked the fate of labeled nucleosomes through DNA replication, and established that nucleosomes present at a locus remembered their position during DNA replication. The replication-associated histone chaperones Dpb3 and Mcm2 were essential for nucleosome position memory, and in the absence of both Dpb3 and Mcm2 histone chaperone activity, nucleosomes did not remember their position. Using the same approach, we tested the model that transcription results in retrograde transposition of nucleosomes along a transcription unit. We found no evidence of retrograde transposition. Our results suggest that nucleosomes have the capacity to transmit epigenetic memory across mitotic generations with exquisite spatial fidelity.
Eukaryotic chromosomes are composed of nucleosomes which stabilize genomic DNA and participate in gene regulation. Posttranslational modifications of nucleosomes influence and reflect local gene expression (1, 2). Furthermore, nucleosomes are inherited semiconservatively during DNA replication, and each duplicated chromosome carries a complement of nucleosomes, half of which are parental nucleosomes synthesized, deposited, and modified before DNA replication, and half of which are new (3). Because nucleosomes are inherited semiconservatively and because nucleosome posttranslational modifications reflect gene expression, it has been postulated that nucleosomes transmit epigenetic information about gene regulation across generations (4, 5). A requirement of this model is that at least some histones from the proteinaceous core of nucleosomes must be inherited during DNA replication without losing memory of their genomic position.
Although the model that nucleosomes store and transmit information about gene regulation is intuitive and widely accepted, it is directly contradicted by experiments that track nucleosomes through DNA replication in vitro. During replication of chromatinized SV40 in cell extracts and in reconstituted biochemical conditions, nucleosomes do not reoccupy the same DNA sequence after DNA replication that they occupied before DNA replication, suggesting that nucleosomes cannot transmit epigenetic memory through DNA replication (6, 7). To understand whether a nucleosome remembers its position through DNA replication in vivo, we covalently labeled nucleosomes at a defined locus with a biologically benign synthetic chromatin modification, then stopped the deposition of new label and tracked the labeled nucleosomes through DNA replication.
Results
Local, Regulated Biotinylation of Nucleosomes at a Single-Copy TetO Sequence.
We developed fusion proteins that attach the Escherichia coli biotin ligase BirA to the tetracycline repressor TetR. In the same strains, we generated fusion proteins in which each endogenous allele of H3 was fused on its carboxyl terminus to the 15-amino acid AviTag, which can be biotinylated by BirA. We also encoded a single-copy 19 bp Tet operator in the GAL10 coding sequence. Tagged histones were biotinylated only in the presence of BirA, and BirA did not biotinylate native yeast proteins, suggesting that we achieved labeling of tagged histones without affecting biotinylation of endogenously biotinylated proteins (SI Appendix, Fig. S1). Tagged histones did not affect heterochromatin stability, as measured by ability of cells to respond to mating pheromone, a classic measure of heterochromatin integrity and transcriptional regulation in Saccharomyces (SI Appendix, Fig. S2). Thus, the tagged histones recapitulated major features of yeast chromatin biology.
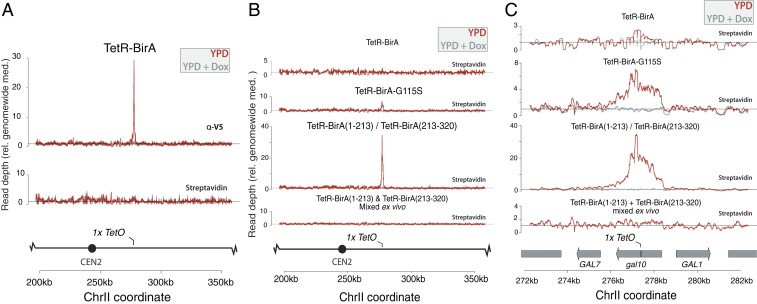
We next asked whether the TetR-BirA fusion protein was correctly localized at the site of the TetO sequence, and whether we could detect enrichment of biotinylated histones at the TetO locus. To ask whether TetR-BirA was recruited to the TetO sequence, we used alleles of TetR-BirA fused to a V5 tag, and performed ChIP-seq on crosslinked chromatin with an anti-V5 agarose resin. In the same extract, we precipitated biotinylated histones with streptavidin-coupled magnetic beads. Although the TetR-BirA protein was recruited to the TetO sequence, high background labeling of H3 genome-wide initially obscured a locus-specific signal (Fig. 1A). To achieve local biotinylation of histones at gal10::TetO, we developed hypomorphic alleles of BirA that reduced background biotinylation without preventing biotinylation at the gal10::TetO locus. We introduced a single-amino acid change in the BirA enzyme to generate a TetR-BirA-G115S allele, which is hypomorphic in E. coli and reduced biotinylation of H3-Avi in yeast (SI Appendix, Fig. S1) (8). The TetR-BirA-G115S fusion protein labeled histones at gal10::TetO ∼7-fold above background with a radius of labeling of ∼4 nucleosomes (Fig. 1 B and C). In addition to developing a hypomorphic allele of BirA, we developed a strategy that relied on a split BirA, in which a portion of BirA (split after position 213) was fused to each of 2 copies of TetR, such that BirA could biotinylate the Avi tag only if the 2 TetR-BirA(s/213) alleles form a heterodimer (9, 10). The split BirA alleles showed reduced bulk histone biotinylation and biotinylated the gal10::TetO locus >30-fold over background, with a radius of labeling of ∼4 nucleosomes (SI Appendix, Fig. S1 and Fig. 1 B and C). Furthermore, growth in medium containing doxycycline, which blocks repressor binding, resulted in no TetR-BirA localization and no local biotin enrichment at the gal10::TetO locus (Fig. 1 B and C). Collectively, these strategies enabled controlled site-specific biotinylation at an edited locus (Fig. 1 B and C).
Fig. 1.
TetR-BirA created a synthetically modified chromatin domain (A) Wild-type TetR-BirA (JRy12392) was enriched at the single-copy gal10::TetO locus in a doxycycline-dependent manner; however, biotinylation was not enriched at the gal10::TetO locus in cells with a wild-type BirA gene. TetR-BirA was tagged with V5, and was detected using ChIP-seq with an anti-V5 agarose resin to precipitate cross-linked chromatin. Biotinylated histones were detected in the same extracts by performing a pulldown with streptavidin-coupled magnetic beads. (B) Comparison of biotinylation signal for alleles of TetR-BirA. TetR-BirA-G115S (JRy12378) showed ∼7-fold labeling over background in haploid cells, TetR-BirA(s/213) (JRy12379) showed ∼35-fold labeling over background in diploid cells. No labeling was detected in mixed extracts expressing either monomer of TetR-BirA(s/213). (C) The same data as (B), plotted at higher resolution. Wild-type TetR-BirA showed minimal biotinylation at gal10::TetO, whereas hypomorphic TetR-BirA-G115S showed ∼7-fold local enrichment for biotinylation and TetR-BirA(s/213) showed ∼35-fold local enrichment for biotinylation at gal10::TetO. In cells bearing either half of the TetR-BirA(s/213) that were mixed ex vivo biotinylation is not observed at gal10::TetO. In each case, the extent of labeling was ∼4 nucleosomes adjacent the TetO sequence, and biotinylation did not accumulate at gal10::TetO if cells are cultured in doxycycline.
TetR-BirA Showed No Specific Off-Target Labeling.
At the gal10::TetO locus, biotinylation was restricted to ∼4 nucleosomes. We asked whether any other loci showed aberrant biotinylation by TetR-BirA. Interestingly, although no other loci were enriched for TetR-BirA binding, a cryptic TetO sequence in the YEL1 promoter was biotinylated in a doxycycline-dependent manner (SI Appendix, Fig. S3). To ask whether site-specific biotinylation could occur in cell extracts ex vivo, we mixed cells expressing either half of the TetR-BirA(s/213) before performing cell lysis and precipitation, using extract from the mixed-cell population, and found no biotinylation at the TetO sequence (Fig. 1 B and C). Thus, locus-specific biotinylation was achieved in vivo.
Labeled Nucleosomes Remembered Their Position through DNA Replication.
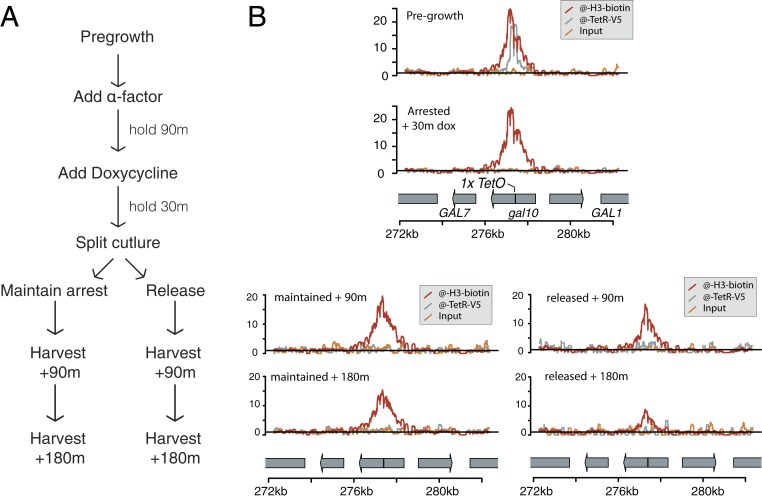
We arrested MATa/mat∆ diploid cells expressing the TetR-BirA(s/213) fusion proteins in G1, using alpha-factor, and then added doxycycline to stop further histone biotinylation at gal10::TetO. We next split the culture and released half the cells into the cell cycle, while holding the other half in G1 (Fig. 2A and SI Appendix, Fig. S4). To isolate the effect of DNA replication on nucleosome position, we cultured strains in glucose medium, in which GAL10 is transcriptionally repressed (11). After a 30-min treatment with doxycycline, TetR-BirA was no longer localized at the TetO sequence, suggesting that there was no new biotinylation of histones at the GAL10 locus after addition of doxycycline (Fig. 2B). After 2 rounds of cell division, the shape of the biotinylated nucleosome density was unchanged, suggesting that histones reoccupied the gal10::TetO locus after DNA replication (Fig. 2B). As expected, the quantitative extent of biotinylation detected at gal10::TetO diminished relative to background biotinylation with each cell division, corresponding to dilution of parentally biotinylated nucleosomes during DNA replication.
Fig. 2.
Histone tracking through DNA replication. (A) Schematic of experimental design. Before the experiment, cells were grown without reaching saturation for at least 8 doublings. (B) For each time and condition, a single sample was harvested, and the purified chromatin was divided and used to precipitate TetR-BirA(s/213) (anti-V5) and biotinylated histone H3-Avi (streptavidin), then sequenced to measure enrichment. Biotinylated histone density did not change in shape, indicating that histones did not move locally along the chromatin fiber; however, the total amount of biotinylated H3-Avi detected (rel. background biotinylation) decreased during an extended G1 arrest or during cell division. Results were representative of 2 biological replicates.
In addition to qualitatively assessing the shape of the nucleosome peak before and after replication, we asked whether nucleosomes rearranged locally by simulating a family of data sets in which we assumed nucleosomes in the arrested sample can each move by 1 position with probability ρ during DNA replication, and then determined which value of ρ results in simulated data that best matched the observed nucleosome positions after a single round of DNA replication. The model ρ = 0, in which nucleosomes do not move during DNA replication, outperformed models in which nucleosomes were assumed to move with ρ > 0 (SI Appendix, Fig. S5). Furthermore, in the absence of DNA replication or transcription, we found that biotinylated histones did not locally reposition at the GAL10 locus (Fig. 2C).
Mcm2 and Dpb3 Were Required for Nucleosome Position-Memory.
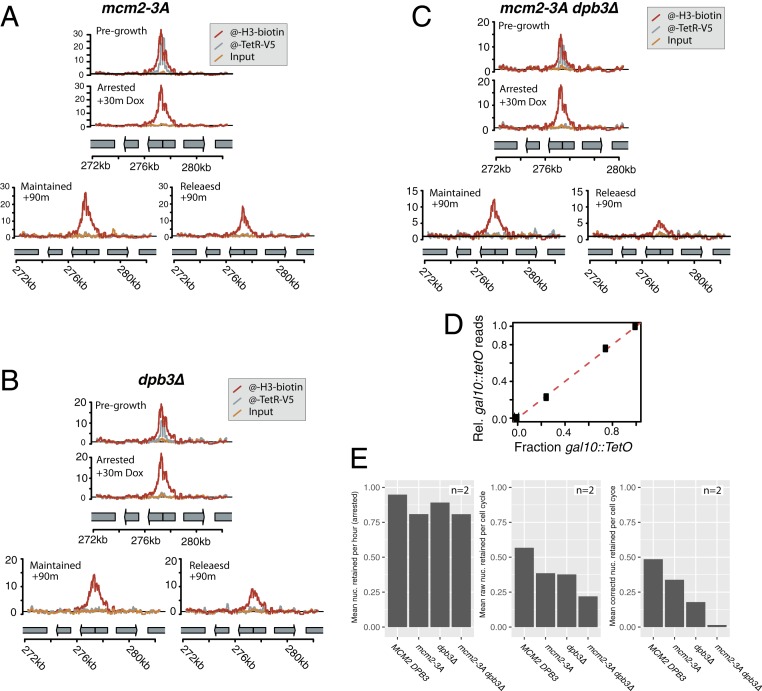
Nucleosome passage across the replication fork is thought to involve chromatin assembly factors including the replication-fork-associated factors Mcm2 and Dpb3, which are each thought to passage parental nucleosomes directly from the parental DNA strand to the lagging and leading strands, respectively (4, 12, 13). We reasoned that if chromatin assembly factors assist in passaging nucleosomes across the replication fork, nucleosomes at the gal10::TetO locus in chromatin assembly mutant strains should show reduced position-memory. In 2 biological replicates, biotinylated histones at the gal10::TetO locus showed no evidence of local movement as a result of DNA replication in dpb3∆ mutant cells, or in mcm2-3A cells, which are deficient in Mcm2 chaperone activity (Fig. 3 and SI Appendix, Figs. S6 and S7). To test whether the number of sequencing reads at the gal10::TetO locus conveyed quantitative information about the fraction of labeled nucleosomes in the experimental cultures, we purified chromatin from samples in which we mixed the experimental TetR-BirA(s/213) gal10::TetO strain in defined ratios with a strain that expressed the split TetR-BirA(s/213) alleles, but did not encode a gal10::TetO target. The number of reads observed that map to the gal10::TetO locus from each mixed culture (per million mapped reads) was directly proportional to the input fraction of cells capable of labeling at gal10::TetO, indicating that the quantitative extent of biotinylation relative to the total number of mapped reads accurately measured the fraction of nucleosomes at the gal10::TetO locus that were biotinylated (Fig. 3D). The total density of labeled nucleosomes retained after replication at the gal10::TetO locus was lower in mcm2-3A and dpb3∆ mutants compared with wild-type, and mcm2-3A dpb3∆ double mutants showed less nucleosome inheritance than either single mutant, consistent with the model that Mcm2 and Dpb3 separately contributed to nucleosome position memory by passaging ancestral histones to the lagging and leading strands, respectively (Fig. 3E) (12, 13). Whereas the density of biotinylated nucleosomes at the gal10::TetO locus decreased through cell division in wild-type and assembly mutant strains, we did not observe any movement of nucleosomes in any of the strains (Fig. 3).
Fig. 3.
Histone tracking through DNA replication in mutants defective in replication-coupled nucleosome chaperone activity. Results plotted are representative of 2 biological replicates. (A) Cells mutant in mcm2-3A (JRy12382) are deficient in lagging-strand nucleosome inheritance; however, there was no shift in nucleosome position among the nucleosomes that were successfully transferred across the replication fork. (B) Cells encoding dpb3∆ (JRy12383) were deficient in leading strand nucleosome inheritance; however, in dpb3∆ mutant cells, the nucleosomes that are inherited during replication were inherited without any local movement of parental nucleosomes. (C) Double-mutant dbp3∆ mcm2-3A (JRy12384) cells showed severely reduced local nucleosome inheritance during DNA replication (D) Nucleosomes were recovered quantitatively from samples that mixed cells encoding gal10::TetO with cells encoding GAL10, indicating that the number of reads (rel. total mapped reads) accurately quantified the number of biotinylated histones at gal10::TetO. The dashed line reflects 1:1 correspondence between the fraction of cells encoding gal10::TetO, and the data are plotted for mixing fractions 0%, 25%, 75%, and 100% gal10::TetO. Both strains in each mixture expressed the TetR-BirA(s/213) monomers. (E) We estimated the rate of nucleosome retention at the gal10::TetO locus by counting the reads whose midpoints map to the 2 nucleosome positions adjacent the gal10::TetO locus. To calculate corrected nucleosome density, we unmixed the raw data to account for cells that did not release from alpha-factor arrest. The data displayed are the average of 2 biological replicates.
Labeled Nucleosomes Did Not Reposition Locally during Transcription.
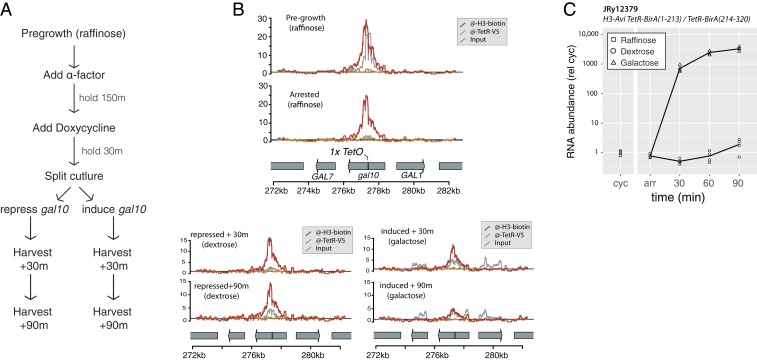
Biochemical analysis of transcription through 227-bp DNA templates results in retrograde nucleosome movement after transcription without dissociation of a nucleosome from DNA (14, 15). On 3-kb templates in vitro, nucleosomes are not displaced by RNA polymerase, and can serve as a ratchet on RNA polymerase to prevent RNA polymerase back-tracking along a DNA template (16). It has been suggested that in vivo, local nucleosome realignment in the wake of RNA polymerase results in concerted retrograde transposition of nucleosomes along a transcription unit during transcription (17). We used the TetR-BirA fusion protein to label H3-avi in the middle of the GAL10 coding sequence to track the position of labeled histones before and after inducing GAL10 transcription (Fig. 4A). Transcription resulted in some loss of labeled nucleosome density from the gal10::TetO locus, consistent with previous observations that transcription evicts some nucleosomes (18–20). However, transcription did not cause any directional nucleosome movement of labeled H3, suggesting that nucleosomes were not moved in either a retrograde or anterograde direction along an ORF during transcription (Fig. 4B and SI Appendix, Fig. S8). We speculate that the reported transcription-dependent movement of nucleosomes along a 227-bp template is constrained in natural chromatin contexts by nucleosomes that occupy upstream DNA, which could prevent propagation of small nucleosome movements in vivo.
Fig. 4.
Histone tracking during transcription. (A) Schematic of the experimental design. Before the experiment, cells were grown without reaching saturation for at least 8 doublings in YP + raffinose. In raffinose, the GAL10 promoter drives low basal transcription. In dextrose, the locus is repressed by Gal80, and in the presence of galactose, transcription is induced 1,000-fold by Gal4. (B) Biotinylated histones did not move locally during transcription; however, biotinylated nucleosome density decreased during transcription (rel. background biotinylation) reflecting removal of nucleosomes during transcription. TetR-BirA (anti-V5) showed ChIP density in induced samples, which reflects a previously characterized ChIP artifact in which highly transcribed genes are enriched in chromatin pulldown fractions (26, 27). The results plotted are representative of 2 biological replicates. (C) Transcription from the gal10::TetO locus was induced >1,000-fold by addition of galactose.
Discussion
Chromatin biochemistry has produced contradictory results with respect to fundamental questions about the physical transactions that occur during DNA replication and RNA transcription. Surprisingly, the published biochemical data are at odds with the field’s conceptual intuition: that chromatin can store and transmit memory (21). In the present study, we designed a synthetic chromatin domain in living cells, and studied the inheritance of the synthetic chromatin domain to understand how the fundamental unit of chromatin, a single nucleosome, is affected during DNA replication and transcription.
To address this question, we developed a strategy to label nucleosomes at a defined locus and then track those nucleosomes through DNA replication. This strategy relied on engineered alleles of the E. coli biotin ligase BirA fused to the DNA-binding TetR protein. Whereas wild-type TetR-BirA did not specifically biotinylate the gal10::TetO target, we developed 2 hypomorphic alleles of TetR-BirA that specifically biotinylated the gal10::TetO target with 7-fold and 35-fold signal relative to background labeling. Whereas ChIP-seq experiments monitoring the location of TetR-BirA did not detect any off-target TetR-BirA binding, streptavidin pulldown sequencing experiments did identify one other TetR-BirA target locus, at the site of a partial TetO sequence. We interpreted this result to mean that the TetR-BirA protein transiently bound the partial TetO sequence, and that the synthetic TetR-BirA-G115S and TetR-BirA(s/213) fusion proteins were capable of converting a transient interaction between TetR and a degenerate TetO sequence into a permanent record of the interaction, despite the interaction being too transient or too weak to be captured by ChIP-seq. This result suggests that naturally occurring nonconsensus transcription factor binding sites might recruit transcription factors transiently, and transient localization might have a biological consequence in some contexts. We propose that hypomorphic BirA fusions to transcription factors or other DNA search proteins might reveal unappreciated transient interactions between transcription factors and DNA that might reflect modes of gene regulation.
In addition to developing a strategy to label a small number of nucleosomes with spatial and temporal control, we applied the technology to understand whether nucleosomes remember their position during DNA replication or transcription. By working at the gal10::TetO locus in Saccharomyces cerevisiae, we could separate the effect of transcription from the effect of replication by growing cells in glucose or galactose to repress or activate transcription as necessary. In the absence of transcription, the histone core particle remembered its position after cell division without detectable local diffusion. Position memory was dependent on the histone chaperones Mcm2 and Dpb3, each of which contributes separately to local nucleosome inheritance. Mutants expressing mcm2-3A or dpb3∆ showed dramatically reduced nucleosome position memory, suggesting that any chromatin-dependent epigenetic process is disrupted in the double mutant. Our results were consistent with the model that Mcm2 and Dpb3 are required to transmit nucleosomes to the lagging and leading strands, respectively, as was suggested based on previous experiments that demonstrated asymmetric inheritance of old histones to the leading and lagging strands in cells expressing mcm2-3A or dpb3∆ (12, 13).
Furthermore, we discovered that during transcription in the absence of DNA replication, labeled H3 proteins could dissociate from their position, but were not translated laterally along the ORF. On the basis of our results, we suggest that previously reported transcription-dependent retrograde transposition of nucleosomes in vitro only occurs over short distances, but is not propagated along an ORF in vivo (14, 15). Whereas previous experiments identified accumulation of older nucleosomes over the 5′ end of ORFs, our data suggest that this phenomenon does not reflect retrograde transposition of nucleosomes, and might instead reflect nonuniform nucleosome turnover along an ORF (17). Alternatively, retrograde movement of nucleosomes could be a rare feature of transcription that only reveals itself when analyzing metagenes that reflect nucleosome occupancy over thousands of ORFs.
Taken together, our data were consistent with the model that the histone core particle can transmit memory across mitotic generations, and is thus a likely vector for epigenetic memory of chromatin state.
Materials and Methods
Yeast Strain Construction.
Strains were constructed using various molecular biology techniques, including standard homology-directed repair or Cas9-mediated homology-directed repair using synthetic gBlocks (IDT) or annealed & extended DNA oligos, as required (22, 23).
Yeast Media, Culture Conditions, and Sample Harvesting.
Yeast were cultured in standard yeast extract peptone (YP) medium with either 2% wt/vol dextrose (YPD) or 2% wt/vol raffinose (YPR), as required for the experiment. Cells were grown in liquid medium shaking at 30 °C.
For DNA replication experiments, cells were grown to a density of 6 × 106 cells/mL in 400 mL YPD, and then arrested with 20 nM alpha-factor. Cell cycle arrest was monitored microscopically, and when >90% of cells were unbudded, each culture was treated with 100 µg/mL doxycycline (Sigma Aldrich D9891) to prevent further TetR localization. After 30 min in doxycycline, each culture was split. Half the culture was treated with an additional 20 nM alpha-factor, and the other half of the culture was treated with 100 µg/mL Streptomyces griseus protease (Sigma Aldrich P5147) to degrade alpha factor and allow reentry into the cell cycle. Next, 1 mL was harvested for flow cytometry, as required for each experiment, and 45 mL of culture corresponding to between 2.7 × 108 and 5.4 × 108 cells were harvested for pulldown experiments.
For transcription-induction experiments, cells were cultured in 400 mL YPR medium overnight to a density of 6 × 106 cells/mL and then arrested with 20 nM alpha-factor. Cell-cycle arrest was monitored microscopically, and when >90% of cells were unbudded, each culture was treated with 100 µg/mL doxycycline (Sigma Aldrich D9891) to prevent further TetR localization. After 30 min in doxycycline, the culture was split. Each half of the culture was treated with YP medium containing either 20% dextrose or 20% galactose, to a final concentration of 2% dextrose or 2% galactose for 90 min. Samples were harvested for flow cytometry and chromatin preparation as described earlier, and samples were harvested for RNA preparation by centrifuging 2 mL culture corresponding to 1.2 × 107 cells, removing the supernatant, and snap-freezing the pellet in liquid nitrogen. Frozen samples were stored at −80 °C until RNA was purified.
Flow Cytometry.
To save samples for flow cytometry, 1 mL culture (∼6 × 106 cells) was pelleted by centrifugation, and the growth medium was removed. Cells were fixed in 80% ethanol and held at 4 °C for up to a week. To perform flow cytometry, 100 µL of fixed cells were transferred to a new tube and pelleted for 2 min at 21,000 rcf. Cells were resuspended in 500 µL 50 mM sodium citrate (pH 7.2) + 0.05% Tween-20 + 10 µg/mL RNase A and incubated overnight at 37 °C. Proteinase K was then added to a final concentration of 20 µg/mL, and cells were incubated at 65 °C for 2 to 4 h. After proteinase K treatment, 500 μL 50 mM sodium citrate (pH 7.2) + 0.05% Tween-20 + 2x SYBR green (Invitrogen; S7563) was added to each sample, and each sample was sonicated at 15% power for 5 s, using a probe sonicator (Branson digital sonifier). Samples were analyzed using a Millipore InCyte benchtop flow cytometer, and data were analyzed and displayed using FlowJo (BD Biosciences). Data were gated to exclude cellular debris and large cell aggregates by gating manually on the interval that extended from ∼1 n inferred DNA content to ∼4 n inferred DNA content.
Immunoblotting.
Cells were grown to a density of ∼6 × 106 cells/mL, and then 5 mL of culture (∼3 × 107 cells) was harvested by centrifugation at 4,200 rcf for 4 min. The cell pellet was resuspended in water and transferred to a 2-mL screw-top tube and then pelleted for 1 min at 21,000 rcf. The supernatant was removed and the pellet was suspended in 1 mL 5% trichloroacetic acid for at least 1 h at 4 °C, then and the cells were pelleted and the supernatant removed. The pellet was resuspended in 2 mL Tris at pH 7.0 and then spun at 21,000 rcf for 1 m and washed with 1 mL acetone. Cells were vortexed thoroughly, although cells do not resuspend well in acetone. Cells were pelleted at 21,000 rcf for 5 min, and the supernatant was carefully removed to avoid disturbing the loose cell pellet. Samples were dried for at least 20 min under vacuum at room temperature to remove residual acetone. After the samples were completely dry, 100 μL acid-washed glass beads and 100 μL protein breaking buffer (5 mM Tris at pH 7.5, 1 mM EDTA, 3 mM DTT) was added to each sample, and samples were disrupted using a FastPrep-24 5G bead beater (MP-Biomedicals) for 40 s at 5 m/s. To each lysate, 50 μL 3x SDS sample buffer was added (188 mM Tris at pH 6.8, 30% glycerol, 150 mM DTT, 6% SDS, 2% beta-mercaptoethanol and bromophenol blue), and samples were incubated in boiling water for 10 min. Boiled samples were cooled on ice and then centrifuged 21,000 rcf for 2 min, and 5 μL of each sample was loaded into a precast Protean Any-KD SDS/PAGE gel (Bio-Rad). The blot was transferred using a TransBlot Turbo (Bio-Rad), then blocked in 10 mL TBS blocking buffer (LiCor Biosciences). The blot was incubated with Abcam ab1791 anti-H3, secondary anti-rabbit 800CW (LiCor 926-32211) and streptaviding-DyLight680 (Thermo Fisher 21848). The immunoblot was imaged using a LiCor infrared fluorescent scanner.
Chromatin Isolation.
To collect samples for pulldown experiments, 45 mL of cells were harvested and fixed in 1% formaldehyde for 15 min at room temperature. Formaldehyde was quenched by adding glycine to 250 mM for 5 min at room temperature. Cells were held on ice until the experiment was complete, and then pelleted and transferred to a 2-mL screw-top tube. Fixed-cell pellets were washed 3 times in 2 mL cold water. Pellets were resuspended in 600 µL FA lysis buffer without EDTA (50 mM Hepes at pH 7.5, 150 mM NaCl, 1% Triton x-100, 0.1% sodium deoxycholate) with 0.1% SDS and 1 mini protease inhibitor tablet (Roche 11836170001) per 15 mL lysis buffer, and 600 µL acid-washed glass beads were added to each tube. Cells were lysed with 4 cycles of 40 s at 10 m/s on a FastPrep-24 5G bead beater (MP-Biomedicals), with 4 min rest on ice after each cycle. After bead-beating, each tube was punctured in the bottom, using a red-hot 18 g hypodermic needle. Each punctured tube was nested inside a 1.5-mL microcentrifuge tube, and was centrifuged for 1 min at 100 rcf to transfer the lysate from the screw-top tube into the microcentrifuge tube. Glass beads were washed once for 1 min with 400 μL FA lysis buffer without EDTA with 0.1% SDS, and centrifuged again at 100 rcf for 1 min to yield 1 mL final lysate. The lysate was centrifuged for 15 min at 18,000 rcf at 4 °C, and the soluble fraction was discarded. The insoluble fraction was washed once (without resuspending) with 1 mL FA lysis buffer without EDTA + 0.1% SDS + protease inhibitors, and then resuspended in 1 mL FA lysis buffer without EDTA + 0.1% SDS + protease inhibitors by sonicating at 30% for 15 s with a probe sonicator, cycling 0.5 s on and then 0.5 s off (Branson digital sonifier). Calcium chloride was added to each tube to a final concentration of 3 mM, and micrococclal nuclease (Worthington Biochemical 9013-53-0) was added at a quantity of 2 units of enzyme per 1.2 × 107 genomes (note this corresponds roughly to 4 units MNase per OD unit of diploid cells). The lysate was incubated 20 min at 37 °C, and then the MNase reaction was stopped by the addition of 10 mM EDTA. The lysate was centrifuged for 15 min at 18,000 rcf at 4 °C. The soluble fraction was moved to a standard microcentrifuge tube, and 450 μL of extract used for pulldown experiments. Input libraries were generated from ∼75 µL of MNase-treated chromatin, corresponding to ∼5 × 107 genomes equivalent of material.
Chromatin Precipitation.
A total of 40 µL (per sample) V5-reactive agarose beads (A7345-1ML) were pelleted by centrifugation at 200 rcf for 1 min and washed once with 500 μL FA lysis buffer + 1 mM EDTA + 0.1% SDS + protease inhibitors + 0.2% BSA (FA-IP buffer). Agarose beads were resuspended in 500 µL FA-IP buffer and mixed with 450 µL MNase-treated chromatin, corresponding to ∼2.4 × 108 genomes, in a 1.5-mL microcentrifuge tube. The slurry was incubated overnight on an end-over-end rotating stand at 4 °C. Agarose beads were collected by centrifugation at 200 rcf for 1 min, and the unbound fraction was aspirated. The bound fraction was washed twice with 1 mL 1x SSC buffer + 0.1% SDS, and twice with 1 mL 0.1x SSC buffer + 0.1% SDS. Each wash was performed at room temperature for 5 min on an end-over-end rotating stand, and beads were collected at each step by centrifugation at 200 rcf for 1 min. The final precipitate was resuspended in 75 µL FA lysis buffer + 10 mM EDTA + 0.1% SDS + protease inhibitors and incubated for 2 h at 70 °C to reverse formaldehyde crosslinks. For streptavidin precipitation, 40 µL (per sample) of streptavidin-coupled Dynabeads MyOne C1 beads (Invitrogen 65001) were separated on a magnet stand and washed once with 500 μL FA lysis buffer + 1 mM EDTA + 0.1% SDS + protease inhibitors + 0.2% BSA (FA-IP buffer). Magnetic beads were resuspended in 500 µL FA-IP buffer and mixed with 450 µL of MNase-treated chromatin, corresponding to ∼2.4 × 108 genomes, in a 1.5-mL microcentrifuge tube. The slurry was incubated overnight on an end-over-end rotating stand at 4 °C. Similar results were obtained with incubation times as short as 1 h, and with bead volumes as little as 10 µL. Magnetic beads were collected on a magnetic separation stand, and the unbound fraction was aspirated. The bound fraction was washed twice with 500 mM LiCl 1 mM EDTA 1% Nonidet P-40 1% NaDOC, and 3 times with TE + 3% SDS. Each wash was performed at room temperature for 5 min on an end-over-end rotating stand, and beads were collected at each step by magnetic separation. The final precipitate was resuspended in 75 µL FA lysis buffer + 10 mM EDTA + 0.1% SDS + protease inhibitors and incubated for 2 h at 70 °C to reverse formaldehyde crosslinks. Each precipitation reaction was cleaned using a Qiagen DNA Clean & Concentrate kit, and then prepared for sequencing.
Library Preparation and Sequencing.
Sequencing libraries were generated using the commercial NEBNext Ultra II DNA library preparation kit (E7645L). Samples were indexed and amplified with 12 cycles of PCR, using i500-series and i700-series indexing primers, and sequenced either with an Illumina MiniSeq or with an Illumina HiSeq4000.
Sequencing Data Manipulation and Presentation.
Raw reads were mapped using Bowtie2 against a SacCer3-derived reference genome that had been edited to include the relevant engineered alleles (24, 25). Only paired-end reads with nondiscordant alignments and an insert size between 150 and 200 bp were mapped. The resulting alignment was analyzed using Python, and visualized using R. To make descriptive figures of chromatin pulldown results, we wrote Python code to integrate all the inferred abundance of each base in the sequenced library (not just the bases mapped to the reference genome). We scaled data by dividing by the genome-wide median coverage for plotting.
To test whether the data were compatible with local nucleosome movement, we permuted the arrested nucleosome midpoint positions by assuming each nucleosome midpoint on the interval ChrII:[276,003: 278,602] moves left or right by 1 nucleosome interval with a probability ρ. We modeled a typical nucleosome interval as a binomial (200,0.81) centered on 162 bp, and we asked which value of ρ resulted in simulated data that minimized the loss function, where the loss function was defined as Euclidean distance between the simulated and observed nucleosome positions in 10 bp nonoverlapping bins along the interval ChrII:[276,003: 278,602] corresponding to the gal10 ORF +/− 250 bp. The result was robust to the bin size between 5 and 45 bp.
To calculate the turnover rate of nucleosomes at the gal10::TetO locus, we computed the midpoint of each read and calculated the number of read midpoints that fell on the interval ChrII:[277050, 277400]. This interval reflected the 2 nucleosome positions closest to the TetO sequence, where biotinylation was strongest. We estimated the sequencing background for each sample as the average of 10,000 permutation tests, in which we sampled 350 continuous bases randomly from the genome and calculated the number of read midpoints that occur over each 350-bp interval. We corrected for background by subtracting each sample’s calculated background from the signal at the GAL10 locus and divided each value by the density of the corresponding arrested sample, immediately before the culture was split by condition. Because the cell-cycle arrest and release were not 100% efficient, we unmixed the signals in cycling conditions by subtracting the signal inferred to have originated from cells that did not re-enter the cell cycle. We did this by solving the formula (observed density) = (fraction G1-stuck) × (arrested density at the corresponding time) + 2 × (fraction G2/M stuck) × (arrested density at the corresponding time) + (fraction correctly cycling) × (cycling density). Sequencing data and processed data files are available on NCBI as GSE135102.
RNA Preparation and RT-qPCR.
A total of 5 mL of cells corresponding to 3 × 107 cells were harvested at each point, and pellets were frozen in liquid nitrogen and then stored at −80 °C for RNA RT-qPCR. RNA was purified using an RNeasy kit, with in-matrix DNase digestion (Qiagen). For each sample, 2 µg total RNA was reversed transcribed using anchored oligo-dT primer and SuperScript III reverse transcriptase (Invitrogen 18080093). cDNA was quantified by quantitative PCR using Dynamo HS SYBR Green master mix (Thermo F410L), with primers specific for the gal10::TetO mRNA (F: GGC TAT GAA AAT GAG GAA GGG; R: CAC AGT ATA CTG TAT GGT TAC C) or for ALG9 (F: CGT TGC CAT GTT GTT GTA TG; R: GCC AGC CTA GTA TAC TAG CC). cDNA abundance was normalized to the same-sample ALG9 abundance and was plotted using R and GGplot2.
Supplementary Material
Acknowledgments
We thank Davis Goodnight for performing early proof of concept experiments, Deborah Thurtle-Schmidt for early experimental design advice and the entire Rine laboratory for spirited discussions. We also thank Doug Koshland and Xavier Darzacq for technical advice and for providing critical feedback on the manuscript. This work was supported by NIGMS 5R01-GM031105-37 and NIGMS 5R01GM120374-03.
Footnotes
The authors declare no conflict of interest.
Data deposition: All raw DNA sequencing reads and bedGraphs of the mapped and filtered sequencing read coverage have been deposited in the Gene Expression Omnibus (accession no. GSE135102).
See Commentary on page 20254.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911943116/-/DCSupplemental.
References
- 1.Lai W. K. M., Pugh B. F., Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister A. J., Kouzarides T., Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson V., Deposition of newly synthesized histones: Hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry 27, 2109–2120 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Serra-Cardona A., Zhang Z., Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends Biochem. Sci. 43, 136–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufman P. D., Rando O. J., Chromatin as a potential carrier of heritable information. Curr. Opin. Cell Biol. 22, 284–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madamba E. V., Berthet E. B., Francis N. J., Inheritance of histones H3 and H4 during DNA replication in vitro. Cell Rep. 21, 1361–1374 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Gruss C., Wu J., Koller T., Sogo J. M., Disruption of the nucleosomes at the replication fork. EMBO J. 12, 4533–4545 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakravartty V., Cronan J. E., The wing of a winged helix-turn-helix transcription factor organizes the active site of BirA, a bifunctional repressor/ligase. J. Biol. Chem. 288, 36029–36039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eginton C., Cressman W. J., Bachas S., Wade H., Beckett D., Allosteric coupling via distant disorder-to-order transitions. J. Mol. Biol. 427, 1695–1704 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Orth P., Schnappinger D., Hillen W., Saenger W., Hinrichs W., Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7, 215–219 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Malik S., Durairaj G., Bhaumik S. R., Mechanisms of antisense transcription initiation from the 3′ end of the GAL10 coding sequence in vivo. Mol. Cell. Biol. 33, 3549–3567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petryk N., et al. , MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Yu C., et al. , A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Studitsky V. M., Clark D. J., Felsenfeld G., A histone octamer can step around a transcribing polymerase without leaving the template. Cell 76, 371–382 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Studitsky V. M., Kassavetis G. A., Geiduschek E. P., Felsenfeld G., Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science 278, 1960–1963 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Hodges C., Bintu L., Lubkowska L., Kashlev M., Bustamante C., Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325, 626–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radman-Livaja M., et al. , Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 9, e1001075 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dion M. F., et al. , Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Rufiange A., Jacques P.-E., Bhat W., Robert F., Nourani A., Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Lee C. K., Shibata Y., Rao B., Strahl B. D., Lieb J. D., Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36, 900–905 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S., Greally J. M., Epigenetics, cellular memory and gene regulation. Curr. Biol. 26, R644–R648 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Ryan O. W., Poddar S., Cate J. H. D., Crispr–cas9 genome engineering in Saccharomyces cerevisiae cells. Cold Spring Harb. Protoc. 2016 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Amberg D. C., Burke D. J., Strathern J. N., Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, 2005 Edition (Cold Spring Harbor, 2005). [Google Scholar]
- 24.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engel S. R., et al. , The reference genome sequence of Saccharomyces cerevisiae: Then and now. G3 (Bethesda) 4, 389–398 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teytelman L., Thurtle D. M., Rine J., van Oudenaarden A., Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 18602–18607 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park D., Lee Y., Bhupindersingh G., Iyer V. R., Widespread misinterpretable ChIP-seq bias in yeast. PLoS One 8, e83506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.