Abstract
Systemic analysis of stress-induced transcription in the cyanobacterium Synechocystis sp. strain PCC 6803 identifies a number of genes as being induced in response to most abiotic stressors (heat, osmotic, saline, acid stress, strong light, and ultraviolet radiation). Genes for heat-shock proteins (HSPs) are activated by all these stresses and form a group that universally responds to all environmental changes. The functions of universal triggers of stress responses in cyanobacteria can be performed by reactive oxygen species (ROS), in particular H2O2, as well as changes in the redox potential of the components of the photosynthetic electron transport chain. The double mutant of Synechocystis sp. PCC 6803 (katG/tpx, or sll1987/sll0755), which is defective in antioxidant enzymes catalase (KatG) and thioredoxin peroxidase (Tpx), cannot grow in the presence of exogenous hydrogen peroxide (H2O2); and it is extremely sensitive to low concentrations of H2O2, especially under conditions of cold stress. Experiments on this mutant demonstrate that H2O2 is involved in regulation of gene expression that responds to a decrease in ambient temperature, and affects both the perception and the signal transduction of cold stress. In addition, they suggest that formation of ROS largely depends on the physical state of the membranes such as fluidity or viscosity. In cyanobacteria, an increase in membrane turnover leads to a decrease in the formation of ROS and an increase in resistance to cold stress. Therefore: (1) H2O2 is the universal trigger of stress responses in cyanobacterial cells; (2) ROS formation (in particular, H2O2) depends on the physical properties of both cytoplasmic and thylakoid membranes; (3) The destructive effect of H2O2 is reduced by increasing of fluidity of biological membranes.
Keywords: cyanobacteria, membrane fluidity, ROS, stress responses, universal triggers
1. Introduction
Cyanobacteria are oxygenic phototrophic prokaryotes that represent the most ancient group of organisms able to perform oxygenic photosynthesis. Cyanobacterial survival and successful function under stress both depend on their capability to adapt to environmental changes. Recent progress made in cyanobacterial molecular signaling research has led to our improved understanding of both basic mechanisms of perception and transduction of stress signals as well as evolution of certain regulatory systems. Such knowledge is useful for related fields, e.g., antimicrobial drug design or stress research in food and industrial crops. With the growing interest in biofuel production in cyanobacteria, which often involves modification of both genetic structure and growth conditions of natural cyanobacterial strains, the response and adaptation of these organisms to multiple internal and external stress factors is of a special interest.
Earlier studies [1] show that cyanobacterial cellular response to various types of abiotic stress (heat shock, oxidative stress, salt stress, hyperosmotic stress, high-intensity illumination, ultraviolet (UV) irradiation) is universal, and includes a transient decrease of cytoplasm volume [2], a decrease in photosynthetic activity [3], and an increase in respiration [4]. In addition, reverse genetics and transcriptome analysis performed with DNA microarray and RNA-seq technologies, suggest that different types of stress might not only cause similar cellular changes, but also induce expression of the same sets of genes in cyanobacteria [1], or activate multisensory proteins that can perceive abiotic stresses regardless of their nature [5,6]. For example, inactivation of a sensory histidine kinase, Hik33, alters cell responses to all cold, salt, hyperosmotic, and oxidative stresses in the cyanobacterium Synechocystis sp. PCC 6803 (hereafter referred to as Synechocystis) [7]. Another histidine kinase, Hik31, is involved in regulation of autotrophic growth [8], responses to light [9,10], and resistance to divalent cations (Cd2+, Cu2+, and Zn2+) [11,12]. Histidine kinase Hik34 acts as a negative transcriptional regulator during heat shock [13], a positive regulator under salt or hyperosmotic stress, and an autoregulator under oxidative stress [14].
These observations suggest that different stressors may generate some universal intracellular signal(s) that triggers adaptive cell responses. Here we review the attempts to identify those signals by systematic analysis of cyanobacterial stress transcriptomes, and the efforts to evaluate the hypothesis of universal signals that trigger stress responses in cyanobacteria.
Unfortunately, there is a limited set of genome-wide data on other cyanobacterial strains subjected to abiotic stressors. Transcriptomic analyses are limited to some Anabaena and Synechococcus strains subjected to ion or nitrogen limitations. Such nutrient limitation data are incomparable to the data on temperature, light, salt or osmotic stress. Therefore, here we focus on the model strain Synechocystis keeping in mind that the discovered patterns can be attributed to other cyanobacterial and bacterial species.
2. Systemic Analysis of Cyanobacterial Stress Transcriptomes
Synechocystis is a model organism used in cyanobacterial research, especially in the stress response field. In general, optimal growth of Synechocystis occurs at temperatures of 30–35 °C, under continuous illumination of 70–120 μmol quanta m−2·s−1, and final CO2 concentration in bubbling air of 1%–1.5%. Various shifts from the optimal conditions result in stress exposure, under which the gene expression is widely researched.
2.1. Genes Induced by Heat Stress
In Synechocystis, heat stress is caused by exposure to 42–45 °C. Synechocystis cells are able to survive a 24-h incubation at 44 °C, pointing to their high thermal resistance [4].
Heat stress has been shown to transiently induce approximately 80 genes with the induction factor (IF) ≥ 2 within the first 15–30 min [13,15,16] (Table 1). When a cell culture is heat stressed for a longer period, in most cases, no increase in transcription is detectable. Many genes are activated just within few minutes from the beginning of the stress treatment, and then, within 30–60 min, their transcription declines and returns to the baseline level recorded before stress exposure [13,15].
Table 1.
Stress-induced genes of Synechocystis sp. PCC 6803: heat stress and other stressors.
| Abiotic Stressors | Redox | ROS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | Gene | Protein | Function | Heat | Salt | Osmo | Light | UV-B | pH | DBMIB | DCMU | H2O2 |
| slr1285 | hik34 | Hik34 | Sensor histidine kinase Hik34 | + | + | + | + | + | + | + | ||
| sll0306 | sigB | SigB | Sigma factor B of RNA polymerase | + | + | + | + | + | + | + | ||
| slr2075 | groES | GroES | Heat-shock protein (HSP) 10 kDa co-chaperonin | + | + | + | + | + | + | |||
| slr2076 | groEL1 | GroEL1 | HSP 60 kDa chaperonin 1 | + | + | + | + | + | + | |||
| sll0416 | groEL2 | GroEL2 | HSP 60 kDa chaperonin 2 | + | + | + | + | |||||
| sll1514 | hspA | HspA | HSP 17 kDa | + | + | + | + | + | + | + | + | |
| sll0170 | dnaK2 | DnaK2 | HSP 70 kDa | + | + | + | + | + | + | + | + | |
| slr0093 | dnaJ | DnaJ | HSP 40 kDa | + | + | + | + | + | + | + | ||
| sll0430 | htpG | HtpG | HSP 90 kDa | + | + | + | + | + | + | |||
| slr1641 | clpB1 | ClpB1 | HSP 100 kDa chaperone | + | + | + | + | + | + | + | ||
| slr1204 | htrA | HtrA | Serine protease | + | + | + | + | + | + | + | ||
| slr0008 | ctpA | CtpA | C-terminal processing protease | + | + | + | + | + | ||||
| sll1621 | sll1621 | PrxA | Peroxiredoxin | + | + | + | + | + | + | |||
| slr0095 | smtA | SmtA | S-adenosyl methionine methyltransferase | + | + | + | + | + | ||||
| slr1674 | slr1674 | Slr1674 | Thermoprotector protein of PS II | + | + | + | + | + | + | + | ||
| slr1963 | ocpA | OCP | Orange carotenoid-binding protein | + | + | + | + | + | ||||
| slr1512 | sbtA | SbtA | Na-dependent bicarnonate transporter | + | + | + | ||||||
| slr1516 | sodB | SodB | Superoxide dismutase | + | + | + | + | + | + | |||
| sll0528 | sll0528 | Sll0528 | Site-2-protease | + | + | + | + | + | + | + | + | |
| slr1675 | hypA1 | HypA1 | Hydrogenase maturation factor HypA1 | + | + | + | + | + | + | + | + | |
| ssl3044 | ssl3044 | Fdx | Ferredoxin | + | + | + | + | + | + | + | + | |
| slr1687 | nblB1 | NblB1 | Phycobilisome degradation protein | + | + | + | + | + | + | + | + | + |
| sll0939 | sll0939 | Sll0939 | Low pH resistance protein | + | + | + | + | + | + | + | + | |
| slr0967 | slr0967 | Slr0967 | Low pH resistance protein | + | + | + | + | + | + | + | + | |
| slr1686 | slr1686 | Unknown function | + | + | + | + | + | |||||
| slr1603 | slr1603 | Unknown function | + | + | + | + | + | + | ||||
| sll1853 | sll1853 | Unknown function | + | + | + | |||||||
| sll0846 | sll0846 | Unknown function | + | + | + | + | + | + | + | + | ||
| sll1884 | sll1884 | Unknown function | + | + | + | + | + | |||||
| sll1009* | frpC* | FrpC | Membrane Fe-regulated protein | + | ||||||||
| sll1501* | cbiA* | CbiA | Cobalamine biosynthesis protein | + | ||||||||
| sll0441* | sll0441* | Unknown function | + | |||||||||
| sll1892* | sll1892* | Unknown function | + | |||||||||
| slr0670* | slr0670* | Unknown function | + | |||||||||
| sll0982* | sll0982* | Unknown function | + | |||||||||
| slr1127* | slr1127* | Unknown function | + | |||||||||
* Genes induced only by heat stress. ORF—open reading frame. Heat—heat stress (44 °C for 15–20 min); Salt—salt stress (0.5 M NaCl for 15–20 min); Osmo—hyperosmotic stress (0.5 M sorbitol for 15–20 min); Light—change of light intensity (from 20 to 300 µmol quanta/m2 sec for 30 min); UV-B—ultraviolet (UV-B) light for 30 min; pH—low pH ~4.0 (30 min); DBMIB—10 µM 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone (30 min); DCMU—10 µM 3-(3,4-dichlorphenyl)-1,1-dimethylurea (30 min); H2O2—250 µM hydrogen peroxide (20 min).
The list of genes for the heat shock proteins (HSPs) includes hspA, groES, groEL1, groEL2, dnaJ, htpG, dnaK2, and clpB1 (chaperones). These genes demonstrate the highest transcriptional upregulation, together with htrA (protease), sigB and sigD (RNA polymerase σ factors), hik34 (heat-sensing histidine kinase), sodB (superoxide dismutase), and several others. Some of the products of these genes tightly interact with each other, for example, RNA polymerase σ factor B (SigB) is necessary for the normal synthesis of the hspA mRNA and cell survival upon a short-term heat [17,18] or salt [19] stress, and the activity of SigB is controlled by Hik34 [13]. SigD regulates the expression of several genes essential for adaptation to oxidative stress induced by high NaCl concentrations [20], and its activity is controlled by a pair of histidine kinase Hik33 and response regulator Rre31 [19].
Apart from the high temperature, the transcription of HSP genes is induced by hyperosmotic [21] or salt stress [19,22,23], strong light [24], UV-B [25], oxidative stress [14,26], and low pH [27,28]. Only 7 out of 80 genes demonstrate a specific response to heat shock (Table 1, labeled with asterisk). The function of more than a half of genes induced by heat shock is still unknown. Therefore, one cannot assume that HSP genes are specific heat shock responders; since their expression changes after a variety of stress factors, HSPs (and their genes, respectively) are rather considered “general stress proteins” (GSPs) [5,6]. Hence, hereafter we will refer to known HSPs as GSPs.
2.2. Genes Induced by Strong Light and Ultraviolet-B (UV-B)
Similarly to heat shock, strong light (here, a shift from optimal intensity of 20 to 400 μmol photons/m2 s) causes a rapid (within 15–20 min) but transient increase in genes transcription. More than 100 genes are induced with IF ≥ 2, including GSP genes: hspA, clpB1, dnaK2, htpG, groES, groEL1, and groEL2; hli (high light inducible) family; genes for NADPH+ dehydrogenase subunit (ndh); and genes for the components of CO2-concentrating mechanism (ccm), and ribulose-1,5-bisphosphate carboxylase (RuBisCO) [24,29]. Transcription of several genes, such as groESL and groEL2, remains at high level even after several hours of exposure to strong light.
Strong light stimulates production of reactive oxygen species (ROS) which are expected to upregulate expression of genes involved in neutralizing ROS. However, out of several ROS-neutralizing genes only superoxide dismutase gene (sodB) and, to a certain extent, glutathione peroxidase gene (gpx1) are induced by strong light. In contrast, transcription of catalase (katG) and thioredoxin peroxidase (tpx) genes have not been found to change during strong light stress [23,26].
UV-B upregulates nearly 50 genes with IF ≥ 2 [24] including genes for GSPs, a precursor of the D1 protein of PS II (psbA2 и psbA3), proteases involved in D1 maturation and degradation (ctpA и ftsH), nblB for a phycobilisome disassembly protein [28], hik34, sigB, and sigD (Table 1).
2.3. Genes Induced by Salt and Hyperosmotic Stress
Salt stress (0.5 M NaCl for 15-20 min) induces more than 100 genes in Synechocystis with IF > 4 [19,22], including genes for GSPs, such as HtrA protease (htrA), sigB and sigD, and superoxide dismutase (sodB). Specifically, NaCl induces genes for ABC-type transporters (cbiQ, norM, and ycf85), quinone synthesis (menB, menG, and ubiH), biotin (birA) and riboflavin (ribF) metabolism, NAD biosynthesis (nadC), protein acetylation (rimI), protease HhoB, translation initiation factor (infC), and some genes with unknown functions [5,22,23].
Hyperosmotic stress (0.5 M sorbitol for 15–20 min) induces a similar set of non-specific stress-response genes (GSPs, sig, etc.) [21]. Such a uniform stress response is explained by a decrease in a cell volume due to rapid water outflow from the cytoplasm during the initial stage of both stresses [30,31]. Only few genes respond specifically to the sorbitol treatment; these include genes for phosphate (pstS) and nitrate/nitrite (ntrABC) transporters, lipid metabolism (sll0208 and sll1377), periplasmic protease (hhoA) and several genes with unknown functions [5,21,32].
2.4. Genes Induced by Cold Shock and Other Stressors
At the early stages of temperature stress, the genes whose transcription is induced by cold are different from those induced by heat (Figure 1). Expression of about 100 genes (Table 2) increases more than twice after lowering the ambient temperature to 22 °C for 30 min [15,33]. These genes belong to several major operational groups, as summarized in [34]:
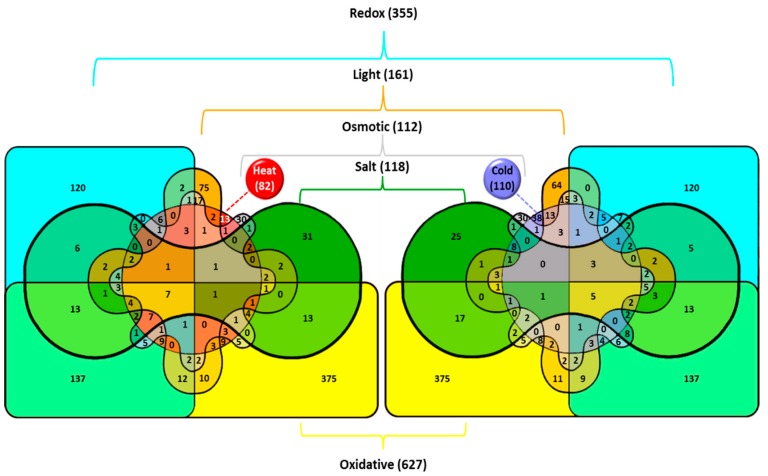
Figure 1.
Venn diagrams illustrating the effects of various stresses on the induction of gene transcription in the cyanobacterium Synechocystis. Total number of genes upregulated in particular stress is shown in parentheses. The number of genes induced by the particular stress or by several stress types is shown without parentheses. Venn diagrams were generated with InteractiVenn at http://www.interactivenn.net. Reproduced from [5] with permission.
Table 2.
Stress-induced genes of Synechocystis sp. PCC 6803: cold stress and other stressors.
| ORF | Gene | Protein | Function | Abiotic stressor | Redox | ROS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cold | Salt | Osmo | Light | UV-B | pH | DBMIB | DCMU | H2O2 | ||||
| sll0790 | hik31 | Hik31 | Two-component sensor histidine kinase | + | + | |||||||
| slr1594 | rre5 | Rre5 | Response regulator | + | + | + | ||||||
| slr0083 | crhR | CrhR | RNA helicase | + | + | + | + | + | ||||
| sll0517 | rbpA1 | RbpA1 | RNA binding protein A1 | + | + | |||||||
| sll0533 | tig | Tig | Ribosome trigger factor | + | + | + | ||||||
| sll1096 | rpsL | Rps12 | 30S ribosomal protein S12 | + | + | + | ||||||
| slr0082 | rimO | RimO | Methyltransferase of ribosomal protein S12 | + | + | + | + | + | ||||
| slr1105 | typA | TypA | GTP-binding protein TypA/BipA | + | + | + | + | + | ||||
| slr1512 | sbtA | SbtA1 | Na-dependent bicarbonate transporter | + | + | + | ||||||
| sll1541 | syc2 | Syc2 | Carotenoid oxygenase | + | + | + | + | + | + | |||
| slr1291 | ndhD2 | NdhD2 | NADH dehydrogenase subunit 4 | + | + | + | + | + | ||||
| ssl2542 | hliA | HliA | High light inducible protein | + | + | + | + | + | + | + | ||
| ssr2595 | hliB | HliB | High light inducible protein | + | + | + | + | + | + | + | + | |
| slr1544 | lilA | LilA | Light-harvesting protein LilA | + | + | + | + | + | + | + | + | + |
| slr1687 | nblB | NblB | Phycocyanobilin lyase NblB | + | + | + | + | + | + | + | + | + |
| sll1742 | nusG | NusG | Transcription antitermination protein | + | + | + | + | + | ||||
| sll1818 | rpoA | RpoA | RNA polymerase alpha subunit σ70 | + | + | + | ||||||
| sll20121 | sigD 1 | SigD1 | RNA polymerase sigma factor SigD | + | + | + | + | + | + | + | + | |
| ssl30441 | ssl3044 1 | Fdx1 | Ferredoxin | + | + | + | + | + | + | + | + | |
| slr1350 | desA | DesA | Δ12 fatty acid desaturase | + | + | |||||||
| sll1441 | desB | DesB | ω3 fatty acid desaturase | + | + | |||||||
| slr1992 | gpx2 | Gpx2 | Hydroperoxy fatty acid reductase | + | + | + | ||||||
| sll1483 | sll1483 | Salt-induced periplasmic protein | + | + | + | + | + | + | + | + | + | |
| sll0157 | sll0157 | Zn-dependent hydrolase | + | + | + | + | + | |||||
| sll1863 | sll1863 | Unknown protein | + | + | + | + | ||||||
| sll1862 | sll1862 | Unknown protein | + | + | + | + | ||||||
| sll1853 | sll1853 | Unknown protein | + | + | + | |||||||
| slr0551 | slr0551 | Unknown protein | + | + | + | + | + | + | ||||
| slr0959 | slr0959 | Unknown protein | + | + | + | + | ||||||
| slr1686 | slr1686 | Unknown protein | + | + | + | + | + | |||||
| slr1687 | slr1687 | Unknown protein | + | + | + | + | + | |||||
ORF—open reading frame; Cold—cold stress (22°C for 30 min); Salt—salt stress (0.5 M NaCl for 15–20 min); Osmo—hyperosmotic stress (0.5 M sorbitol for 15–20 min); Light—strong light (a shift from 20 to 300 μmol photons/m2 s); UV—exposure to UV-B for 30 min; pH—lower pH from 8.0 to 4.0 for 30 min; DBMIB—10 μM 2,5-dibromo-6-isopropyl-3-methyl-1,4-benzoquinone; DCMU—10 μM 3-(3,4-dichlorophenyl)-1,1-dimethylure; H2O2—0.25 mM hydrogen peroxide. Reproduced from [5] with permission.
(1) signal perception and transduction;
regulatory genes for cold stress are limited to two histidine kinases (light-regulated Hik3 [35] and multifunctional Hik31 [8,9,10,11,12]), two genes for the DNA-binding transcriptional regulators (rre5 and sfsA) [36], and a gene for the RNA polymerase σ-factor sigD. Hik31 is activated by Cu2+, released as a result of oxidation of plastocyanin by photosynthetically producing electrons, and triggers the connected regulatory cascade [11,12]. Rre5 is a PatA-subfamily response regulator, which is involved in CO2 uptake and is associated pH homeostasis [34]. Transcriptional factor SfsA regulates sugar catabolism [37], and SigD is involved in light-dependent redox regulation of transcription [18,38].
(2) transcription and translation;
genes from this group encode proteins that prevent issues with transcription and translation caused by cold. For example, a hindrance such as antisense transcription is reduced with the assistance of NusG (a transcriptional terminator) [39], ribosome protein misfolding is prevented by Tig (a chaperone trigger factor), and stalled ribosomes are rescued with the assistance of SmpB [40]. Interestingly, an RNA helicase CrhR from this group participates in redox regulation of the plastoquinone (PQ) pool [41,42].
(3) photosynthesis and respiration—these genes will be discussed further;
(4) cell wall and membrane maintenance;
three of four genes for the fatty acid desaturases (FADs) induced by cold treatment are the key enzymes modulating cold-dependent changes in membrane fluidity. In cyanobacteria, enzymatic dehydrogenation occurs in fatty acids (FAs) of glycerolipids. Lowering of ambient temperature slows down many biosynthetic processes, including synthesis of FA, forcing modification of already existing fatty acids in membrane lipids to adapt to changes in fluidity. Such modification includes desaturation by three fatty acid desaturation enzymes, which add double bonds to FA thus increasing fluidity previously lowered by cold [43]. Evidently, the transcription of genes encoding these FADs (desA for Δ12-desaturase, desB for ω3-desaturase, and desD for Δ6-desaturase) is induced by cold stress [44]. In addition, RbpA3 secures the stability of mRNA of these genes [45].
(5) cofactors and nucleotide metabolism, other metabolic functions;
(6) genes with unspecified functions.
It should be noted that some cold-induced genes strictly depend on light, and are never induced in the dark: hliA and hliB [46,47] ycf39 for a PS II assembly factor [48], lilA [49], and several genes encoding proteins involved in redox-sensitive regulation: ndhD2, pedR and gshB [50,51,52].
While more than 100 genes induce their transcription in response to cold, only 30 of them are cold-specific and do not respond to any other stresses. Together with genes of unspecified functions, this group includes genes that encode proteins involved in transcription and translation (rbpA-family, smpB, rplT, pfbB), methylation (mbpA), and metabolism (cbiF, ams1).
The other two thirds of cold-induced genes react to additional two or more abiotic stresses (Table 2).
The induction of the expression of the same genes by different stressors suggests the existence of some common, universal stress components. In photosynthetic cells or organisms, such components are hypothesized to be related to changes in oxidative state of cellular components. The mechanism of their action would include either electric or chemical signaling, specifically by reactive oxygen species [53].
2.5. Redox Regulation and Oxidative Stress
The redox status of a cell is important for DNA synthesis, gene expression, and enzymatic activity. The redox homeostasis is mainly maintained by thiol-containing molecules such as glutathione, thioredoxins, glutaredoxins, and peroxiredoxins. Stress-induced changes in the redox status of thylakoid membranes directly affect the function of the photosynthetic electron transport chain (ETC).
The majority of the GSP genes are regulated, among other stresses, by the redox status of the plastoquinone (PQ) pool. The status changes as electrons are transferred through the ETC. Such changes are experimentally achieved with the chemicals that interrupt electron flow either upstream (DCMU) or downstream (DBMIB) of the PQ pool. Alternatively, strong light [54] or acidification [27] may alter the QB redox status that affects gene transcription. For example, the reduction of PQ by DBMIB causes the induction of the GSP genes [12].
Expression of GSP genes is also induced by reactive oxygen species (ROS), which are formed during stress and change the oxidative state of a cell. Of all various forms of ROS, summarized in [52], hydrogen peroxide (H2O2) is considered the most important in oxidative stress due to its reasonable lifetime allowing it to perform regulatory functions.
H2O2 in characterized by relatively long lifetime, and is moderately reactive. However, it might get reduced to form hydroxyl radical via a Fenton reaction and, thus, become destructive for cells. Oxidative stress (0.25 mM H2O2) in a liquid culture of Synechocystis induces transcription of ~100 genes with IF ≥ 3 [14], among which there are GSP genes. Similarly to heat and other stresses, ROS also induce genes that are necessary for stress neutralization: genes encoding thioredoxin (trx), peroxiredoxin (aphC), and catalase (katG). H2O2 specifically induces transcription of perR (slr1738) for the transcriptional regulator PerR [38] that is directly regulated by hydrogen peroxide via oxidation of His37 and His91 residues, which leads to dissociation of PerR-DNA complex [55].
Overall analysis of cyanobacterial stress transcriptome suggests that H2O2 and redox changes in ETC (specifically in PQ redox state), commonly present in most abiotic stresses, are the universal triggers of stress responses in photosynthetic bacteria. Further, we discuss experimental evidence to support this hypothesis.
3. The Redox State of Plastoquinone (PQ) Depends on Membrane Fluidity
Temperature fluctuations cause significant changes in membrane fluidity, which in turn (1) activates membrane receptors or (2) affects membrane-related metabolic processes.
Transmembrane receptors that perceive cold stress in bacteria are histidine kinases (Hiks). Hiks sense a change in thickness of the cytoplasmic membrane due to changes in its viscosity [56]. In cyanobacteria, about a half of the cold-sensitive genes are controlled by the red-light-dependent transmembrane histidine kinase Hik33 [47,57], which also partially controls the reactions to hyperosmotic, salt, and oxidative stress [1]. In general, the cold-altered membrane fluidity can be restored via FA desaturation by FADs, a process similar in all eubacteria and plants.
Photosynthesis and respiration are two major metabolic processes located in membranes thus affected by its physical state. Structural and functional organization of photosynthetic apparatus in cyanobacteria is similar to that in higher plants. In cyanobacteria, however, water is not the only electron donor. Some substrates are oxidized by respiratory ETC enzymes, which interact with the photosynthetic ETC via PQ. The redox state of PQ determines the activity of photosynthetic ETC and redistribution of energy between PS II and PS I. Since interaction of PQ with ETC components is a diffusion-driven process, the fluidity of thylakoid membranes is a key factor defining the efficiency of these interactions. In turn, the membrane fluidity depends on the proportion of saturated and (poly)unsaturated FAs that are produced by FADs [43]. X-ray crystallography of cyanobacterial PS II and PS I clearly indicate the presence of a significant amount of lipids in photosynthetic reaction centers that may participate in structuring and regulation of electron transfer [58].
Synechocystis cell culture grown at its optimal temperature (30–35 °C), is able to adapt to cold (20–22 °C) by adjusting its membrane fluidity via induction of several FADs which in turn increase the amount of unsaturated fatty acids in membrane lipids.
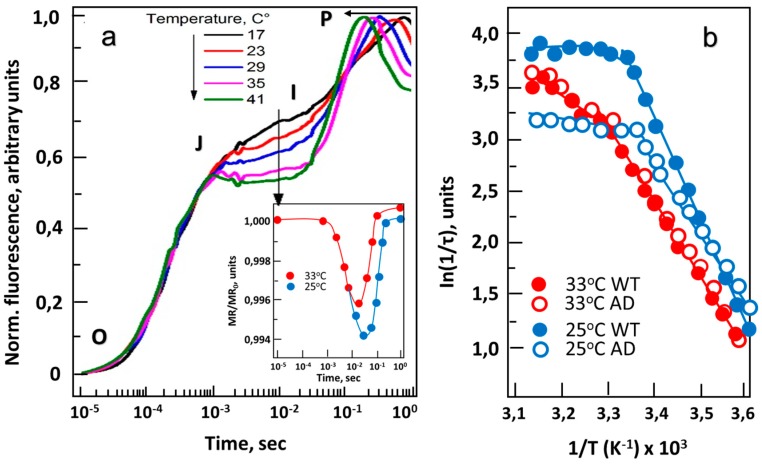
Assuming that differences in membrane fluidity affect the diffusion rate of PQ and, accordingly, the electron transfer rate at the donor side of PS I, one would expect both to change under cold stress. Evidence suggests this hypothesis to be true [59]. In the experiment where redox activity of photosystems was measured at different temperatures using modulated reflection (MR), it was observed that initially (100 µs–10 ms), the reduction rate of P700+ does not depend on temperature in a range of 5–45 °C, but strongly depends on the density of photon flux (Figure 2a), indicating that MR is controlled by photo-activated formation of P700+. However, later (10−2–10 s) the MR signal strongly depends on temperature, but not on the photon flux density (Figure 2a). This means that the reduction of P700+ depends on the rate of oxidation of PQ by cytochrome b6f complex, which is a diffusion-driven process. In addition, during cold treatment at 25 °C, the reduction of P700+ occurs much faster in wild-type (WT) than in mutant cells defective in genes for cold-induced FADs (Figure 2b), since the mutant is not able to compensate for membrane rigidification caused by a decrease in temperature [59].
Figure 2.
The effect of temperature on chlorophyll fluorescence induction in Synechocystis. (a) Wild-type (WT) strain. Characteristic induction curves obtained at 17, 23, 29, 35 and 41 °C; letters O, J, I and P indicate different stages of plastoquinone (PQ) reduction, according to standard nomenclature. Insert panel: the effect of temperature on kinetics of modulated reflection (MR) at 820 nm obtained at 25 and 32 °C. (b) Arrhenius plots of P700+ reduction rates of wild-type strain (WT, solid symbols) and desA−/desD− mutant (AD; open symbols) at 33 °C (red circles) and 25 °C (blue circles). Cell cultures were dark-adapted for 5 min prior to MR measurements. MR changes were induced by array of red (627 ± 10 nm) light-emitting diodes (LEDs) delivering 3000 µmol photons/m2 sec of actinic light to a sample [59].
At optimal temperature of 32 °C, wild type and double-mutant (desA¯/desD¯) Synechocystis with inactivated Δ12- and Δ6-FADs (hereafter – AD cell culture) show similar reduction rates of P700+. However, at 25 °C, AD cell culture had significantly lower reduction rates of P700+ compared to WT cells (Figure 2b), demonstrating the importance of unsaturated FAs for regulating ETC under cold stress [57]. It turns out that at low temperatures, regulation of the redox state of the PQ pool strongly depends on the fluidity of the membrane controlled by cold-induced Δ12- and Δ6-FADs.
The correlation between FAs unsaturation and membrane fluidity have been established by different approaches including differential scanning calorimetry [60], Fourier transform [15] and Raman [59] spectroscopy, measurements of steady-state fluorescence anisotropy with 1,6-diphenyl-1,3,5-hexatriene (DPH) [46] and other lipid-phase intercalating probes. All experimental evidence suggests significant difference in fluidity of thylakoid and cytoplasmic membranes between wild-type and AD cell cultures. Thus, the oxidation rate and diffusion coefficient of PQ, as well as the exchange rate of electrons between the PQ and PS II, all depend on membrane fluidity, which is mainly regulated by the FADs.
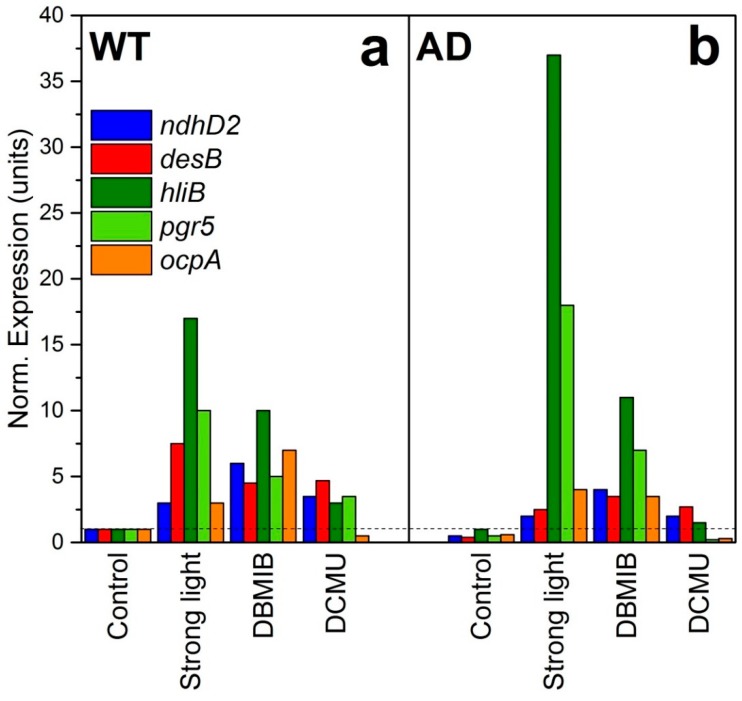
Consequently, fluidity-dependent changes in redox potential of PQ should trigger cellular responses to cold stress. To confirm this hypothesis, redox changes in PQ pool have been stimulated at optimal growth temperature by the ETC inhibitors, such as 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU or diuron, an oxidizer of PQ) and dibromotiminoquinone (DBMIB—the reducer of PQ). Diuron blocks the transfer of electrons from PS II, which leads to the oxidation of the following ETC components, including PQ. DBMIB acts in the region of the Qo site of cytochrome b6f and inhibits electron transfer to Cyt b6f, thus, reducing PQ. The experimental treatments with DCMU and DBMIB were conducted in WT and AD cells (with rigidified membranes) grown at optimal temperature (32 °C). Simultaneously, expression of several cold-responsive genes, including ndhD2 (NADH-dehydrogenase) and desB (terminal ω3-FAD) has been measured [59]. The expression of these two genes is controlled by Hik33 and also depends on both light and membrane fluidity. It was found, that at optimal temperature of 33 °C, the ndhD2 gene is induced by strong light and DCMU in both WT and AD cell cultures, but induction in AD is markedly lower. Treatment with DBMIB induced expression of ndhD2 and desB to rather a similar extent in both WT and AD cells. The results suggest that redox-dependent transcription of these two genes is less sensitive to PQ oxidation by strong light and DCMU in cells with rigidified membranes, whereas DBMIB-induced PQ reduction does not produce a significant difference between these two strains (Figure 3).
Figure 3.
Gene expression in WT (A) and desA−/desD− (=AD) (B) cells grown at 32°C under illumination of 70 µmol photons/m2 sec and exposed for 30 min to strong high light (2500 µmol photons/m2 sec), or treated with 10 µM 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) or 10 µM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). Reproduced from [59] with permission.
Taking into consideration that cold-treated WT cells are able to compensate for the PS II dependent reduction of PQ and maintain the H+ gradient via the respiratory ETC, while AD mutant cells are not, one may expect the relevant differences in cellular stress responses.
Besides ndhD2 and desB, expression of other stress-responsive genes was affected by PQ redox state. HliB and pgr5 genes were highly induced in AD cells, and their transcription was regulated by PQ reduction (Figure 3). Both are shown to be induced by strong light and cold shock; HliB is associated with PSI trimers and protects it under stress conditions [46], and pgr5 encodes an analog of a higher plants proton gradient regulation 5 protein (Prg5) with a controversial function [61]. Expression of another gene, ocpA, was induced by DBMIB, but to lower extent in AD cells to compare to WT cells. Orange carotenoid protein (OCP) participates in non-photochemical quenching in cyanobacterial cells. Overall, the evidence suggests that PQ reduction affects the transcription of genes related to stress regulation (Figure 3).
Apparently, oxidation and reduction of plastoquinone caused by changes in temperature-dependent fluidity of membranes affect transcription of a vast number of stress genes. The redox state of plastoquinone must work as a key signal turning on the expression of FADs and consequent desaturation of fatty acids in membranes. Change of PQ oxidative state is a good candidate for a role of a common component connecting light, cold or other stresses.
An alternative experiment, where instead of membrane rigidification via targeted FAD mutations, various alcohols were applied to cells as membrane “fluidizers”, was also performed earlier [62]. Expression of GSP and FAD genes expression was measured. Hexanol (hexan-1-ol, C6) was found to have the strongest fluidizing effect on membranes among all alcohols (C1-C9) studied. It was found that the effectiveness of hexanol correlates with both duration of exposure and concentration of alcohol in the environment. The spectrum of hexanol-inducible genes generally coincides with the spectrum of genes induced by aromatic benzyl alcohol or heat stress [62]. This suggests again [63,64] that a change in the membrane fluidity may serve as a primary signal for triggering stress responses, probably through a change in the redox potential of the quinone pool. This, in turn, may lead to emergence of long-lived ROS in the form of hydrogen peroxide, which can serve as a universal chemical trigger for launching stress-protective systems.
4. H2O2 is Involved in Regulation of Cold Stress Responses
To test the hypothesis on H2O2 as a universal trigger of stress responses, a double-mutant of Synechocystis, simultaneously defective in two antioxidant enzymes, catalase-peroxidase (KatG) and thioredoxin-peroxidase (Tpx) [65] was used to study transcription under oxidative stress [66]. The mutant was necessary to perform such analysis since due to a rapid detoxification of peroxide under normal conditions, its effect is not traceable in the wild-type cyanobacterium [66].
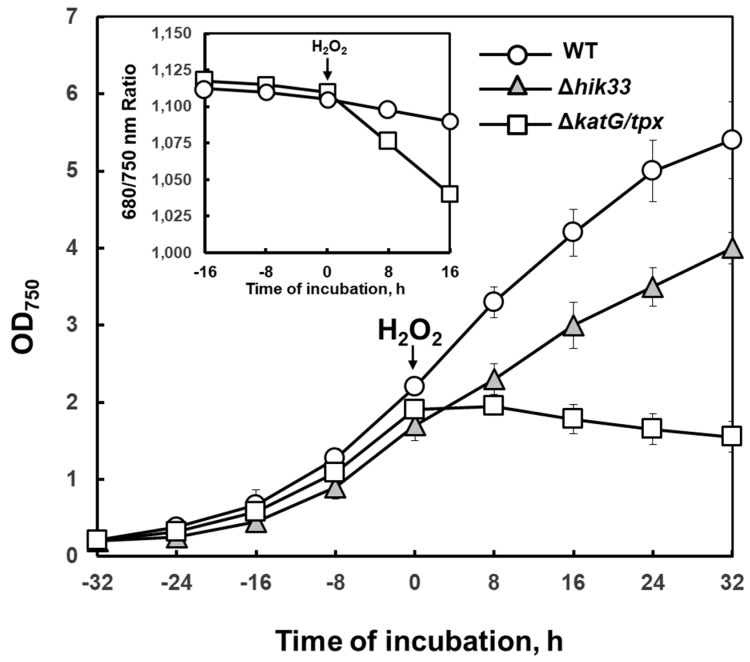
It was shown that such mutant has a delayed detoxification mechanism, which gives plenty of time to detect changes in gene expression. Unlike WT, mutant cells cannot grow in the presence of exogenous H2O2. The katG/tpx mutant is extremely sensitive to low concentrations of H2O2, especially under low temperature stress (Figure 4).
Figure 4.
The effect of exogenous hydrogen peroxide on Synechocystis growth at 32 °C. Point 0: H2O2 was added to a final concentration of 0.25 mM. Optical density measured at 750 nm reflects cell growth. Insert panel: ratio of absorbance at 680/750 nm, characteristic of the relative content of chlorophyll in cells. Reproduced from [66] with permission.
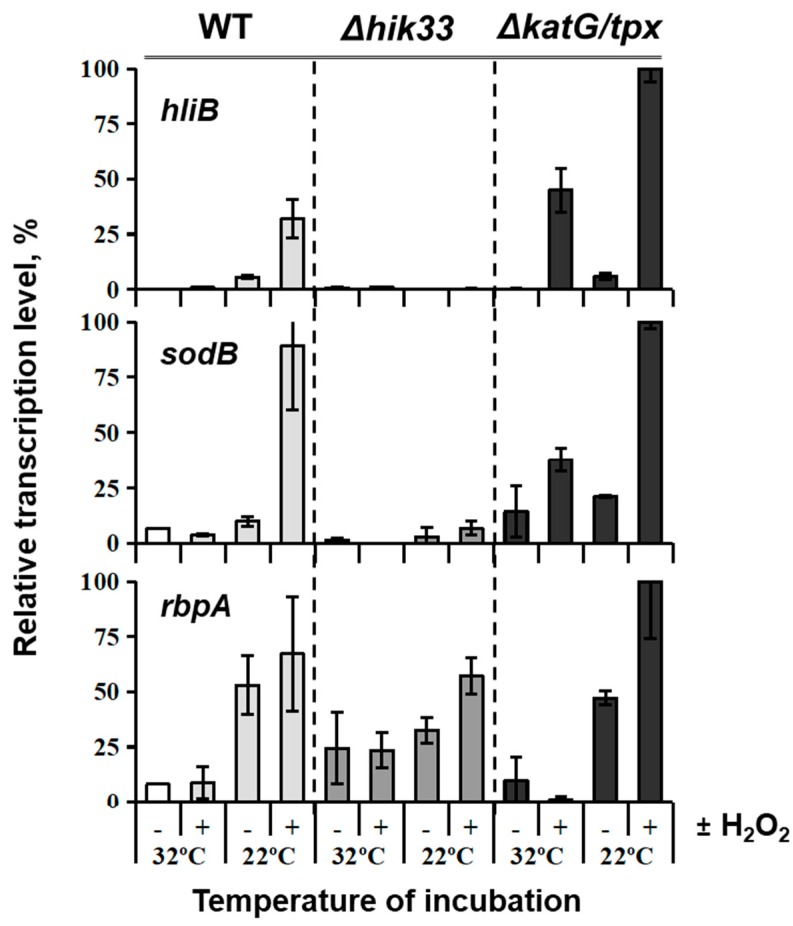
Subjected to both treatments with H2O2 and low temperatures, use of the double-mutant managed to demonstrate that peroxide is involved in regulation of genes induced by cold (Figure 5) in a Hik33-dependent manner [66]. This connection was made based on the fact that cold-inducible Hik33-regulated genes, hliB and sodB, were stimulated by H2O2 in a wild type strain, while in the Δhik33 mutant these genes were not induced by cold anymore despite the presence or absence of hydrogen peroxide. Hik33-independent rbpA, however, served as control in this experiment and responded to cold stress uniformly in both WT and mutant [66]. Apparently, Hik33 is sensitive to ROS such as H2O2, which explains its general involvement in the sensing of stresses where reactive oxygen species are present, with light being the major stimulant of ROS formation.
Figure 5.
Analysis of stress-inducible gene transcription in Synechocystis strains. Wild-type (WT), Δhik33 mutant, and ΔkatG/tpx double mutant strains were grown at 32 °C (control), then exposed to 22 °C for 30 min in the presence (+) or absence (−) of H2O2 at a final concentration of 0.25 mM. Control samples (32 °C) were also subjected to 0.25 mM H2O2. Transcription of Hik33-dependent (hliB, and sodB) and Hik33-independent (rbpA) genes was analyzed. Amounts of transcripts were assessed by qRT-PCR, normalized to those of the housekeeping gene rnpB, and expressed in % of the maximum value of gene expression in all samples [66].
An additional conformation of this statement recently came from experimental studies on RpaB, a cognate response regulator for Hik33, which is shown to interact with ~150 target light-regulated promoters [67]. RpaB is suggested to pair with Hik33 to sense ROS, controlling expression of genes involved in diverse photosynthetic functions, e.g., photoprotection, cyclic electron flow and state transitions, photorespiration, etc. Similarly to Hik33, RpaB controls responses to strong light, cold stress, and iron deficiency [7,66,67].
In Synechocystis, an interaction between H2O2 and either regulatory Hiks [14], transcription factors [50,51,67], or σ-factors of RNA polymerase [68] has been demonstrated. Two important participants in oxidative stress response have been identified as non-essential group 2 σ-factors, SigB and SigD (see Section 2 of this article). Both of them belong to GSPs. Cells that lack SigB and SigD fail to acclimate to high light, singlet oxygen and H2O2, while the overproduction of these σ-factors leads to superior growth of cells under those stress conditions [68].
The question of the exact mechanism of H2O2 action is still being discussed. Presumably, it acts on proteins as an oxidant, affecting their folding and activity [69,70,71], starting multiple protective pathways. Interestingly, Hik33 of Synechocystis does not contain amino acid residues that might be oxidized by H2O2, rather it contains specific light-dependent electron-transport domains PAS and HAMP that might participate in redox-regulated signal transduction [72,73], that are likely to interact with PQ which in turn is sensitive to H2O2.
5. Conclusions
Despite the significant amount of research on cyanobacterial stress response, certain aspects of the latter have remained unclear until the present day. A considerable amount of data on stress-induced transcriptome in Synechocystis suggest that there is no simple relationship between one stress type, one stress sensor and one specific gene or a group of genes changing expression in response. The fact that one sensor can perceive signals from multiple stresses, and that expression of certain groups of genes is not specific to one type of stress, has been puzzling researchers for a long period of time. In certain stresses, such as cold, hyperosmotic and salt stress, a common component whose action results in a comparable cellular response can be identified relatively easily as a decrease in cell volume and rigidification of the membrane. Here, we suggest reactive oxygen species, and specifically H2O2, to be another common component interconnecting several types of abiotic stresses. In summary, we conclude that: (1) H2O2 and redox status of PQ are the universal components of various stresses that trigger similar responses in cyanobacterial cells; (2) ROS formation (in particular, H2O2) depends on the physical properties of both cytoplasmic and thylakoid membranes; (3) the destructive effect of H2O2 is reduced by increasing of fluidity of biological membranes.
Author Contributions
K.S.M. performed the experiments and analyzed the data, M.S. analyzed the data and wrote the article, M.A.S. performed bioinformatic analysis, D.A.L. generated the idea and wrote the article. All authors contributed to data analysis and writing of the manuscript.
Funding
This research was funded by the Russian Science Foundation (grant nos. 19-74-10100 to K.S.M. and 14-24-00020 to D.A.L.).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Los D.A., Zorina A., Sinetova M., Kryazhov S., Mironov K., Zinchenko V.V. Stress sensors and signal transducers in cyanobacteria. Sensors. 2010;10:2386–2415. doi: 10.3390/s100302386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachin D., Nazarenko L.V., Mironov K.S., Pisareva T., Allakhverdiev S.I., Los D.A. Mechanosensitive ion channel MscL controls ionic fluxes during cold and heat stress in Synechocystis. FEMS Microbiol. Lett. 2015;362:fnv090. doi: 10.1093/femsle/fnv090. [DOI] [PubMed] [Google Scholar]
- 3.Los D.A., Mironov K.S., Allakhverdiev S.I. Regulatory role of membrane fluidity in gene expression and physiological functions. Photosynth. Res. 2013;116:489–509. doi: 10.1007/s11120-013-9823-4. [DOI] [PubMed] [Google Scholar]
- 4.Červený J., Sinetova M.A., Zavřel T., Los D.A. Mechanisms of high temperature resistance of Synechocystis sp. PCC 6803: An impact of histidine kinase 34. Life. 2015;5:676–699. doi: 10.3390/life5010676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinetova M.A., Los D.A. Systemic analysis of transcriptomics of Synechocystis: Common stress genes and their universal triggers. Mol. BioSyst. 2016;12:3254–3258. doi: 10.1039/C6MB00551A. [DOI] [PubMed] [Google Scholar]
- 6.Sinetova M.A., Los D.A. Lessons from cyanobacterial transcriptomics: Universal genes and triggers of stress responses. Mol. Biol. (Moscow) 2016;50:685–694. doi: 10.1134/S0026893316040117. [DOI] [PubMed] [Google Scholar]
- 7.Murata N., Los D.A. Histidine kinase Hik33 is an important participant in cold signal transduction in cyanobacteria. Physiol. Plant. 2006;126:17–27. doi: 10.1111/j.1399-3054.2006.00608.x. [DOI] [Google Scholar]
- 8.Kahlon S., Beeri K., Ohkawa H., Hihara Y., Murik O., Suzuki I., Ogawa T., Kaplan A. A putative sensor kinase, Hik31, is involved in the response of Synechocystis sp. strain PCC 6803 to the presence of glucose. Microbiology. 2006;152:647–655. doi: 10.1099/mic.0.28510-0. [DOI] [PubMed] [Google Scholar]
- 9.Nagarajan S., Sherman D.M., Shawhttp I., Sherman L.A. Functions of the duplicated hik31 operons in central metabolism and responses to light, dark, and carbon sources in Synechocystis sp. strain PCC 6803. J. Bacteriol. 2012;194:448–459. doi: 10.1128/JB.06207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagarajan S., Srivastava S., Sherman L.A. Essential role of the plasmid hik31 operon in regulating central metabolism in the dark in Synechocystis sp. PCC 6803. Mol. Microbiol. 2014;91:79–97. doi: 10.1111/mmi.12442. [DOI] [PubMed] [Google Scholar]
- 11.Giner-Lamia J., López-Maury L., Reyes J.C., Florencio F.J. The CopRS two-component system is responsible for resistance to copper in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2012;159:1806–1818. doi: 10.1104/pp.112.200659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giner-Lamia J., López-Maury L., Florencio F.J. Global transcriptional profiles of the copper responses in the cyanobacterium Synechocystis sp. PCC 6803. PLoS ONE. 2014;9:e108912. doi: 10.1371/journal.pone.0108912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki I., Kanesaki Y., Hayashi H., Hall J.J., Simon W.J., Slabas A.R., Murata N. The histidine kinase Hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis. Plant Physiol. 2005;138:1409–1421. doi: 10.1104/pp.104.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanesaki Y., Yamamoto H., Paithoonrangsarid K., Shumskaya M., Suzuki I., Hayashi H., Murata N. Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J. 2007;49:313–324. doi: 10.1111/j.1365-313X.2006.02959.x. [DOI] [PubMed] [Google Scholar]
- 15.Inaba M., Suzuki I., Szalontai B., Kanesaki Y., Los D.A., Hayashi H., Murata N. Genetically-engineered rigidification of membrane lipids enhances the cold inducibility of gene expression in Synechocystis. J. Biol. Chem. 2003;278:12191–12198. doi: 10.1074/jbc.M212204200. [DOI] [PubMed] [Google Scholar]
- 16.Prakash J.S.S., Sinetova M., Kupriyanova E., Zorina A., Suzuki I., Murata N., Los D.A. DNA supercoiling regulates the stress-inducible expression of genes in the cyanobacterium. Mol. BioSyst. 2009;5:1904–1912. doi: 10.1039/b903022k. [DOI] [PubMed] [Google Scholar]
- 17.Imamura S., Asayama M., Takahashi H., Tanaka K., Takahashi H., Shirai M. Antagonistic dark/light-induced SigB/SigD, group 2 sigma factors, expression through redox potential and their roles in cyanobacteria. FEBS Lett. 2003;554:357–362. doi: 10.1016/S0014-5793(03)01188-8. [DOI] [PubMed] [Google Scholar]
- 18.Tuominen I., Pollari M., Tyystjärvi E., Tyystjärvi T. The SigB sigma factor mediates high-temperature responses in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 2006;580:319–323. doi: 10.1016/j.febslet.2005.11.082. [DOI] [PubMed] [Google Scholar]
- 19.Shumskaya M.A., Paithoonrangsarid K., Kanesaki Y., Los D.A., Zinchenko V.V., Tanticharoen M., Suzuki I., Murata N. Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in Synechocystis. J. Biol. Chem. 2005;280:21531–21538. doi: 10.1074/jbc.M412174200. [DOI] [PubMed] [Google Scholar]
- 20.Pollari M., Gunnelius L., Tuominen I., Ruotsalainen V., Tyystjarvi E., Salminen T., Tyystjarvi T. Characterization of single and double inactivation strains reveals new physiological roles for group 2 sigma factors in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2008;147:1994–2005. doi: 10.1104/pp.108.122713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paithoonrangsarid K., Shoumskaya M.A., Kanesaki Y., Satoh S., Tabata S., Los D.A., Zinchenko V.V., Hayashi H., Tanticharoen M., Suzuki I., et al. Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J. Biol. Chem. 2004;279:53078–53086. doi: 10.1074/jbc.M410162200. [DOI] [PubMed] [Google Scholar]
- 22.Marin K., Kanesaki Y., Los D.A., Murata N., Suzuki I., Hagemann M. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 2004;136:3290–3300. doi: 10.1104/pp.104.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagemann M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev. 2011;35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 24.Hihara Y., Kamei A., Kanehisa M., Kaplan A., Ikeuchi M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001;13:793–806. doi: 10.1105/tpc.13.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L., McCluskey M.P., Ni H., LaRossa R.A. Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J. Bacteriol. 2003;184:6845–6858. doi: 10.1128/JB.184.24.6845-6858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hihara Y., Sonoike K., Kanehisa M., Ikeuchi M. DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 2003;185:1719–1725. doi: 10.1128/JB.185.5.1719-1725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohta H., Shibata Y., Haseyama Y., Yoshino Y., Suzuki T., Kagasawa T., Kamei A., Ikeuchi M., Enami I. Identification of genes expressed in response to acid stress in Synechocystis sp. PCC 6803 using DNA microarrays. Photosynth. Res. 2005;84:225–230. doi: 10.1007/s11120-004-7761-x. [DOI] [PubMed] [Google Scholar]
- 28.Uchiyama J., Asakura R., Kimura M., Moriyama A., Tahara H., Kobayashi Y., Kubo Y., Yoshihara T., Ohta H. Slr0967 and Sll0939 induced by the SphR response regulator in Synechocystis sp. PCC 6803 are essential for growth under acid stress conditions. Biochim. Biophys. Acta. 2012;1817:1270–1276. doi: 10.1016/j.bbabio.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Hsiao H.Y., He Q., Van Waasbergen L.G., Grossman A.R. Control of photosynthetic and high-light-responsive genes by the histidine kinase DspA: Negative and positive regulation and interactions between signal transduction pathways. J. Bacteriol. 2004;186:3882–3888. doi: 10.1128/JB.186.12.3882-3888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiguzov A., Lyukevich A.A., Allakhverdiev S.I., Sergeyenko T.V., Suzuki I., Murata N., Los D.A. Osmotic shrinkage of cells of Synechocystis sp. PCC 6803 by water efflux via aquaporins regulates osmostress-inducible gene expression. Microbiology. 2005;151:447–455. doi: 10.1099/mic.0.27530-0. [DOI] [PubMed] [Google Scholar]
- 31.Sinetova M.A., Mironov K.S., Mustardy L., Shapiguzov A., Bachin D., Allakhverdiev S.I., Los D.A. Aquaporin-deficient mutant of Synechocystis is sensitive to salt and high-light stress. J. Photochem. Photobiol. B. Biol. 2015;152:377–382. doi: 10.1016/j.jphotobiol.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Kanesaki Y., Suzuki I., Allakhverdiev S.I., Mikami K., Murata N. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 2002;290:339–348. doi: 10.1006/bbrc.2001.6201. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki I., Kanesaki Y., Mikami K., Kanehisa M., Murata N. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 2001;40:235–244. doi: 10.1046/j.1365-2958.2001.02379.x. [DOI] [PubMed] [Google Scholar]
- 34.Sinetova M.A., Los D.A. New insights in cyanobacterial cold stress responses: Genes, sensors, and molecular triggers. Biochim. Biophys. Acta. 2016;1860:2391–2403. doi: 10.1016/j.bbagen.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 35.Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J.A., et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H.L., Postier B.L., Burnap R.L. Alterations in global 5 patterns of gene expression in Synechocystis sp. PCC 6803 in response to 6 inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004;279:5739–5751. doi: 10.1074/jbc.M311336200. [DOI] [PubMed] [Google Scholar]
- 37.Steczkiewicz K., Muszewska A., Knizewski L., Rychlewski L., Ginalski1 K. Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 2012;40:7016–7045. doi: 10.1093/nar/gks382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., Singh A.K., McIntyre L.M., Sherman L.A. Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 2004;186:3331–3345. doi: 10.1128/JB.186.11.3331-3345.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peters J.M., Mooney R.A., Grass J.A., Jessen E.D., Tran F., Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–2633. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karzai A.W., Susskind M.M., Sauer R.T. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sireesha K., Radharani B., Krishna P.S., Sreedhar N., Subramanyam R., Mohanty P., Prakash J.S. RNA helicase, CrhR is indispensable for the energy redistribution and the regulation of photosystem stoichiometry at low temperature in Synechocystis sp. PCC6803. Biochim. Biophys. Acta. 2012;1817:1525–1536. doi: 10.1016/j.bbabio.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Georg J., Rosana A.R.R., Chamot D., Migur A., Hess W.R., Owttrim G.W. Inactivation of the RNA helicase CrhR impacts a specific subset of the transcriptome in the cyanobacterium Synechocystis sp. PCC 6803. RNA Biol. 2019;24:1–10. doi: 10.1080/15476286.2019.1621622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Los D.A., Murata N. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta. 1998;1394:3–15. doi: 10.1016/S0005-2760(98)00091-5. [DOI] [PubMed] [Google Scholar]
- 44.Los D.A., Ray M.K., Murata N. Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol. Microbiol. 1997;25:1167–1175. doi: 10.1046/j.1365-2958.1997.5641912.x. [DOI] [PubMed] [Google Scholar]
- 45.Tang Q., Tan X., Xu X. Effects of a type-II RNA-binding protein on fatty acid composition in Synechocystis sp. PCC 6803. Chin. Sci. Bull. 2010;55:2416–2421. doi: 10.1007/s11434-010-3254-9. [DOI] [Google Scholar]
- 46.He Q., Dolganov N., Bjorkman O., Grossman A.R. The high light-inducible polypeptides in Synechocystis PCC6803. Expression and function in high light. J. Biol. Chem. 2001;276:306–314. doi: 10.1074/jbc.M008686200. [DOI] [PubMed] [Google Scholar]
- 47.Mironov K.S., Sidorov R.A., Trofimova M.S., Bedbenov V.S., Tsydendambaev V.D., Allakhverdiev S.I., Los D.A. Light-dependent cold-induced fatty acid unsaturation, changes in membrane fluidity, and alterations in gene expression in Synechocystis. Biochim. Biophys. Acta. 2012;1817:1352–1359. doi: 10.1016/j.bbabio.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 48.Chidgey J.W., Linhartová M., Komenda J., Jackson P.J., Dickman M.J., Canniffe D.P., Koník P., Pilný J., Hunter C.N., Sobotka R. A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell. 2014;26:1267–1279. doi: 10.1105/tpc.114.124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kufryk G., Hernandez-Prieto M.A., Kieselbach T., Miranda H., Vermaas W., Funk C. Association of small CAB-like proteins (SCPs) of Synechocystis sp. PCC 6803 with photosystem II. Photosynth. Res. 2008;95:135–145. doi: 10.1007/s11120-007-9244-3. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura K., Hihara Y. Photon flux density-dependent gene expression in Synechocystis sp. PCC 6803 is regulated by a small redox-responsive, LuxR-type regulator. J. Biol. Chem. 2006;281:36758–36766. doi: 10.1074/jbc.M606797200. [DOI] [PubMed] [Google Scholar]
- 51.Horiuchi M., Nakamura K., Kojima K., Nishiyama Y., Hatakeyama W., Hisabori T., Hihara Y. The PedR transcriptional regulator interacts with thioredoxin to connect photosynthesis with gene expression in cyanobacteria. Biochem. J. 2010;431:135–140. doi: 10.1042/BJ20100789. [DOI] [PubMed] [Google Scholar]
- 52.Cameron J.C., Pakrasi H.B. Essential role of glutathione in acclimation to environmental and redox perturbations in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2010;154:1672–1685. doi: 10.1104/pp.110.162990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmitt F.-J., Renger G., Friedrich T., Kreslavski V.D., Zharmukhamedov S.K., Los D.A., Kuznetsov V.V., Allakhverdiev S.I. Reactive oxygen species: Re-evaluation of generation, monitoring and role in stress-signaling in phototrophic organisms. Biochim. Biophys. Acta. 2014;1837:835–848. doi: 10.1016/j.bbabio.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Singh A.K., Li H., Sherman L.A. Microarray analysis and redox control of gene expression in the cyanobacterium Synechocystis sp. PCC 6803. Physiol. Plant. 2004;120:27–35. doi: 10.1111/j.0031-9317.2004.0232.x. [DOI] [PubMed] [Google Scholar]
- 55.Lee J.W., Helmann J.D. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 56.Saita E., Albanesi D., de Mendoza D. Sensing membrane thickness: Lessons learned from cold stress. Biochim. Biophys. Acta. 2016;1861:837–846. doi: 10.1016/j.bbalip.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Mironov K.S., Sidorov R.A., Kreslavski V.D., Bedbenov V.S., Tsydendambaev V.D., Los D.A. Cold-induced gene expression and ω3 fatty acid unsaturation is controlled by red light in Synechocystis. J. Photochem. Photobiol. B Biol. 2014;137:84–88. doi: 10.1016/j.jphotobiol.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 58.Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 59.Maksimov E.G., Mironov K.S., Trofimova M.S., Nechaeva N.L., Todorenko D.A., Klementiev K.E., Tsoraev G.V., Tyutyaev E.V., Zorina A.A., Feduraev P.V., et al. Membrane fluidity controls redox-regulated cold stress responses in cyanobacteria. Photosynth. Res. 2017;133:215–223. doi: 10.1007/s11120-017-0337-3. [DOI] [PubMed] [Google Scholar]
- 60.Tasaka Y., Gombos Z., Nishiyama Y., Mohanty P., Ohba T., Ohki K., Murata N. Targeted mutagenesis of acyl-lipid desaturases in Synechocystis: Evidence for the important roles of polyunsaturated membrane lipids in growth, respiration and photosynthesis. EMBO J. 1996;15:6416–6425. doi: 10.1002/j.1460-2075.1996.tb01033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allahverdiyeva Y., Mustila H., Ermakova M., Bersanini L., Richaud P., Ajlani G., Battchikova N., Cournac L., Aro E.M. Flavodiiron proteins Flv1 and Flv3 enable cyanobacterial growth and photosynthesis under fluctuating light. Proc. Natl. Acad. Sci. USA. 2013;110:4111–4116. doi: 10.1073/pnas.1221194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mironov K.S., Shumskaya M., Sidorov R.A., Trofimova M.S., Los D.A. Membrane physical state and stress regulation in Synechocystis: Fluidizing alcohols repress fatty acid desaturation. Plant J. 2018;96:1007–1017. doi: 10.1111/tpj.14086. [DOI] [PubMed] [Google Scholar]
- 63.Vigh L., Los D.A., Horváth I., Murata N. The primary signal in the biological perception of temperature: Pd-catalyzed hydrogenation of membrane lipids stimulated the expression of the desA gene in Synechocystis PCC6803. Proc. Natl. Acad. Sci. USA. 1993;90:9090–9094. doi: 10.1073/pnas.90.19.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horváth I., Glatz A., Nakamoto H., Mishkind M.L., Munnik T., Saidi Y., Goloubinoff P., Harwood J.L., Vigh L. Heat shock response in photosynthetic organisms: Membrane and lipid connections. Prog Lipid Res. 2012;51:208–220. doi: 10.1016/j.plipres.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 65.Yamamoto H., Miyake C., Dietz K.J., Tomizawa K., Murata N., Yokota A. Thioredoxin peroxidase in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 1999;447:269–273. doi: 10.1016/S0014-5793(99)00309-9. [DOI] [PubMed] [Google Scholar]
- 66.Fedurayev P.V., Mironov K.S., Gabrielyan D.A., Bedbenov V.S., Zorina A.A., Shumskaya M., Los D.A. Hydrogen peroxide participates in perception and transduction of cold stress signal in Synechocystis. Plant Cell Physiol. 2018;59:1255–1264. doi: 10.1093/pcp/pcy067. [DOI] [PubMed] [Google Scholar]
- 67.Kujirai J., Nanba S., Kadowaki T., Oka Y., Nishiyama Y., Hayashi Y., Arai M., Hihara Y. Interaction of the GntR-family transcription factor Sll1961 with thioredoxin in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2018;8:6666. doi: 10.1038/s41598-018-25077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hakkila K., Valev D., Antal T., Tyystjävi E., Tyystjävi T. Group 2 sigma factors are central regulators of oxidative stress acclimation in cyanobacteria. Plant Cell Physiol. 2019;60:436–447. doi: 10.1093/pcp/pcy221. [DOI] [PubMed] [Google Scholar]
- 69.Marinho H.S., Real C., Cyrne L., Soares H., Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kojima K., Oshita M., Nanjo Y., Kasai K., Tozawa Y., Hayashi H., Nishiyama Y. Oxidation of elongation factor G inhibits the synthesis of the D1 protein of photosystem II. Mol. Microbiol. 2007;65:936–947. doi: 10.1111/j.1365-2958.2007.05836.x. [DOI] [PubMed] [Google Scholar]
- 71.Yutthanasirikul R., Nagano T., Jimbo H., Hihara Y., Kanamori T., Ueda T., Haruyama T., Konno H., Yoshida K., Hisabori T., et al. Oxidation of a cysteine residue in elongation factor EF-Tu reversibly inhibits translation in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2016;291:5860–5870. doi: 10.1074/jbc.M115.706424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khursigara C.M., Wu X., Zhang P., Lefman J., Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. USA. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sporer A.J., Kahl L.J., Price-Whelan A., Dietrich L.E.P. Redox-based regulation of bacterial development and behavior. Annu. Rev. Biochem. 2017;86:777–797. doi: 10.1146/annurev-biochem-061516-044453. [DOI] [PubMed] [Google Scholar]