Abstract
Obligate intracellular bacteria belonging to the genus Anaplasma spp. are responsible for causing a hemolytic disease called anaplasmosis in animals, as well as in humans. This study was aimed at the molecular identification and genetic analysis of responsible causative agents of anaplasmosis beyond those already reported. A survey was performed during July and August 2018 in the Jhang District, Punjab, Pakistan. Four hundred and fifty blood samples from asymptomatic, tick-infested cattle were collected on FTA cards and tested for the Anaplasma spp. presence using nested-polymerase chain reaction (PCR) methods. The 16S ribosomal RNA gene sequences generated from the positive samples were used for genetic analysis of Anaplasma spp. The nested-PCR results showed the presence of two Anaplasma spp. with an overall prevalence rate of 10.44%, where the prevalence of A. bovis and A. phagocytophilum was 7.78% and 2.66%, respectively. The study portrayed new molecular data on the prevalence of Anaplasma spp. in the studied cattle population, indicating a potential threat to the human population as well.
Keywords: Anaplasma bovis, Anaplasma phagocytophilum, nested-PCR, bovine, Pakistan
1. Introduction
Being an agricultural country, the economy of Pakistan gets a real boost from its livestock industry, with a share of 58.92% in the agriculture sector and a contribution of almost 11.11% during the 2017–2018 economic year, as far as total gross domestic product is concerned. In Pakistan, there are currently 84.9 million heads of buffalo and cattle present, sharing 96.80% of the total milk gross production; whereas 96.03% and 50.56% of the total milk and meat, respectively, is consumed by the human population [http://www.finance.gov.pk/survey_1617.html]. Despite being an integral part of the economy, and despite the fact that a major proportion of the human population of Pakistan is involved in the livestock industry, detailed information regarding disease prevalence and its prevention, management practices and control strategies is lacking.
Because of the developing pathogenicity in farm animals and to a lesser extent in people, among other Rickettsiales, the genus Anaplasma demandeds special attention. A variety of the Anaplasma species, which is a gram negative bacteria possessing an obligate intracellular nature, is responsible for causing anaplasmosis, a hemolytic tick transmitted disease, in humans and animals. It has a wide distribution in the temperate, subtropical and tropical regions of the world [1]. The disease is continuously becoming a serious concern for the animal breeding system, as the infection puts an additional burden on veterinary care by reducing the body weight of animals, decreasing milk production, and frequently causing abortions leading to death [2,3,4,5].
The Anaplasma genus is comprised of six species that exhibit versatility in cell tropism and in the preferential selection of hosts [6,7]. The red blood cells of cattle and wild ruminants are chosen by A. centrale and A. marginale as a site of infection, while small ruminants presenting the same cells to be infected are encountered by A. ovis. Anaplasma bovis causing anaplasmosis, targets small mammals and ruminants, which results in the infection of monocytes. Infection is prevalent in different regions of the world with a variable prevalence rate of 3.94 to 39.80% and 9 to 15% in domestic ruminants and wild cervids (Sika deer and Red deer), respectively. The prevalence rate is dependent on the type of species infected and the diagnostic method used [8,9,10,11,12]. Having zoonotic potential, A. phagocytophilum preferentially tends to reside and infect neutrophil granulocytes causing granulocytic anaplasmosis in a range of hosts including horses, ruminants, dogs and humans. The organism has been characterized in different regions including Asia, the Americas, Africa and Europe [13,14,15,16,17,18,19]. Age, immune status and the host’s exposure to tick vectors are attributed to its prevalence in different regions [http://www.cdc.gov/anaplasmosis/]. Unique tropism is shown by the A. platys bacteria appears in dog platelets and is an etiological agent for infectious canine cyclic thrombocytopenia. Out of six species of the Anaplasma genus, five of them specifically look for domestic and wild ruminants to serve as hosts for them [7,13,14,15,16,17,18,19].
From Pakistan, a report is available on the distribution of A. marginale and A. centrale in cattle and buffaloes from one district of Sindh Province using Giemsa’s stained blood smears, but the study lacks sequence analysis [20]. Whereas some reports using molecular diagnostic approaches have also been made. A study using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) has been carried out reporting one Anaplasma spp. (A. marginale) only in two provinces of Pakistan [21]. Recently, a PCR-based investigation reported A. marginale in the northern areas of Pakistan [22]. To date, however, there is no report on the prevalence of A. bovis and A. phagocytophilum in bovine from Pakistan, even with the use of microscopic and molecular diagnostic tools.
PCR has been characterized as the gold standard diagnostic approach for anaplasmosis [23] but it has not been used preferentially as a diagnostic tool in most Anaplasma-related epidemiological studies in Pakistan. This nested-PCR based study reveals the first 16S ribosomal RNA based evidence of the two Anaplasma spp. viz; A. bovis, and a zoonotic pathogen, A. phagocytophilum, in bovine in the Jhang District, Punjab, Pakistan.
2. Results
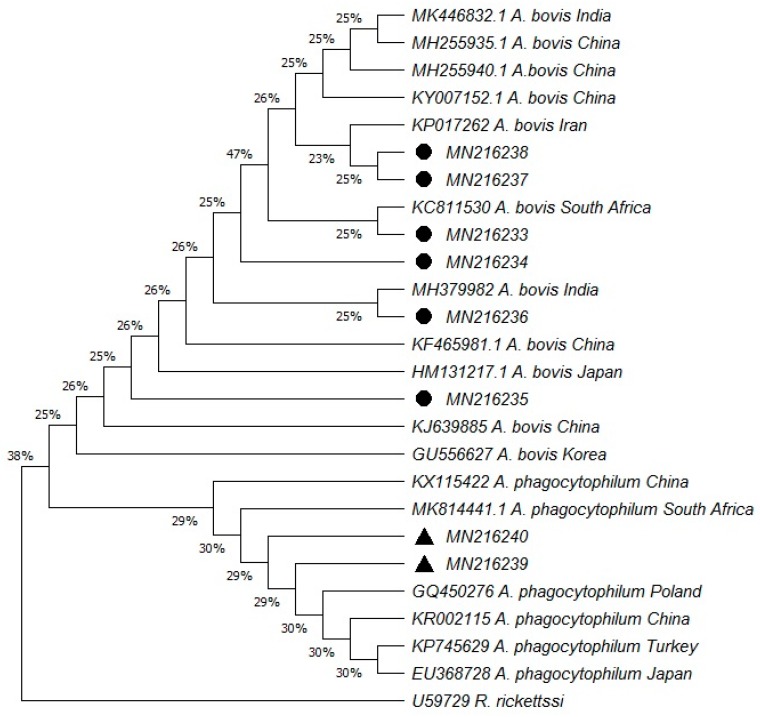
Out of 450 bovine blood samples, 47 samples (10.44%) were positive for Anaplasma infection. The overall prevalence rate observed for A. bovis was higher than that of A. phagocytphilum, which was 7.78% and 2.66%, respectively. Sequencing results of the 16S rRNA gene from the positively detected, randomly selected PCR products confirmed the presence of the Anaplasma infection in the screened samples that were correctly amplified earlier. The tree constructed on the basis of the 16S rRNA gene explains the phylogenetic relationship (Figure 1).
Figure 1.
Phylogenetic analysis of the sequences of the 16S rRNA gene using the neighbor-joining method. The optimal tree with the sum of branch length = 1.36582012 is shown. The evolutionary distances were computed using the maximum composite likelihood method. The pathogens identified in the present study are marked in bold, where circles and triangles indicate A. bovis and A. phagocytophilum, respectively.
Nucleotide sequence accession numbers. Accession numbers received from NCBI GenBank for 16S rRNA gene sequences of A. phagocytophilum are MN216239 and MN216240, while the 16S rRNA gene sequence of A. bovis is MN216233-38.
3. Discussion
Bovine anaplasmosis caused by different Anaplasma spp. is highly endemic in different developing countries [24]. In Pakistan, rural communities commonly fulfill their domestic and commercial needs from small-holder cattle farming systems. However, farmers have also shifted towards commercial dairy farming by adopting modern techniques and importing exotic cattle breeds (Bos taurus). While exotic cattle breeds have a greater milk yield potential, they are also at a higher risk of getting ticks and tick borne infections, with a mortality rate more than double of that compared to local breeds (Bos indicus).
Ticks have been characterized as the major vectors of Anaplasma spp., particularly belonging to the genera Ixodes, Amblyomma, Rhipicephalus and Dermacentor. Susceptibility of the animal population towards Anaplasma infection is attributed with the distribution and infestation of ticks [25]. Although bovine anaplasmosis exhibits major limitations to the livestock production system, only a few studies provide limited information about bovine anaplasmosis in Pakistan. Most of the exisiting studies rely only on conventional microscopy with low sensitivity and specificity. As far as it could have been ascertained in Pakistan, A. bovis has not been detected as an etiological agent of the bovine anaplasmosis and only A. marginale has been identified as the major cause of infection from Southern Punjab and Khyber Pakhtunkhwa provinces with prevalence rates of 17% and 18.33% respectively [21,22].
With the objective of defining the spectrum of potential causative agents of bovine anaplasmosis in Pakistan, the designed PCR-based study was successful in diagnosing the Anaplasma infection, even in the carrier animals. Use of a PCR tool based on 16S rRNA gene amplification as a preferential method was established for Anaplasma spp. detection in carrier animals [26,27,28,29].
This study confers the use of 16S rRNA based nested-PCR in the subclinical diagnosis of the bovine anaplasmosis as it revealed the presence of A. bovis and A. phagocytophilum in apparently asymptomatic animals with an overall prevalence rate of 10.44%. The obtained sequences showed 98–100% identity to the Anaplasma reference sequences. The retrieved sequences of A. bovis and A. phagocytophilum isolates using BLAST query for phylogenetic analysis presented themselves to be highly homologous with NCBI reference sequences. Phylogenetic analysis of A. bovis revealed maximum identity with the sequences previously reported in China, India, Iran, Japan and South Africa. While sequences from A. phagocytophilum were obtained from positive samples and were seen to be closely related with the sequences previously reported from South Africa and Poland (Figure 1).
Our findings were dissimilar to an investigation that reported 61% prevalence of Anaplasma spp. in Karachi and its adjacent areas [25]. The difference of experimental outcomes regarding Anaplasma infection in the two different studies is presumably attributable to the environmental compatibility of Karachi, a coastal city with high relative humidity and moderate climate which supports tick infestation and the different tools used for the pathogen detection [30]. By using conventional microscopy, Anaplasma was detected with a very high prevalence rate of 80%, but with no specific differential distribution and evidence of A. bovis and A. phagocytophilum [31]. Another study made in the neighboring country, Iran, reported 22.22% prevalence rate of Anaplasma infection in the cattle population, without species differentiation [32]. Likewise, an investigation claimed the presence of only A. marginale with 68.75% prevalence in bovine in Punjab State of India [33]. Anaplasma bovis and A. phagocytophilum have been identified in Xinjiang, China in cattle with 4.80% and 6.40% prevalence, respectively [11]. In contrast, other researchers described the prevalence of Anaplasma infection ranging between 4–12% without establishing A. bovis and A. phagocytophilum as potential pathogens for the infection in Pakistan, where only the conventional tools for detection were used [34,35].
4. Methods
4.1. Sample Collection
In the present study, blood samples were collected randomly on Whatman FTATM Classic Cards from 450 tick-infested but asymptomatic cattle during July and August 2018 from Jhang District in Punjab, Pakistan. Blood was collected from the jugular vein using 10 ml disposable syringes and transferred to the Whatman® FTA cards (GE Healthcare Limited, Buckinghamshire HP7 9NA, UK) and allowed to air-dry. Sample collection and animal treatments complied with the Animal Ethics Procedures and Guidelines and was approved by the Animal Ethics Committee of the Punjab Livestock Department, Pakistan. The FTA cards were shipped to Lanzhou Veterinary Research Institute at the Chinese Academy of Agricultural Sciences Lanzhou, China for further processing.
4.2. DNA Extraction
Genomic DNA was extracted from the FTA cards using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA concentration was determined with a Nano-Drop 2000 spectrophotometer (Nanodrop Technologies®, Wilmington, DE, USA). DNA was stored at −20 °C until further analysis.
4.3. PCR Amplification
Nested PCR was carried out to detect Anaplasma infection from the collected bovine samples. During the first round, genomic DNA from field blood samples was amplified using the primers EE1 and EE2 [36]. The PCR products were used as templates for the second round using the A. bovis specific primers AB1f and AB1r, which generate a product of 551 bp, and the A. phagocytophilum specific primers SSAP2f and SSAP2r, which generate a product of 641 bp [13]. The reactions were performed in a final volume of 50 µL, containing 1.0 mM of each primer, 5 µL of PCR buffer, 4 µL of deoxynucleoside triphosphates, 0.25 µL of TaKaRa Taq (5 µ/mL) (TaKaRa, China), and 1 µL of DNA sample. Reactions were conducted in an automated DNA C1000 Thermal Cycler (Bio-Rad, Beijing, China). For the EE1 and EE2 primers, the cycling conditions were denaturation for 4 min at 94 °C, followed by 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 30 s. The annealing temperature (62 °C) was stepped down four times by 2 °C every two cycles. The final annealing temperature used was 54 °C for 28 cycles, followed by a final extension for 5 min at 72 °C. For the nested PCR, 2 µL of the product from the first amplification was used for amplification with specific primers; the amplification consisted of 40 cycles, each of 1 min at 94 °C, 1 min at 55 °C, and 1 min at 72 °C. Cattle genomic DNA and distilled water were used as negative and blank controls, respectively. The PCR products were subjected to electrophoresis on 1% agarose gel containing 0.5 g/ml ethidium bromide and visualized under UV light.
DNA Sequencing and Data Analysis
Positive PCR products amplified by primers SSAP2f/2r and AB1f/AB1r were excised from the gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen, USA). The DNA fragments were cloned into pGEM-T vector (Promega, Madison, WI). Escherichia coli Trans 5α (TaKaRa, China) was transformed and plasmid DNA from the selected clones was identified using PCR with the set of primers T7 (5’-TAATACGACTCACTATAG GG-3’) and SP6 (5’-ATTTAGGTGACACTATAG-3’) to verify the presence of correct inserts in selected clones and then sequenced by Sangon Biotech Company (Shanghai, China). The obtained sequences were analyzed by a BLAST search in GenBank for determining the accuracy of the PCR method.
4.4. Phylogenetic Analysis
For genotyping, obtained sequences of A. phagocytophilum and A. bovis were aligned using the MegAlign component of the DNAStar software program (Version 4.0 DNAStar, Madison, USA). After alignment with related Anaplasma spp. 16S rDNA sequences retrieved from GenBank, parts of the cloning vector region were removed manually. The resulting sequences were then submitted to the GenBank database. A phylogenetic tree was generated based on the cloned sequences and the related Anaplasma spp. 16S rDNA sequences in GenBank by using the neighbor-joining method [37].
5. Conclusions
The present study provides the first evidence of A. bovis and A. phagocytophilum as potential causative agents of bovine anaplasmosis in Pakistan; of which the latter alarms for its own health significance. A comprehensive molecular epidemiological investigation is required for appropriate disease mapping in the country which can help devise control strategies for ticks and tick-transmitted diseases of livestock and public health significance.
Abbreviations
| PCR | polymerase chain reaction |
| DNA | deoxyribonucleic acid |
Author Contributions
H.Y. and Z.L. designed this study and critically revised the manuscript; M.S.S. participated in sample collection; N.I., M.U.M., J.Y., Q.N. and G.G. performed the experiments, data analysis, and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was financially supported by the National Key R&D Program of China (2017YFD0501200, 2016YFC1202000); 973 Program (2015CB150300); NSFC (No:31402189, No:31372432); ASTIP, FRIP (2014ZL010), CAAS; NBCIS (CARS-38); Jiangsu Co-Innovation Center for the Prevention and Control of Important Animal Infectious Disease and Zoonose, and the State Key Laboratory of Veterinary Etiological Biology Projects.
Conflicts of Interest
The authors declare no conflict of interest.
Ethics Approval and Consent to Participate
Animal treatments and sample preparation complied with the Animal Ethics Procedures and Guidelines, and was approved by the Animal Ethics Committee of Punjab Livestock Department, Pakistan.
Availability of Data and Materials
Sequences submitted in the GenBank database under accession numbers are as follows: A. phagocytophilum: MN216239 and MN216240; and A. bovis: MN216233, MN216234, MN216235, MN216236, MN216237 and MN216238.
References
- 1.Kocan K.M., de la Fuente J., Guglielmone A.A., Melendez R.D. Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin. Microbiol. Rev. 2003;16:698–712. doi: 10.1128/CMR.16.4.698-712.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sainz A., Amusategui I., Tesouro M.A. Ehrlichia platys infection and disease in dogs in Spain. J. Vet. Diagn. Investig. 1999;11:382–384. doi: 10.1177/104063879901100419. [DOI] [PubMed] [Google Scholar]
- 3.Melendez R.D. Future perspectives on veterinary hemoparasite research in the tropics at the start of this century. Ann. N. Y. Acad. Sci. 2000;916:253–258. doi: 10.1111/j.1749-6632.2000.tb05297.x. [DOI] [PubMed] [Google Scholar]
- 4.Stuen S., Bergstrom K., Palmer E. Reduced weight gain due to subclinical Anaplasma phagocytophilum (formerly Ehrlichia phagocytophila) infection. Exp. Appl. Acarol. 2002;28:209–215. doi: 10.1023/A:1025350517733. [DOI] [PubMed] [Google Scholar]
- 5.Stuen S., Nevland S., Moum T. Fatal cases of Tick-borne fever (TBF) in sheep caused by several 16S rRNA gene variants of Anaplasma phagocytophilum. Ann. N. Y. Acad. Sci. 2003;990:433–434. doi: 10.1111/j.1749-6632.2003.tb07407.x. [DOI] [PubMed] [Google Scholar]
- 6.Dumler J.S., Barbet A.F., Bekker C.P., Dasch G.A., Palmer G.H., Ray S.C., Rikihisa Y., Rurangirwa F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ’HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 7.Rar V., Golovljova I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: Pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect. Genet. Evol. 2011;11:1842–1861. doi: 10.1016/j.meegid.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Ceci L., Iarussi F., Greco B., Lacinio R., Fornelli S., Carelli G. Retrospective study of haemoparasites in cattle in Southern Italy by reverse line blot hybridization. J. Vet. Med. Sci. 2014;76:869–875. doi: 10.1292/jvms.13-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belkahia H., Said M.B., Alberti A., Abdi K., Issaoui Z., Hattab D., Gharbi M., Messadi L. First molecular survey and novel genetic variants’ identification of Anaplasma marginale, A. centrale and A. bovis in cattle from Tunisia. Infect. Genet. Evol. 2015;34:361–371. doi: 10.1016/j.meegid.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Chen Z., Liu Z., Liu J., Yang J., Li Q., Li Y., Luo J., Yin H. Molecular Survey of Anaplasma and Ehrlichia of Red Deer and Sika Deer in Gansu, China in 2013. Transbound Emerg. Dis. 2016;63:e228–e236. doi: 10.1111/tbed.12335. [DOI] [PubMed] [Google Scholar]
- 11.Yang J., Li Y., Liu Z., Liu J., Niu Q., Ren Q., Chen Z., Guan G., Luo J., Yin H. Molecular detection and characterization of Anaplasma spp. in sheep and cattle from Xinjiang, northwest China. Parasites Vectors. 2015;8:108. doi: 10.1186/s13071-015-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Njiiri N.E., Bronsvoort B.M., Collins N.E., Steyn H.C., Troskie M., Vorster I., Thumbi S.M., Sibeko K.P., Jennings A., van Wyk I.C., et al. The epidemiology of tick-borne haemoparasites as determined by the reverse line blot hybridization assay in an intensively studied cohort of calves in western Kenya. Vet. Parasitol. 2015;210:69–76. doi: 10.1016/j.vetpar.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawahara M., Rikihisa Y., Lin Q., Isogai E., Tahara K., Itagaki A., Hiramitsu Y., Tajima T. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl. Environ. Microbiol. 2006;72:1102–1109. doi: 10.1128/AEM.72.2.1102-1109.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M’Ghirbi Y., Yaich H., Ghorbel A., Bouattour A. Anaplasma phagocytophilum in horses and ticks in Tunisia. Parasites Vectors. 2012;5:180. doi: 10.1186/1756-3305-5-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang J.G., Kim H.C., Choi C.Y., Nam H.Y., Chae H.Y., Chong S.T., Klein T.A., Ko S., Chae J.S. Molecular detection of Anaplasma, Bartonella, and Borrelia species in ticks collected from migratory birds from Hong-do Island, Republic of Korea. Vectors Borne Zoonotic. Dis. 2013;13:215–225. doi: 10.1089/vbz.2012.1149. [DOI] [PubMed] [Google Scholar]
- 16.Djiba M.L., Mediannikov O., Mbengue M., Thiongane Y., Molez J.F., Seck M.T., Fenollar F., Raoult D., Ndiaye M. Survey of Anaplasmataceae bacteria in sheep from Senegal. Trop. Anim. Health. Prod. 2013;45:1557–1561. doi: 10.1007/s11250-013-0399-y. [DOI] [PubMed] [Google Scholar]
- 17.Stuen S., Pettersen K.S., Granquist E.G., Bergstrom K., Bown K.J., Birtles R.J. Anaplasma phagocytophilum variants in sympatric red deer (Cervus elaphus) and sheep in southern Norway. Ticks Tick Borne Dis. 2013;4:197–201. doi: 10.1016/j.ttbdis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Borthakur S., Deka D., Bhattacharjee K., Sarmah P. Seroprevalence of canine dirofilariosis, granulocytic anaplasmosis and lyme borreliosis of public health importance in dogs from India’s North East. Vet. World. 2014;7:665–667. doi: 10.14202/vetworld.2014.665-667. [DOI] [Google Scholar]
- 19.Razzaq F., Khosa T., Ahmad S., Hussain M., Saeed Z., Khan M., Shaikh R., Ali M., Iqbal F. Prevalence of Anaplasma phagocytophilum in horses from Southern Punjab (Pakistan) Trop. Biomed. 2015;32:233–239. [PubMed] [Google Scholar]
- 20.Rajput Z.I., Hu S.H., Arijo A.G., Habib M., Khalid M. Comparative study of Anaplasma parasites in tick carrying buffaloes and cattle. J. Zhejiang Univ. Sci. B. 2005;6:1057–1062. doi: 10.1631/jzus.2005.B1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashraf Q.U., Khan A.U., Khattak R.M., Ali M., Shaikh R.S., Ali M., Iqbal F. A report on the high prevalence of Anaplasma sp. in buffaloes from two provinces in Pakistan. Ticks Tick Borne Dis. 2013;4:395–398. doi: 10.1016/j.ttbdis.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Farooqi S.H., Ijaz M., Rashid M.I., Nabi H., Islam S., Aqib A.I., Hussain K., Khan A., Rizvi S.N.B., Mahmood S., et al. Molecular epidemiology of bovine anaplasmosis in Khyber Pakhtunkhwa, Pakistan. Trop. Anim. Health Prod. 2018;50:1591–1598. doi: 10.1007/s11250-018-1599-2. [DOI] [PubMed] [Google Scholar]
- 23.De Echaide S.T., Bono M.F., Lugaresi C., Aguirre N., Mangold A., Moretta R., Farber M., Mondillo C. Detection of antibodies against Anaplasma marginale in milk using a recombinant MSP5 indirect ELISA. Vet. Microbiol. 2005;106:287–292. doi: 10.1016/j.vetmic.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez S.D., Garcia Ortiz M.A., Jimenez Ocampo R., Vega y Murguia C.A. Molecular epidemiology of bovine anaplasmosis with a particular focus in Mexico. Infect. Genet. Evol. 2009;9:1092–1101. doi: 10.1016/j.meegid.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Haider M., Bilqees F. Anaplasmosis in certain mammals in Karachi and adjoin areas. Proc. Parasitol. 1988;6:85–88. [Google Scholar]
- 26.Molad T., Mazuz M., Fleiderovitz L., Fish L., Savitsky I., Krigel Y., Leibovitz B., Molloy J., Jongejan F., Shkap V. Molecular and serological detection of A. centrale-and A. marginale-infected cattle grazing within an endemic area. Vet. Microbiol. 2006;113:55–62. doi: 10.1016/j.vetmic.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Bekker C.P., De Vos S., Taoufik A., Sparagano O.A., Jongejan F. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichiaruminantium in Amblyomma variegatum ticks by reverse line blot hybridization. Vet. Microbiol. 2002;89:223–238. doi: 10.1016/S0378-1135(02)00179-7. [DOI] [PubMed] [Google Scholar]
- 28.Carelli G., Decaro N., Lorusso A., Elia G., Lorusso E., Mari V., Ceci L., Buonavoglia C. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet. Microbiol. 2007;124:107–114. doi: 10.1016/j.vetmic.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 29.Noaman V., Shayan P., Amininia N. Molecular diagnostic of Anaplasma marginale in carrier cattle. Iran. J. Parasitol. 2009;4:26–33. [Google Scholar]
- 30.Shahnawaz S., Ali M., Aslam M.A., Fatima R., Chaudhry Z.I., Hassan M.U., Ali M., Iqbal F. A study on the prevalence of a tick-transmitted pathogen, Theileria annulata, and hematological profile of cattle from Southern Punjab (Pakistan) Parasitol. Res. 2011;109:1155–1160. doi: 10.1007/s00436-011-2360-1. [DOI] [PubMed] [Google Scholar]
- 31.Khan M., Zahoor A., Jahangir M., Mirza M.A. Prevalence of blood parasites in cattle and buffaloes. Pak. Vet. J. 2004;24:193–194. [Google Scholar]
- 32.Hosseini-Vasoukolaei N., Oshaghi M.A., Shayan P., Vatandoost H., Babamahmoudi F., Yaghoobi-Ershadi M.R., Telmadarraiy Z., Mohtarami F. Anaplasma Infection in Ticks, Livestock and Human in Ghaemshahr, Mazandaran Province, Iran. J. Arthropod. Borne Dis. 2014;8:204–211. [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma A., Singla L.D., Kaur P., Bal M.S. PCR and ELISA vis-a-vis microscopy for detection of bovine anaplasmosis: A study on associated risk of an upcoming problem in North India. Sci. World J. 2015;2015:352519. doi: 10.1155/2015/352519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atif F.A., Khan M.S., Iqbal H.J., Arshad G.M., Ashraf E., Ullah S. Prevalence of Anaplasma marginale, Babesia bigemina and Theileria annulata infections among cattle in Sargodha District, Pakistan. Afr. J. Agric. Res. 2012;7:302–3307. [Google Scholar]
- 35.Sajid M., Siddique R., Khan S., Zafar I., Khan M. Prevalence and risk factors of anaplasmosis in cattle and buffalo populations of district Khanewal, Punjab, Pakistan. Glob. Vet. 2014;12:146–153. [Google Scholar]
- 36.Barlough J.E., Madigan J.E., DeRock E., Bigornia L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus) Vet. Parasitol. 1996;63:319–329. doi: 10.1016/0304-4017(95)00904-3. [DOI] [PubMed] [Google Scholar]
- 37.Saitou N., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences submitted in the GenBank database under accession numbers are as follows: A. phagocytophilum: MN216239 and MN216240; and A. bovis: MN216233, MN216234, MN216235, MN216236, MN216237 and MN216238.