Significance
Genomic DNA must be compacted to fit in the cell but must simultaneously remain accessible for transcription. While genome organization influences gene expression, the impact of transcription on genome organization remains to be understood. The process of “loop extrusion” is central to chromosome organization. In bacteria, the condensin SMC complex performs extrusion by translocating along arms of the chromosome. During translocation, condensins encounter bulky transcription machinery, which could interfere with loop extrusion. This work investigates the interplay between 2 important active processes; it demonstrates that extruding SMCs can efficiently bypass transcribing RNA polymerases within mere seconds, thus allowing spatial organization of the transcriptionally active genome, and predicts that extruding SMC complexes have a mechanism to bypass other bulky DNA-bound obstacles.
Keywords: condensin, transcription, loop extrusion, SMC, chromosome conformation capture
Abstract
To separate replicated sister chromatids during mitosis, eukaryotes and prokaryotes have structural maintenance of chromosome (SMC) condensin complexes that were recently shown to organize chromosomes by a process known as DNA loop extrusion. In rapidly dividing bacterial cells, the process of separating sister chromatids occurs concomitantly with ongoing transcription. How transcription interferes with the condensin loop-extrusion process is largely unexplored, but recent experiments have shown that sites of high transcription may directionally affect condensin loop extrusion. We quantitatively investigate different mechanisms of interaction between condensin and elongating RNA polymerases (RNAPs) and find that RNAPs are likely steric barriers that can push and interact with condensins. Supported by chromosome conformation capture and chromatin immunoprecipitation for cells after transcription inhibition and RNAP degradation, we argue that translocating condensins must bypass transcribing RNAPs within ∼1 to 2 s of an encounter at rRNA genes and within ∼10 s at protein-coding genes. Thus, while individual RNAPs have little effect on the progress of loop extrusion, long, highly transcribed operons can significantly impede the extrusion process. Our data and quantitative models further suggest that bacterial condensin loop extrusion occurs by 2 independent, uncoupled motor activities; the motors translocate on DNA in opposing directions and function together to enlarge chromosomal loops, each independently bypassing steric barriers in their path. Our study provides a quantitative link between transcription and 3D genome organization and proposes a mechanism of interactions between SMC complexes and elongating transcription machinery relevant from bacteria to higher eukaryotes.
The structural maintenance of chromosome (SMC) complexes are an evolutionarily conserved family of protein complexes, including condensin, cohesin, SMCHD1, Smc5/6, and others, present in most organisms from eubacteria to humans (1). These proteins are involved in processes as diverse as DNA damage repair, sister chromatid cohesion, and organization of mitotic and interphase chromosomes. SMC complexes are characterized by a 3-part ring, composed of a dimer of SMC subunits, each with an ATPase domain and a long coiled-coil domain; a kleisin linker which closes the ring; and accessory proteins which bind to the linker to perform specific functions, depending on the organism (1).
Recent in vivo studies have provided evidence that SMC complexes have a motor activity, allowing them to translocate processively on a chromatin fiber and perform active chromatin reorganization by loop extrusion. In the proposed loop-extrusion mechanism (2–8), SMC complexes (or oligomers of SMC complexes) bind to DNA at a single site; bridge 2 flanking DNA segments, forming a loop; and then progressively enlarge the loop by translocating away from the loading site. Thus, loop extrusion is thought to result from the activity of 2 connected motors translocating in opposite directions that expand a DNA loop.
Single-molecule studies provide support for this or a similar mechanism by demonstrating that budding yeast condensin SMCs are mechano-chemical motors that can translocate along DNA (9), extrude DNA loops (10), and actively compact DNA (11–13). While the molecular details of this process are yet to be fully understood (14, 15), loop extrusion appears to be a mechanism that can explain a wealth of chromosomal phenomena in eukaryotes and bacteria.
In eukaryotes, during mitosis, loop extrusion by condensin can explain the compaction and resolution of sister chromatids and may underlie the formation of arrays of loops and nested loops central to mitotic chromosomes. Evidence also suggests that loop extrusion by cohesin SMCs underlies the formation of chromosomal domains during interphase (16–20).
In bacteria, condensin SMC complexes help resolve newly replicated origins and appear to do so by juxtaposing the left and right chromosome arms of the newly replicated sister chromosomes. Amazingly, DNA juxtaposition extends from origin to terminus, generating a single 4-Mb “loop” (7, 21, 22). Studies using chromosome conformation capture (Hi-C) and chromatin immunoprecipitation combined with deep sequencing (ChIP-seq) have shown that condensins are preferentially loaded onto chromosomes via ParB proteins bound at parS sites located primarily adjacent to the origin of replication (23–25). Once loaded, condensins progress away from the parS sites (8, 26) along both chromosomal arms, thus juxtaposing them (7, 21), resulting in the characteristic “X”-shaped pattern on the Hi-C maps of many bacteria (27). In vivo experiments recently showed that translocation by condensin is an active process (8, 28): Condensin complexes travel processively and bidirectionally away from the parS loading site, in a manner that appears to be ATP-dependent, at speeds exceeding 800 bp/s (29). This active juxtaposition of chromosome arms by the bacterial condensin SMC condensin complex suggests a mechanism of loop extrusion in bacteria (7, 8, 28, 30) nearly identical to the proposed loop-extrusion process in eukaryotes.
Beyond their function in directly shaping spatial chromosome structure, SMCs can potentially also respond to various signals, allowing the reorganization of chromosomes in response. As an example, recent studies provide strong evidence that transcription can affect genome structure and SMC action (31–36). It remains unknown, however, how an SMC loop extruder interacts with the transcription machinery and how these nanometer-scale interactions affect global chromosome structure.
Here, we study the effect of transcription on chromosome structure by developing models of condensin dynamics and validating them using experimental data. Central to these models is the hypothesis that the speed of condensin translocation is affected by transcription, depending on the relative orientation of genes and the direction of extrusion. We propose that once a condensin encounters an actively transcribed gene, it slows down due to interactions with the transcription machinery, with the slowing down being greater if condensin and RNA polymerase (RNAP) meet in a head-to-head versus head-to-tail interaction.
Our models predict condensin juxtaposition trajectories that are in excellent quantitative agreement with Hi-C data for wild-type and engineered bacterial strains where the condensin loading site has been moved to different genome positions. Our analysis further supports the idea that loop extrusion by bacterial condensins is mediated by at least 2 independently acting and uncoupled motor activities. To understand the molecular mechanisms that underlie the directional effect of slow transcription (∼40–80 bp/s) on the much faster condensin translocation (∼800 bp/s), we develop a mechanistic “moving-barriers” model for interactions of SMC complexes with transcription machinery. The analytical solution of the stochastic “moving-barriers” model allows us to integrate diverse experimental data to predict chromosome structure arising through the interplay of loop extrusion and transcription. We find strong evidence that SMC molecules can bypass elongating RNAPs, impeding their translocation within 2 s of an encounter at rRNA operons and within 10 s at protein-coding operons. This finding has important implications for understanding the mode of DNA translocation by condensins and their ability to overcome steric barriers. We also investigated changes in DNA juxtaposition following transcription inhibition and acute RNAP degradation. Our analysis revealed that both transcription-dependent and -independent effects impacted condensins’ genome-wide chromosome juxtaposition activity. Our quantitative models of transcription–condensin interactions tested on bacterial data have widespread implications for chromosome organization and can provide a framework to study the effect of transcription on chromosome organization in higher organisms
Materials and Methods
Hi-C and ChIP-seq were performed as described (7) for Bacillus subtilis PY79 cells grown to midexponential phase (SI Appendix, Materials and Methods). For sources of data present in all figures, refer to SI Appendix, Tables S1–S17. For data processing, see SI Appendix, Data Processing. Simulations and theory are described in SI Appendix, sections 3–6. Source codes are available at https://github.com/hbbrandao/bacterialSMCtrajectories. Data are deposited in the Gene Expression Omnibus (accession no. GSE117854).
Results
Predicting Condensin’s Spatiotemporal Trajectory from Gene Directions and Positions.
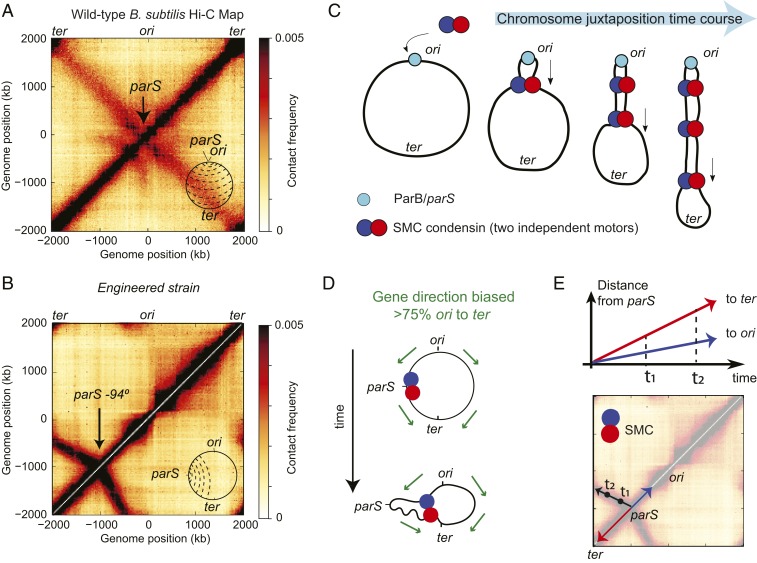
In the Hi-C map of wild-type B. subtilis, as reported (7, 21), contacts from DNA segments close in the linear genome sequence, as in all organisms, give rise to the primary interaction diagonal, which extends from the bottom left to top right of the map (see Fig. 1A and SI Appendix, Tables S2–S14 for sources of data in all figures). A secondary diagonal, which runs perpendicular to the primary, typical of many bacterial Hi-C interaction maps (7, 22, 27), indicates a symmetric juxtaposition of 2 chromosome arms about the parS sites located next to the origin of replication (ori) (Fig. 1 A, Inset). However, engineered strains of B. subtilis, in which all of the endogenous parS sites are deleted, and a single site is inserted at other positions, reveal different shapes of the Hi-C secondary diagonal. In such strains, the wild-type secondary diagonal is missing and is replaced with a diagonal emanating from the new parS location (7, 8). Without exception, these new interaction signatures are tilted or curved away from the ori (Fig. 1B). These curved diagonals represent an asymmetry in interactions between DNA flanking these displaced parS sites. In all cases, larger tracks of terminus-proximal DNA interact with shorter tracks of origin-proximal DNA. In the loop-extrusion model, chromosome juxtaposition occurs by 2 motor activities of condensin translocating away from the parS site (Fig. 1C); at the molecular level, the secondary diagonal, visible by Hi-C, arises from individual trajectories of condensin’s loop-extrusion motors, averaged over a population of cells. Thus, in the context of this model, a curved diagonal suggests that at certain loci, one of the loop-extruding motors translocates more slowly that the other one (Fig. 1D).
Fig. 1.
The posited role of transcription in shaping asymmetric SMC translocation rates. (A) Hi-C map of a wild-type B. subtilis PY79. A, Inset depicts the juxtaposition of chromosome arms (dashed lines) centered on the ori to ter axis. (B) Hi-C map of a B. subtilis strain (see SI Appendix for strain tables) with a single parS site at −94° which has a secondary diagonal that points biasedly away from the ori. (C) Loop-extrusion model schematic depicting the active juxtaposition of chromosome arms performed by the B. subtilis condensin loop-extruding complex. (D) Gene directions point biasedly toward the ter (green arrows); for SMC motors translocating toward the ori (blue), there will be increased frequencies of head-on collisions with transcripts as compared to SMCs translocating toward the ter (red). (E) Interpretation of Hi-C: Condensin translocates bidirectionally from the parS site juxtaposing flanking DNA; motion toward the ter is faster than toward the ori, resulting in the asymmetric secondary diagonal.
Recalling that over 75% of genes in B. subtilis are co-oriented with replication (37), we posited that transcription could account for the tilt of the secondary diagonal by slowing down condensin translocation (Fig. 1D). Since condensin translocation toward the ori will be more frequently opposing transcription, the increased numbers of “head-to-head” encounters of condensin with RNAP potentially lead to a slower overall translocation rate for ori-oriented condensins. This results in a gene-direction-based effect on condensin speed (Fig. 1E). This hypothesis is supported by recent experimental evidence in Caulobacter crescentus and B. subtilis, where the relative orientations of genes to condensin-loading sites have been altered (8, 28).
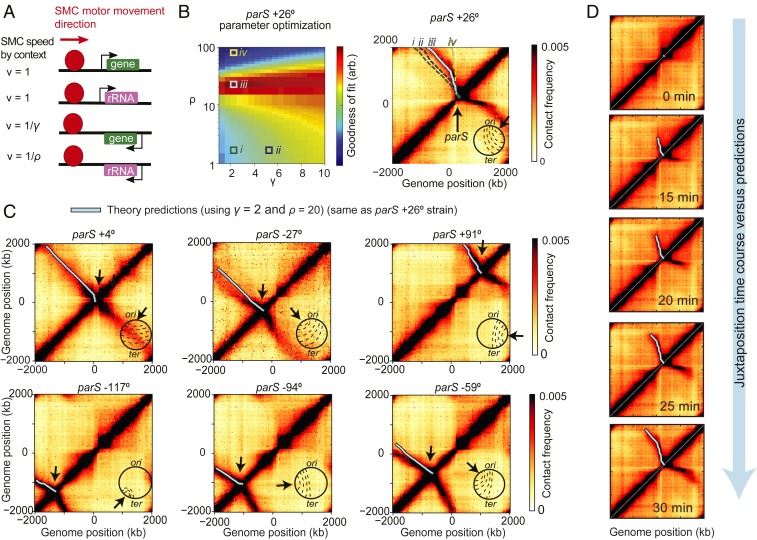
To test whether the interplay between condensin translocation and transcription can shape chromosome structure, we developed a model where condensin trajectories can be predicted based solely on gene locations and orientations (Fig. 2A). In this model, condensins form a loop-extrusion complex that has 2 motors, each translocating independently and deterministically along the DNA with the maximum speed vmax; the complex begins at a single point (the parS site), and each motor progresses in opposing directions with the following rules: When a motor encounters a gene, its instantaneous speed v is changed such that
The value γ is interpreted as the fold increase in time required for a condensin motor to traverse a gene against the direction of transcription. ρ is similarly interpreted, but reserved for the highly transcribed rRNA operons which are found at 7 distinct loci in B. subtilis PY79. Below, we generalize this model to incorporate locus-specific rates of transcription. Crucially, we assumed that speeds of condensin motors in the same extrusion complex were independent of each other—i.e., if one motor is slowed down, the other continues unaffected at its own speed; we revisit this assumption later. As the 2 motors move away from the parS site, they bring together flanking DNA, generating the Hi-C secondary diagonal (Fig. 1 C and D). By computing the displacement from parS versus the time of each motor, base pair by base pair, using gene position and direction data (SI Appendix, section 3.1), it was possible to trace the expected extrusion complex trajectory parametrized by time on the Hi-C map (Fig. 1E); the trajectory depends on the values of γ and ρ as shown (Fig. 2 B, Right).
Fig. 2.
Model of SMC translocation based on gene position and orientation. (A) Rules for the model of condensin-translocation rates based on gene orientation. (B) A parameter sweep of the model shows the agreement with Hi-C data as a function of γ and ρ parameters (Left); illustrative example trajectories (i–iv) are superimposed on the Hi-C map to show how juxtaposition traces change as a function of these parameters (Right). (C) Examples of using the parS+26° strain-specific optimum parameters (γ = 2 and ρ = 20) (see SI Appendix, Fig. S4 for globally optimum trajectories) to predict the juxtaposition trajectories of other engineered B. subtilis strains. (D) The model of condensin translocation using only gene positions and orientations captures the spatiotemporal behavior of Hi-C secondary diagonal formation during a time-course Hi-C experiment where condensin loading was induced at t = 0 min.
We hypothesized that if transcriptional interference with loop extrusion is a universal phenomenon which depends largely on gene orientation, then a single set of the parameters γ, ρ might predict the shapes of secondary diagonals in different engineered bacterial strains with ectopic condensin loading sites. By sweeping over parameter values and comparing predicted extrusion traces to Hi-C experiments, we can determine the best values of γ and ρ (SI Appendix, Fig. S1A). For example, we can use the best values of γ and ρ found for one strain—i.e., parS+26° (Fig. 2B)—to adequately predict the condensin trajectories in 9 other strains (Fig. 2C and SI Appendix, Fig. S1B). Moreover, and importantly, we found that the optimal solutions for γ and ρ across strains have similar values (SI Appendix, Fig. S1A). Combining parameter-fit values of the different strains (SI Appendix, Fig. S2A), we found that a single set of parameters (γ = 3.5, ρ = 20) provides the overall best predictive power for the secondary diagonals and extrusion traces (SI Appendix, Fig. S2B) and resembles the strain-specific optimum trajectories (SI Appendix, Fig. S1A).
To validate the temporal aspect of our above model of condensin loop extrusion, we tested whether it agreed with time-course Hi-C of chromosome juxtaposition (8). In the time-course experiments, loading of condensin at the parS site was induced at time t = 0 min, and the progression of the juxtaposition front was monitored by Hi-C over 5-min time intervals. Using the average juxtaposition rate of 800 bp/s measured previously (8), we calibrated the relative condensin speeds into absolute speeds (SI Appendix, section 4.1). In our model, knowing the average condensin speed, vavg, we inferred the maximum speed, vmax, using
where N is the number of base pairs of a genome arm, and i is the relative translocation time to move across a locus (where ηi = γ,ρ if the base pair, ηi, belongs to a gene or rRNA locus that is oriented opposite to condensin’s translocation, and ηi =1 otherwise). Plotting the model predictions made for each arm using the 2 globally optimal parameter values γ = 3.5, ρ = 20, known experimental values for vavg = 800 bp/s, N = 2 × 106 (base pairs), we found excellent quantitative agreement with the time course of experimental Hi-C (Fig. 2D and SI Appendix, Fig. S3) and inferred that the maximum speed of condensin is vmax ≈ 1,500 ± 200 bp/s.
Thus, this minimal model which uses only gene positions and orientations agrees well with experimental data; it captures the major aspects of the Hi-C secondary diagonals and suggests that transcription orientation is a key factor in controlling the speed of extrusion. However, the model does not establish a direct link between condensin translocation and the process of transcription. Accordingly, it remains a possibility that other DNA motifs or processes, correlated with gene orientations, influence condensin speeds (38). This calls for more direct experimental tests for the role of genes and transcription. Nonetheless, we can conclude from this analysis that, either via transcription or other mechanisms, origin to terminus sequence biases strongly influence condensin translocation and alter chromosome organization in a predictable and universal way.
Bidirectional Condensin Translocation Is Performed by 2 Independent Motor Activities.
The model above and previous analyses (8) suggest that B. subtilis condensin complexes bidirectionally translocate along chromosomal arms, enlarging chromosomal loops by 2 independent motor activities. We sought to rigorously test whether this assumption of independence is necessary and the degree to which it holds true. We modified the model such that the instantaneous waiting times for each locus (ηi = γ,ρ) were partially correlated between the 2 motors, thereby breaking the assumption of independent translocation for each motor. Correlation was quantified by the mixing parameter f (f = 0 for independent motion, f = 1 for fully correlated motion; see SI Appendix, section 3.2 for details).
As done previously, we swept the parameters γ, ρ for various fixed values of f in the mixing model (ranging from f = 0 to f = 1) to obtain goodness-of-fit values (SI Appendix, Fig. S4A). Intriguingly, the model with f = 0 (independent translocation) had the highest overall goodness-of-fit value (SI Appendix, Fig. S4B) and exhibited visually better predictions (SI Appendix, Fig. S5). Thus, our model strongly suggests that the translocation process occurs via 2 independent motor activities, which do not sense impediments to their counterpart in the other translocation direction. This finding argues that dimerization (or oligomerization) of condensin may be required to obtain 2 distinct motor activities, consistent with previous experimental evidence and theoretical models (8, 14, 39, 40), or that a single 1-sided condensin dynamically switches directions of translocation, leading to apparent “2-sided” extrusion, with effectively independent motor activities (41).
Transcription Slows Down the Condensin Translocation Rate at Highly Transcribed Genes.
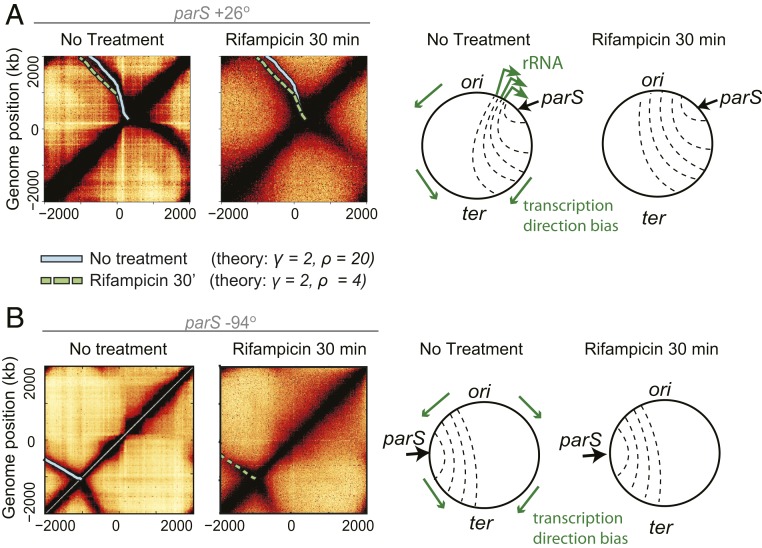
To directly test the effect of transcription on condensin translocation, we studied how transcription inhibition affects chromosome structure, as assayed by Hi-C using the small molecule rifampicin. Rifampicin is a well-characterized transcription inhibitor that prevents the transition from transcription initiation to elongation, but does not prevent binding of RNAP to DNA (42); moreover, RNAP molecules that have started to synthesize RNA will continue to synthesize their RNA until they reach the transcription termination site (43, 44). Residual transcription elongation after treatment with rifampicin is shown to have a genome-wide half-life of ∼6 min in Escherichia coli (45), so after 30 min of rifampicin treatment, the estimated remaining elongation is below 1% of untreated levels.
As shown previously, in a strain with a parS site close to a large cluster of rRNA operons (parS+26°), adding rifampicin to exponentially growing cells for 30 min results in a partial straightening out of the secondary diagonal (8, 28) (Fig. 3A). We reasoned that if transcription elongation shapes the overall tilt in the secondary diagonal, our quantitative model (Fig. 2A) should reveal a decrease in both the γ and ρ values following transcription inhibition.
Fig. 3.
Transcription-dependent and -independent features of SMC-mediated chromosome juxtaposition. (A) Hi-C data from previous experiments [Wang et al. (8)] showing the effect of transcription inhibition by rifampicin for 30 min on chromosome arm juxtaposition; superimposed SMC translocation model trajectories (solid and dashed lines) suggest a transcription-elongation independent effect on asymmetric SMC trajectories (i.e., the factor γ is unchanged before and after treatment; SI Appendix, Fig. S6A); schematic representations of the changes to chromosome juxtaposition are shown on the right. (B) Hi-C before and after rifampicin treatment experiments for a strain (parS+94°) with condensin loading far from the highly transcribed rRNA clusters of the other strain (parS+26°); the unchanging secondary diagonal angle confirms a transcription-independent effect.
We fit the published data on transcription inhibition to obtain the best-fit values for γ, ρ before and after treatment (SI Appendix, Fig. S6A). To our surprise, we found that γ = 2, ρ = 4.5 best describe the data after treatment and γ = 2, ρ = 20 before treatment (Fig. 3A and SI Appendix, Fig. S6A). This suggests that the overall tilt (“baseline asymmetry”) in the Hi-C secondary diagonal away from rRNA loci, captured by our parameter γ, is largely independent of transcription elongation (i.e., γ = 2 before and after treatment). Conversely, ρ, which quantifies the slowdown of condensin translocation going head-to-head with transcription at rRNA loci, largely depends on elongation.
To independently investigate this observation, we performed additional transcription-inhibition experiments using a strain with a parS site at the −94° position. In this strain, the parS site is far from the highly transcribed rRNA loci that affect condensin movement in the former strain (parS+26°). Consistent with the effect suggested by our model, we observed virtually no changes to the angle of the Hi-C secondary diagonal in the newly tested strain after 10 min (SI Appendix, Fig. S6B) or 30 min (Fig. 3B) of rifampicin treatment. This suggests that protein-coding genes (non-rRNA) have little effect on the speed of condensin translocation. These observations and the partial, but not complete, straightening out of the secondary diagonal in the parS+26° experiments led us to consider 2 possible models: Either nontranscribing RNAPs (e.g., trapped at transcription start sites by rifampicin) are directional barriers to condensin translocation, akin to CTCFs as directional barriers to cohesin in eukaryotes (4), or an RNAP-independent mechanism generates the “baseline asymmetry” of chromosome juxtaposition at loci outside of rRNA operons.
To differentiate between these the 2 models, we analyzed changes to chromosome structure following the degradation of RNAP. We reasoned that degrading RNAP will have no effect on the secondary diagonal tilt if the mechanism that generates the asymmetric juxtaposition is RNAP-independent. We generated a strain in which the sole copy of the β′ (beta prime) subunit of RNAP was fused to yellow fluorescent protein (YFP) and an SsrA tag (β′-YFP-SsrA) and could be conditionally targeted for degradation (46) (SI Appendix, Fig. S7). We induced degradation of the β′ fusion in a strain with a parS site at the −59° position and monitored the levels of protein over 90 min; β′ levels dropped to 5% of their initial value, as assayed via quantitative immunoblotting (SI Appendix, Fig. S7C) and imaging of β′-YFP-SsrA fluorescence in single cells (SI Appendix, Fig. S7B). Strikingly, a time-course Hi-C after induction of RNAP degradation revealed only a minor change in the tilt of the secondary diagonal (SI Appendix, Fig. S7A). As with rifampicin treatment, the most significant changes to the Hi-C maps manifested as a “blurring” of Hi-C features along the main diagonal and the disappearance of high-intensity spots along the secondary diagonal (SI Appendix, Fig. S8A). After RNAP degradation for 90 min, contact probability at short distances (<200 kb) decayed more quickly as compared to normal growth conditions (SI Appendix, Fig. S9A) and was indistinguishable from DNA contact probabilities of cells treated for 30 min with rifampicin (SI Appendix, Fig. S9B). Thus, RNAP and transcription are necessary for creating the texture in the Hi-C maps, which results in increased DNA–DNA contacts within ∼200 kb of separation; however, it does not strongly affect the overall secondary diagonal tilt. Altogether, the degradation of RNAP experiments rule out the role of paused RNAPs in establishing the “baseline asymmetry” of chromosome juxtaposition; this further indicates that the condensin translocation slowdown toward the ori (at non-rRNA loci) is largely independent of RNAP.
In hindsight, we understand that our phenomenological model (Fig. 2A) worked so well because in B. subtilis, gene density is high and homogeneous (SI Appendix, Fig. S8B), and there are relatively few highly transcribed genes (47); the average RNAP density is 0.1 RNAP/kb for most genes (48), in contrast with ∼10 RNAP/kb for rRNA genes (48) in our growth conditions (SI Appendix, section 4.2). Thus, the parameter γ reflects a systematic ori to ter bias in the condensin translocation speed (i.e., the “baseline asymmetry”) which correlates with gene direction but is largely RNAP-independent. We posit that such a bias may come from the process of DNA replication and will be a topic of future study. In contrast to γ, however, the parameter ρ, which reflects condensin slowdown at rRNA loci, does depend strongly on transcription. We thus chose to focus on understanding how the parameter ρ emerges from the process of transcription. Accordingly, we tested mechanistic models to help explain how highly transcribed genes become directional barriers to condensin translocation.
The Moving-Barriers Mechanism of Condensin–Transcription Interactions.
Plausible mechanisms of SMC and RNAP interaction must solve the following puzzle: How can condensin’s effective speed of translocation [measured at >800 bp/s via Hi-C and ChIP-seq (8)] be so strongly attenuated (>20-fold) by RNAP transcription [which moves at 40–90 bp/s (49)], depending only on the relative orientation of the 2 processes?
Previous studies have suggested that a passive SMC ring can be pushed by transcribing RNAP (34, 50). This idea emerged from the observation that cohesin SMCs are enriched at sites of convergent transcription in yeast (50, 51) and in mammalian cells (35); this is further supported by other experiments demonstrating localization of cohesin mediated by transcription in cells (32) and in vitro (52). The central assumption of the models is that transcription machinery forms an impermeable moving barrier to the SMC ring, pushing it along the direction of transcription.
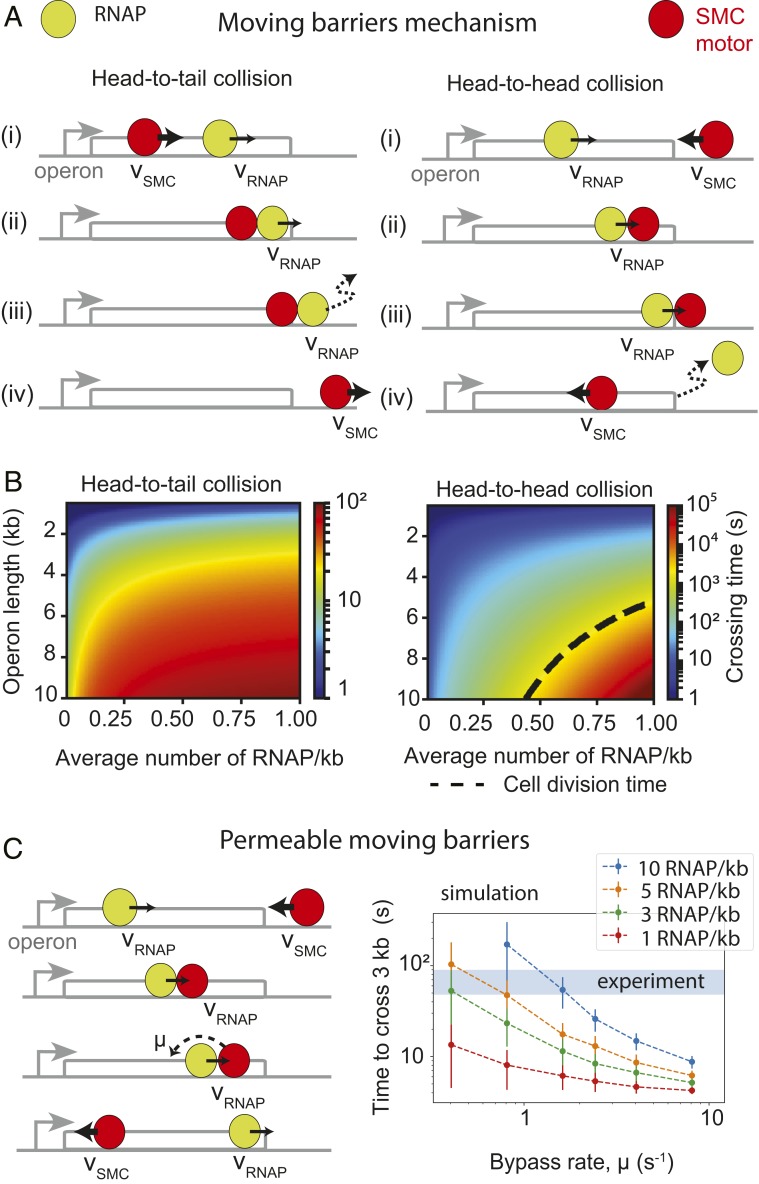
By combining the moving-barrier idea with active translocation by condensin, we can intuitively explain the directional effect of RNAP elongation on SMC translocation at highly expressed genes and the emergence of the direction parameter, ρ. In the case of head-to-tail interactions between condensin and RNAP (Fig. 4 A, Left), a condensin motor translocates at a high rate (e.g., ∼1,500 bp/s) until it encounters a transcribing RNAP moving in the same direction at a much lower speed [e.g., 80 bp/s as measured in E. coli (49)]. Since RNAP is assumed to be an impermeable barrier (we generalize this later to allow for partial permeability), when condensin encounters an RNAP, it slows down its translocation rate to match the RNAP until the end of the operon. Dissociation of RNAP at the end of an operon allows the condensin to continue translocating at its original high speed. In contrast, in the case of head-to-head interactions (Fig. 4 A, Right), when a translocating condensin encounters a transcribing RNAP, condensin is stalled and pushed back to the transcription termination site [condensin has a very low stall force measured in vitro (10) compared to RNAP (53)]. Once RNAP dissociates at the transcription termination site, condensin is left to attempt crossing the operon again; the condensin will only successfully cross the operon if no RNAPs are encountered during its run through the operon. In this “moving-barriers” model, the relative directional slowing down of condensin arises from the fact that multiple attempts may be required for the condensin to successfully cross an operon in a head-to-head orientation.
Fig. 4.
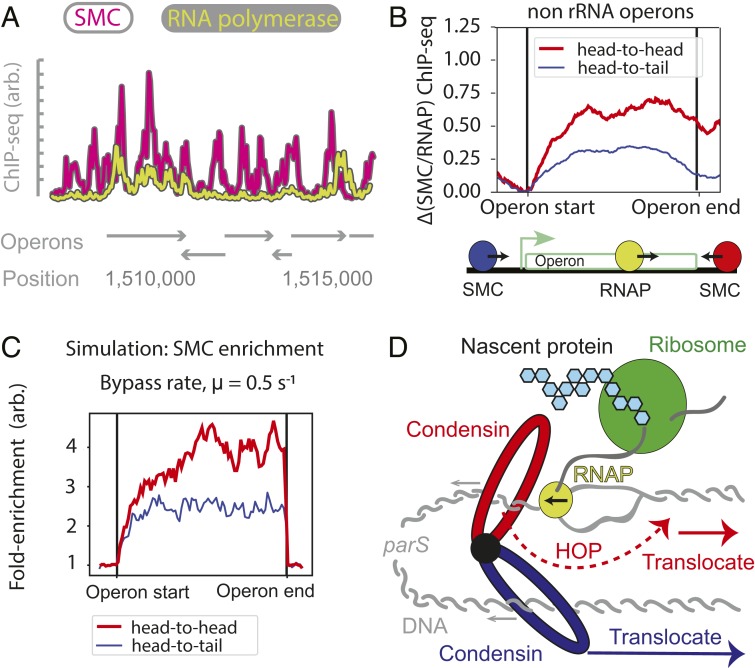
Mechanistic models of extrusion-transcription interference. (A) The “moving-barriers” model of condensin transcription interactions: Transcription complexes are posited as impermeable barriers to condensin movement, which results in different translocation dynamics when crossing operons in the co-oriented or convergent fashion. Condensin translocates (i) at its native speed (vSMC) until (ii) it reaches a slowly moving RNAP, then (iii) it proceeds in the direction and at the speed of RNAP (vRNAP) until (iv) RNAP reaches the end of the operon, whereby condensin continues translocation at its original speed and direction. (B) Analytically computed average times to cross an operon for each of the head-to-tail (Left) and head-to-head (Right) cases as a function of operon length and RNAP density. Despite similar “rules,” in the head-to-tail case (B, Left), the SMC always reaches the end of the operon, whereas in the head-to-head case (B, Right), a successful traversal by SMC may occur only if no RNAP is encountered within a cell-division time (dashed line). (C) Extension of the “moving-barriers” model allowing for condensin to bypass transcribing RNAP (Left, schematic); simulations of the locus-crossing times with varying permeability (“bypass”) rates and RNAP densities (Right); blue region indicates the experimentally estimated time for condensin to cross a 3-kb rRNA gene locus (with density ∼10 RNAP/kb).
The moving-barriers concept can be incorporated into a quantitative model, allowing us to compute the parameter ρ. To obtain theoretical estimates for the times to cross a locus in the head-to-head versus head-to-tail cases, we solved the moving-barriers model analytically (Fig. 4B) (SI Appendix, section 5). The ratios of the calculated head-to-head to baseline operon crossing times gave us a theoretical local value of ρ as a function of operon lengths and RNAP density and produced a strong directional effect with ρ >> 1, as desired. However, for characteristic rRNA operon lengths (10 kb) and average rRNA locus densities (∼10 RNAP/kb), the calculated directionality parameter produced values of ρ > 104 (SI Appendix, Fig. S9A), far exceeding the sought range of ρ ≈ 20–100 (SI Appendix, Fig. S1A). These calculations raise the possibility that translocating condensins can somehow bypass (“hop over”) elongating RNAP.
As a consistency check, we investigated whether the moving-barriers model would support the observations that transcription inhibition (Fig. 4B) and RNAP degradation (SI Appendix, Fig. S7A) resulted in only a minor change to the tilt of the secondary diagonal. Using the moving-barriers model, we calculated the relative contribution to our parameter γ due to transcription elongation at regular operons (i.e., non-rRNA). For operons of length ∼3 kb, and average RNAP densities of ∼0.1 RNAP/kb, the ratio of head-to-head versus head-to-tail crossing times was ∼1.3 (SI Appendix, section 5). The ∼1.3-fold relative slowdown suggests that active transcription can only account for a γ value up to ∼1.3. Since γ is found to be between 2 and 7 (SI Appendix, Fig. S1A), this suggests that transcription elongation can contribute only up to 30% of the observed tilt. If condensins can bypass RNAPs (as we will see below), then the upper limit of 30% will be further reduced. Thus, the model agrees with the apparent lack of change in the secondary diagonal tilt at non-rRNA loci. Interestingly, this calculated fold increase in crossing times between the head-to-tail and head-to-head encounters agrees well with other recent experimental results (28). In C. crescentus, the enrichment of SMCs within genes was up to 1.4-fold larger, depending on whether the genes transcribed “against” or “with” the direction of condensin translocation (see figure 5 in ref. 28). This suggests that the moving-barriers concept is not only applicable to B. subtilis genes, but is a general feature of SMC interactions with transcription in other organisms.
Translocating Condensins Can Efficiently Bypass Sites of Active Transcription.
To study the possibility that translocating condensin can bypass transcribing RNAP, we generalized our moving-barriers model by introducing a finite permeability to the barrier. In the permeable moving-barriers model, condensins that are hindered by a transcription complex can bypass it with a characteristic rate, µ (Fig. 4 C, Left and SI Appendix, Fig. S10A), i.e., pausing at each RNAP for on average 1/µ s. The limiting case µ → 0 s−1 is the impermeable-barriers model (Fig. 4A), and the limit µ → ∞ s−1 is where condensins do not interact with RNAP at all.
We studied the model analytically and performed 1D simulations of the RNAP and condensin translocation with varying permeability (or bypass) rates, µ, and computed the average times for condensin to cross operons of various lengths (1–10 kb). We searched for permeability rates which would reproduce rRNA head-to-head and head-to-tail locus crossing times measured by Hi-C as well as the parameter ρ ≈ 20. Time-course Hi-C data indicated that to cross the clusters of rRNA operons near the ori (e.g., parS+26° strain), it takes <1 min for condensins traveling along the direction of transcription (SI Appendix, Fig. S3A) and between 8 and 15 min against transcription (SI Appendix, Fig. S3B). Assuming RNAP densities of ∼10 RNAP/kb, we found from simulations that the permeability rate µ ∼ 0.8–1.6 s−1 was most consistent with the experimental data on rRNA locus-crossing times and µ ∼ 0.6–1.7 s−1 from the analytical model (Fig. 4C, SI Appendix, Fig. S10B, and SI Appendix, section 6.2). Reassuringly, this range of rates also reproduced the value ρ ≈ 20 required to reproduce condensin traces in Hi-C data (SI Appendix, Fig. S10B). Together, these results suggest that in crossing the rRNA locus, condensins stall for 0.5–2 s at each elongating RNAP molecule before bypassing it. Curiously, the rate at which SMCs bypass RNAP molecules (0.5–1.6 s−1) is very close to the rate of ATP consumption by SMCs (∼0.7 ± 0.1 ATP/condensin/s) (29), suggesting that a few (∼1 to 5) ATP cycles (or SMC “step”) are required for condensin to bypass an elongating transcription complex. Interestingly, these estimates suggest that most genes (due to low levels of transcription compared to rRNA genes and high permeability values) will not significantly slow down condensin translocation; this is consistent with the small effect of transcription inhibition on the “baseline asymmetry” of chromosome juxtaposition (i.e., parameter γ).
We next sought to independently validate the permeability-rate estimates measured above. The analytical formulation of the permeable moving-barriers model makes several predictions. While most genes will not measurably change the overall condensin trajectories visible by Hi-C, they will leave signatures in SMC accumulation patterns measurable by ChIP-seq. We made 3 predictions: First, there will be a positive correlation between RNAP and SMC ChIP-seq signals. Second, the model predicted a nonuniform SMC accumulation pattern within operon bodies: Wherever RNAP accumulates within a gene body, our theory predicted that SMCs will also accumulate. Additionally, even for uniform RNAP distributions, simulations of head-to-head encounters suggested that for high RNAP densities and inferred permeability rates (e.g., 10 RNAP/kb and µ = 0.8 s−1), there will be a strong accumulation of SMC at transcription termination sites, whereas for lower RNAP densities and similar rates (e.g., 1 RNAP/kb and µ = 0.8 s−1), the SMC distribution will be more uniform (SI Appendix, Fig. S10C); the pattern and strength of accumulation is a function of the permeability rate, RNAP density, and distance from the transcription start site (SI Appendix, Fig. S10C). Third, and most importantly, the model predicted a stronger ChIP-seq enrichment for condensins crossing operons in the head-to-head versus head-to-tail directions (SI Appendix, Fig. S10 B and C).
Thus, to test the permeable moving-barriers model, we performed ChIP-seq for RNAP and condensin. As expected, we found a strong positive correlation between RNAP and SMC ChIP-seq signals (SI Appendix, Fig. S11A) (Pearson correlation coefficient, R = 0.51, P < 10−28). Then, visualizing SMC and RNAP ChIP-seq signals alongside genome annotations, we found that wherever RNAP accumulated, SMC also accumulated, consistent with the analytical theory (Fig. 5A). Interestingly, the reverse was not always true (Fig. 5A); this suggests that RNAPs can be barriers to translocating condensins, but that other DNA-bound proteins may also be barriers; consequently, this makes our estimates of the permeability rates lower bounds on the true rates. Lastly, to probe for the gene-body- and gene-direction-dependent SMC accumulation, we performed an aggregate analysis of SMC accumulation within operons. We found that SMC accumulated 2 times more strongly in operons where the transcription direction opposed condensin’s translocation direction (Fig. 5B and SI Appendix, Fig. S11B), consistent with our simulations (Fig. 5C) and analytical model (SI Appendix, sections 6.3–6.7). The simulations with a range µ ∼0.1–0.8 s−1 were in the best agreement with the SMC ChIP-seq data using estimated numbers of RNAP (∼1–3 RNAPs per transcription burst) (ref. 54; Fig. 5C and SI Appendix, Fig. S10B); the analytical model suggested µ ∼ 0.12–0.36 s−1 for the same values (SI Appendix, section 6.6). Thus, 2 independent sets of data (derived from Hi-C and ChIP-seq) produced similar results within the permeable moving-barrier model framework for rRNA loci and protein-coding loci. Together, these analyses lend strong support for the idea that elongating RNAPs can push translocating condensins and support our findings that condensins bypass the elongating RNAPs with high efficiencies (Fig. 5D).
Fig. 5.
Evidence for the “permeable moving barriers” model of condensin–transcription interactions. (A) Comparison of SMC ChIP-seq tracks (anti-SMC) with RNAP tracks (anti-GFP and RpoC-GFP) showing that RNAP colocalizes with SMC, but SMC peaks may occur without RNAP; operon locations are shown. (B) Fold difference of ChIP-seq signal for SMC tracks normalized by RNAP tracks; average signal is shown for all genes of length up to length 1 kb, separated by the direction of SMC translocation relative to transcription direction (SI Appendix, section 1.3). (C) Simulation SMC ChIP-seq for 1-kb gene, demonstrating the differential accumulation of SMC within the gene body for the “moving barriers with permeable boundaries” model; a permeability rate of 0.5 s−1 well describes the ∼2-fold change in experimentally observed head-to-tail versus head-to-head SMC accumulation. (D) Summary model: Condensins (possibly oligomers) translocate away from the parS loading site by 2 independent (blue and red) motor activities; condensin motors can bypass steric barriers (like transcription machinery or other DNA-bound proteins) which are of similar size or larger than the condensin lumen; while condensin attempts to bypass a steric barrier, it may be “pushed” by other translocating factors like RNAP, leading to transcription-dependent SMC translocation rates.
Discussion
Collectively, our analysis leads to the permeable moving-barriers model (Fig. 5D). The model, which is quantitative in nature, makes several yet untested predictions. It suggests that by increasing the density of transcribing RNAPs beyond a critical value, actively transcribed loci will become directionally impermeable to condensin translocation within physiological time scales. For example, it is highly improbable for a condensin to cross a 10-kb operon with a density of ∼20 RNAP/kb in the head-to-head orientation within ∼35 min (or one cell division time), but it will bypass the same operon in the head-to-tail orientation within 30 s. Prior experimental observations have demonstrated that condensin translocation speed can be directionally slowed down by the rRNA locus (8, 28) and can result in a gene-direction-dependent build-up of SMC at other highly expressed operons (28, 55), but the impermeability of a transcribed locus to condensin has not been shown. A Hi-C experiment whereby the transcription levels are increased at a specific locus may test the prediction of a directionally impermeable locus, and our quantitative analyses can provide a guide to estimate conditions (based on transcript length and RNAP density) when a locus can totally block condensin translocation.
The permeable moving-barriers model posits that transcription elongation is the key driver of the directional slowing down of condensin by RNAP. One can hypothesize that RNAP has a directional effect on condensin translocation, like the effect of CTCF proteins on cohesin in eukaryotes (4). The similar effects of rifampicin treatment (where some RNAPs remain stalled at promoters) and RNAP degradation on the bacterial Hi-C maps rule out the possibility that RNAPs are strong “CTCF-like” barriers to condensin translocation. However, we cannot rule out the possibility of a weak “CTCF-like” effect by RNAP on condensin speed. To rigorously test this assumption of our model, future experiments will have to decouple the effects of density of RNAP in genes from the physical process of RNAP elongation. This may involve performing Hi-C on cells treated with drugs that stall RNAP elongation while preserving the RNAP density.
Predictions of the permeable moving-barriers mechanism can also be tested by single-molecule experiments similar to those that observed ATP-dependent condensin translocation (9) and loop extrusion (10) in vitro. For example, in head-to-head “collision” types of experiments, bypassing of elongating RNAP by condensin, and pushing/pausing of condensins for the predicted ∼1 to 2 s may be measured; however, while observing the directional effect on a single RNAP on condensin could be difficult, our model suggests that a train of transcribing RNAPs will better help measure the permeability value (i.e., the rate at which condensin bypasses DNA-bound obstacles like RNAP). Our analyses of SMC ChIP-seq data and estimates of the permeability rates (i.e., µ ∼ 0.1–0.8 s−1 for protein-coding sequences versus µ ∼ 0.8–1.6 s−1 for rRNA loci) also raise the interesting possibility that different types of genes (i.e., coding versus noncoding) may have different permeabilities. The lower permeability rate in protein-coding genes could arise from a higher steric hindrance imposed by ribosomes that bind to nascent RNA during transcription in protein coding, but not rRNA genes. This can be tested in vitro by attaching fluorophores or beads of different sizes to a transcribing RNAP or by increasing the length of the transcribed RNA (see below), potentially achieving different rates of permeability.
The permeable moving-barriers mechanism proposed here for bacteria may be a general mechanism with implications in eukaryotes as well. There is growing evidence that transcription can affect cohesin SMC localization in yeast (50) and mammalian cells (32, 35) and can potentially interfere with the process of loop extrusion by cohesin. Although the size of isolated eukaryotic RNAPs are comparable to their bacterial counterparts [∼10–15 nm in diameter (56)], sizes of whole elongating transcription complexes in eukaryotes can be considerably larger. A typical 1.5- to 3-kb bacterial operon generates RNA of physical dimensions up to ∼60 × 30 × 10 nm (57). Together with coupled transcription and translation machinery, we estimate that the transcription complex will have a globular diameter of 45–60 nm (SI Appendix, section 6.8), which could be just small enough to pass through the SMC lumen (Fig. 5D). In comparison, the average human gene is ∼27 kb (58), increasing the linear dimensions of the RNA molecule by ∼2-fold (see SI Appendix, section 6.8 for further details). While eukaryotes do not have coupled transcription and translation, there is transcription-coupled splicing, and the spliceosome is of similar size to the ribosome (58), making the whole complex of a globular diameter > 70 nm. Future experiments will be needed to test whether SMCs can bypass steric barriers larger than what fits through their lumen. Of particular interest, a recent preprint has shown that purified yeast condensins that extrude loops on DNA can bypass one another with short pauses of ∼7 s (59); this is similar to the time we measured for bacterial condensins to bypass elongating transcripts. If eukaryotic RNAPs turn out as permeable to SMC as bacterial RNAPs, transcription in eukaryotes may not have a major effect on chromosome organization by loop extrusion, as compared to other molecules that specifically (and directionally) impede loop extrusion, like CTCF (20, 60). Nevertheless, since SMC and CTCF interactions in eukaryotes play a regulatory role in gene expression (61), it is of interest to explore the potential role of SMC–transcription interactions in gene regulation in both bacteria and eukaryotes.
The remarkable ability of loop-extruding condensin SMCs to bypass large elongating transcription complexes in bacteria is important from both molecular and evolutionary standpoints. While the molecular mechanisms of loop extrusion by SMCs remain to be elucidated, our results suggest that to overcome large steric barriers, either: 1) a sufficiently large opening (large enough to fit the entire transcription complex) emerges in the SMC complex lumen during an SMC ATPase cycle, and SMC loop extrusion proceeds by maintaining DNA in a topological embrace [with mechanisms like “inchworm,” “pumping,” “segment-capture,” and “shackled-walker” (9, 14, 25, 62–64)]; or 2) the SMC rings transiently open (65), disengage the topological embrace of the DNA, and reengage after passing the steric barrier [closer to the “walker”/“rock-climber” models (39, 40)].
Furthermore, our work suggests a possible link between the SMCs’ ATP hydrolysis rate and the rate at which the SMC complex can bypass sites of active transcription or other obstacles. We found that condensins pause for about 1 (or a few) ATP hydrolysis cycles before bypassing RNAP. This suggests that the rate of bypassing will differ between types of SMC complexes, depending on their respective ATP hydrolysis rates. For instance, yeast cohesin SMCs are shown to have lower ATP hydrolysis rates in vitro of <0.2 ATP/cohesin/s (66, 67), whereas yeast condensins have higher rates of ∼1.5 ATP/condensin/s (9) compared to B. subtilis SMC’s 0.7 ATP/condensin/s (29).
Another surprising result is that loop extrusion activity occurred by 2 effectively uncoupled motor activities in vivo; i.e., occlusion of 1 motor did not affect the translocation of the other. This could suggest that linked dimers of condensins each separately perform directional translocation (thus loop extrusion) (8, 40). We note, however, that this does not preclude the possibility that a single SMC complex performs the 2 motor activities: For instance, an anchored “1-sided” SMC extruder can alternate the anchoring site and the “DNA reeling arm,” effectively performing 2-sided extrusion with uncoupled kinetics (41); this will be a topic of future study.
Irrespective of the molecular details, the ability to overcome steric barriers is likely an important evolutionary adaptation for SMCs to organize chromosomes (e.g., to resolve sister chromatids in bacteria and to form domains and compacted genomes in eukaryotes) largely unobstructed by active transcription. Permeability of RNAPs to loop extrusion also suggests that SMCs should be able to effectively bypass other large steric barriers, such as nucleosomes in eukaryotes [consistent with experimental evidence, where nucleosome depletion does not affect condensin’s ability to form a mitotic chromosome (68) and a recent preprint showing that human condensins can bypass nucleosome-bound DNA (69)], as well as long plectonemes (22, 31) and other DNA-bound proteins [as suggested for the SMC homolog MukBEF (70)] in bacteria. It remains to be seen how bigger molecular complexes—e.g., replication machinery—can interfere with the process of loop extrusion.
In summary, our analyses suggest that bacterial condensin’s loop-extrusion activity occurs by 2 effectively independent and uncoupled motor activities in vivo. Further, it appears that 2 major processes may be at play in shaping the genome-wide condensin trajectories and, hence, chromosome organization. The first is a transcription-independent mechanism that slows down loop extrusion when condensin proceeds toward the origin. The second is a transcription-elongation-dependent effect at highly transcribed loci like rRNA operons. Most crucially, our models with their inferred parameters show how SMC translocation speeds can vary as they progress through the genome; we show that the speed of extrusion is slowed down by interactions with transcription machinery and depends on the relative directions of transcription and extrusion. Our permeable moving-barrier models show that trains of RNAPs can serve as directional barriers to extrusion, with individual RNAPs having only a modest effect on translocating condensin, by pausing and pushing it back for a mere ∼2 s at rRNA loci and ∼10 s at protein-coding loci (Fig. 5D). In all, our work provides a quantitative and predictive framework to study the dynamics of SMC complexes and their interactions with other translocating DNA-bound complexes in vivo and in vitro.
Supplementary Material
Acknowledgments
We thank Johannes Nuebler, Ed Banigan, John Marko, Stuart Sevier, Jean-Benoit Lalanne, Maxim Imakaev, Geoff Fudenberg, Anders Hansen, and Stephan Gruber for helpful insights and suggestions and/or feedback on the manuscript. We are especially grateful to Tung Le for sharing data and discussions. H.B.B. was supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarships–Doctoral fellowship. This work was supported by NIH Grants GM086466 and GM073831 (to D.Z.R.) and GM114190 (to L.A.M.); and start-up funds from Indiana University (X.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.M.-R. is a guest editor invited by the Editorial Board.
Data deposition: Source codes have been made available at https://github.com/hbbrandao/bacterialSMCtrajectories. Data have been deposited at the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE117854).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907009116/-/DCSupplemental.
References
- 1.Hirano T., Condensin-based chromosome organization from bacteria to vertebrates. Cell 164, 847–857 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Alipour E., Marko J. F., Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 40, 11202–11212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fudenberg G., et al. , Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanborn A. L., et al. , Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E6456–E6465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riggs A. D., DNA methylation and late replication probably aid cell memory, and type I DNA reeling could aid chromosome folding and enhancer function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326, 285–297 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Nasmyth K., Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673–745 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Wang X., et al. , Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29, 1661–1675 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Brandão H. B., Le T. B. K., Laub M. T., Rudner D. Z., Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 355, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terakawa T., et al. , The condensin complex is a mechanochemical motor that translocates along DNA. Science 358, 672–676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganji M., et al. , Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eeftens J. M., et al. , Real-time detection of condensin-driven DNA compaction reveals a multistep binding mechanism. EMBO J. 36, 3448–3457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keenholtz R. A., et al. , Oligomerization and ATP stimulate condensin-mediated DNA compaction. Sci. Rep. 7, 14279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H., Loparo J. J., Multistep assembly of DNA condensation clusters by SMC. Nat. Commun. 7, 10200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marko J. F., De Los Rios P., Barducci A., Gruber S., DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res. 47, 6956–6972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fudenberg G., Abdennur N., Imakaev M., Goloborodko A., Mirny L. A., Emerging evidence of chromosome folding by loop extrusion. Cold Spring Harb. Symp. Quant. Biol. 82, 45–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gassler J., et al. , A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J. 36, 3600–3618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haarhuis J. H. I., et al. , The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao S. S. P., et al. , Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzer W., et al. , Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wutz G., et al. , Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marbouty M., et al. , Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 59, 588–602 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Le T. B. K., Imakaev M. V., Mirny L. A., Laub M. T., High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342, 731–734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber S., Errington J., Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137, 685–696 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Sullivan N. L., Marquis K. A., Rudner D. Z., Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137, 697–707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilhelm L., et al. , SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4, e06659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minnen A., et al. , Control of Smc coiled coil architecture by the ATPase heads facilitates targeting to chromosomal ParB/parS and release onto flanking DNA. Cell Rep. 14, 2003–2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marbouty M., et al. , Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. eLife 3, e03318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran N. T., Laub M. T., Le T. B. K., SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 20, 2057–2071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., et al. , In vivo evidence for ATPase-dependent DNA translocation by the Bacillus subtilis SMC condensin complex. Mol. Cell 71, 841–847.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miermans C. A., Broedersz C. P., Bacterial chromosome organization by collective dynamics of SMC condensins. J. R. Soc. Interface 15, 20180495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le T. B. K., Laub M. T., Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. EMBO J. 35, 1582–1595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S., et al. , Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowley M. J., et al. , Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlmann F., SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Busslinger G. A., et al. , Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544, 503–507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowley M. J., et al. , Condensin II counteracts cohesin and RNA polymerase II in the establishment of 3D chromatin organization. Cell Rep. 26, 2890–2903.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merrikh H., Zhang Y., Grossman A. D., Wang J. D., Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 10, 449–458 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Touzain F., Petit M.-A., Schbath S., El Karoui M., DNA motifs that sculpt the bacterial chromosome. Nat. Rev. Microbiol. 9, 15–26 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Badrinarayanan A., Reyes-Lamothe R., Uphoff S., Leake M. C., Sherratt D. J., In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338, 528–531 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zawadzka K., et al. , MukB ATPases are regulated independently by the N- and C-terminal domains of MukF kleisin. eLife 7, e31522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banigan E. J., Mirny L. A., Limits of chromosome compaction by loop-extruding motors. Phys. Rev. X 9, 031007 (2019). [Google Scholar]
- 42.Mosteller R. D., Yanofsky C., Transcription of the tryptophan operon in Escherichia coli: Rifampicin as an inhibitor of initiation. J. Mol. Biol. 48, 525–531 (1970). [DOI] [PubMed] [Google Scholar]
- 43.Pato M. L., von Meyenburg K., Residual RNA synthesis in E. coli after inhibition of initiation of transcription by rifampicin. Cold Spring Harb. Symp. Quant. Biol. 35, 497–504 (1970). [Google Scholar]
- 44.Villain-Guillot P., Bastide L., Gualtieri M., Leonetti J. P., Progress in targeting bacterial transcription. Drug Discov. Today 12, 200–208 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Chen H., Shiroguchi K., Ge H., Xie X. S., Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 11, 781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffith K. L., Grossman A. D., Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol. Microbiol. 70, 1012–1025 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolas P., et al. , Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Klumpp S., Hwa T., Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 20245–20250 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel U., Jensen K. F., The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 176, 2807–2813 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lengronne A., et al. , Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573–578 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizuguchi T., et al. , Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 516, 432–435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson I. F., et al. , Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 35, 2671–2685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M. D., et al. , Force and velocity measured for single molecules of RNA polymerase. Science 282, 902–907 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Golding I., Paulsson J., Zawilski S. M., Cox E. C., Real-time kinetics of gene activity in individual bacteria. Cell 123, 1025–1036 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Vazquez Nunez R., Ruiz Avila L. B., Gruber S., Transient DNA occupancy of the SMC interarm space in prokaryotic condensin. Mol. Cell 75, 209–223.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milo R., Phillips R., Cell Biology by the Numbers (Garland Science, Taylor & Francis Group, LLC, Abindgon, UK, 2016). [Google Scholar]
- 57.Gopal A., Zhou Z. H., Knobler C. M., Gelbart W. M., Visualizing large RNA molecules in solution. RNA 18, 284–299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson J. D., et al. , Molecular Biology of the Gene (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, ed. 7, 2014). [Google Scholar]
- 59.Kim E., Kerssemakers J., Shaltiel I. A., Haering C. H., Dekker C., DNA-loop extruding condensin complexes can traverse one another. bioRxiv:10.1101/682864 (26 June 2019). [DOI] [PubMed]
- 60.Nora E. P., et al. , Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–944.e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Y., et al. , CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diebold-Durand M.-L., et al. , Structure of full-length SMC and rearrangements required for chromosome organization. Mol. Cell 67, 334–347.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bürmann F., et al. , A folded conformation of MukBEF and cohesin. Nat. Struct. Mol. Biol. 26, 227–236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nichols M. H., Corces V. G., A tethered-inchworm model of SMC DNA translocation. Nat. Struct. Mol. Biol. 25, 906–910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eeftens J. M., et al. , Condensin Smc2-Smc4 dimers are flexible and dynamic. Cell Rep. 14, 1813–1818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murayama Y., Uhlmann F., Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505, 367–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murayama Y., Uhlmann F., DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell 163, 1628–1640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shintomi K., et al. , Mitotic chromosome assembly despite nucleosome depletion in Xenopus egg extracts. Science 356, 1284–1287 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Kong M., et al. , Human condensin I and II drive extensive ATP-dependent compaction of nucleosome–bound DNA. bioRxiv:10.1101/683540 (27 June 2019). [DOI] [PMC free article] [PubMed]
- 70.Lioy V. S., et al. , Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172, 771–783.e18 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.