Significance
The study addresses a basic immunology topic with considerable clinical relevance, namely the nature of the pathogenic T helper cells that drive autoimmune disorders like multiple sclerosis (MS) and inflammatory arthritis. Using mouse MS, we identify precursors of the encephalitogenic/pathogenic T cells based on their dependence on the protease inhibitor SerpinB1. Newly identified signature genes reveal the unusual nature of these T cells that combine (i) inflammatory cytokine secretion, (ii) a cytolytic system, and (iii) extreme rapid proliferation. We demonstrate that their survival/expansion depends on SerpinB1 and involves regulation of a proliferation-associated protease-mediated cell suicide mechanism. Importantly, we discovered that cell surface CXCR6 is an exquisite marker of pathogenic T helper cells and demonstrated that anti-CXCR6 treatment has potential to prevent or mitigate MS.
Keywords: serpins, multiple sclerosis, inflammatory arthritis, pathogenic T helper cells, autoimmune
Abstract
SerpinB1, a protease inhibitor and neutrophil survival factor, was recently linked with IL-17–expressing T cells. Here, we show that serpinB1 (Sb1) is dramatically induced in a subset of effector CD4 cells in experimental autoimmune encephalomyelitis (EAE). Despite normal T cell priming, Sb1−/− mice are resistant to EAE with a paucity of T helper (TH) cells that produce two or more of the cytokines, IFNγ, GM-CSF, and IL-17. These multiple cytokine-producing CD4 cells proliferate extremely rapidly; highly express the cytolytic granule proteins perforin-A, granzyme C (GzmC), and GzmA and surface receptors IL-23R, IL-7Rα, and IL-1R1; and can be identified by the surface marker CXCR6. In Sb1−/− mice, CXCR6+ TH cells are generated but fail to expand due to enhanced granule protease-mediated mitochondrial damage leading to suicidal cell death. Finally, anti-CXCR6 antibody treatment, like Sb1 deletion, dramatically reverts EAE, strongly indicating that the CXCR6+ T cells are the drivers of encephalitis.
Multiple sclerosis (MS) and murine experimental autoimmune encephalomyelitis (EAE) are chronic demyelinating disorders of the central nervous system driven by self-reactive TH cells (1). The disease-inducing autoimmune T cells, which are present at low numbers in the periphery and as expanded populations in the central nervous system (CNS), were initially thought to be TH1 cells because disease is abrogated by deletion of the interleukin (IL)-12 subunit p40 (2, 3). With the discoveries that p40 is also a subunit of IL-23 and IL-23 plays a pivotal role in mediating disease (4–7), MS was reinterpreted as TH17-driven (8, 9). More recent studies established that TH17 cells themselves are not pathogenic but are converted in vivo under the priming of myeloid cell-derived IL-1β and IL-23 into pathogenic (encephalitogenic) TH cells, the true drivers of disease (4, 10–14). These cells produce interferon (IFN)γ and granulocyte-macrophage colony-stimulating factor (GM-CSF) (10, 15–18), the latter required for encephalitogenicity (16–18). Despite the importance of the encephalitogenic TH cells, little else is known about their nature or the factors and pathway that drive their development.
In cytolytic CD8 cells and NK cells, powerful granule serine proteases that are regulated by endogenous inhibitors called serpins play pivotal roles in immune surveillance against tumors and viral infection while simultaneously maintaining immune homeostasis (19–26). Whether analogous granzyme-serpin regulation exists in CD4 cells is not known. SerpinB1 (Sb1), previously called MNEI (monocyte/neutrophil elastase inhibitor), is an ancestral member of the superfamily of serpins (SERine Protease Inhibitors). It is a highly efficient inhibitor of elastinolytic and chymotryptic proteases that has been best studied in neutrophils (27–31). For example, in bacterial lung infection, Sb1 protects against inflammatory tissue injury and neutrophil death, and in naïve mice, preserves the bone marrow reserve of mature neutrophils by restricting spontaneous cell death mediated by the granule serine proteases cathepsin G and proteinase-3 (32–35). Recently, we and others demonstrated that Sb1 selectively restricts expansion of IL-17–expressing γδ T cells (36) and NK T cells (37), findings that led us to study adaptive Th cell development where we identified Sb1 as a signature gene of TH17 cells (38).
Here, we report that Sb1 expression is required for optimal development of paralysis in MOG-immunized mice. We identified a highly selective subset of IFNγ- and GM-CSF–expressing IL17+ Sb1-dependent CD4 cells in the periphery of immunized mice at onset of disease. We describe here isolation of these Sb1-dependent primed T cells and their molecular and functional signature and developmental pathway in EAE, and we demonstrate that these are the T helper cells responsible for disease.
Results
Sb1 Is Highly Expressed in TH Cells in EAE.
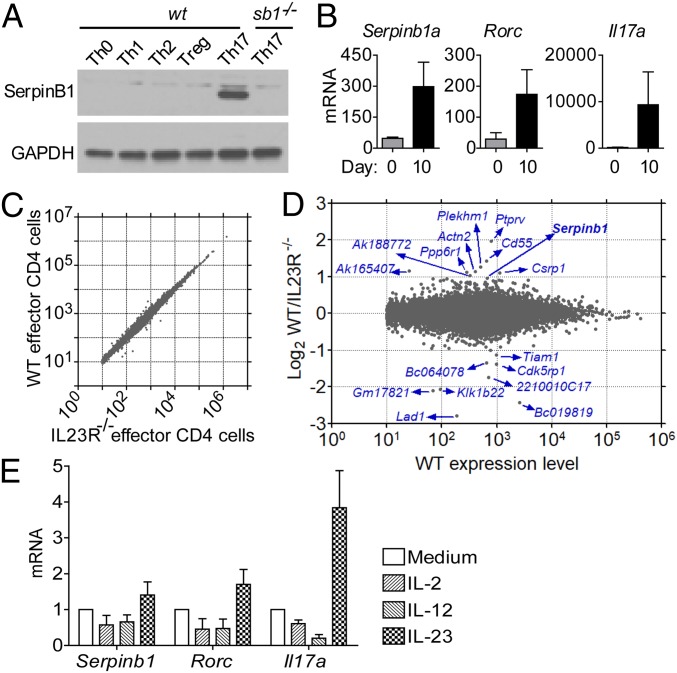
Previously, we identified Sb1 as preferentially expressed in TH17 cells by studying 129S6 strain mouse cells in vitro. In preparation for working with the EAE model, we polarized naïve CD4 cells of C57BL/6 mice to TH17 cells driven by IL-6 and TGFβ (Fig. 1A and SI Appendix, Fig. S1A) and we confirmed select expression of Sb1, consistent with previous findings (38) (Fig. 1A). We then showed that Sb1 is also dramatically up-regulated in vivo along with Rorc and Il17a in effector (CD44hi) CD4 cells during EAE development (Fig. 1B). To date, the factors controlling expression of Sb1 in TH cells are unknown. Online gene arrays for mice deleted for the Th17 inducer serine/threonine protein kinase (Sgk)-1 (39) revealed that Sb1 is among the top down-regulated genes in IL23-stimulated Sgk1-deficient TH17 cells, suggesting a correlation between Il23r and Sb1 in TH17 cells. To investigate this putative link, we generated mice with Il23r deleted in CD4 cells (Il23rΔCD4). We then compared the transcriptome of wt and Il23rΔCD4 effector (CD44hiCD62Llo) CD4 cells from the lymph node (LN) of MOG-immunized mixed chimeric mice at onset of EAE. Surprisingly, wt and Il23rΔCD4 effector CD4 cells were not very different at the transcriptome level (Fig. 1C), and only few genes were decreased more than 2fold in Il23rΔCD4 compared with wt cells (Fig. 1D). Among the prominent genes with skewed expression and known immune function, we found Sb1, confirming a critical role of IL-23R signals in inducing or maintaining Sb1 expression in effector CD4 cells at the onset of EAE. To investigate the effects of IL-23 on TH17 cells and Sb1, we returned to the IL-6/TGFβ in vitro differentiation system. Adding IL-23 did not further increase Sb1 expression (SI Appendix, Fig. S1B); however, on restimulation in a two-stage protocol, the addition of IL-23 in the second stage maintained expression of Sb1, Rorc, and Il17 (Fig. 1E).
Fig. 1.
Serpinb1a (Sb1) is highly expressed in TH17 cells and in TH cells in EAE. (A) Sb1 expression in wt T cell subsets differentiated in vitro and analyzed by Western blot. Data are representative of five experiments. (B) qRT-PCR analysis of CD44+ (effector) CD4 cells isolated from LN of naïve mice (day 0) and MOG/CFA immunized mice at onset of EAE (day 10). Expression levels were normalized relative to CD44neg (naïve) CD4 cells isolated from LN of naïve mice. Depicted data are mean ± SEM for pooled cells of 3 to 9 mice per genotype in three experiments. (C and D) RNA-sequencing (seq) analysis. Mixed chimeric mice (CD45.1 wt/CD45.2 Il23rΔCD4) were immunized with MOG/CFA to induce EAE. On day 9, effector (CD44hiCD62Llo) CD4 cells were sorted from draining LNs. (C) Gene expression in wt and Il23rΔCD4 effector CD4 cells. Data are mean of five replicates with 3 to 4 chimeric mice per replicate. (D) Top hits with identities. (E) IL-23 treatment maintains expression of Sb1, Rorc, and Il17a in Th17 cells. In vitro-differentiated TH17 cells were maintained in IL-2 for 2 d and restimulated with anti-CD3/CD28 and the indicated cytokine for 24 h and then analyzed by qRT-PCR. Data are mean ± SEM of three experiments.
EAE Amelioration Due to Deficiency of Sb1 in CD4 Cells.
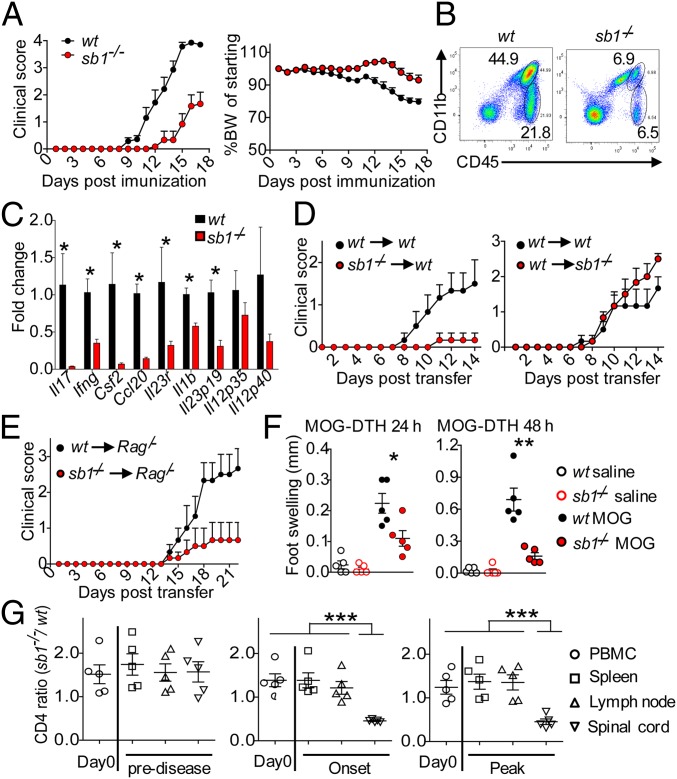
To determine whether the expression of Sb1 affects the encephalitogenic TH cells, we induced EAE in Sb1−/− mice. Compared to the severe encephalomyelitis that developed in MOG-immunized wt mice, Sb1−/− mice exhibited delayed and ameliorated disease (Fig. 2A). Fewer leukocytes, both lymphocytes and myeloid cells, infiltrated the spinal cord (Fig. 2B). The deficit of cells was reflected at the mRNA level in the decrease of TH and myeloid cell cytokines (Fig. 2C). Because Sb1 is expressed in multiple cells and very prominently in myeloid cells, we performed adoptive transfer studies to identify the cellular source of Sb1 that controls encephalitogenicity. We found that, compared to wt T cells, Sb1−/− T cells of immunized mice were less encephalitogenic. Moreover, disease was not ameliorated in the reciprocal experiment (Fig. 2D), solidifying the notion that Sb1 influences the pathogenic potential of T cells. In a complementary model, naïve CD4 cells were transferred into Rag1/− mice prior to immunization. Clinical disease was attenuated in mice receiving Sb1−/− CD4 cells compared to mice receiving wt CD4 cells, and immune cell accumulation in spinal cord was blunted (Fig. 2E). Further, comparison of the delayed-type hypersensitivity (DTH) response of MOG-immunized wt and Sb1−/− mice showed that T cell priming was already impaired in the periphery in Sb1−/− mice (Fig. 2F). Finally, a mixed chimeric mouse model revealed that the ratio of Sb1−/− to wt CD4 cells in the periphery did not change following MOG immunization, but the Sb1−/− to wt CD4 cell ratio decreased in the spinal cord (Fig. 2G), a phenotype that largely replicates that of wt:Il23r-deficient mixed chimeric mice (11). The cumulative findings indicate that attenuation of encephalomyelitis and paucity of immune cells in the spinal cord of Sb1−/− mice are due to Sb1 absence in CD4 cells.
Fig. 2.
CD4 cell autonomous deficiency of Sb1 ameliorates EAE. (A–C) Wt and Sb1−/− mice were immunized with MOG/CFA to induce EAE. (A) Mean clinical score (Left) and body weight (Right) of wt (n = 13) and Sb1−/− (n = 14) mice. Experiment was repeated more than 5 times with the same pattern. (B) Spinal cord infiltrates on day 10 analyzed by FACS. n = 4–5 mice in each genotype, shown are representative of five experiments. (C) Relative gene expression of spinal infiltrates analyzed by qRT-PCR. Data represent mean of four biological replicates, each with pooled cells from 2 to 3 mice per genotype. (D) Adoptive transfer EAE. Wt or Sb1−/− T cells from MOG-immunized mice were expanded ex vivo and transferred to naive wt or Sb1−/− recipients. Mean clinical scores for 6 mice each genotype. (E) Naive CD4 cell transfer EAE. Wt or Sb1−/− naïve CD4 cells were transferred to Rag1/− mice, which were then MOG-immunized to induce EAE. Mean clinical scores for 6 mice each genotype. (F) DTH response of wt and Sb1−/− mice to challenge in the footpad with MOG or vehicle on day 6 after MOG immunization. (G) Ratio of Sb1−/− to wt CD4 cells in active EAE in chimeric mice. Symbols indicate individual mice. Data are representative of two (D, F, and G) or three (E) experiments. Error bars indicate ±SEM, *P < 0.05; **P < 0.01 by Student’s t test (C and F); ***P < 0.001 by one-way ANOVA (G).
Sb1 Controls IFNγ+ and GM-CSF+CD4 Cells during CD4 Cell Priming.
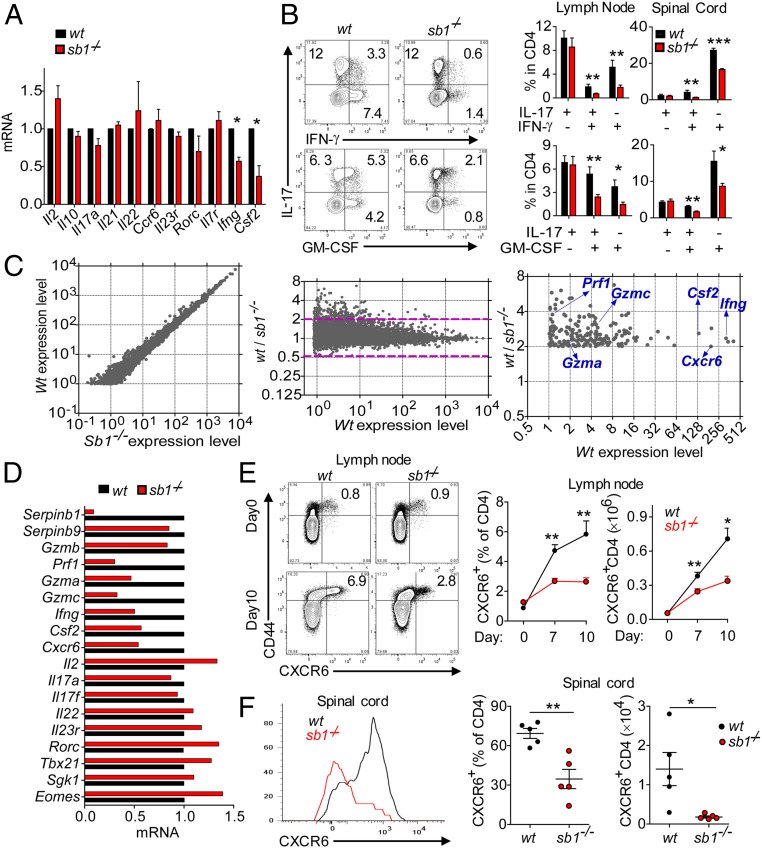
To determine what causes the deficit of spinal cord T cells, we examined draining LN CD4 cells at disease onset. No differences were found between Sb1−/− and wt mice in immune cell counts or frequencies of effector (CD44+) CD4 T cells, T regulatory cells, or CD4 cells expressing CCR2 or CCR6, which are thought to be important for early infiltration of the CNS (40) (SI Appendix, Fig. S2 A–D). There were no differences between the genotypes in recall properties, IL-17 production, responsiveness to IL-23, up-regulation of IL-1R1, TH17 metabolic enzymes, expression of integrins including VLA4 and LFA1, and expression of myeloid cytokines (SI Appendix, Fig. S2 E–J). Moreover, Il23r and many other genes generally associated with TH17 cells are expressed at normal levels in Sb1−/− effector CD4 cells (Fig. 3A). However, expression of Csf2 and Ifng, encoding GM-CSF and IFNγ, respectively, was decreased in LN of Sb1−/− effector CD4 cells compared to corresponding wt cells (Fig. 3A).
Fig. 3.
Decreased frequency of IFNγ+ and GM-CSF+ CD4 cells in LN of Sb1−/− mice provided the key to signature of pathogenic TH cells. (A and B) IFNγ+- and GM-CSF+ CD4 cells at the onset of EAE. (A) Relative gene expression of effector CD4 cells determined by qRT-PCR. Depicted data are mean ± SEM for pooled cells of 3 to 5 mice per genotype in three experiments. (B) Cytokine profiles analyzed by FACS. (Left) Representative contour plots of LN CD4 cells. (Right) Cumulative frequencies for LN and spinal cord CD4 cells. Data for 5 mice per genotype are representative of five experiments. (C) RNA-seq analysis. RNA of wt and Sb1−/− LN effector CD4 cells harvested at disease onset and incubated with P+I. Depicted (Left and Center) are the 9,650 genes with expression levels (FPKM) >1.0. Area above the dashed lines in Center depicts the 258 genes with Sb1−/− expression relative to wt decreased by >2.0-fold. Identities are indicated for the verified genes (Right). (D) Verification by qRT-PCR. Depicted data are representative of two cell isolates analyzed after P+I stimulation. (E and F) CXCR6 expression on CD4 cells of MOG-immunized wt and Sb1−/− mice. Depicted are representative plots (Left) and mean frequencies (Center) and absolute cell numbers (Right) for LNs at indicated days and spinal cord on day 14 (peak disease) (F). Data for 3 to 6 mice per time point per experiment are representative of two experiments. Symbols in F indicate individual mice; error bars represent ±SEM, *P < 0.05; **P < 0.01, ***P < 0.001 by Student’s t test.
To determine whether the decreased expression of Csf2 and Ifng represents decreased cytokine per cell or fewer cytokine-expressing cells, LN and spinal cord CD4 cells were examined by flow cytometry. The frequencies of IL-17 single positive (SP) cells were not different in Sb1−/− and wt mice; however, the frequencies of cytokine double positive (DP) (IL17+IFNγ+, IL17+GM-CSF+) cells as well as GM-CSF SP and IFNγ SP cells were decreased in LNs and spinal cord of Sb1−/− mice (Fig. 3B). TH cells that produce multiple cytokines have been previously described in affected organs of patients with autoimmunity (41, 42), and GM-CSF+ cells are known to be essential for autoimmune neural inflammation (16–18). In the LN of wt and sb1−/− mice, the absolute numbers of cytokine-producing cells reflected the frequency patterns, but in the spinal cord, the absolute numbers of all Sb1−/− CD4 cells were greatly decreased (SI Appendix, Fig. S3A). In MOG-immunized mixed bone marrow chimeras, frequencies of most cytokine DP Sb1−/− CD4 cells were skewed downward (SI Appendix, Fig. S3B). Cumulatively, the findings support the concept that encephalitogenic TH cells, identifiable by production of GM-CSF and IFNγ, are expanded already in the LN of MOG-immunized mice of both genotypes, but the frequency of these cells is decreased in Sb1−/− mice.
Signature Genes Identified for Sb1-Dependent TH Cells in EAE.
Next, we aimed to identify genes that confer encephalitogenic properties to TH cells through Sb1. We profiled the transcriptome of Sb1−/− and wt effector CD4 cells isolated from LN at disease onset, anticipating that other encephalitogenity-conferring genes would be decreased along with Csf2 and Ifng among Sb1−/− effector CD4 cells. Of 9,650 expressed genes, 258 genes were decreased >2fold in Sb1−/− compared with wt cells, and no genes were increased >2fold (Fig. 3C). From among the decreased genes, we selected a subset with immune-related functions for further study. Those verified by qRT-PCR are Ifng and Csf2, as expected, and also Gzmc (GzmC), Gzma (GzmA), and Prf1 (perforin A), which are components of cytotoxic granules (Fig. 3D). Of note, cathepsin L, which promotes differentiation of TH17 cells and is inhibited by Sb1 (38), was not among the genes underexpressed in Sb1−/− effector CD4 cells. The skewed genes included the chemokine receptor Cxcr6 encoding CXCR6 (43), which was verified by both qRT-PCR and flow cytometry.
CXCR6 Marks Sb1-Dependent Encephalitogenic TH Cells.
We next quantified CD4 cells expressing CXCR6 as a function of time during EAE development in wt mice. CXCR6+ CD4 cells, which comprised <1% of CD4 cells in naïve mice, increased after MOG immunization to ∼6% in the LN (Fig. 3E) and constituted the major population (∼70%) in the spinal cord at peak of disease (Fig. 3F). CXCR6+CD4 cells also increased in frequency in LN of Sb1−/− mice postimmunization, but not to the same extent as in wt mice, and failed to accumulate in the Sb1−/− spinal cord. The deficit of CXCR6+CD4 cells in Sb1−/− mice can be best appreciated by comparing absolute cell numbers in the spinal cord (Fig. 3 E and F, Right).
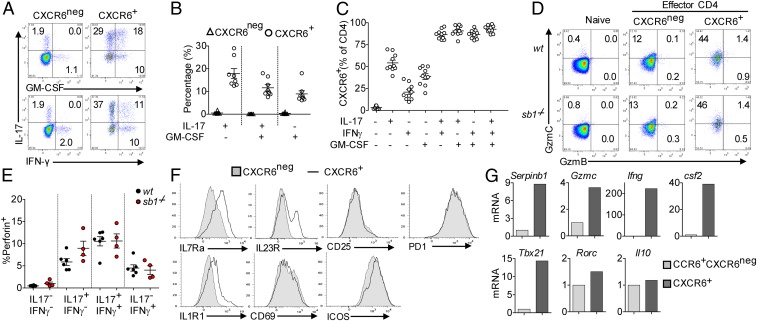
Combined analysis of CXCR6 and cytokines showed that essentially all LN CD4 cells that produce two or more of the cytokines IL-17, GM-CSF, and IFNγ were CXCR6+, as were half of IL-17 SP, a third of GM-CSF-SP, and a smaller fraction of IFNγ-SP cells (Fig. 4 A–C). Strikingly, GzmC, but not GzmB, was preferentially expressed in CXCR6+CD4 cells (Fig. 4D). Concomitantly, perforin-A expression, which was negligible in cytokineneg CD4 cells, was increased in IL-17 SP cells and further increased in IL17/IFNγ DP cells (Fig. 4E). On a “per cell” basis, the content of GzmC and perforin were not different between the genotypes. Except for Csf2 and Ifng, the signature genes Gzmc, Gzma, Prf1, and Cxcr6 identified here for in vivo-generated encephalitogenic TH cells differ from the signature genes of pathogenic TH17 cells generated in vitro (44). Compared with CXCR6neg effector CD4 cells, CXCR6+ effector CD4 cells had increased surface expression of IL7Ra, IL23R, and IL1R1, but not PD-1, ICOS, CD69, and CD25 (Fig. 4F). Compared with conventional TH17 cells (CCR6+CXCR6negCD44+CD4 cells), CXCR6+CD44+CD4 cells in EAE showed increased Sb1, Gzmc, Tbx21, Csf2, and Ifng expression but comparable levels of Rorc and Il10 (Fig. 4G). These findings strongly suggest that the CXCR6+ Sb1-dependent CD4 cells are the TH17-derived encephalitogenic TH cells in EAE.
Fig. 4.
Pathogenic TH cells are marked by CXCR6 and produce multiple cytokines and express GzmC and perforin. MOG-immunized wt mice were killed at onset of EAE, and LN cells were analyzed. (A–C) CXCR6 expression on cytokine-producing CD4 cells. (A) Representative dot plots. (B) Cytokine frequency when gated on CXCR6neg or CXCR6+ CD4 cells. (C) CXCR6 frequency when gated on different cytokine-producing CD4 cells. Cumulative data are from three experiments; symbols indicate individual mice. (D) GzmC and GzmB expression in naïve, CXCR6neg-, and CXCR6+-effector CD4 cells analyzed by FACS. Depicted are representative data of four experiments. (E) Perforin expression in cytokine-producing cells detected by FACS. Symbols indicate individual mice. Data are representative of two experiments. (F) Histograms of IL-7Ra, IL-23R, IL-1R1, CD25, ICOS, PD-1, and CD69 on CXCR6neg and CXCR6+ effector CD4 cells. Depicted data are from pooled cells of 5 to 9 mice wt mice per experiment and are representative of two experiments. (G) Relative gene expression of CCR6+CXCR6neg- and CXCR6+-effector CD4 cells analyzed by qRT-PCR; data are representative of two experiments.
Verification of Encephalitogenic Function of CXCR6+ CD4 Cells.
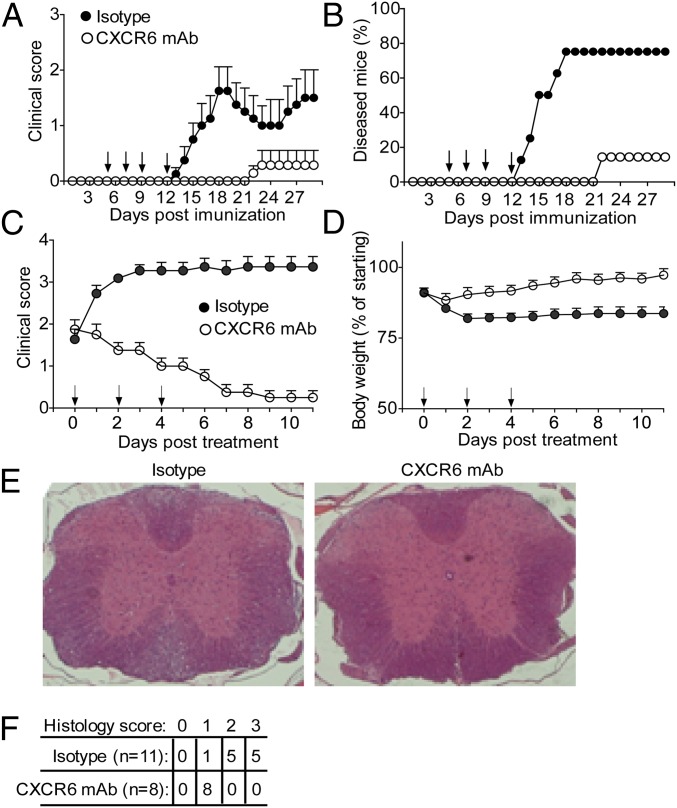
To test the apparent role of CXCR6-marked CD4 cells as mediator of pathogenicity in EAE, we used a cell depletion strategy. A previous study found that disease is not altered in MOG-immunized mice on deletion of CXCR6, indicating that the CXCR6 molecule itself is not required for disease (45). We therefore hypothesized that a CXCR6-directed therapy might be used to deplete encephalitogenic TH cells. In feasibility studies, MOG-immunized wt mice received a single dose of anti-CXCR6 mAb at disease onset and LN cells were examined 24 h later. FACS showed that the GM-CSF/IFNγ DP and GM-CSF SP CD4 cells were decreased in anti-CXCR6–treated mice compared with isotype-treated mice (SI Appendix, Fig. S4A), suggesting successful targeting of CXCR6+CD4 cells. Treating immunized mice with four doses of anti-CXCR6 mAb starting before appearance of symptoms (“prevention protocol”) largely abrogated clinical disease (Fig. 5 A and B), and fewer lymphocytes and myeloid cells infiltrated the spinal cord (SI Appendix, Fig. S4B). Moreover, delivering anti-CXCR6 mAb after appearance of symptoms (“therapeutic protocol”) reversed the clinical score to baseline (Fig. 5C and Movies S1–S5), prevented body weight loss (Fig. 5D), decreased leukocyte accumulation (Fig. 5E), and dramatically diminished the histology score (Fig. 5F).
Fig. 5.
Anti-CXCR6 treatment prevents EAE and reverses established disease. Wt mice were immunized with MOG and treated with anti-mouse CXCR6 mAb or isotype control at the days indicated by arrows. (A and B) Disease prevention protocol. Clinical score (mean ± SEM) (A) and disease frequency (n = 8 per group) (B). One diseased mouse in the isotype-treated group recovered spontaneously on day 22. (C–E) Therapeutic protocol. Clinical score (C) and body weight (D). Data are mean ± SEM. (E) Histology. Representative spinal cord sections on day 11 of therapeutic treatment stained with haemotoxylin and eosin. (F) Histopathology scores, which measure inflammation and degeneration.
CXCR6 Identifies Pathogenic TH Cells in Different Autoimmune Disorders.
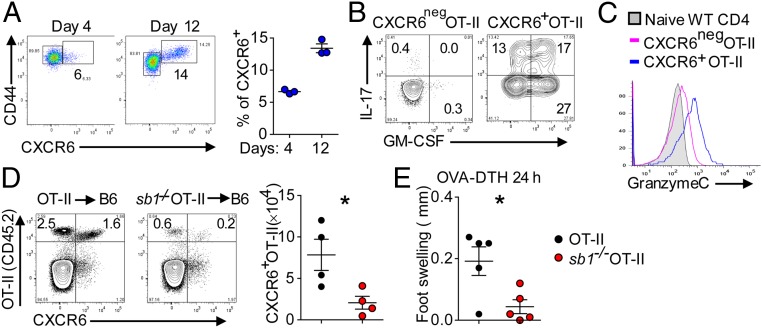
CXCR6 also marks an expanded population of CD4 cells expressing multiple cytokines and GzmC in mice adoptively transferred with OT-II cells and immunized with ovalbumin peptide (OVA) (Fig. 6 A–C). In the absence of Sb1, the expanded population of CXCR6+OT-II cells was largely abrogated (Fig. 6D), and pathogenic function of these cells was lacking as indicated by decreased footpad swelling on OVA challenge in the footpad (DTH response) (Fig. 6E). A blunted DTH response was seen also in MOG-immunized Sb1−/− mice challenged in the footpad with MOG peptide (Fig. 2F).
Fig. 6.
CXCR6 expression on OT-II cells. (A–C) OT-II cell transfer studies. Naïve OT-II cells (CD45.2) were transferred into naïve congenic CD45.1 mice, and the mice were immunized with OVA/CFA. (A) CXCR6 expression on LN OT-II cells on days 4 and 12 after immunization. (Left) Representative contour plots. (Right) Cell frequencies. (B) Cytokine profile of CXCR6neg and CXCR6+ OT-II cells on day 12 analyzed by FACS. (C) Histogram of GzmC expression. (D and E) Sb1−/− OT-II transfer studies. Naïve wt OT-II and Sb1−/− OT-II cells were separately transferred as in A, and the mice were immunized with OVA/CFA. (D) CXCR6-expressing wt and Sb1−/− OT-II cells in LN on day 10. (Left) Representative contour plots. (Right) Mean number of cells. (E) OVA-induced DTH (footpad swelling) in mice transferred with wt or Sb1−/− OT-II cells and challenged in the footpad. Symbols represent individual mice. Data are representative of three (A) and two (B–E) experiments. *P < 0.05 by Student’s t test.
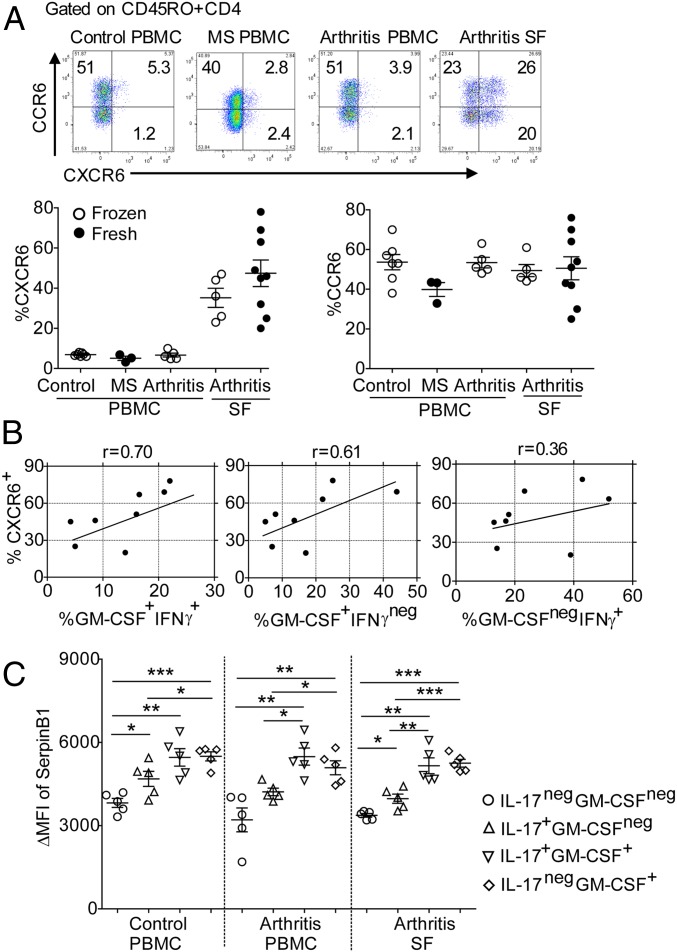
We also evaluated human CXCR6+ CD4 cells in synovial fluid (SF) cells of inflammatory arthritis patients and, for comparison, the corresponding cells in peripheral blood of control individuals, MS patients, and inflammatory arthritis patients. CXCR6+CD4 cells were highly enriched only in arthritis patient synovial fluid and not in peripheral blood samples (Fig. 7A). In contrast, no disease association was noted for CCR6+ CD4 cells (Fig. 7A). The proportions of CXCR6+CD4 cells in synovial fluids correlated well with the proportions of GM-CSF/IFNγ DP and GM-CSF SP cells, but not with IFNγ SP cells (Fig. 7B). Thus, in both mouse and human autoimmune disorders, CXCR6 identifies CD4 cells that produce multiple key pathogenic cytokines and are enriched in inflamed tissues. Consistent with the dependence of the CXCR6+ cells on Sb1 expression in mice, levels of human SerpinB1 (SB1) were higher in CD45RO+CD4 cells compared with CD45RA+ cells (SI Appendix, Fig. S5 A and B). SB1 levels were increased also in IL-17+ CD4 cells compared with cytokineneg CD4 cells and further increased in GM-CSF/IL-17 DP, IFNγ/GM-CSF DP, and GM-CSF SP cells (Fig. 7C and SI Appendix, Fig. S5 C and D). These subset-specific SB1 differences were largely independent of the source of CD4 cells (Fig. 7C and SI Appendix, Fig. S5D). The finding that CD4 cell subsets have characteristic SB1 levels, whether from an inflammatory site or peripheral blood, is consistent with the dominant role of cell number rather than phenotypic aberrancies in determining the pathogenicity of CXCR6+ cells.
Fig. 7.
CXCR6, cytokines, and SerpinB1 expression in SF CD4 cells of patients with inflammatory arthritis. (A) CXCR6 and CCR6 expression: (Upper) Representative contour plots. (Lower Left) Cumulative frequencies of CXCR6+ cells in CD45RO+CD4 cells of peripheral blood of control individuals and MS patients and both peripheral blood and synovial fluid of inflammatory arthritis patients. (Lower Right) Cumulative frequencies of CCR6+ cells in the same populations. (B) Pearson’s correlation coefficients for frequency of CXCR6+ cells and indicated cytokine-expressing cells. Because the cytokine-producing cell incubation with P+I cause CXCR6 to be down-regulated, the results of separate assays were used to determine correlation coefficients. (C) Sb1 expression in the indicated cytokine-producing CD4 cells. Intracellular cytokines and Sb1 (clone ELA-5) were stained after 4-h stimulation with P+I and analyzed by FACS. Symbols indicate individual patients. *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test.
Sb1 Controls the Longevity of CXCR6+ Encephalitogenic TH Cells.
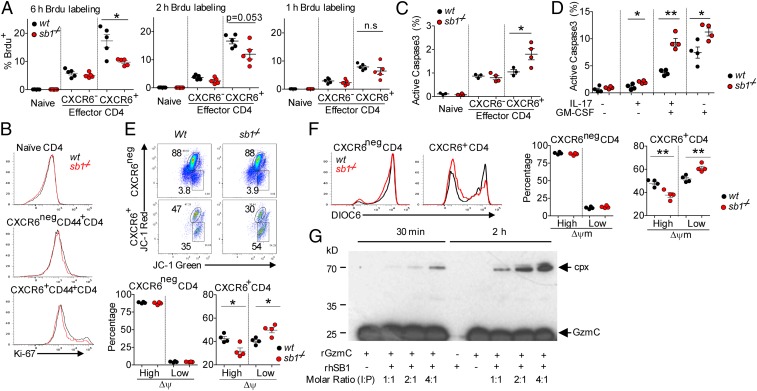
Having established molecular and functional features of the Sb1-dependent CXCR6+CD4 cells, we then sought to account for their deficiency in Sb1−/− mice. We injected MOG-immunized wt and Sb1−/− mice at disease onset with the thymidine analog bromodeoxyuridine (BrdU) to label proliferating cells. Analysis of LN cells 6 h later revealed that (i) CXCR6+CD4 cells have higher BrdU labeling than CXCR6neg cells, suggesting that CXCR6+ cells have a high proliferation rate and (ii) the frequency of BrdU+ Sb1−/− CXCR6+ cells was decreased compared with that of BrdU+ wt CXCR6+ cells. Because the frequency of cells labeled with BrdU after a fixed time span can be affected by both cell death as well as cell proliferation, we shortened the labeling time to minimize effects of cell death. After 2 h, the frequency of BrdU+ wt cells was unchanged, but the deficit of BrdU+ Sb1−/− CXCR6+CD4 cells was largely diminished, and at 1 h the frequency of BrdU+ Sb1−/− CXCR6+CD4 cells was not different from corresponding wt cells, indicating that Sb1−/− and wt CXCR6+CD4 cells proliferate at the same rate (Fig. 8A). Further studies comparing the two genotypes for staining with Ki-67, a nuclear marker of recently proliferated cells, provided verifying evidence that proliferation of CXCR6+CD4 cells is rapid and is not different between Sb1−/− and wt mice (Fig. 8B).
Fig. 8.
CXCR6+ TH cells of Sb1−/− mice are subject to enhanced cell death during robust proliferation. Wt and Sb1−/− mice were immunized with MOG/CFA to induce EAE. (A) Frequency of BrdU+ population in LN CD4 cells quantified by FACS. Data are representative of 2 to 3 experiments. (B) Ki-67 expression of LN CD4 cells at disease onset. Depicted histograms are representative of 7 mice per genotype in two experiments. (C) Active caspase-3 staining of freshly isolated LN CD4 cells at disease onset. (D) Active caspase-3 of cytokine-producing cells. LN cells were stimulated with P+I for 2.5 h and stained for cytokines and active caspase-3. Depicted data are representative of two to three experiments. (E and F) Mitochondrial membrane potential (Δψm) of CXCR6neg and CXCR6+ wt and Sb1−/− CD4 cells at onset of EAE. (E) Δψm measured with the mitochondrial dye JC-1. Representative dot plots (Upper) and Cumulative frequencies (Lower). (F) Δψm measured with the mitochondrial dye DiOC6. (Left) Histograms. (Right) Cumulative frequencies of CXCR6neg and CXCR6+ CD4 cells. (E and F) Data in each are representative of two experiments with 5 mice per genotype. (A and C–F) Symbols represent individual mice. (G) Recombinant human SerpinB1 (rhSB1) forms an inhibitory complex with rGzmC. Western blot stained with rabbit anti-GzmC. Arrows indicate GzmC (Glu193Gly) at 26 kDa and the covalent SB1–GzmC complex (cpx) at 66 kDa. SB1, detected in a parallel protein-stained gel, migrates at 42 kDa. Data are representative of three experiments. *P < 0.05, **P < 0.01 by Student’s t test.
To examine cell death, freshly isolated LN cells were stained for active caspase-3. Active caspase-3+ cells, although few in number, were significantly increased among Sb1−/− CXCR6+CD4 cells compared to wt ones (Fig. 8C). Because dead cells bearing active caspase-3 are rapidly removed in vivo, we repeated the measurement after stimulating the cells ex vivo, conditions less favorable to dead cell removal. After ex vivo stimulation, the excess of active caspase-3+ Sb1−/− cells over wt cells was substantial, especially for IL-17/GM-CSF DP and GM-CSF SP cells (Fig. 8D).
We then considered whether the Sb1-dependent CD4 cells are subject to self-inflicted cell death as occurs in other granule-containing cells such as NK cells, CD8 cells, and neutrophils (22, 34, 35, 46). In this mechanism, high-level activation or stress causes permeabilization of granule membranes allowing granzymes to leak into the cytoplasm (47). GzmB, a serine protease released in cytolytic CD8 cells and NK cells, can induce cell suicide, but this is opposed by the cytoprotective inhibitor Serpinb9 (Sb9). In neutrophils, cell death is mediated by the azurophil granule proteases cathepsin G and proteinase-3 (PR3) and opposed by Sb1, which irreversibly inactivates these serine proteases (34, 35).
Because loss of mitochondrial membrane potential (Δψm) is an early and irreversible step of this intrinsic death process (48), we used mitochondrial dyes to measure Δψm at disease onset. The dye JC-1 forms red-fluorescing aggregates in mitochondria at high Δψm and green-fluorescing monomers at low Δψm. More than 80% of CXCR6neg CD4 cells of wt and Sb1−/− mice had red fluorescence, indicating intact mitochondria. In contrast, a substantial percentage of wt CXCR6+CD4 cells and an even greater percentage of Sb1−/− CXCR6+CD4 cells had green-fluorescing JC-1, indicating mitochondrial damage and irreversible commitment to cell death (Fig. 8E). We also used the unrelated mitochondrial probe DiOC6, where high fluorescence of wt and Sb1−/− CXCR6neg CD4 cell indicated intact mitochondria. However, a substantial percentage of wt CXCR6+CD4 cells and an even greater percentage of Sb1−/− CXCR6+CD4 cells had dim fluorescence (low Δψm), indicating damaged mitochondria and irreversible commitment to cell death (Fig. 8F). Overall, the findings indicate that Sb1, by preventing cell suicide, determines whether sufficient CXCR6+CD4 cells survive to form an expanded population capable of implementing pathogenesis.
Further study will be required to fully document the death process and identify the Sb1- inhibitable protease (or proteases) responsible for the death of CXCR6+CD4 cells, but the expression data suggest GzmC, a serine protease that has a cytolytic efficiency comparable to GzmB and acts via a cell death pathway involving direct mitochondrial damage (49). We found that GzmC, a chymotryptase, is directly inhibited by Sb1 as indicated by the covalent complex formed on incubating rGzmC with human rSB1 (Fig. 8G). Neither GzmA, a tryptase, nor GzmB, an aspase, can be inhibited by Sb1 (27).
Discussion
Here, we report that the protease inhibitor Sb1 is expressed at the onset of EAE in a subset of peripheral effector CD4 cells that we subsequently identified as the encephalitogenic T cells. We found further that Sb1 is required for survival and expansion but not generation of these cells. On deletion of Sb1, encephalitogenic T cells do not accumulate in the CNS of immunized mice, and disease is substantially ameliorated. Findings from transcriptomics attesting to the unusual nature of these TH cells include the newly identified signature genes GzmC, GzmA, and PrfA and the previously documented Csf2 and Ifng. These TH cells are distinguished also by the presence of cytolytic granules along with the previously documented secretion system for multiple cytokines. Important also is the finding that CXCR6, a chemokine receptor, is suitable as a cell surface marker of the Sb1-dependent encephalitogenic TH cells. Having a marker that identifies the truly encephalitogenic T cells in EAE paves the way for design of novel therapy for human MS.
It is generally accepted that the function of CD4 cells in MS and related autoimmune disorders is not fully explained by the action of polarized TH1 or TH17 cells, but rather by cells generated through a not-yet-characterized encephalitogenic program initiated and maintained by IL1β and IL23 (reviewed in ref. 50). A link of Sb1 with the encephalitogenic program was strongly suggested by finding indistinguishable phenotypes for Sb1-deficient and Il23r-deficient mice (Figs. 1 and 2 and ref. 11). The cumulative findings for these mice suggest that Sb1 functions downstream of IL-23 to regulate the encephalitogenic program, which has at its core function the successful expansion of a select subset of primed TH cells. In the program, Sb1 restricts a proliferation-associated granule protease-mediated mitochondrial damage/suicidal death pathway and, thus, is crucial for survival and expansion of the select T helper cells that constitute the encephalitogenic population. Altogether, the findings describe encephalitogenic TH cells as cells that produce multiple pathogenic cytokines especially GM-CSF; proliferate rapidly; rely on Sb1 to survive during rapid expansion; express cytotoxic granule components perforin A, GzmA, and GzmC; and are marked by CXCR6.
TH cells expressing most of the features of encephalitogenic TH cells, specifically CXCR6+, multiple cytokines, granzymes, pathogenic function, and IL23 dependence, were found in the OT-II transfer model of DTH, indicating that the disease-inducing TH cells described here are not limited to autoimmune neuroinflammation.
TH cells with similarities to murine Sb1-dependent encephalitogenic TH cells have been reported previously in autoimmune disorders. The first were the IL-17/IFNγ DP CD4 cells noted in the gut of Crohn’s disease patients (41) and later in brain tissue of MS patients (51). In MS, myelin-reactive cytokine-producing CD4 cell clones were characterized as IL-17/GM-CSF DP, GM-CSF SP, and IFNγ SP (42, 52), a pattern similar to murine encephalitogenic TH cells. It is now appreciated that IL-17/IFNγ DP CD4 cells, known as TH1/TH17 and TH17/TH1 cells, and also a subset of IFNγ SP CD4 cells called nonclassic TH1 cells, are not TH1 cells but rather are derived from TH17 cells (15, 53).
An earlier study found SFs of inflammatory arthritis patients enriched in CXCR6+ CD4 cells that produce IFNγ and, on that basis, were reported as TH1 cells (54). This led to the notion that CXCR6 marks inflammatory TH1 cells at tissue sites. We show here that the CD4 cells marked by CXCR6 in inflammatory arthritis SF are enriched in cytokine DP (GM-CSF/IL-17 and GM-CSF/IFNγ) cells (Fig. 7), suggesting their relatedness to the TH17-derived encephalitogenic TH cells in murine EAE. Relatedness is suggested also for IL17/IFNγ DP cells that emerge in an IL-23–dependent fashion in murine inflammatory bowel disease (55). In a T cell transfer model of chronic colitis, pathogenic CD4 cells marked by CXCR6 include IL-17/IFNγ DP cells along with predominant IL-17 SP and IFNγ SP cells (56); poor proliferation characterizes these cells and thus distinguishes them from the rapidly proliferating CXCR6+ TH cells in EAE. Lastly, encephalitogenic CXCR6+ TH cells have at least one feature, cytotoxic granules, in common with CD4+ cytolytic T cells (CD4+ CTL) that provide, e.g., antiviral protection. Recent work showed that progression of disease in MS patients correlates with the density of circulating CD4+ CTL (57).
To determine how Sb1 regulates the density of encephalitogenic TH cells in EAE mice, we evaluated cell proliferation and cell death. Multiple approaches to proliferation including in vivo BrdU labeling showed that the proliferation rate for encephalitogenic TH cells is not different in Sb1−/− and wt mice. Cell death quantitation was challenging because dead cells are rapidly removed in vivo. The most definitive experiments involved quantifying cells in the process of dying, i.e., cells irreversibly committed to death due to mitochondrial damage (48), a process induced by leakage of cytotoxic granule contents (22). This approach demonstrated (i) robust ongoing death of wt encephalitogenic TH cells occurring concurrent with robust proliferation and (ii) further increase of dying encephalitogenic TH cells in mice lacking Sb1. The cumulative findings indicate that the extent of expansion of CXCR6+ TH cell subset in EAE and, hence, their encephalitogenicity is the net result of simultaneous robust proliferation and robust cell death, the latter restricted by Sb1 and increased in its absence. The factors driving evolution of this inherently inefficient cell expansion mechanism are unknown, but we speculate that they reflect the biological need for highly potent cell populations to be tightly and irreversibly regulated.
Of note, the Sb1-mediated mechanism proposed here as the basis of the IL-1β and IL-23–driven TH cell encephalitogenic program, although new for CD4 cells, is not unique, but rather is analogous to the mechanisms by which Sb9 controls expansion and retraction of human and mouse populations of activated CD8 cytolytic cells and NK cells (22, 46). In a similar program, Sb1 reacts stoichiometrically with endogenous granule serine proteases cathepsin G and proteinase-3 to control neutrophil survival (34, 35). Recently, Sb1 together with Serpinb6 (Sb6) were shown to restrict cathepsin G-mediated death of neutrophils and monocytes and to prevent proteolytic release of inflammatory cytokines (58).
Limitations stemming from the use of a mouse model for mechanistic study of the TH cell encephalitogenic process are mitigated somewhat by related findings for synovial fluid TH cells in human inflammatory arthritis. Further study will be required to identify the Sb1-inhibitable granule protease or proteases that cause cell suicide of these highly differentiated and highly activated CXCR6+ CD4 cells. We identified the murine serine protease granzyme C as a likely candidate but were unable to be specific because of limitations of reagents and because that region of the genome is greatly expanded in mice comprising Gzm-C, Gzm-F, Gzm-L, Gzm-N, Gzm-G, Gzm-D, and Gzm-E (59). In contrast, there is one counterpart gene in humans, GZM-H–encoding granzyme H, a serine protease, which is rapidly and stoichiometrically inhibited by SB1 (60).
Finally, apparent depletion of CXCR6+ cells by anti-CXCR6 treatment of immunized mice prevented the development of clinical disease and decreased the accumulation of cytokine-producing TH cells in the spinal cord and reversed or ameliorated clinical symptoms in diseased mice. These findings indicate that the Sb1-dependent multifunctional cells described here indeed mediate encephalitogenicity in EAE. They suggest that therapies to regulate Sb1 levels or, more realistically, strategies to deplete CXCR6-marked TH cells hold promise for mitigating autoimmune disorders such as MS.
Materials and Methods
Described here is a summary; details are provided in SI Appendix, SI Material and Methods. Animal studies were approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital or the cantonal veterinary office of Zurich. To induce EAE, wt and sb1−/− mice in the C57BL/6 background were injected with MOG35–55 emulsified with complete Freund’s adjuvant followed by 200 ng of pertussis toxin on days 0 and 2. Discarded synovial fluid specimens were obtained from patients with inflammatory arthritis undergoing diagnostic and/or therapeutic arthrocentesis for active joint inflammation, and research on these specimens was conducted under approved Institutional Review Board (IRB) protocols 2007P002441 (Brigham and Women’s Hospital) and S09-10-0557 (Boston Children’s Hospital). A preliminary report of some of these findings has appeared in abstract form (61).
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism 4. The data were analyzed by Student’s t test, unpaired and paired, or one-way ANOVA. P values ≤0.05 were considered significant.
Footnote/Note Added in Proof.
While this manuscript was in review, a publication appeared coauthored by one of us (B.B.) that characterized GM-CSF+ CD4 cells in EAE and included verifying evidence that IL-1β- and IL-23–driven GM-CSF+ CD4 cells are marked by cell surface CXCR6 (62).
Supplementary Material
Acknowledgments
We thank Timothy Ley, Charaf Benarafa, Sheng Xiao, Susana Camposano, Norma Gerard, Gregory Keras, and Catherine Brownstein of the Intellectual and Developmental Disabilities Research Center Molecular Genetics Core Facility and Roderick Bronson of the Dana–Farber Cancer Institute Rodent Histopathology for reagents, animals, sample curation, advice and the use of equipment, and Heinz Remold, Susanna Remold, Andrew Croxford, Mazier Divangahi, and Ruben Martinez Barricarte for review of the manuscript. We thank the Functional Genomic Center Zurich for NGS analysis. The work was supported by NIH Grants R21 AI117440 (to E.R.-O.), RO1 AR065538 (to P.A.N.), KO8 AR073339 (to L.A.H.), and K08 AR072791 (to D.K.) and Swiss National Science Foundation Grants 316030_150768 and 310030_146130 (to B.B.). Synovial fluid samples were collected through an infrastructure supported by NIH Grant P30 AR070253 Joint Biology Consortium (to P.A.N. and L.A.H.).
Footnotes
Conflict of interest statement: L.H. and E.R.-O. filed a provisional patent related to the findings of this study and have equity ownership and are on the Scientific Advisory Board of the recently launched Edelweiss Immune, Inc. The other authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905762116/-/DCSupplemental.
References
- 1.Compston A., Coles A., Multiple sclerosis. Lancet 372, 1502–1517 (2008). [DOI] [PubMed] [Google Scholar]
- 2.El-behi M., Rostami A., Ciric B., Current views on the roles of Th1 and Th17 cells in experimental autoimmune encephalomyelitis. J. Neuroimmune Pharmacol. 5, 189–197 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal B. M., Shevach E. M., IL-12 unmasks latent autoimmune disease in resistant mice. J. Exp. Med. 184, 771–775 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cua D. J., et al. , Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421, 744–748 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Segal B. M., Dwyer B. K., Shevach E. M., An interleukin (IL)-10/IL-12 immunoregulatory circuit controls susceptibility to autoimmune disease. J. Exp. Med. 187, 537–546 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langrish C. L., et al. , IL-12 and IL-23: Master regulators of innate and adaptive immunity. Immunol. Rev. 202, 96–105 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Langrish C. L., et al. , IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201, 233–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haak S., et al. , IL-17A and IL-17F do not contribute vitally to autoimmune neuro-inflammation in mice. J. Clin. Invest. 119, 61–69 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komiyama Y., et al. , IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J. Immunol. 177, 566–573 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Ghoreschi K., et al. , Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGeachy M. J., et al. , The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10, 314–324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutton C., Brereton C., Keogh B., Mills K. H., Lavelle E. C., A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. J. Exp. Med. 203, 1685–1691 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronchi F., et al. , Experimental priming of encephalitogenic Th1/Th17 cells requires pertussis toxin-driven IL-1β production by myeloid cells. Nat. Commun. 7, 11541 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung Y., et al. , Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 30, 576–587 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirota K., et al. , Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12, 255–263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codarri L., et al. , RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12, 560–567 (2011). [DOI] [PubMed] [Google Scholar]
- 17.El-Behi M., et al. , The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12, 568–575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McQualter J. L., et al. , Granulocyte macrophage colony-stimulating factor: A new putative therapeutic target in multiple sclerosis. J. Exp. Med. 194, 873–882 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun J., et al. , A cytosolic granzyme B inhibitor related to the viral apoptotic regulator cytokine response modifier A is present in cytotoxic lymphocytes. J. Biol. Chem. 271, 27802–27809 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Phillips T., et al. , A role for the granzyme B inhibitor serine protease inhibitor 6 in CD8+ memory cell homeostasis. J. Immunol. 173, 3801–3809 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Zhang M., et al. , Serine protease inhibitor 6 protects cytotoxic T cells from self-inflicted injury by ensuring the integrity of cytotoxic granules. Immunity 24, 451–461 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Bird C. H., et al. , Selective regulation of apoptosis: The cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 18, 6387–6398 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bird C. H., et al. , The granzyme B-Serpinb9 axis controls the fate of lymphocytes after lysosomal stress. Cell Death Differ. 21, 876–887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangan M. S., et al. , A pro-survival role for the intracellular granzyme B inhibitor Serpinb9 in natural killer cells during poxvirus infection. Immunol. Cell Biol. 95, 884–894 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Kaiserman D., Bird P. I., Control of granzymes by serpins. Cell Death Differ. 17, 586–595 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Ashton-Rickardt P. G., Serine protease inhibitors and cytotoxic T lymphocytes. Immunol. Rev. 235, 147–158 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Cooley J., Takayama T. K., Shapiro S. D., Schechter N. M., Remold-O’Donnell E., The serpin MNEI inhibits elastase-like and chymotrypsin-like serine proteases through efficient reactions at two active sites. Biochemistry 40, 15762–15770 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Remold-O’Donnell E., Chin J., Alberts M., Sequence and molecular characterization of human monocyte/neutrophil elastase inhibitor. Proc. Natl. Acad. Sci. U.S.A. 89, 5635–5639 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng W., Silverman G. A., Remold-O’Donnell E., Structure and sequence of human M/NEI (monocyte/neutrophil elastase inhibitor), an Ov-serpin family gene. Gene 213, 179–187 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Benarafa C., Cooley J., Zeng W., Bird P. I., Remold-O’Donnell E., Characterization of four murine homologs of the human ov-serpin monocyte neutrophil elastase inhibitor MNEI (SERPINB1). J. Biol. Chem. 277, 42028–42033 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Benarafa C., Remold-O’Donnell E., The ovalbumin serpins revisited: Perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 102, 11367–11372 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benarafa C., et al. , SerpinB1 protects the mature neutrophil reserve in the bone marrow. J. Leukoc. Biol. 90, 21–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benarafa C., Priebe G. P., Remold-O’Donnell E., The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection. J. Exp. Med. 204, 1901–1909 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baumann M., Pham C. T., Benarafa C., SerpinB1 is critical for neutrophil survival through cell-autonomous inhibition of cathepsin G. Blood 121, 3900–3907, S1–S6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loison F., et al. , Proteinase 3-dependent caspase-3 cleavage modulates neutrophil death and inflammation. J. Clin. Invest. 124, 4445–4458 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao P., Hou L., Farley K., Sundrud M. S., Remold-O’Donnell E., SerpinB1 regulates homeostatic expansion of IL-17+ γδ and CD4+ Th17 cells. J. Leukoc. Biol. 95, 521–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Georgiev H., Ravens I., Benarafa C., Förster R., Bernhardt G., Distinct gene expression patterns correlate with developmental and functional traits of iNKT subsets. Nat. Commun. 7, 13116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou L., et al. , The protease cathepsin L regulates Th17 cell differentiation. J. Autoimmun. 65, 56–63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C., et al. , Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reboldi A., et al. , C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat. Immunol. 10, 514–523 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Annunziato F., et al. , Phenotypic and functional features of human Th17 cells. J. Exp. Med. 204, 1849–1861 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Y., et al. , Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci. Transl. Med. 7, 287ra74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng H. K., Unutmaz D., KewalRamani V. N., Littman D. R., Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388, 296–300 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Lee Y., et al. , Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J. V., et al. , Two-photon laser scanning microscopy imaging of intact spinal cord and cerebral cortex reveals requirement for CXCR6 and neuroinflammation in immune cell infiltration of cortical injury sites. J. Immunol. Methods 352, 89–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashton-Rickardt P. G., An emerging role for Serine Protease Inhibitors in T lymphocyte immunity and beyond. Immunol. Lett. 152, 65–76 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Boya P., Kroemer G., Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Zamzami N., et al. , Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J. Exp. Med. 181, 1661–1672 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson H., Scorrano L., Korsmeyer S. J., Ley T. J., Cell death induced by granzyme C. Blood 101, 3093–3101 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Becher B., Segal B. M., T(H)17 cytokines in autoimmune neuro-inflammation. Curr. Opin. Immunol. 23, 707–712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kebir H., et al. , Preferential recruitment of interferon-gamma-expressing TH17 cells in multiple sclerosis. Ann. Neurol. 66, 390–402 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Restorick S. M., et al. , CCR6+ Th cells in the cerebrospinal fluid of persons with multiple sclerosis are dominated by pathogenic non-classic Th1 cells and GM-CSF-only-secreting Th cells. Brain Behav. Immun. 64, 71–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazzoni A., et al. , Demethylation of the RORC2 and IL17A in human CD4+ T lymphocytes defines Th17 origin of nonclassic Th1 cells. J. Immunol. 194, 3116–3126 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Kim C. H., et al. , Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J. Clin. Invest. 107, 595–601 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahern P. P., et al. , Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33, 279–288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mandai Y., et al. , Distinct roles for CXCR6(+) and CXCR6(−) CD4(+) T cells in the pathogenesis of chronic colitis. PLoS One 8, e65488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peeters L. M., et al. , Cytotoxic CD4+ T cells drive multiple sclerosis progression. Front. Immunol. 8, 1160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burgener S. S., et al. , Cathepsin G inhibition by Serpinb1 and Serpinb6 prevents programmed necrosis in neutrophils and monocytes and reduces GSDMD-driven inflammation. Cell Rep. 27, 3646–3656.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grossman W. J., et al. , The orphan granzymes of humans and mice. Curr. Opin. Immunol. 15, 544–552 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Wang L., et al. , Identification of SERPINB1 as a physiological inhibitor of human granzyme H. J. Immunol. 190, 1319–1330 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Hou L., et al. , SerpinB1 deficiency ameliorates experimental autoimmune encephalomyelitis (abstract). J Immunol 196, 1-S58.13 (2016). [Google Scholar]
- 62.Komuczki J., et al. , Fate-mapping of GM-CSF expression identifies a discrete subset of inflammation-driving T helper cells regulated by cytokines IL-23 and IL-1beta. Immunity 50, 1289–1304.e6 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.