Significance
While it is well established that heavily O-glycosylated mucins are essential for normal gastrointestinal function, it is not known how polypeptide N-acetylgalactosaminyl transferase (GalNAc-T12)–specific O-glycans may be contributing. Here we use structural, computational, and biochemical methods to show how the GALNT12 mutations present in patients with colorectal cancer disrupt the enzymatic function of GalNAc-T12. This information will help identify specific biological substrates of GalNAc-T12 and serve to inform our understanding of their functions, including their potential contributions to cancer initiation and progression.
Keywords: GalNAc-Ts, mucin-type O-glycosylation, colorectal cancer, substrate selectivity, enzyme catalysis
Abstract
Polypeptide N-acetylgalactosaminyl transferases (GalNAc-Ts) initiate mucin type O-glycosylation by catalyzing the transfer of N-acetylgalactosamine (GalNAc) to Ser or Thr on a protein substrate. Inactive and partially active variants of the isoenzyme GalNAc-T12 are present in subsets of patients with colorectal cancer, and several of these variants alter nonconserved residues with unknown functions. While previous biochemical studies have demonstrated that GalNAc-T12 selects for peptide and glycopeptide substrates through unique interactions with its catalytic and lectin domains, the molecular basis for this distinct substrate selectivity remains elusive. Here we examine the molecular basis of the activity and substrate selectivity of GalNAc-T12. The X-ray crystal structure of GalNAc-T12 in complex with a di-glycosylated peptide substrate reveals how a nonconserved GalNAc binding pocket in the GalNAc-T12 catalytic domain dictates its unique substrate selectivity. In addition, the structure provides insight into how colorectal cancer mutations disrupt the activity of GalNAc-T12 and illustrates how the rules dictating GalNAc-T12 function are distinct from those for other GalNAc-Ts.
The mucus layer in the gastrointestinal (GI) epithelium that protects underlying organs from infection and physical and chemical damage (1–4) contains heavily O-glycosylated mucin proteins. Colorectal cancer (CRC) is characterized by abnormal glycosylation states of mucins that can promote the dysregulation of the microbiota in the GI tract and lead to infection and inflammation (5–8). Mucin-type O-glycosylation results in the addition of N-acetylgalactosamine (GalNAc) to Ser or Thr to yield GalNAc-α-1-O-Ser/Thr (Tn antigen), which can be further modified stepwise to yield an array of heterogeneous glycan structures (9–11). The process is initiated by a family of UDP-GalNAc polypeptide N-acetylgalactosaminyl-transferases (GalNAc-Ts) (E.C. 2.4.1.41) that are conserved across metazoan, with 20 human isoenzymes identified thus far (9). GalNAc-Ts are Golgi-bound, type II transmembrane, Mn2+-dependent enzymes belonging to CAZy family 27 (9). The luminal portion of the enzymes includes the GT-A type catalytic domain that contains an active site consisting of Mn2+ coordinated by the canonical Asp-His-His residues, water, and the sugar donor UDP-GalNAc. A catalytic flexible loop becomes stabilized and adopts a closed “active” conformation upon peptide substrate and UDP-GalNAc binding (12, 13). A flexible linker (∼10 aa) connects the catalytic domain to a C-terminal ricin B-type lectin domain (carbohydrate-binding module group 13 in the CAZy database) containing 3 tandem repeats termed α, β, and γ that can each potentially bind a sugar if the canonical sugar-binding residues are present.
Human GalNAc-T12 is highly expressed in GI organs, with the highest expression in the colon (14). However, its expression is diminished in colon cancer cell lines and tissues from patients with CRC (15). Over the last decade, several studies in patients with CRC have led to the characterization of somatic and germ line mutations that reduce the in vitro enzymatic activity of GalNAc-T12 (16–18). Many mutations occur in conserved residues, while a distinct subset alters nonconserved residues, suggesting the presence of GalNAc-T12–specific features (SI Appendix, Table S1).
Although an association between GalNAc-T12 and CRC has been demonstrated, the substrates and downstream effects of GalNAc-T12 deactivation are not known. Generally, GalNAc-T substrate identification is a challenge due to the absence of a strong consensus sequence or substrate recognition motif. Instead, GalNAc-Ts select for substrates through interactions with a limited set of preferred residues and extant GalNAcs on a peptide substrate. For instance, a Pro located +3 amino acids C-terminal to an acceptor Ser/Thr on a peptide substrate enhances enzymatic activity through interactions with a conserved pocket in the GalNAc-T catalytic domain (13, 19, 20). In GalNAc-T4, T7, T10, and T12, extant GalNAcs at short distances from the acceptor Ser/Thr on a peptide likely interact with regions in the catalytic domain (20). Furthermore, nearly all GalNAc-Ts utilize their lectin domain to recognize an extant GalNAc located 5 to 17 residues from the acceptor Ser/Thr on a substrate (20, 21). It was recently shown that the Drosophila homolog PGANT9 recognizes substrates via electrostatic, GalNAc-independent interactions with the lectin domain (22). By utilizing these different activities, each GalNAc-T selects its substrates through a unique combination of catalytic and/or lectin domain interactions with nonglycosylated and/or previously glycosylated peptide substrates (20, 21, 23–25).
GalNAc-T12 is unusual among the GalNAc-T family members because its activity is enhanced through interactions between both an N-terminal (−17 to 5 residues) GalNAc and a C-terminal (+3 residues) GalNAc on peptide substrates using its lectin and catalytic domains, respectively. This dual glycopeptide activity suggests that GalNAc-T12 likely prefers to interact with and modify densely glycosylated substrates. Here we characterize the molecular basis of the unique functions of GalNAc-T12. The X-ray crystal structure of GalNAc-T12 in complex with a di-glycosylated peptide substrate reveals how a subset of CRC mutations deactivate the enzyme and shows that GalNAc-T12 uses a distinct mechanism to align its active site and stabilize its catalytic flexible loop in the closed “active” conformation. The structure also confirms that lectin domain binding to GalNAc on a substrate occurs in a conserved pocket and reveals the nonconserved residues that constitute the unique catalytic domain GalNAc-binding pocket.
Results
Structure of GalNAc-T12 Bound to Di-Glycosylated Peptide, UDP, and Mn2+.
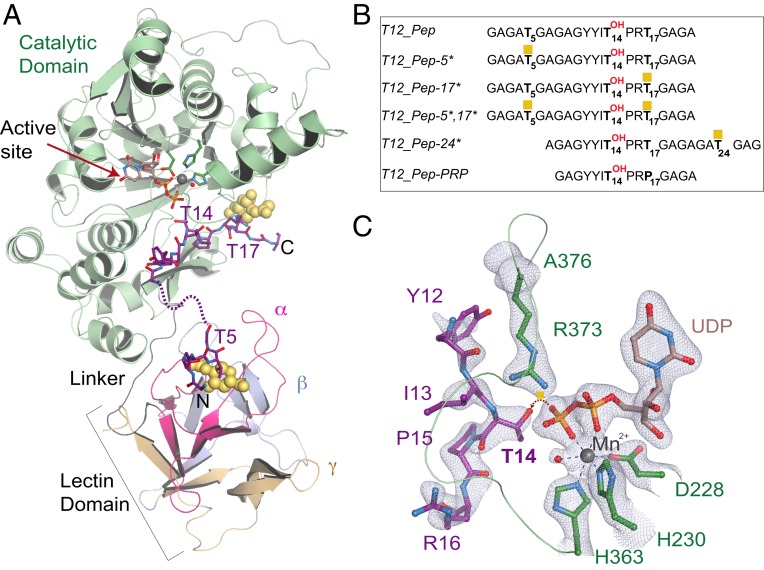
We solved the X-ray crystal structure of human GalNAc-T12 bound to a model di-glycosylated peptide termed T12_Pep-5*,17*, UDP, and Mn2+ at 2.0-Å resolution (Fig. 1A and Table 1) (26). The di-glycosylated peptide sequence (Fig. 1B) is based on a previously optimized GalNAc-T12 peptide, T12_Pep (17), containing residues that uniquely enhance GalNAc-T12 activity, including Tyr at positions −3 and −2 (Y11 and Y12) N-terminal to the acceptor Thr at position 14 (T14) (SI Appendix, Fig. S1A) and Arg at position +2 (R16) C-terminal to T14 (20, 23). The ternary complex crystallizes in the P1 space group with 2 complexes per asymmetric unit. The catalytic domain of GalNAc-T12 adopts the characteristic GT-A fold and is connected to the C-terminal lectin domain by a flexible linker (Fig. 1A). T12_Pep-5*,17* is positioned with amino acids 1 to 6 containing Thr5-O-GalNAc in the lectin domain. Amino acids 7 to 11 (AGAGY) are located between the 2 domains and are disordered. Weak electron density for Y11 on the peptide suggests that this residue is dynamic and makes transient interactions with the enzyme. Thus, how it enhances enzymatic activity is unclear from the structure. The remaining T12_Pep-5*,17* amino acids 12 to 21, including the acceptor T14 and Thr17-O-GalNAc, are ordered and bound to the catalytic domain (Fig. 1A). The active site is correctly aligned in the product-bound state with clear density for UDP; the canonical active site residues D228, H230, and H363; and a water coordinating the active site Mn2+ (Fig. 1C). The acceptor T14 is adjacent to UDP in the active site and correctly oriented for accepting GalNAc, and the catalytic flexible loop (H363-A376) is ordered and in the closed, “active” conformation (Fig. 1C).
Fig. 1.
The structure of the GalNAc-T12 ternary complex. (A) Structure of GalNAc-T12 in complex with UDP (light brown), Mn2+ (gray), and T12_Pep-5*,17* di-glycosylated peptide substrate (purple, with the disordered region of the peptide shown as purple dashes). The catalytic domain (green) contains the active site (red arrow). The lectin domain tandem repeats are labeled α (magenta), β (light blue), and γ (beige). GalNAc (yellow spheres) at the N terminus is bound to a pocket in the α repeat of the lectin domain, and GalNAc at the C terminus is bound to the catalytic domain. (B) Optimized GalNAc-T12 peptides used in this study, with GalNAc shown as yellow squares. (C) The active site of GalNAc-T12 superposed over an Fo-Fc omit map (light blue) contoured at 2.5 σ contains the peptide amino acids 12 to 16 (purple), UDP (light brown), and active site residues D228, H230, and H363 (dark green) coordinating Mn2+ (gray). T14 on the peptide is positioned for GalNAc transfer, as indicated by the dashed red arrow. The catalytic loop from A376 to H363 includes CRC residue R373 (dark green).
Table 1.
Data collection and refinement statistics, GalNAc-T12/UDP/Mn2+/di-glycosylated peptide (PDB ID code 6PXU)
| Parameter | Value |
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c, Å | 72.8, 73.1, 74.4 |
| a, b, γ, ° | 113.1, 100.5, 108.2 |
| Resolution, Å* | 19.98–2.01 (2.08–2.01) |
| Rpim* | 0.100 (0.903) |
| I/σI* | 7.5 (0.95) |
| Completeness, %* | 97.3 (89.5) |
| Redundancy* | 3.8 (3.1) |
| No. of unique reflections* | 81,588 (3,795) |
| Refinement | |
| Rwork/Rfree | 17.1/21.9 |
| No. of atoms | |
| Protein | 8,756 |
| Mn2+ | 4 |
| UDP | 50 |
| Peptide/GalNAc | 195/56 |
| Water/solvent† | 562/30 |
| B-factors | |
| Average | 42.0 |
| Macromolecules | 41.9 |
| Solvent‡ | 42.6 |
| Ligands | 48.0 |
| Rmsd | |
| Bond lengths, Å | 0.007 |
| Bond angles, ° | 0.98 |
Data in the highest-resolution shell is shown in parentheses.
Ethylene glycol and glycerol.
Water, ethylene glycol, and glycerol.
The CRC Variant GalNAc-T12R373H Reveals the Role of Active Site Residue Arg373.
We mapped the CRC-associated mutations to the structure of GalNAc-T12 (SI Appendix, Fig. S1B and Table S1). Several conserved mutations likely destabilize the GalNAc-T12 tertiary structure. Another subset of mutations alter residues that are poorly conserved, including R373, which is located in the catalytic flexible loop (Fig. 1C). A sequence alignment of the human GalNAc-T family catalytic flexible loop region reveals that R373 only occurs at that position in GalNAc-T12 and its closest homolog GalNAc-T4 (Fig. 2A) (12, 27). Since R373 is distant from extant glycans on the peptide substrate, we predicted that the effect of an R373H mutation on activity would be independent of GalNAc binding and tested the activity of GalNAc-T12R373H against the unglycosylated substrate T12_Pep (Fig. 1B). GalNAc-T12R373H is ∼10-fold less active than GalNAc-T12WT, similar to previous results showing diminished activity of GalNAc-T12R373H compared to GalNAc-T12WT against a Muc5Ac peptide substrate (Fig. 2B) (18).
Fig. 2.
Nonconserved residues in the catalytic flexible loop of GalNAc-T12. (A) P366, A369, and R373 are uniquely found within the catalytic flexible loop of GalNAc-T12 and in the corresponding positions of GalNAc-T4 (salmon). (B) The CRC variant GalNAc-T12R373H is less active against the GalNAc-T12–specific peptide T12_Pep compared with the wild-type enzyme. A P366R mutation further reduces the activity of GalNAc-T12R373H. (C) Active site comparison of GalNAc-T12 bound to T12_Pep substrate (dark colors) to GalNAc-T2 bound to EA2 mucin peptide (PDB ID code 2FFU; light colors). GalNAc-T12-R373 interacts with UDP, the backbone carbonyl of T12_Pep residue Y12, and the backbone amide of T12_Pep acceptor residue T14 through indirect interactions with water (encircled red sphere). GalNAc-T2-H365 superimposes over GalNAc-T12-R373 and similarly interacts with the peptide backbone of EA2 and the β-phosphate of UDP. GalNAc-T2 contains an additional R362 corresponding to GalNAc-T12-P366 that indirectly coordinates UDP through interactions with water (encircled salmon sphere). (D) The catalytic flexible loops of GalNAc-T12 (dark green) and GalNAc-T4 (salmon) adopt a similar conformation that is distinct from the conformation of the GalNAc-T2 loop (light cyan). GalNAc-T12-K367 is positioned to make CH–π bonds with a neighboring W262. Peptide residue R16 interacts with the catalytic flexible loop. The pink conformation makes backbone interactions with P366, and the purple conformation makes indirect interactions with F365 and W262.
We examined the interactions between R373 and the surrounding residues in the structure and found that R373 interacts with the peptide backbone carbonyl of Y12, the β-phosphate of UDP, and a water that is hydrogen-bonded to the acceptor T14 backbone amide (Fig. 2C). R373 also makes cation π interactions with W335 to stabilize the conserved WGGE motif that interacts with the β-phosphate of UDP (SI Appendix, Fig. S1C) (28). The WGGE motif residue E338 is critical for GalNAc-T activity and is positioned to interact with the GalNAc moiety of UDP-GalNAc based on a comparison with the structure of GalNAc-T10 containing an active site- bound GalNAc (SI Appendix, Fig. S1C) (29). Overall, the structure suggests that R373 promotes catalysis by aligning the active site and correctly orienting the acceptor T14 and UDP-GalNAc for transfer. Since R373 is not conserved, its interaction with the backbone carbonyl of Y12 is unique and could partially explain why Tyr at position −2 of the GalNAc-T12–optimized substrate enhances enzymatic activity. A structural comparison of GalNAc-T12 to GalNAc-T2 bound to UDP and an EA2 mucin peptide (Protein Data Bank [PDB] ID code 2FFU) (13) reveals that GalNAc-T2-F369 corresponding to GalNAc-T12-R373 in the sequence alignment does not contact EA2 or UDP (Fig. 2C). Instead, GalNAc-T2-H365 superimposes with GalNAc-T12-R373 in the structure and similarly coordinates the EA2 backbone and a UDP-interacting water molecule.
GalNAc-T12 Uses a Unique Mechanism for Catalytic Flexible Loop Stabilization.
The structural comparison between GalNAc-T12 and GalNAc-T2 further reveals a nonconserved P366 in the GalNAc-T12 catalytic flexible loop that corresponds to a conserved R362 in GalNAc-T2 (Fig. 2 A and C). GalNAc-T2-R362 makes indirect interactions with UDP but is also critical for GalNAc-T2 activity, because it makes CH–π interactions with a conserved GalNAc-T2-F104 (corresponding to GalNAc-T12-I103) that stabilizes its catalytic flexible loop in the closed “active” conformation (30). However, since GalNAc-T12 does not contain Arg at position 366 or Phe at position 103, it must use distinct mechanisms to stabilize its catalytic flexible loop. The structure shows that a nonconserved W262 is positioned to make CH–π interactions with catalytic flexible loop residue K367 (Fig. 2D and SI Appendix, Fig. S2). In GalNAc-T2, the corresponding K363 is oriented in the opposite direction to K367, and GalNAc-T2-M258 superimposes with W262 (Fig. 2D), indicating that this mechanism of loop stabilization is unique to GalNAc-T12 and GalNAc-T4. The conformational rigidity of a Pro at position 366 strongly influences the loop conformation and could stabilize the loop orientation for optimal CH–π interactions between K367 and W262. Indeed, the activity of GalNAc-T12R373H/P366R is ∼1.5-fold less than GalNAc-T12R373H, suggesting that a change from Pro to Arg in the catalytic flexible loop further destabilizes the enzyme (Fig. 2B).
Interestingly, R16 (at the +2 position) in the peptide substrate that uniquely enhances the enzymatic activity of GalNAc-T12 (20) interacts with the catalytic flexible loop in each crystallographic complex (Fig. 2D). In 1 complex, R16 coordinates a water that interacts with the backbone carbonyl of I260, the backbone amide of W262, and the backbone carbonyl of loop residue F365. In the second complex, R16 interacts with the backbone carbonyl of P366. Thus, the GalNAc-T12–specific preference for Arg at position +2 of the peptide is associated with its unique residues P366 and W262. The slightly decreased activity observed for GalNAc-T12R373H/P366R could also be attributed to the introduction of positive charges in the loop resulting in unfavorable electrostatic interactions with Arg16. Interestingly, GalNAc-T4 has a similar catalytic flexible loop conformation as GalNAc-T12 but does not have a preference for Arg at position +2 (20). A nonconserved R368 adjacent to GalNAc-T4-K368 (corresponding to GalNAc-T12-K367) increases the positive charge density in the GalNAc-T4 catalytic flexible loop that could result in unfavorable electrostatic interactions with a peptide containing Arg at the +2 position (Fig. 2D).
The Lectin Domain GalNAc-Binding Pocket Is Conserved.
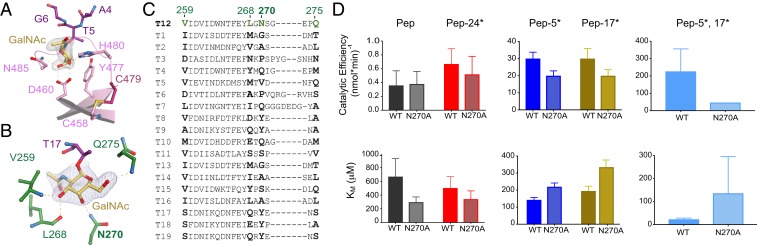
Thr5-O-GalNAc binds to the α repeat sugar-binding pocket of the GalNAc-T12 lectin domain that consists of D460, Y477, H480, and N485 (Fig. 3A and SI Appendix, Fig. S3A). A structural superposition of the GalNAc-T12 and GalNAc-T2 α repeats containing GalNAc reveals a similar side chain configuration (SI Appendix, Fig. S3B), verifying that GalNAc binding is structurally conserved. A deactivating CRC mutation, C479F, occurs in the α repeat and prevents disulfide bond formation between C479 (adjacent to H480 of the GalNAc-binding pocket) and C458 to disrupt the tertiary structure of the enzyme (Fig. 3A). Although the GalNAc-binding pockets of GalNAc-T12 and GalNAc-T2 are conserved, their lectin domains are not superimposable when their catalytic domains are aligned, placing the α repeat on opposite faces of the lectin domain (SI Appendix, Fig. S3C). A similar reorientation occurs in GalNAc-T4 as the result of the poorly conserved linker region that connects the catalytic and lectin domains (27).
Fig. 3.
GalNAc recognition by GalNAc-T12. (A) The lectin domain α repeat-binding pocket of GalNAc-T12 showing GalNAc (yellow) superimposed over an Fo-Fc omit map contoured at 3 σ hydrogen bonding to side chains of conserved residues (pink). The CRC variant C479F destabilizes the GalNAc-binding pocket by disrupting the disulfide between C479 and C458 that is adjacent to the α-binding pocket. (B) Catalytic domain-binding pocket of GalNAc-T12 showing GalNAc (yellow) superimposed over an Fo-Fc omit map contoured at 3 σ hydrogen bonding to backbone atoms of nonconserved residues (dark green) with the exception of N270, which makes side chain interactions. (C) Sequence alignment showing that the catalytic GalNAc-binding residues in GalNAc-T12 are not conserved in human GalNAc-Ts. (D) GalNAc-T12WT and GalNAc-T12N270A enzyme activity derived from the kinetic plots in SI Appendix, Figs. S3D and S4A. Catalytic efficiency (Vmax/KM) is reduced with N270A, verifying that this pocket is important for GalNAc binding and activity due to increases in KM values of the glycopeptide substrates.
To probe GalNAc lectin domain binding, we compared the activity of unglycosylated T12_Pep to the monoglycosylated substrates T12_Pep-5* (N-terminal to acceptor T14) and T12_Pep-24* (C-terminal to T14) (SI Appendix, Fig. S3D). As expected, glycosylation of T12_Pep and T12_Pep-24* (C-terminal GalNAc) is less than that seen with T12_Pep-5* (N-terminal GalNAc), verifying a preference for lectin domain recognition of GalNAc in the N-terminal region of a glycopeptide (20).
The Unique Catalytic Domain GalNAc-Binding Pocket of GalNAc-T12.
The catalytic domain GalNAc-binding pocket interacts with Thr17-O-GalNAc and consists of hydrogen-bonding interactions with the backbone atoms of V259, L268, and Q275 (Fig. 3B). V259 is semiconserved, while L268 and Q275 are not conserved (Fig. 3C). Interestingly, the only side chain interaction occurs between GalNAc and N270, the least conserved residue in that pocket (Fig. 3 B and C). To probe catalytic domain GalNAc binding, we compared the activity of the unglycosylated T12_Pep substrate with that of a monoglycosylated substrate T12_Pep-17* and observed activity enhancements with T12_Pep-17* (Fig. 3D and SI Appendix, Fig. S3D), verifying the proposed role of the GalNAc-T12 catalytic domain in GalNAc binding and substrate selectivity (20). Interestingly, enhancement of the catalytic domain-binding substrate increased the Vmax to a similar degree as the lectin domain-binding substrate. Although T12_Pep-17* has an ∼25% higher Vmax, it also has a slightly higher KM than T12_Pep-5*; thus, the catalytic efficiencies (Vmax/KM) of both monoglycosylated peptide substrates are essentially the same (Fig. 3D and SI Appendix, Table S2). This is in contrast to GalNAc-T4, in which catalytic domain-mediated activity is significantly lower than lectin domain-mediated activity (12). This is likely due to the different catalytic domain GalNAc-binding modes; in GalNAc-T4, GalNAc has minimal interactions with surface residues, while in GalNAc-T12, GalNAc has extensive interactions with residues in the pocket.
To examine the effects of simultaneous lectin and catalytic glycopeptide binding, we assessed the activity of GalNAc-T12 toward a di-glycosylated substrate (T12_Pep-5*,17*) (Fig. 3D and SI Appendix, Fig. S3D and Table S2). We found that GalNAc-T12 is most active at low substrate concentrations, with an apparent Vmax identical to that of T12_Pep-17*, but, more interestingly, has an apparent KM that is ∼6- to 9-fold lower than that of either monoglycosylated substrate. This is due to the synergistic binding of both GalNAcs of the di-glycosylated peptide to GalNAc-T12. Intriguingly, the activity of the transferase is inhibited at higher concentrations of T12_Pep-5*,17* (Ki ∼70 to 80 μM), suggesting that tight binding of GalNAc-T12 to the di-glycosylated peptide may indeed inhibit product release at high substrate concentrations (SI Appendix, Fig. S3D and Table S2). Alternatively, it is possible that at high substrate concentrations, nonproductive binding of the glycosylated Thr residues to incorrect binding sites (i.e., Thr*5 at the catalytic domain and Thr*17 at the lectin domain) could lead to reduced activities, as was recently proposed for GalNAc-T4 (12).
N270 Is a Nonconserved Residue That Is Important for Binding GalNAc.
We tested GalNAc-T12N270A against our substrates and found that its Vmax values were nearly identical to those of wild-type GalNAc-T12, while its KM values against the glycopeptide substrates T12_Pep-5*, T12_Pep-17*, and T12_Pep-5*,17* were all significantly higher than those of the wild-type enzyme (Fig. 3D and SI Appendix, Fig. S4A and Table S2). On this basis, the catalytic efficiency of the mutant is reduced to approximately two-thirds for the monoglycosylated peptide substrates and to approximately one-fifth for the di-glycosylated peptide substrate (Fig. 3D and SI Appendix, Table S2). Interestingly, the activity of the mutant against the unglycosylated peptide shows an apparent ∼2-fold decrease in its KM, which doubles its catalytic efficiency. This may be consistent with the structure showing that N270A changes a hydrophilic side chain to a hydrophobic side chain that subsequently could better interact with the T17 methyl group on a small movement of the loop containing N270A (Fig. 3B).
To assess the sugar specificity of the binding pocket, we modeled in galactose, mannose, and glucose (SI Appendix, Fig. S4B). Galactose makes similar contacts as GalNAc and likely binds with comparable affinity, since the N-acetylamino group of GalNAc is not directly contacting the enzyme. With mannose, C4-OH appears to be 3.8 Å from N270, while with glucose, C4-OH is ∼5 Å away from N270. Based on these distances, we predict a slightly decreased affinity for mannose in the pocket and weak interactions with glucose (SI Appendix, Fig. S3B).
The GalNAc-T12 preference for extant GalNAc in the +3 position overlaps with a well-characterized preference for Pro at the +3 position for GalNAc-Ts (13, 19, 20, 23). Kinetic studies indeed show that GalNAc-T12 has an ∼8-fold higher preference for Pro over Thr at the +3 position (i.e., ∼TPRP∼ vs. ∼TPRT∼) and an ∼15-fold preference for a GalNAc (∼TPRT*∼) at this position (SI Appendix, Fig. S4C). A structural alignment of Pro pockets of GalNAc-T2, -T4, and -T12 shows that residues in the 3 enzymes are well aligned despite the presence of GalNAc in the GalNAc-T12 structure (SI Appendix, Fig. S4D). However, the GalNAc-T2 equivalent of the GalNAc-T12-N270 residue (GalNAc-T2-A266) cannot form appropriate side chain interactions with GalNAc, while for GalNAc-T4, the equivalent Q269 side chain would clash with GalNAc (SI Appendix, Fig. S4D). GalNAc would similarly not likely be accommodated in the hydrophobic Pro pocket of GalNAc-T1, PGANT9, or T10 (SI Appendix, Fig. S4E).
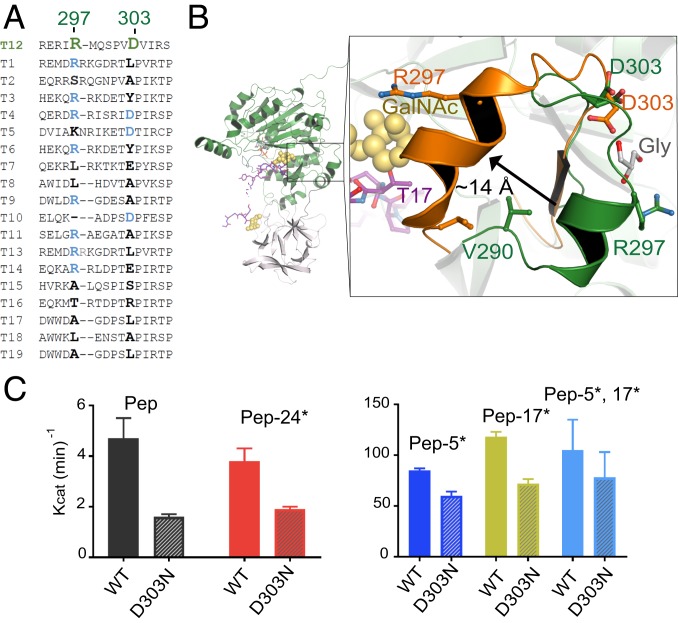
CRC-Associated Residues D303 and R297 Are Located in a Semiconserved Loop.
GalNAc-T12D303N and GalNAc-T12R297W occur widely among CRC patients (17, 18). D303 and R297 are located in a semiconserved loop within the catalytic domain and interact with glycerol (Fig. 4 A and B). Another CRC mutation, V290F, introduces a bulky hydrophobic side chain in the loop that would clash with nearby residues and destabilize that region of the enzyme (Fig. 4B and SI Appendix, Fig. S5A). GalNAc-T12R297W is nearly inactive against the Muc5Ac peptide and T12_Pep, while GalNAc-T12D303N retains partial activity against both peptides (SI Appendix, Fig. S5B) (18). Thus, we used D303N to assess the function of the loop and performed a detailed kinetic analysis of the GalNAc-T12D303N mutant to determine whether its peptide or glycopeptide activities were differentially altered. As with previous findings, GalNAc-T12D303N is ∼50% less active than GalNAc-T12WT against T12_Pep and monoglycosylated peptide substrates (Fig. 4C and SI Appendix, Fig. S5C and Table S2). The decreased activity of GalNAc-T12D303N is less pronounced in the presence of T12_Pep-5*,17*. This may be due to the synergy of di-glycosylated peptide binding by GalNAc-T12D303N, where GalNAc-binding sites in both the catalytic and lectin domains are not likely to be altered by the mutations. These results indicate that GalNAc-T12D303N does not have significantly altered activities against glycopeptide substrates compared with the unglycosylated substrate, and that the role of the semiconserved loop in modulating activity is independent of extant GalNAc binding.
Fig. 4.
Effect of CRC-associated mutations D303N and R297W on the function of GalNAc-T12. (A) Sequence alignment highlighting the CRC-associated residues R297 and D303. (B) Both residues are located on a semiconserved loop near the C terminus of the peptide substrate (purple, GalNAc in yellow spheres) and are coordinating glycerol (gray). The crystal structure loop (dark green) is superimposed over 2 of the low-scoring loop conformations from computational studies (orange) and depicts a movement of ∼14 Å that repositions the loop at the C terminus of the peptide substrate. (C) Activity assay verifying that the CRC-associated variant D303N has lower activity than wild-type GalNAc-T12 against all the T12_Pep substrates.
How CRC mutations alter the function of the loop containing D303 and R297 is unclear from the crystal structure, since the loop is ∼25 Å from the active site and does not have extensive interactions with surrounding amino acids. We performed loop simulations that suggest alternate low-energy conformations, including 1 where the loop undergoes an ∼14-Å movement that significantly changes the spatial position of R297 (Fig. 4B). Thus, it is possible that the loop may be influencing the activity of the enzyme through transient movements that are not observed in the static GalNAc-T12 structure. Due to the presence of bound glycerol, we conducted docking simulations that predicted potential sugar-binding pockets with interaction energies comparable to those of known GalNAc-binding pockets. We found that GalNAc is more likely to interact with the back side of the loop, where glycerol is bound in the structure (Fig. 4B and SI Appendix, Fig. S5 D–F and Table S3). Interestingly, the docking simulations are consistent with kinetic results showing that GalNAc-T12 binds GalNAc efficiently in its lectin and catalytic domain, while GalNAc-T4 binds GalNAc efficiently in its lectin domain but not its catalytic domain (SI Appendix, Fig. S5F) (12). However, since extant GalNAcs on a substrate do not alter the activity of GalNAc-T12D303N, a role for sugar binding in the loop is currently not clear.
Discussion
The fundamental challenge in understanding how GalNAc-T dysfunction may potentially contribute to cancer initiation and/or progression has been determining how each member in this large family of enzymes uniquely recognizes and modifies substrates. In this study, we investigated the unique functions of the glycopeptide-preferring family member GalNAc-T12, in which mutations resulting in the reduction or loss of activity are found in a subset of patients with CRC. We found that the clinical variant GalNAc-T12R373H alters the nonconserved catalytic loop residue R373 that aligns the active site for catalysis. The related isoenzyme GalNAc-T4 also contains an Arg at that position, where it appears to have a similar role in coordinating substrate recognition (12). The data further reveal a unique catalytic flexible loop stabilization that occurs through interactions between a conserved K367 in the loop and a nearby nonconserved W262. The loop conformation is influenced by the nonconserved residue P366, which also interacts with R16 at the +2 position of the peptide, which was uniquely shown to enhance the activity of GalNAc-T12. Overall, these results highlight the distinct mechanisms and interplay between unique and conserved residues used by different family members to modulate activity.
The ternary structure verifies that GalNAc-T12 binds to extant GalNAc on a substrate in the α repeat of its lectin domain and GalNAc at the +3 position through a nonconserved pocket in the catalytic domain and explains how the +3 position is unique to GalNAc-T12. Peptides with Pro in the +3 position have been shown to enhance enzymatic activity for most of the isoenzymes, including GalNAc-T12 (20, 23), yet GalNAc-T12 prefers GalNAc over Pro. This is unsurprising, given that our kinetic studies show that GalNAc at specific positions on the peptide substrate greatly enhances the activity of GalNAc-T12, suggesting that even if the enzyme had a sequence preference, it still would function more efficiently as a glycopeptide-preferring enzyme.
Finally, the structure of GalNAc-T12 shows that the CRC mutations V290F, D303N, and R297W occur on a semiconserved loop that that binds to glycerol. The importance of this loop is presently not clear from the structure, but computational simulations suggest that it could adopt various conformations. Thus, it is possible that this loop is mobile in solution and undergoes dynamic conformational changes to influence function by, for instance, potentially folding over to help position substrates in the active site or changing conformation to stabilize transient states of the enzyme during catalysis. The loop glycerol-binding pocket could play a role in GalNAc or sugar binding, although previous studies and these data show that GalNAc-T12 does not have a preference for extant GalNAcs at the C-terminal position of a peptide (20). While this remains to be further investigated, the structure reveals a functionally important region in the catalytic domain unique to both GalNAc-T12 and GalNAc-T4. Overall, we anticipate that the structure and supporting biochemical data will provide a starting point for designing in vivo experiments for discovering the biological substrate(s) of GalNAc-T12 and potentially explain how aberrant glycosylation states of its substrate in patients with CRC contribute to cancer progression.
Methods
In brief, His6-TEV-GalNAc-T1239–581 was cloned from the template GALNT12 (NM_024642.4) into pPICZα A for secreted protein expression in Pichia pastoris. GalNAc-T12 mutants were constructed by site-directed mutagenesis (SI Appendix, Table S4). Proteins were purified on a HisTrap HP column, followed by treatment with TEV protease to remove the His6 tag (SI Appendix, Table S5). Crystals of GalNAc-T12 bound to T12_Pep-5*,17*, Mn2+, and UDP formed in 0.2 M NaCl and 20% PEG 3350 (wt/vol), and the structure was solved by molecular replacement. Enzyme kinetics were measured by quantifying the transfer of either [3H]-GalNAc or [14C]-GalNAc to peptide substrates. Loop structure predictions were conducted using Rosetta KIC, and GalNAc docking was done using RosettaLigand. The methodology is described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Kelly Ten Hagen for helpful discussions and for critically reading and editing the manuscript; Dr. Wei Yang (National Institute of Diabetes and Digestive and Kidney Diseases) for providing access to equipment and editing the manuscript; Dr. Yang Gao (National Institute of Diabetes and Digestive and Kidney Diseases) for editing the manuscript; the beamline staff at the Advanced Photon Source for assistance; and XingJie Pan and Tanja Kortemme for providing access to the latest Rosetta loop closure algorithms. This research was supported by NIH Grants 1-ZIA-DE000739–05 (to L.A.T.) and R01 GM113534 (to T.A.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Structure coordinates and X-ray diffraction data have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6PXU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902211116/-/DCSupplemental.
References
- 1.Bergstrom K., et al. , Defective intestinal mucin-type O-glycosylation causes spontaneous colitis-associated cancer in mice. Gastroenterology 151, 152–164.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duarte H. O., et al. , Mucin-type O-glycosylation in gastric carcinogenesis. Biomolecules 6, E33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kudelka M. R., Ju T., Heimburg-Molinaro J., Cummings R. D., Simple sugars to complex disease—Mucin-type O-glycans in cancer. Adv. Cancer Res. 126, 53–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niv Y., Rokkas T., Mucin expression in colorectal cancer (CRC): Systematic review and meta-analysis. J. Clin. Gastroenterol. 53, 434–440 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Alexander J. L., et al. , Colorectal carcinogenesis: An archetype of gut microbiota–host interaction. Ecancermedicalscience 12, 865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahmus J. D., Kotler D. L., Kastenberg D. M., Kistler C. A., The gut microbiome and colorectal cancer: A review of bacterial pathogenesis. J. Gastrointest. Oncol. 9, 769–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly D., Yang L., Pei Z., Gut microbiota, fusobacteria, and colorectal cancer. Diseases 6, E109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucas C., Barnich N., Nguyen H. T. T., Microbiota, inflammation and colorectal cancer. Int. J. Mol. Sci. 18, E1310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett E. P., et al. , Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ten Hagen K. G., Fritz T. A., Tabak L. A., All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13, 1R–16R (2003). [DOI] [PubMed] [Google Scholar]
- 11.Tian E., Ten Hagen K. G., Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj. J. 26, 325–334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Las Rivas M., et al. , Structural and mechanistic insights into the catalytic domain-mediated short-range glycosylation preferences of GalNAc-T4. ACS Cent. Sci. 4, 1274–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fritz T. A., Raman J., Tabak L. A., Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-2. J. Biol. Chem. 281, 8613–8619 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Fagerberg L., et al. , Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J. M., Chen H. L., Wang G. M., Zhang Y. K., Narimatsu H., Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-12 in gastric and colonic cancer cell lines and in human colorectal cancer. Oncology 67, 271–276 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Clarke E., et al. , Inherited deleterious variants in GALNT12 are associated with CRC susceptibility. Hum. Mutat. 33, 1056–1058 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Evans D. R., et al. , Evidence for GALNT12 as a moderate penetrance gene for colorectal cancer. Hum. Mutat. 39, 1092–1101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guda K., et al. , Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc. Natl. Acad. Sci. U.S.A. 106, 12921–12925 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F., et al. , The small molecule luteolin inhibits N-acetyl-α-galactosaminyltransferases and reduces mucin-type O-glycosylation of amyloid precursor protein. J. Biol. Chem. 292, 21304–21319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Revoredo L., et al. , Mucin-type O-glycosylation is controlled by short- and long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. Glycobiology 26, 360–376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerken T. A., et al. , The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J. Biol. Chem. 288, 19900–19914 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji S., et al. , A molecular switch orchestrates enzyme specificity and secretory granule morphology. Nat. Commun. 9, 3508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerken T. A., et al. , Emerging paradigms for the initiation of mucin-type protein O-glycosylation by the polypeptide GalNAc transferase family of glycosyltransferases. J. Biol. Chem. 286, 14493–14507 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milac A. L., Buchete N. V., Fritz T. A., Hummer G., Tabak L. A., Substrate-induced conformational changes and dynamics of UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase-2. J. Mol. Biol. 373, 439–451 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman J., et al. , The catalytic and lectin domains of UDP-GalNAc: Polypeptide α-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J. Biol. Chem. 283, 22942–22951 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samara N. L., Fernandez A. J., Crystal structure of human GalNAc-T12 bound to a diglycosylated peptide, Mn2+, and UDP. Protein Data Bank. http://www.rcsb.org/pdb/results/results.do?tabtoshow=Unreleased&qrid=D30FC3B2. Deposited 30 July 2019.
- 27.de Las Rivas M., et al. , The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences. Nat. Commun. 8, 1959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagen F. K., Hazes B., Raffo R., deSa D., Tabak L. A., Structure-function analysis of the UDP-N-acetyl-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase. Essential residues lie in a predicted active site cleft resembling a lactose repressor fold. J. Biol. Chem. 274, 6797–6803 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Kubota T., et al. , Structural basis of carbohydrate transfer activity by human UDP-GalNAc: Polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10). J. Mol. Biol. 359, 708–727 (2006). [DOI] [PubMed] [Google Scholar]
- 30.de Las Rivas M., et al. , Structural analysis of a GalNAc-T2 mutant reveals an induced-fit catalytic mechanism for GalNAc-Ts. Chemistry 24, 8382–8392 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.