Significance
Horizontal gene transfer (HGT) is the movement of genetic material between organisms other than by reproduction, which plays an important role in bacterial evolution. Often, mobile genetic elements such as plasmids are involved in HGT. In this study, we present phylogenetic, biogeographic, and functional analyses of a previously unrecognized plasmid that is found with 100% sequence identity in multiple distinct bacterial genera obtained from geographically separated locations. This is the only known instance where actual nucleotide identity and not only high synteny has been described for plasmids in environmental organisms. Furthermore, we provide experimental evidence for the potential of this plasmid to be transmitted across bacterial orders, thereby increasing our understanding of evolution and microbial niche adaptation in the environment.
Keywords: horizontal gene transfer, Roseobacter group, RepL-type plasmid, chromate resistance
Abstract
Horizontal gene transfer (HGT) plays an important role in bacterial evolution and serves as a driving force for bacterial diversity and versatility. HGT events often involve mobile genetic elements like plasmids, which can promote their own dissemination by associating with adaptive traits in the gene pool of the so-called mobilome. Novel traits that evolve through HGT can therefore lead to the exploitation of new ecological niches, prompting an adaptive radiation of bacterial species. In this study, we present phylogenetic, biogeographic, and functional analyses of a previously unrecognized RepL-type plasmid found in diverse members of the marine Roseobacter group across the globe. Noteworthy, 100% identical plasmids were detected in phylogenetically and geographically distant bacteria, revealing a so-far overlooked, but environmentally highly relevant vector for HGT. The genomic and functional characterization of this plasmid showed a completely conserved backbone dedicated to replication, stability, and mobilization as well as an interchangeable gene cassette with highly diverse, but recurring motifs. The majority of the latter appear to be involved in mechanisms coping with toxins and/or pollutants in the marine environment. Furthermore, we provide experimental evidence that the plasmid has the potential to be transmitted across bacterial orders, thereby increasing our understanding of evolution and microbial niche adaptation in the environment.
Horizontal gene transfer (HGT) events allow prokaryotes to exploit novel traits and habitats and thereby influence the role and impact of bacterial communities in changing environments (1). Common vectors of HGT are gene transfer agents (GTAs) (2) and mobilizable or conjugative plasmids (3), whose transfer depends on type IV secretion systems (T4SSs) (3–5). GTAs can efficiently package and deliver genomic DNA between bacteria via a phage-like transduction process. However, the size of DNA fragments is severely restricted (<5 kbp) and packaging is nonrandom depending on GC content and the genomic source region (6). Plasmids, on the other hand, have been shown to potentially transfer large coherent gene clusters between distant relatives (5, 7, 8). Plasmid transfer is mediated by dedicated T4SSs, encoded on the same replicon in the case of conjugative plasmids, or elsewhere within the genome in the case of mobilizable plasmids. However, the number and types of different plasmids that can stably coexist within a cell are restricted by “plasmid incompatibility,” which is defined by the similarity of the corresponding replicases (9, 10). In contrast to the anthropogenic spread of antibiotic resistance plasmids across the globe by, e.g., antibiotic misuse and overuse, very little is known about the extent of natural plasmid transfer in the environment.
HGT vectors are widespread among members of the Roseobacter group (11), which are diverse and abundant marine Rhodobacteraceae (Alphaproteobacteria). They are key players in the global cycles of carbon and sulfur (12, 13) and are known for their functional and metabolic diversity. Due to their remarkable genome plasticity (14–16) and potential for HGT they are a good model for studying niche adaptations in marine habitats (11, 17). As exemplified for Roseobacter group members, it is suspected that the phylogenetically scattered occurrence of photosynthesis among Proteobacteria is essentially caused by plasmid transfer (7, 10, 18). Furthermore, the recent discovery of 2 largely syntenic plasmids in phylogenetically distant Roseobacter members retraces the horizontal transfer and individual evolution of plasmids in marine habitats (5).
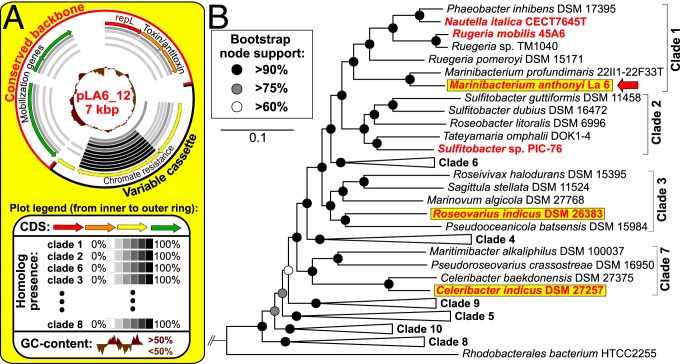
Recently, addition of the rare earth element lanthanum to methanol enrichments of coastal seawater from the United Kingdom facilitated the isolation of a new methylotroph within the Roseobacter group: Marinibacterium anthonyi La 6 (19). This bacterium contains an unusually large genome and unique genetic adaptations, clearly distinguishing it from its closest phylogenetic neighbors. In order to understand its genome plasticity, we performed gap closure and circularization of the replicons by PacBio long-read DNA sequencing. When analyzing these replicons with respect to current knowledge on Roseobacter mobile elements (9, 10, 20), we found a RepL-type (21, 22) plasmid, designated pLA6_12, representing an additional and previously unrecognized roseobacterial compatibility group.
Most interestingly, this plasmid is found with 100% sequence identity in multiple distinct genera obtained from geographically separated locations. This is the only known instance where actual nucleotide identity, and not only high plasmid synteny, has been observed for extrachromosomal replicons in environmental organisms of distinct genera. This likely reflects very recent widespread HGT events. The genomic and functional characterization of this plasmid revealed a completely conserved backbone, with 92 to 100% sequence identity in all observed cases, dedicated to replication, stability, and mobilization as well as an interchangeable gene cassette with diverse, but globally recurring motifs involved in mechanisms coping with toxins and/or pollutants.
Furthermore, we provide experimental evidence for the potential of this plasmid to be transmitted across bacterial orders. We also verified the functionality of the encoded toxin resistance operon after transfer to new hosts, thereby exemplifying the environmental and evolutionary relevance of this plasmid as a potent vector for HGT. This study documents the only known instance of identical plasmids bridging very large phylogenetic and geographic distances in natural ecosystems worldwide.
Results
The Replicons of M. anthonyi La 6.
Sequencing, gap closure, circularization, and reassembly of M. anthonyi La 6 yielded a total genome size of 6.8 Mbp, which is the largest genome reported for Rhodobacteraceae. This difference from the initially estimated size of the draft genome (7.2 Mbp) (19) is the result of improved repeat resolution mediated by PacBio long-read DNA sequencing. The total number of replicons in this strain was exceptionally high, consisting of 1 5.6-Mbp chromosome and 12 extrachromosomal replicons (ECRs; pLA6_01 to pLA6_12), ranging from 7 to 184 kbp in size. Based on a relative synonymous codon usage (RSCU) analysis, which allows the comparison of all replicons of a completely sequenced genome (10), these ECRs could be further classified as 5 chromids with a chromosome-like genetic imprint, considered to be essential parts of the native genome (10, 23), and 7 “dispensable” plasmids (SI Appendix, Fig. S1). Six of those plasmids encode a T4SS or plasmid mobility genes, reflecting the potential of HGT via conjugation. One of the chromids, pLA6_08, is susceptible to loss after cultivation in complex medium, thereby seemingly contradicting the usually essential nature of chromids (23). However, functional annotation revealed that most genes on pLA6_08 are involved in uptake, degradation, and utilization of various carbon compounds. They are therefore presumably essential in natural oligotrophic marine habitats, but dispensable under artificial nutrient-rich laboratory conditions. For the sake of completeness, both the wild-type strain and the natural plasmid-“curing” mutant of M. anthonyi La 6 have now been deposited at the German Collection of Microorganisms and Cell Cultures (DSM) (wild type: DSM 107130; ΔpLA6_08: DSM 104755). Another ECR, the T4SS-containing plasmid pLA6_07, is ambiguously positioned basal to chromids, but also close to plasmids by RSCU-based clustering (SI Appendix, Figs. S1 and S2), possibly illustrating its ongoing transition from a plasmid to a chromid due to the formation and resolution of cointegrates (23). For further discussion of the M. anthonyi La 6 ECRs, pLA_01 to 11, please refer to SI Appendix, section 3).
With 1 exception, all ECRs of M. anthonyi La 6 harbor well-characterized replicases associated with the 4 most abundant plasmid types in Rhodobacteraceae, i.e., RepA, RepB, RepABC, and DnaA-like (9, 20). However, the smallest plasmid, pLA6_12, with a size of 7,053 bp encodes a solitary RepL replication initiation protein and lacks the parAB partitioning operon otherwise typical for Rhodobacteraceae (Fig. 1A). Among all sequenced Rhodobacteraceae, the occurrence of RepL-type plasmids is very scarce and none of them have been characterized. In addition, this type of plasmid has never been reported within the Roseobacter group. Further comparative analyses revealed the presence of this exact plasmid, in the form of replicons sharing 100% nucleotide identity with pLA6_12 over their entire length, in 2 other Roseobacter group members belonging to phylogenetically distinct clades, i.e., Roseovarius indicus DSM 26383 and Celeribacter indicus DSM 27257 (Fig. 1B and SI Appendix, Fig. S3). This is therefore the only known instance of identical plasmids naturally occurring in different genera. Recently, Kothari et al. (24) described high sequence similarities between plasmids found in multiple adjacent groundwater wells. However, although the sampled wells were in very close proximity to each other, no absolutely identical plasmids were shared. In contrast, the 3 strains harboring the 7,053 bp pLA6_12 plasmid were independently isolated from widespread geographic locations in the North Atlantic and the Indian Ocean (19, 25, 26). For M. anthonyi La 6, the presence of pLa6_12 could be additionally verified by DNA isolation, library preparation, and sequencing in 2 different laboratories (United Kingdom and Germany). Furthermore, 3 additional homologous plasmids sharing 92 to 94% nucleotide identity over a conserved 4.5-kbp backbone region encoding the replicase RepL, a toxin–antitoxin system and a plasmid mobility gene cluster were found in 3 other genera of the Roseobacter group (Fig. 1B). This broad phylogenetic distribution reflects the relevance of this plasmid type for ongoing HGT events within marine bacteria.
Fig. 1.
(A) Circular visualization of the pLA6_12 plasmid and (B) phylogenomic maximum likelihood tree of Roseobacter group members. Multi locus sequence alignment is based on 582 orthologous proteins with 182,693 amino acid positions. Strains with identical RepL-type plasmids are highlighted in yellow. Roseobacter group members containing a RepL-type plasmid with a backbone >92% identical to those of pLA6_12 are marked in red. Clades are shown partially collapsed. A noncollapsed version is given in SI Appendix, Fig. S3.
Variety and Biogeography of pLA6_12-Like Plasmids.
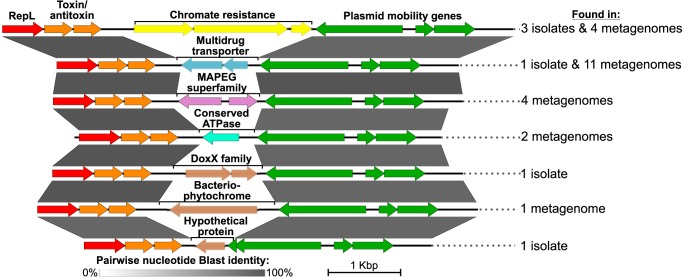
RepL-type plasmids can be found in a highly diverse range of bacterial phyla (SI Appendix, Fig. S4). Background information and a detailed overview is given in SI Appendix, section 1. However, comparative analyses indicated that the occurrence of ECRs with the conserved backbone of pLA6_12 is restricted to the Roseobacter group. Their replicase RepL shares an amino acid identity of at least 97%, whereas the most closely related RepL-type plasmids lacking the conserved backbone exhibit only 75% amino acid identity (SI Appendix, Table S1). Accordingly, we employed an identity cutoff of 95% in subsequent screenings of public metagenome databases to find further homologs of this replicon. The database search yielded 21 completely circularized metagenomic replicons sharing the highly conserved pLA6_12 backbone region and a variable integration cassette (Fig. 2 and SI Appendix, Fig. S5). In total, we identified 7 distinct integration cassette motifs, 4 of which are recurring in multiple samples and/or isolates of members of the Roseobacter group worldwide. Nineteen additional metagenome fragments contained a complete pLA6_12-like repL gene, but were of insufficient length to cover the full backbone and integration cassette. The 3 nonrecurring motifs comprised a putative bacteriophytochrome, a DoxX-family protein of unknown function, and a hypothetical protein, respectively. The recurring motifs found in multiple metagenomic pLA6_12-like replicons included 1) conserved ATPases; 2) MAPEG (“membrane-associated proteins in eicosanoid and glutathione metabolism”) superfamily proteins (27); 3) multidrug transporters; and, most interestingly, 4) chromate resistance gene clusters. Global nucleotide alignments showed that these chromate resistance plasmids display sequence dissimilarities compared to the corresponding replicons of M. anthonyi La 6, R. indicus B108, and C. indicus P73 (SI Appendix, Fig. S6). However, most of these differences are located within the variable cassette region, thereby supporting the assumption that pLA6_12 is a highly conserved and potent natural vector for HGT. Considering the small size of all detected pLA6_12-like plasmids, which range from 4.8 to 7 kbp, it is likely that functional integration cassettes are restricted in their maximal size and thus limited to relatively compact gene clusters.
Fig. 2.
Linear representation of pLA6_12-like plasmids with different integration cassette motifs in isolates and environmental samples. Alignment visualization was produced using EasyFig. Pairwise Blast identities are indicated by gray shading showing a conserved backbone region. Only 1 representative is shown for each integration motif. The number of instances the motif was found in isolates and/or metagenomes is given on the Right. A detailed overview of all complete plasmid instances found for each integration motif is given in SI Appendix, Fig. S5.
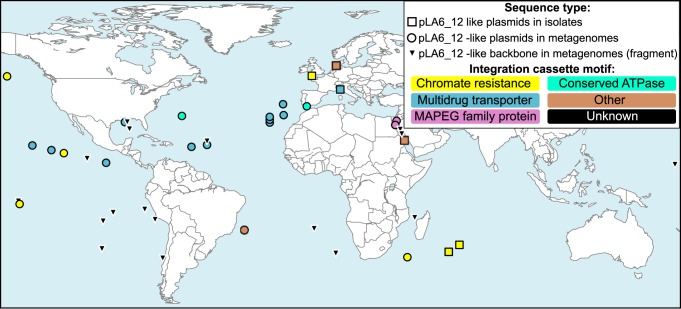
The biogeography of the detected RepL-type plasmids indicates a worldwide distribution with representatives found in the Indian, North and South Atlantic, the North and South Pacific Oceans, as well as the North, Baltic, Red, and Mediterranean Seas (Fig. 3). Extensive metagenome and reference genome analyses provided no evidence for its presence in limnic or terrestrial habitats, which is in accordance with the observation that all isolates with pLA6_12-like plasmids represent marine Rhodobacteraceae (SI Appendix, Table S1). Apart from MAPEG-family proteins (27), which were only found in Red Sea samples, no clear regional preference for specific integration cassette motifs could be observed (Fig. 3). Therefore, this plasmid appears to be abundant in all marine environments, mediating HGT across vast geographic and large phylogenetic distances (Figs. 1 and 3). Its strict limitation to marine habitats, but detection at numerous distant locations via independent sampling and sequencing approaches, emphasizes the significance of our finding.
Fig. 3.
Biogeography of pLA6_12-like plasmids. Sampling locations of Roseobacter group isolates are shown in squares. Locations where metagenomes have been obtained are shown in circles and triangles. Integration cassette motifs are indicated by a color code consistent with Fig. 2. pLA6_12-like plasmids were found exclusively in marine environments, but scattered across the world. Black triangles represent metagenome fragments, which encode pLA6_12-like repL genes, but are too incomplete to allow conclusions on the respective integration cassette.
Functional Analyses of the pLA6_12 Backbone.
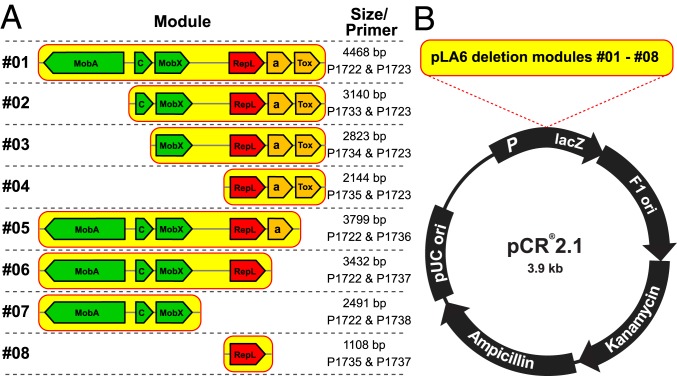
For this study, a shuttle plasmid for genetic engineering in Escherichia coli and functional tests in Roseobacter group members was created by cloning the conserved pLA6_12 backbone consisting of 6 genes (repL, mobACX, and 2 toxin–antitoxin genes) into the commercial cloning vector pCR2.1 (Fig. 4). The model organisms Phaeobacter inhibens DSM 17395 and Dinoroseobacter shibae DSM 16493 were chosen as test strains for plasmid transformation and conjugation experiments (8, 20, 28, 29). The kanamycin resistance gene of the shuttle plasmid served as a selection marker.
Fig. 4.
Deletion series of the conserved backbone from pLA6_12. (A) Module #01 with the complete pLA6_12 backbone and modules #02 to #08, representing stepwise deletions of each of the 6 backbone genes mobACX, repL, and the toxin/antitoxin system genes (here designated “Tox” and “a,” respectively), were amplified from pLA6_12 via specific PCRs. The size of each module and the primers used for its respective creation are listed to the Right. The respective primer sequences are given in SI Appendix, Table S4. (B) Each module was individually cloned into the commercially available E. coli vector pCR2.1 encoding kanamycin resistance as a universal selective marker.
Plasmid functionality.
The functionality of RepL shuttle-plasmid constructs was investigated in P. inhibens by monitoring plasmid replication in the novel host (30). It was shown that module #01 (Fig. 4), comprising the complete conserved pLA6_12 backbone, is successfully replicated in this host (SI Appendix, Figs. S7 and S8), thereby expanding the known natural host range of pLA6_12 to the genus Phaeobacter. Furthermore, a series of deletion mutants of each backbone gene (modules #02 to #07; Fig. 4) was established and their respective functional role was investigated. The experiments showed that all genes except repL are dispensable for plasmid replication, illustrating the essential role of the replicase RepL (SI Appendix, Figs. S7 and S9). Finally, it was established that a solitary repL gene is sufficient for replication (module #08) and thus can be regarded as the “functional heart” of the pLA6_12 plasmid (28).
Plasmid stability.
Initial plasmid stability tests of the complete conserved backbone (module #01) showed that all 150 investigated P. inhibens colonies maintained their RepL construct after 14 h of cultivation in complex medium without kanamycin (30) (SI Appendix, Fig. S8). Long-term stability assays furthermore revealed that none of 50 tested colonies lost their RepL plasmid construct during starvation after 7 d of incubation (SI Appendix, Fig. S9). This indicates a remarkable stability of pLA6_12, especially in the light of spontaneous chromid loss (pLA6_08) in M. anthonyi La 6 (see above) and compared to some unstable RepABC-type plasmids (30).
Deletion of the mobility gene cluster (module #04) also did not affect plasmid stability in Phaeobacter; however, removal of the presumed toxin–antitoxin operon (module #06) correlated with plasmid loss in 16% of 50 tested colonies. The experimental outcome confirmed the annotated function of this operon as a postsegregational killing mechanism (31). Since 1 of its genes is homologous to toxin proteins and the other contains an N-terminal helix-turn-helix domain that is characteristic of antitoxins (32), we conclude that it represents a type II toxin–antitoxin system. Interestingly, deletion of all backbone genes except repL (module #08) even resulted in a more frequent plasmid loss (68%), indicting a lower stability than the deletion of only the toxin–antitoxin system (module #06; SI Appendix, Fig. S9). This difference is indicative of additional regulatory elements in deleted noncoding regions, possibly influencing plasmid copy number.
Plasmid mobilization.
The function of the mobility gene cluster was tested in a conjugation experiment between 2 distinct Roseobacter group members. Since pLA6_12 does not encode its own T4SS for the formation of conjugative pili, D. shibae DSM 16493, which possesses 2 T4SS-encoding plasmids (28), was chosen as donor and electroporated with the complete-backbone shuttle vector (module #01, Fig. 4). The P. inhibens DSM 17395 mutant Δ262 (Materials and Methods), which bears gentamicin resistance as selective marker and is easily distinguishable from the brightly pink D. shibae (33) due to its distinct white color, was chosen as recipient. The conjugation experiment, performed according to Patzelt et al. (8), followed by selection for plasmid-encoded kanamycin resistance as well as the recipient-specific gentamicin resistance, yielded hundreds of positive transconjugants. The successful conjugal transfer of the shuttle vector was validated by PCR assays (SI Appendix, Fig. S10). Accordingly, we could show that the mobACX mobilization module of pLA6_12 is functional and mediates conjugation of this plasmid across genus borders.
Host range.
The presence of pLA6_12-like plasmids in 6 distinct genera covering 4 distinct clades of the Roseobacter group (Fig. 1), as well as its exclusiveness to marine habitats (Fig. 3), indicates a general functionality within these bacteria. In accordance with these observations, extensive searches in public sequence databases have not yet revealed any bacterial strains outside the Roseobacter group that contain a pLA6_12-like plasmid. However, as recently shown for RepABC-type plasmids (30), this does not necessarily mean that the natural host range of pLA6_12 is restricted to the Roseobacter group. Accordingly, we investigated if the pLA6_12 backbone is also functional in selected representatives of 3 other alphaproteobacterial orders (Rhizobiales, Rhodospirillales, and Sphingomonadales). We could confirm stable replication of the pLA6_12-like vector construct in the pathogenic rhizobium Agrobacterium tumefaciens (SI Appendix, Fig. S11), but none of the other tested bacteria. Therefore, we could show that its host range spans at least 2 different alphaproteobacterial orders, i.e., Rhodobacterales and Rhizobiales. Further investigations will be necessary to reveal whether this plasmid is also stable in other bacterial orders, or even phyla as indicated by the presence of homologous RepL replicases in taxonomically highly diverse lineages (SI Appendix, Fig. S4).
Functional Analyses of the pLA6_12 Integration Cassette.
The majority of recurring integration cassette motifs of pLA6_12-like ECRs appear to be involved in mechanisms coping with toxins and/or pollutants in the marine environment. This is in concordance with previous studies, which describe heavy metal resistance genes to be among the most frequently found phenotypic modules carried by bacterial plasmids (24, 34, 35). As these functions can be encoded in relatively compact gene clusters, this may represent insert selection bias, especially when considering the small size of all pLA6_12-like plasmids ranging from 4.8 to 7 kbp. Alternatively, this distribution may also reflect a high impact of toxins and/or pollutants on environmental communities. The MAPEG superfamily (36) comprises membrane proteins involved in eicosanoid and glutathione metabolism and may provide protection against xenobiotics and/or oxidative stress (36, 37). The term “multidrug transporter,” which represents the most frequently detected motif in metagenomic samples (SI Appendix, Fig. S5), refers to diverse efflux systems that provide protection against a range of toxins and antibiotics (38). ChrBAC chromate resistance gene clusters, as found in M. anthonyi La 6 as well as 6 other isolates and metagenome samples, provide protection against hexavalent chromium (CrO42−) (39). This highly toxic and carcinogenic pollutant is imported into the bacterial cytoplasm by the sulfate (SO42−) uptake pathway and reduced to lower oxidation states [Cr(V) and Cr(III)] by which free radicals are formed that cause oxidative stress (40).
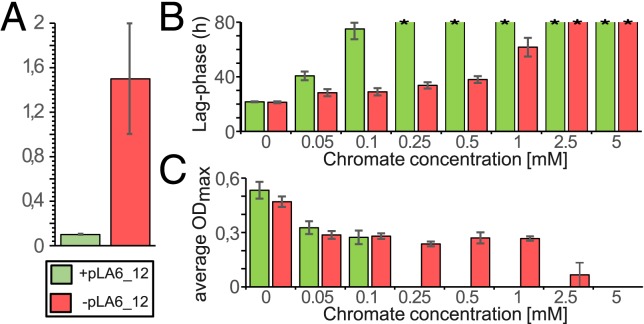
ChrA is an energy-dependent efflux transporter for cytoplasmic chromate (41); ChrB acts as a chromium-sensitive regulator of the chr operon (39); and ChrC is a superoxide dismutase for the detoxification of free radicals (42). Homologs of ChrA that share >59% sequence identity can also be found on the chromosome of M. anthonyi La 6 and in 39 of 45 analyzed Roseobacter group members (Dataset S1). While this distribution indicates a general relevance of chromate resistance for marine habitats, in silico prediction of a bacterial transporter’s actual substrate(s) is problematic and needs to be verified experimentally. Furthermore, it is questionable whether the solitary chromosomal ChrA homolog could provide an equivalent protection against chromate compared to a complete, plasmid encoded chrABC operon. We therefore investigated the susceptibility of the pLA6_12-bearing strain C. indicus DSM 27257 to hexavalent chromium with growth experiments in defined sea water medium (SWM) and compared it with the D. shibae DSM 16493 and P. inhibens DSM 17395 ∆65/3, which both lack this plasmid (28, 29). Similar tests for M. anthonyi La 6 and R. indicus DSM 26383 were not possible, due to poor growth of these strains in SWM. The results show that P. inhibens and D. shibae are more severely affected by chromate than C. indicus (SI Appendix, Fig. S12), with an increased lag phase of up to 41 h occurring at 0.1 mM, and no detectable growth beyond these concentrations. In contrast, C. indicus was still able to grow at 2 mM chromate, indicating an at least 1 order of magnitude increased chromate tolerance. In order to verify whether this tolerance was actually conveyed by the pLA6_12 encoded chromate resistance operon, plasmids were extracted from C. indicus DSM 27257 and transformed into P. inhibens DSM 17395 ∆65/3. Selection on SWM plates with 0.5 mM chromate, exceeding the 0.1-mM growth limit of the original P. inhibens strain, resulted in resistant colonies harboring the pLA6_12 plasmid (SI Appendix, Figs. S13 and S14). In subsequent growth experiments, pLA6_12-containing P. inhibens transformants displayed a chromate tolerance of at least 1 mM, which is a 10-fold increase compared to the original strain and close to the natural chromate tolerance of C. indicus (Fig. 5 and SI Appendix, Fig. S15). Furthermore, extremely long lag phases of more than 61 h were observed at 0.1-mM chromate concentration for the original P. inhibens ∆65/3 strain, which dropped down to 28 h for the pLA6_12-containing transformants. These results reflect a higher efficiency of the chrBAC operon compared to any indigeneous mechanisms like the putative chromosomal ChrA homolog, due perhaps to the detoxification by the plasmid-specific superoxide dismutase ChrC. However, the effect may also be partly influenced by a “gene dosage effect” based on its extrachromosomal localization. In conclusion, our experiments showed the functionality of the chromate resistance plasmid pLA6_12 in 1) the genuine host and 2) transformants of our model organism P. inhibens, thereby simulating the feasibility and benefit of its horizontal transfer in chromate-contaminated habitats (43).
Fig. 5.
Influence of pLA6_12 on the chromate tolerance of P. inhibens DSM 17395 ∆65/3. Comparative growth experiments of P. inhibens Δ65/3 (lacking pLA6_12) and the transformant P. inhibens Δ65/3+pLA6_12 were performed with chromate (K2CrO4) concentrations between 0 and 5 mM. (A) Maximum chromate tolerance was found to be 10 to 20× higher in strains containing pLA6_12. (B and C) Susceptibility or resistance to chromate manifested in form of changes in lag-phase duration and maximum optical density. The length of the lag phase was determined by the “threshold hour” (Ht), at which OD600 exceeded a predefined threshold of 0.03. *, infinity. The corresponding growth curves are shown in SI Appendix, Fig. S15.
Discussion
The main discovery of this study is the identification of an absolutely conserved RepL plasmid in 3 distantly related genera of Rhodobacteraceae. So far, no absolutely identical plasmids shared between phylogenetically distinct genera have previously been reported. In order to rule out the possibility that such instances may have been present, but simply overlooked in the past, we searched and compared publicly available sequence databases (44, 45) (SI Appendix, section 2 and Table S3). We found only 3 occurrences of identical plasmids shared by different taxa above species level, all of which occurred in strictly clinical settings. However, such settings represent a specialized habitat, since pathogens are constantly (re)introduced and subjected to artificially high selective pressure via antibiotics and disinfectants (46, 47). Our current study is therefore the first time 100% identical plasmids shared beyond genus borders were ever observed in natural environmental settings. In the postgenomics era, this is highly noteworthy because experimental host range tests have been a common tool in plasmid biology for decades.
Despite its ubiquitous global distribution, the mobilizable RepL-type plasmid pLA6_12 had previously been overlooked in the Roseobacter group. This potent HGT vector displays an extraordinary degree of conservation and transports different integration cassette motifs across vast geographic as well as phylogenetic distances. Consequently, it acts as an indicator for very recent global HGT events, reflecting a higher connectivity of the pangenome of this bacterial group than previously assumed. It is likely that those very small pLa6_12-like plasmids can be more efficiently mobilized than larger ones. The majority of the observed integration cassettes confer resistance to toxins and xenobiotics, which might reflect rapid adaptations to the respective pollutants in marine habitats. We exemplified this by experimentally validating the function of the chromate transporter gene cluster in the pLA6_12 plasmid of M. anthonyi La 6. This gene cluster conferred a 10 to 20× increase in chromate resistance of up to 2.5 mM, which is about a factor of 1,000 higher than the estimated average chromium concentration in open ocean waters (48, 49). It therefore should ensure the survival of its host bacterium in heavily contaminated habitats. However, the replication and maintenance of plasmids are a metabolic burden for the host bacterium (50) and the related fitness costs likely result in a gradual outcompeting of the plasmid-containing strains in the absence of chromium. Accordingly, we expect a higher frequency of this plasmid type in marine habitats with higher concentrations of this heavy metal. Furthermore, in light of the broad range variability of the RepL cassettes (Fig. 2), the observed presence of pLA6_12-like plasmids in the marine pangenome probably assures the quick adaptability of Roseobacter to changing concentrations of pollutants. It may be assumed that such an extreme tolerance is not necessarily relevant under most environmental conditions, but is precisely what is to be expected from an accessory gene. Any genes required to adapt to permanent changes in environmental conditions would be expected to eventually become part of the respective core genome of the organism.
The pLA6_12-like plasmid type seems to be restricted to marine habitats, as we specifically searched terrestric (and limnic) metagenomes and found no instance of this plasmid type there. Nevertheless, surface currents such as the Gulf Stream are reported to reach 0.4 to 1.2 m/s (51), making them conceivable dispersal mechanisms. Furthermore, ballast waters transported in freighters could also be a significant factor in speeding up dispersal.
The scattered phylogenetic and biogeographical distribution (Figs. 1 and 3) of this feature, combined with its localization on a plasmid and its deviating codon usage (SI Appendix, Figs. S1 and S2) clearly illustrate its general dispensability but nonetheless frequent intergenus HGT. Such horizontally transferred accessory genes are usually not present in every member of a microbial community, but still benefit the whole population, i.e., when adaptations to sudden variable extreme conditions are required (52, 53). The global distribution and exceptional conservation of pLA6_12-like plasmids therefore likely demonstrate recent adaptations to globally increasing frequencies of regional and temporal spikes in toxin concentrations from anthropogenic pollution (54, 55). In this regard, it seems to pose a striking analogy to the role of antibiotic resistance plasmids for multidrug-resistant hospital pathogens (56, 57).
Furthermore, the shuttle vectors constructed in this study possess a high biotechnological potential for genetic manipulations and heterologous gene expression in Alphaproteobacteria due to their broad host range and stability. Although the mobilization of the plasmid depends on the presence of a separate conjugative plasmid harboring a functional T4SSs for the formation of the conjugation pilus within the same bacterial host cell, this does not diminish its applicability for facilitating gene transfer between Roseobacter group members: Such systems are highly common within this group. Our study therefore illustrates the potential impact of this recently discovered vector on HGT-based adaptation mechanisms in bacterial communities worldwide. In conclusion, pLA6_12-like plasmids are globetrotting conjugational hitchhikers, likely playing an important role in bacterial evolution.
Materials and Methods
The updated M. anthonyi La 6 genome assembly is available under GenBank deposit (NSDV00000000), as well as under the individual accession nos. CP031585–CP031597.
For a detailed description of Gap closure as well as the selection and processing of reference genomes, please refer to SI Appendix, SM1 and 2.
Construction and Transformation of Shuttle Vectors and Deletion Modules.
A series of deletion modules of the pLa6_12 backbone was created by amplifying different backbone regions via specific PCR (Fig. 4A and SI Appendix, Table S4) and ligating them into pCR2.1 (Thermo Fisher Scientific; Fig. 4B). This vector encodes a kanamycin resistance gene as a selective marker and does not replicate in Alphaproteobacteria (30). Electro-competent E. coli New England Biolabs (NEB) Turbo cells were used for cloning. Initial PCR amplification was performed with the Phusion High-Fidelity DNA polymerase (NEB) while the conventional OneTaq DNA polymerase without proof-reading activity (NEB) was used for subsequent tailing with a single adenosine residue at the 3′ end. All modules were sequenced via primer walking, thereby excluding any PCR error that might impede the outcome of the physiological experiments. For details on functionality and stability tests based on these modules, please refer to SI Appendix, SM3.
Extraction of pLa6_12 and Transformation into P. inhibens.
The complete set of plasmids including pLa6_12 was isolated from C. indicus DSM 27257 with the Qiagen large-construct kit (Qiagen, Germany) and electroporated into competent cells of P. inhibens DSM 17395 mutant Δ65/3 (20, 58). The transformants were plated on SWM agar plates supplemented with 0.5 mM potassium chromate, thereby using increased chromate tolerance as a selective marker. Presence of the plasmid pLa6_12 in the transformants was additionally verified via specific PCR.
Chromate Tolerance Tests.
Growth experiments were performed in a defined SWM, modified from Zech et al. (59) by supplying only half the original concentration of trace elements. SWM was furthermore supplemented with 15 mM succinate and final potassium chromate concentrations ranging between 0 and 20 mM. Growth was monitored in 24-well plates for 96 h, using the “Infinite 200 Pro” microplate reader (Tecan Trading AG, Switzerland). In order to compare fluctuations in lag-phase length, we introduced a “threshold hour” (Ht) metric, as the timepoint at which OD600 cell density exceeded a defined threshold of 0.03. Growth tests were performed for the type strains of C. indicus DSM 27257 and D. shibae DSM 16493. In the case of P. inhibens, the plasmid curing mutant ∆65/3 (20, 58) of strain DSM 17395 was used instead of the wild type. This mutant is lacking its 65-kbp biofilm plasmid (60), thus preventing attachment to the surface of the test plates, thereby allowing reliable measurement of culture growth.
Supplementary Material
Acknowledgments
We thank Pascal Bartling for bioinformatic support, and Sandra Hacke as well as Simone Severitt for excellent technical assistance. We would also like to thank Ulrich Nübel (DSMZ) for helpful discussions. This work was partially supported by the Transregional Collaborative Research Center “Roseobacter” (Transregio TRR 51) of the Deutsche Forschungsgemeinschaft and by Gordon and Betty Moore Foundation Grant GBMF3303. We would also like to acknowledge support by the state of Baden-Württemberg through High Performance Computing, Data Intensive Computing and Large Scale Scientific Data Management in Baden-Württemberg (bwHPC).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905878116/-/DCSupplemental.
References
- 1.Koonin E. V., Horizontal gene transfer: Essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. F1000 Res. 5, 1805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lang A. S., Zhaxybayeva O., Beatty J. T., Gene transfer agents: Phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsay J. P., et al. , An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob. Genet. Elements 6, e1208317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallden K., Rivera-Calzada A., Waksman G., Type IV secretion systems: Versatility and diversity in function. Cell. Microbiol. 12, 1203–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petersen J., Wagner-Döbler I., Plasmid transfer in the ocean–A case study from the Roseobacter group. Front. Microbiol. 8, 1350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomasch J., et al. , Packaging of Dinoroseobacter shibae DNA into gene transfer agent particles is not random. Genome Biol. Evol. 10, 359–369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkmann H., Göker M., Koblížek M., Wagner-Döbler I., Petersen J., Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae. ISME J. 12, 1994–2010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patzelt D., et al. , Gene flow across genus barriers–Conjugation of Dinoroseobacter shibae’s 191-kb killer plasmid into Phaeobacter inhibens and AHL-mediated expression of type IV secretion systems. Front. Microbiol. 7, 742 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen J., Phylogeny and compatibility: Plasmid classification in the genomics era. Arch. Microbiol. 193, 313–321 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Petersen J., Frank O., Göker M., Pradella S., Extrachromosomal, extraordinary and essential–The plasmids of the Roseobacter clade. Appl. Microbiol. Biotechnol. 97, 2805–2815 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Luo H., Moran M. A., Evolutionary ecology of the marine Roseobacter clade. Microbiol. Mol. Biol. Rev. 78, 573–587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner-Döbler I., Biebl H., Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Moran M. A., González J. M., Kiene R. P., Linking a bacterial taxon to sulfur cycling in the sea: Studies of the marine Roseobacter group. Geomicrobiol. J. 20, 375–388 (2003). [Google Scholar]
- 14.Newton R. J., et al. , Genome characteristics of a generalist marine bacterial lineage. ISME J. 4, 784–798 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Brinkhoff T., Giebel H.-A., Simon M., Diversity, ecology, and genomics of the Roseobacter clade: A short overview. Arch. Microbiol. 189, 531–539 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Vollmers J., et al. , Poles apart: Arctic and Antarctic Octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLoS One 8, e63422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon M., et al. , Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen J., et al. , Think pink: Photosynthesis, plasmids and the Roseobacter clade. Environ. Microbiol. 14, 2661–2672 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Howat A. M., et al. , Comparative genomics and mutational analysis reveals a novel XoxF-utilizing methylotroph in the Roseobacter group isolated from the marine environment. Front. Microbiol. 9, 766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen J., et al. , Origin and evolution of a novel DnaA-like plasmid replication type in Rhodobacterales. Mol. Biol. Evol. 28, 1229–1240 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Kovach M. E., et al. , Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Greated A., Lambertsen L., Williams P. A., Thomas C. M., Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ. Microbiol. 4, 856–871 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Harrison P. W., Lower R. P. J., Kim N. K. D., Young J. P. W., Introducing the bacterial ‘chromid’: Not a chromosome, not a plasmid. Trends Microbiol. 18, 141–148 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Kothari A., et al. , Large circular plasmids from groundwater plasmidomes span multiple incompatibility groups and are enriched in multimetal resistance genes. MBio 10, e02899-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai Q., Cao J., Yuan J., Li F., Shao Z., Celeribacter indicus sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium from deep-sea sediment and reclassification of Huaishuia halophila as Celeribacter halophilus comb. nov. Int. J. Syst. Evol. Microbiol. 64, 4160–4167 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Lai Q., et al. , Roseovarius indicus sp. nov., isolated from deep-sea water of the Indian Ocean. Int. J. Syst. Evol. Microbiol. 61, 2040–2044 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Jakobsson P.-J., Morgenstern R., Mancini J., Forf-Hutchinsen A., Persson B., Membrane-associated proteins in eicosanoid and glutathione metabolism (MAPEG). Am. J. Respir. Crit. Care Med. 161, S20–S24 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Wagner-Döbler I., et al. , The complete genome sequence of the algal symbiont Dinoroseobacter shibae: A hitchhiker’s guide to life in the sea. ISME J. 4, 61–77 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Thole S., et al. , Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J. 6, 2229–2244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartling P., et al. , The composite 259-kb plasmid of Martelella mediterranea DSM 17316T-A natural replicon with functional RepABC modules from Rhodobacteraceae and Rhizobiaceae. Front. Microbiol. 8, 1787 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zielenkiewicz U., Cegłowski P., Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48, 1003–1023 (2001). [PubMed] [Google Scholar]
- 32.Yamaguchi Y., Park J. H., Inouye M., Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45, 61–79 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Biebl H., et al. , Dinoroseobacter shibae gen. nov., sp. nov., a new aerobic phototrophic bacterium isolated from dinoflagellates. Int. J. Syst. Evol. Microbiol. 55, 1089–1096 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Hemme C. L., et al. , Comparative metagenomics reveals impact of contaminants on groundwater microbiomes. Front. Microbiol. 6, 1205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster T. J., Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol. Rev. 47, 361–409 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bresell A., et al. , Bioinformatic and enzymatic characterization of the MAPEG superfamily. FEBS J. 272, 1688–1703 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Allocati N., Federici L., Masulli M., Di Ilio C., Glutathione transferases in bacteria. FEBS J. 276, 58–75 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Putman M., van Veen H. W., Konings W. N., Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64, 672–693 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viti C., Marchi E., Decorosi F., Giovannetti L., Molecular mechanisms of Cr(VI) resistance in bacteria and fungi. FEMS Microbiol. Rev. 38, 633–659 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Ramírez-Díaz M. I., et al. , Mechanisms of bacterial resistance to chromium compounds. Biometals 21, 321–332 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Díaz-Pérez C., Cervantes C., Campos-García J., Julián-Sánchez A., Riveros-Rosas H., Phylogenetic analysis of the chromate ion transporter (CHR) superfamily. FEBS J. 274, 6215–6227 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Juhnke S., Peitzsch N., Hübener N., Grosse C., Nies D. H., New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch. Microbiol. 179, 15–25 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Hausladen D. M., Alexander-Ozinskas A., McClain C., Fendorf S., Hexavalent chromium sources and distribution in California groundwater. Environ. Sci. Technol. 52, 8242–8251 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Brooks L., Kaze M., Sistrom M., A curated, comprehensive database of plasmid sequences. Microbiol. Resour. Announc. 8, e01325-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayers E. W., et al. , Database resources of the national center for biotechnology information. Nucleic Acids Res. 47, D23–D28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbosa T. M., Levy S. B., The impact of antibiotic use on resistance development and persistence. Drug Resist. Updat. 3, 303–311 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Russell A. D., Biocide use and antibiotic resistance: The relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 3, 794–803 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Bonnand P., James R. H., Parkinson I. J., Connelly D. P., Fairchild I. J., The chromium isotopic composition of seawater and marine carbonates. Earth Planet. Sci. Lett. 382, 10–20 (2013). [Google Scholar]
- 49.Geisler C.-D., Schmidt D., An overview of chromium in the marine environment. Dtsch. Hydrogr. Zeitschrift 44, 185–196 (1991). [Google Scholar]
- 50.San Millan A., MacLean R. C., Fitness costs of plasmids: A limit to plasmid transmission. Microbiol. Spectr. 5, MTBP-0016-2017 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Dohan K., Ocean surface currents from satellite data. J. Geophys. Res. Oceans 122, 2647–2651 (2017). [Google Scholar]
- 52.Mira A., Martín-Cuadrado A. B., D’Auria G., Rodríguez-Valera F., The bacterial pan-genome:a new paradigm in microbiology. Int. Microbiol. 13, 45–57 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R., The microbial pan-genome. Curr. Opin. Genet. Dev. 15, 589–594 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Wise J. P., Sr, et al. , A global assessment of chromium pollution using sperm whales (Physeter macrocephalus) as an indicator species. Chemosphere 75, 1461–1467 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Pei Y., Yu Z., Ji J., Khan A., Li X., Microbial community structure and function indicate the severity of chromium contamination of the yellow river. Front. Microbiol. 9, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palmer K. L., Kos V. N., Gilmore M. S., Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 13, 632–639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry J. A., Wright G. D., The antibiotic resistance “mobilome”: Searching for the link between environment and clinic. Front. Microbiol. 4, 138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dower W. J., Miller J. F., Ragsdale C. W., High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zech H., et al. , Growth phase-dependent global protein and metabolite profiles of Phaeobacter gallaeciensis strain DSM 17395, a member of the marine Roseobacter-clade. Proteomics 9, 3677–3697 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Frank O., et al. , Plasmid curing and the loss of grip–the 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst. Appl. Microbiol. 38, 120–127 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.