Significance
Long interspersed element-1s (L1s) encode 2 proteins (ORF1p and ORF2p) that preferentially mobilize (i.e., retrotranspose) their encoding messenger RNA (mRNA) transcript. ORF1p and/or ORF2p can also mobilize other cellular RNAs, including short interspersed elements (SINEs), U6 small nuclear RNA (snRNA), and mRNAs. Here, we demonstrate the RNA ligase RtcB can join U6 snRNA to L1 or other cellular RNAs to create chimeric RNAs; retrotransposition of the resultant chimeric RNAs leads to chimeric pseudogene formation; and chimeric U6/L1 RNAs are part of the transcriptome in multiple human cells. These data suggest RNA ligation contributes to the plasticity of the transcriptome and that the retrotransposition of chimeric RNAs can generate genetic variation in the human genome.
Keywords: LINE-1, retrotransposon, U6 snRNA, RtcB, RNA ligation
Abstract
Long interspersed element-1 (LINE-1 or L1) amplifies via retrotransposition. Active L1s encode 2 proteins (ORF1p and ORF2p) that bind their encoding transcript to promote retrotransposition in cis. The L1-encoded proteins also promote the retrotransposition of small-interspersed element RNAs, noncoding RNAs, and messenger RNAs in trans. Some L1-mediated retrotransposition events consist of a copy of U6 RNA conjoined to a variably 5′-truncated L1, but how U6/L1 chimeras are formed requires elucidation. Here, we report the following: The RNA ligase RtcB can join U6 RNAs ending in a 2′,3′-cyclic phosphate to L1 RNAs containing a 5′-OH in vitro; depletion of endogenous RtcB in HeLa cell extracts reduces U6/L1 RNA ligation efficiency; retrotransposition of U6/L1 RNAs leads to U6/L1 pseudogene formation; and a unique cohort of U6/L1 chimeric RNAs are present in multiple human cell lines. Thus, these data suggest that U6 small nuclear RNA (snRNA) and RtcB participate in the formation of chimeric RNAs and that retrotransposition of chimeric RNA contributes to interindividual genetic variation.
Long interspersed element-1 sequences (LINE-1s or L1s) comprise ∼17% of human DNA and have mobilized by a replicative process termed retrotransposition (1, 2). Most L1s are immobile (3–5); however, an average human genome harbors ∼80 to 100 retrotransposition-competent L1s (RC-L1s) (6–8). Human RC-L1s are ∼6 kilobases (kb) and contain a 5′ untranslated region (UTR) that is followed by 2 open reading frames (ORFs) (ORF1 and ORF2) and a short 3′ UTR that ends in a variable length poly-adenosine [poly(A)] tract (4, 9). ORF1 encodes an ∼40-kDa nucleic acid binding protein (ORF1p) (10–14) that has nucleic acid chaperone activity (14, 15). ORF2 encodes an ∼150-kDa protein (ORF2p) (16–18) that has DNA endonuclease (L1 EN) (19) and reverse transcriptase (L1 RT) (20, 21) activities. ORF1p, ORF2p, and full-length polyadenylated L1 RNA are required for efficient L1 retrotransposition in cis (19, 22, 23).
L1 retrotransposition begins with the transcription of a full-length genomic L1 from an RNA polymerase II promoter that resides within its 5′ UTR (24–26). The bicistronic L1 messenger RNA (mRNA) is exported to the cytoplasm where it undergoes translation (27–29). ORF1p and ORF2p preferentially bind their encoding L1 mRNA by a process termed cis-preference (30, 31), leading to the formation of an L1 ribonucleoprotein particle (RNP) (10, 13, 17, 32, 33). The association of ORF2p with the L1 mRNA poly(A) tail is a critical step in both L1 RNP formation and L1 retrotransposition (23). Components of the L1 RNP then enter the nucleus where a new L1 copy is integrated into genomic DNA by target site primed reverse transcription (TPRT) (19, 34, 35).
U6 small nuclear RNA (snRNA) is a uridine-rich small noncoding RNA that plays an essential role in nuclear intron splicing (36–38). U6 snRNA is the most conserved spliceosomal snRNA (39), is transcribed by RNA polymerase III (40), and has structural and functional similarities to domain V of self-splicing group II introns (41–44). The major form of U6 snRNA terminates in a 5-base poly-uridine [poly(U)] tract that ends in a terminal 2′,3′-cyclic phosphate (45). The 2′,3′-cyclic phosphate is generated posttranscriptionally by the Mpn1 enzyme (46, 47), which is encoded by the U6 snRNA biogenesis phosphodiesterase 1 (USB1) gene. Deletions or mutations in USB1 disrupt U6 snRNA 3′ end processing (46, 47) and are associated with the human genetic disease poikiloderma with neutropenia (48).
The L1 proteins can act in trans to promote the retrotransposition of a variety of cellular RNAs, including, small interspersed element (SINE) RNAs (49–52), noncoding RNAs (53–56), and messenger RNAs (30, 31). L1-mediated retrotransposition events have likely dispersed hundreds of copies of U6 snRNA throughout the human genome (57). Approximately 100 to 200 U6 pseudogenes consist of a copy of U6 fused to either a variably 5′-truncated L1 or a complementary DNA (cDNA) derived from a cellular RNA (53, 55–58), and they contain structural hallmarks that indicate they were formed by L1 retrotransposition [e.g., they end in a poly(A) tail, integrate into an L1 EN consensus cleavage sequence, and are flanked by short, variably sized target-site duplications] (53, 55, 56, 58). Experiments using engineered human L1s suggest that U6/L1 chimeric pseudogenes account for up to 1 out of 15 L1 retrotransposition events in HeLa cells (55) whereas computational analyses strongly suggest that 3 different LINE clades (LINE-1, LINE-2, and RTE) participate in U6/LINE chimeric pseudogene formation (57). Thus, U6/LINE chimeric pseudogene formation appears to be an ancient and ongoing process.
Here, we used genetic, molecular biological, biochemical, and computational approaches to dissect the mechanism of U6/L1 chimeric pseudogene formation. We demonstrate that U6/L1 chimeric RNAs arise independently of L1 retrotransposition and are formed through the ligation of a 2′,3′-cyclic phosphate on the 3′ end of U6 snRNA and 5′-OH on L1 RNA. Biochemical and genetic evidence suggest that the RNA 2′,3′-cyclic phosphate ligase RtcB (59–61) can join U6 RNAs ending in a 2′,3′-cyclic phosphate to both L1 and other mRNAs (e.g., green fluorescent protein [GFP] RNAs) containing a 5′-OH. Finally, we demonstrate that U6/L1 chimeric RNAs are a component of the transcriptome in multiple human cell lines.
Results
U6/L1 RNA Is Generated Independently of L1 Retrotransposition.
Previous hypotheses suggested that U6/L1 chimeric pseudogene formation could occur if ORF2p undergoes a template-switching event from L1 RNA to the 3′ end of U6 snRNA during TPRT (53, 55). However, recent studies have demonstrated that ORF2p exhibits a profound preference for binding, either directly or indirectly, to the 3′ poly(A) tract of L1, Alu, and other cellular RNAs (23, 62), raising the question of how ORF2p would switch templates to an RNA (e.g., U6 snRNA) that ends in a poly(U) tract. We hypothesized that U6 snRNA could be joined to L1 RNA to form a chimeric U6/L1 RNA prior to retrotransposition.
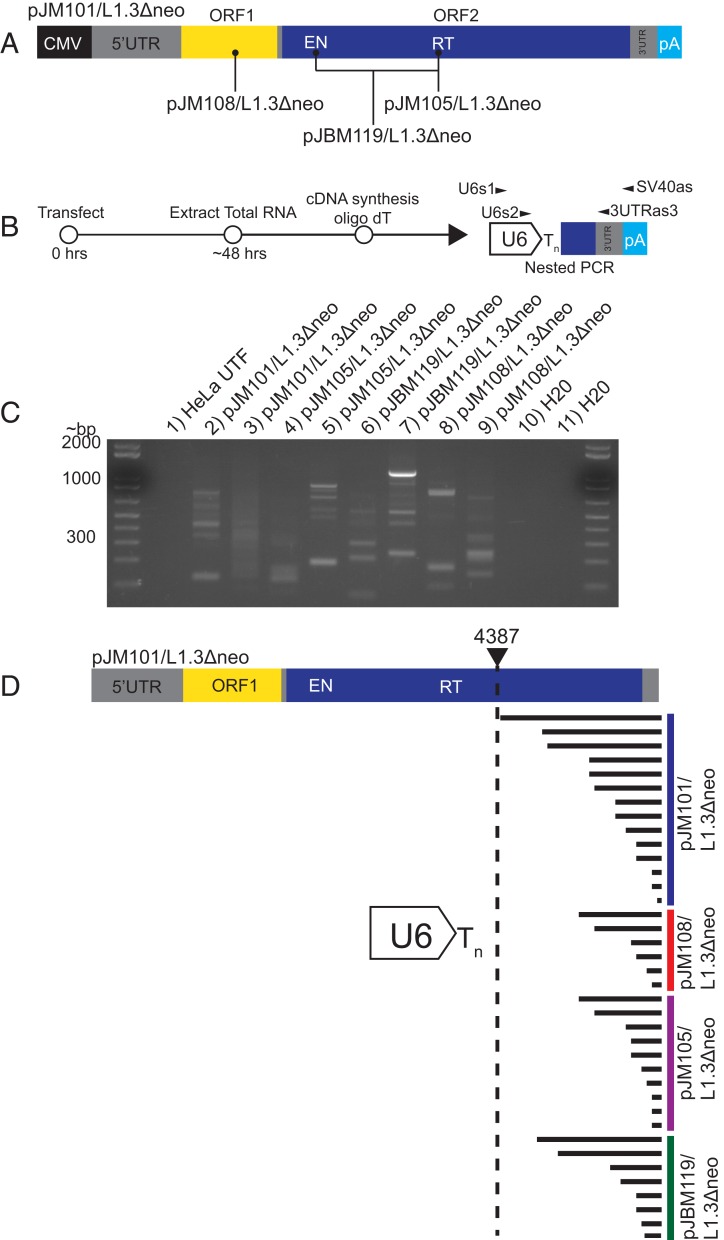
To test the above hypothesis, HeLa-JVM cells were transfected with either a wild-type engineered human L1 expression plasmid (Fig. 1A) (pJM101/L1.3Δneo) or human L1 expression plasmids that contain a nonsense mutation in ORF1 (Fig. 1A) (pJM108/L1.3Δneo) or missense mutations in the L1 EN and/or L1 RT domain of ORF2p that severely inhibit L1 retrotransposition (Fig. 1A) (pJBM119/L1.3Δneo or pJM105/L1.3Δneo) (19, 22, 31). Whole cell RNAs from transfected HeLa cells were subjected to cDNA synthesis using an oligo-dT primer, and the resultant cDNAs were used as templates in nested reverse-transcriptase PCRs (RT-PCRs) using primers complementary to U6 snRNA and the 3′ end of the engineered L1 plasmid (Fig. 1B). To ensure specificity, the outer L1 primer (Fig. 1B) (SV40as) was complementary to a specific sequence within the engineered L1 expression construct. The RT-PCR products were separated on agarose gels, visible DNA fragments were isolated from gels, and the products were characterized using Sanger sequencing (Fig. 1C). RT-PCR products were not detected in control RT-PCR experiments that lacked the reverse transcriptase (SI Appendix, Fig. S1A).
Fig. 1.
Chimeric U6/L1 RNA is generated in HeLa cells transfected with human L1 expression constructs. (A) Schematics of wild-type and mutant L1s. Gray rectangles represent 5′ UTR, inter-ORF space, and 3′ UTR, respectively; yellow rectangle, ORF1; blue rectangle, ORF2. L1s were cloned into the pCEP4 mammalian expression vector. A cytomegalovirus immediate early promoter (CMV, black rectangle) that augments L1 expression and an SV40 polyadenylation signal (light blue rectangle, pA) flank the L1. The pJM101/L1.3Δneo plasmid expresses an active human L1 (L1.3). The pJM108/L1.3Δneo, pJM105/L1.3Δneo, and pJBM119/L1.3Δneo plasmids express versions of L1.3 that contain mutations that render them unable to retrotranspose; the approximate locations of the respective mutations are indicated in the schematic. (B) Rationale of the RT-PCR experiments used to detect U6/L1 chimeric RNAs. HeLa cells were transfected with L1 expression plasmids, total cellular RNA was extracted ∼48 h posttransfection, and cDNAs were synthesized using an oligo-dT primer. Nested PCR was carried out using primers complementary to sequences within U6 and the 3′ end of the L1 construct (U6s1 and SV40as, then U6s2 and 3UTRas3). (C) Results from a representative RT-PCR experiment. The transfected L1 construct is indicated above each lane of the agarose gel image. Each lane contains a single biological replicate. Lane 1, HeLa UTF (untransfected HeLa cells); lanes 2 and 3, HeLa transfected with pJM101/L1.3Δneo; lanes 4 and 5, HeLa transfected with pJM105/L1.3Δneo; lanes 6 and 7, HeLa transfected with pJBM119/L1.3Δneo; lanes 8 and 9, HeLa transfected with pJM108/L1.3Δneo; lanes 10 and 11, H2O PCR controls. Molecular weight standards (in bp) are shown in the first and last gel lanes. At least 3 independent biological replicates were conducted for each transfection condition. (D) Structures of 38 U6/L1 chimeric RNAs found in transfected HeLa cells. U6/L1 RNA chimera sequences contain the 3′ terminus of U6 snRNA cDNA (white arrow) ending in ∼4 to 6 thymidine nucleotides (Tn) conjoined to a variable 5′−truncated L1. A schematic of the full-length L1.3 sequence is represented at the top of the schematic. The horizontal black lines indicate the approximate length of L1 sequence conjoined to the U6 poly(T) tract. The 5′-most U6/L1 junction occurred at L1.3 nucleotide position 4387.
Sequence analyses revealed the presence of U6/L1 chimeric cDNAs in HeLa cells transfected with either the wild-type or mutant L1 expression plasmids (Fig. 1 C, lanes 2 to 9 and D), but not in independent control experiments using untransfected HeLa cells (Fig. 1C, lane 1). The cDNA products typically consisted of the 3′ end of U6 snRNA cDNA ending in ∼4 to 6 thymidines conjoined to a variably 5′-truncated L1 sequence derived from the transfected L1 expression plasmid and closely resembled those of previously characterized genomic U6/L1 pseudogenes (53, 55–58). The constellation of U6/L1 cDNA products generally varied in size between ∼100 and ∼1,000 base pairs, depending upon where the 3′ end of U6 was conjoined to the 5′-truncated L1 sequence. The size and number of cDNA fragments for each transfection condition also varied between independent RT-PCR experiments. Additional analyses revealed that there were not specific sequences within L1 that facilitated U6/L1 chimeric RNA formation (Fig. 1D and SI Appendix, Fig. S1B and Table S1).
BLAT searches (63) revealed that the U6/L1 junction sequences were not present in the human genome reference sequence (HGR/build Grch38), indicating that they did not arise from the transcription of existing genomic U6/L1 pseudogenes. Notably, atypical U6/L1 chimeric cDNAs consisting of 3′-truncated U6 conjoined to a 5′-truncated L1 also were recovered from these experiments. These cDNAs sometimes exhibited microhomologies of 1 to 3 nucleotides (nt) at the U6/L1 cDNA junction and were similar in structure to previously described artifacts encountered in RT-PCR experiments (SI Appendix, Fig. S1C) (64). Thus, U6/L1 chimeric RNAs are generated in transfected HeLa cells independently of L1 retrotransposition.
Purified RtcB Ligates U6 RNA to L1 RNA In Vitro.
We next investigated the mechanism of U6/L1 chimeric RNA formation. The 3′ end of mature U6 snRNA terminates in a 2′,3′-cyclic phosphate (38, 45). During transfer RNA (tRNA) splicing, the tRNA splicing endoribonuclease excises an intron from a subset of tRNA precursor RNAs, generating a 2′,3′-cyclic phosphate and a 5′-OH on the 3′ and 5′ ends of the cleaved tRNA halves, respectively (65). In archaea and animals, the cleaved tRNAs are spliced together by the RNA 2′,3′-cyclic phosphate ligase RtcB (59–61). RtcB joins 2′,3′-cyclic-PO4 and 5′-OH ends via RNA-3′-PO4 and RNA-(3′)pp(5′)G intermediates (66, 67).
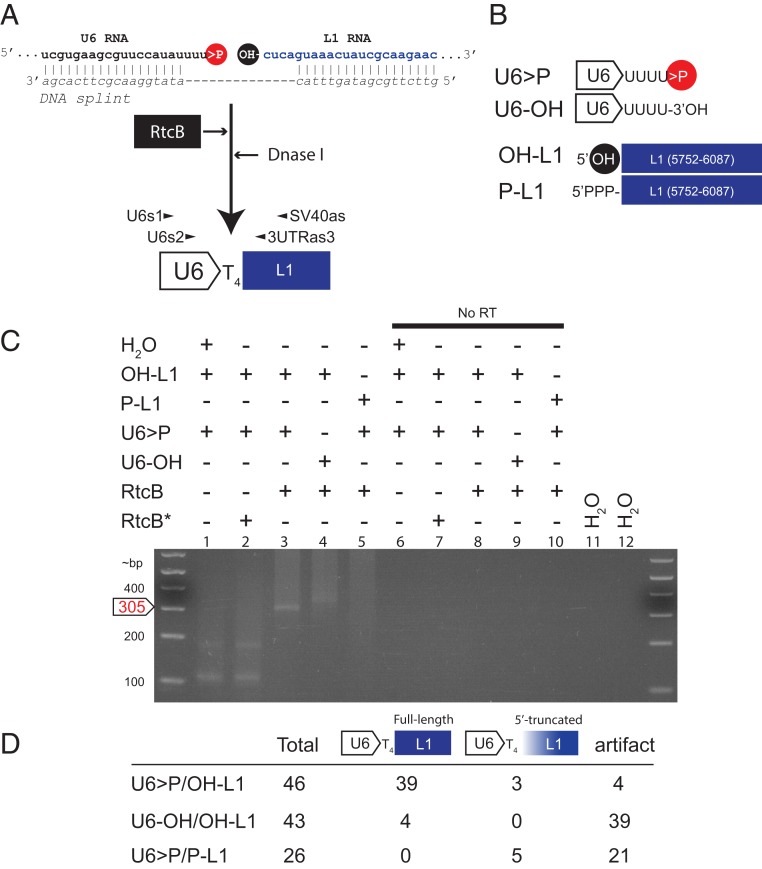
We hypothesized RtcB could ligate U6 snRNA to L1 RNA to generate U6/L1 chimeric RNAs. To test this hypothesis, we used ribozymes to generate a synthetic human U6 snRNA bearing a 2′,3′-cyclic phosphate (herein called U6 > P) and a synthetic L1 RNA fragment containing a 5′-OH (herein called OH-L1) (SI Appendix, Fig. S2 A and B and Methods). We then used a cDNA oligonucleotide as a splint to ensure that the U6 snRNA 2′,3′-cyclic phosphate and L1 5′-OH ends remained in close proximity to one another (Fig. 2A). We reasoned that the DNA oligonucleotide splint would simulate the situation encountered during tRNA splicing where base pairing interactions within and between the tRNA halves stabilize the cleaved tRNA. The resultant RNA/DNA complex was incubated with purified bacterial RtcB (59) and treated with DNase I to remove the oligonucleotide splint. The RNAs were purified, and nested RT-PCR was used to detect U6/L1 chimeric RNA (Fig. 2 A and B).
Fig. 2.
Purified recombinant RtcB ligates U6 RNA to L1 RNA in vitro. (A) The rationale of the U6/L1 in vitro ligation experiment. A synthetic human U6 RNA containing a 2′,3′-cyclic phosphate (>P, red circle) and a synthetic L1 RNA (blue font) containing a 5′-OH (black circle) were generated using a ribozyme-based in vitro transcription reaction. U6 and L1 RNAs were splinted with a cDNA oligonucleotide (DNA splint, italics), and then the resultant RNA/DNA hybrid was incubated with purified RtcB (black rectangle, white font) for 1 h. The reaction was then treated with DNase I, the RNA was purified, and cDNAs were synthesized using the SV40as oligonucleotide primer. RT-PCR reactions using nested primers (U6s1 and SV40as, then U6s2 and 3UTRas3) were used to detect U6/L1 chimeric cDNAs. (B) Schematic representations of the synthetic RNAs used in in vitro experiments. The in vitro transcribed U6 RNA ends in 4 uridine ribonucleotides and contains a 2′,3′-cyclic phosphate (U6 > P) or a 3′-OH (U6-OH). The in vitro transcribed L1 RNA consists of pJM101/L1.3Δneo sequence (nt positions 5752 to 6087) and contains a 5′-OH (OH-L1) or a 5′-triphosphate (P-L1). (C) Results from the in vitro U6/L1 ligation reactions. The constituents of U6/L1 ligation reactions are indicated above each gel lane (+) of the agarose gel image. An asterisk (*) indicates that RtcB was heat treated at 95 °C for 10 min prior to adding it to the reaction. No RT, no RT control; H2O, water PCR controls. DNA size markers (in bp) are shown to the left of the gel image. The predicted position of the 305-bp U6/L1 RT-PCR product is noted on the left side of the gel image (white arrow, red font). (D) Summary of results from product characterization experiments. Column 1, synthetic RNAs used in the reaction; column 2, number of RT-PCR products characterized for each reaction condition; column 3, number of RT-PCR products that correspond to the full-length ligation product; column 4, number of RT-PCR products that contain a variably 5′-truncated L1 sequence; column 5, number of putative RT-PCR artifact products. Each in vitro experiment was repeated 3 independent times and yielded similar results.
Agarose gel analyses revealed that reactions containing the U6 > P and OH-L1 templates yielded the predicted 305-bp U6/L1 cDNA product (Fig. 2C, lane 3), which was not visible in negative controls (Fig. 2C, lanes 1 and 2). DNA sequencing revealed that 39 of 46 (∼85%) U6/L1 cDNA products contained a copy of the 3′ end of a U6 snRNA cDNA ending in 4 thymidine nucleotides precisely conjoined to the 5′-OH of the full-length L1 RNA fragment (Fig. 2D). Three products (∼7%) contained a copy of the 3′ end of a U6 snRNA cDNA ending in 4 thymidine nucleotides conjoined to a 5′-truncated L1 fragment. These products could arise from the ligation of U6 > P to broken or degraded L1 RNA fragments containing a 5′-OH (Fig. 2D and SI Appendix, Table S2). Four RT-PCR products contained either a 3′-truncated U6 snRNA cDNA sequence conjoined to a 5′-truncated L1, a U6 snRNA followed by a partial hepatitis delta virus ribozyme (HDVr) sequence conjoined to a 5′-truncated L1, and/or untemplated nucleotides at the U6/L1 junction. These products also sometimes exhibited a microhomology of 1 to 3 nucleotides at the U6/L1 junction and structurally resemble previously described artifacts generated during RT-PCR (64) (Fig. 2D and SI Appendix, Fig. S2C).
U6/L1 RNA Ligation Requires a 2′,3′-Cyclic Phosphate and a 5′-OH.
To test whether the U6 2′,3′-cyclic phosphate was required for RtcB-mediated U6/L1 RNA ligation, purified RtcB was incubated with a synthetic U6 snRNA that contained a 3′-OH terminus (U6-OH) and an L1 that contained a 5′ hydroxyl end (OH-L1) (Fig. 2 A and B). In contrast to reactions with U6 > P, the 305-bp cDNA diagnostic for bona fide U6/L1 chimeric cDNAs was not overtly visible in reactions containing the U6-OH substrate (Fig. 2C, lane 4). The majority (39 of 43) of recovered cDNA products appeared to be RT-PCR artifacts similar to those described in the preceding paragraph (Fig. 2D and SI Appendix, Fig. S2C). Only 4 RT-PCR products contained a copy of the 3′ end of a U6 snRNA cDNA ending in 4 thymidine nucleotides precisely conjoined to the 5′-OH of the full-length L1 RNA (Fig. 2D).
To test whether an L1 5′-OH terminus was required for RtcB-mediated U6/L1 RNA ligation, we incubated purified RtcB with U6 > P and an L1 RNA fragment containing a 5′-triphosphate (P-L1) (Fig. 2 A and B). The 305-bp cDNA diagnostic for bona fide U6/L1 chimeras was not overtly visible in reactions that contained P-L1 substrate (Fig. 2C, lane 5). We did not recover any full-length U6/L1 chimeric cDNAs (0 of 26) from these reactions; however, 5 of 26 (∼19%) products contained the 3′ end of a U6 snRNA cDNA ending in 4 thymidine nucleotides conjoined to a 5′-truncated P-L1 (Fig. 2D and SI Appendix, Table S2). As above, these products could result from the ligation of U6 > P to broken or degraded L1 RNAs containing a 5′-OH. Thus, efficient RtcB-mediated ligation in vitro requires a U6 snRNA ending with a 2′,3′-cyclic phosphate and an L1 RNA that contains a 5′-OH end.
U6 snRNA and L1 RNA Are Ligated in HeLa Cell Nuclear Extracts.
We next tested whether U6 and L1 RNA could be ligated in cell free extracts. Briefly, we incubated U6 > P and OH-L1 RNA in HeLa cell nuclear extracts and then used the same nested RT-PCR strategy used in the in vitro assay to detect U6/L1 chimeric cDNAs (Fig. 2 A and B and Methods). Of note, we did not use a DNA oligonucleotide splint in these assays as endogenous RNase H activity may result in the degradation of the complementary regions of the U6 > P and OH-L1 RNAs. Control Western blots confirmed the presence of RtcB in HeLa nuclear extracts (Fig. 3D and SI Appendix, Fig. S3A).
Fig. 3.
HeLa cell nuclear extracts mediate the ligation of U6 and L1 RNAs. (A) Results from U6/L1 ligation reactions using HeLa cell nuclear extracts. The ligated U6/L1 RNA was purified from ligation reactions and analyzed using RT-PCR and agarose gel electrophoresis. The constituents of U6/L1 ligation reactions are indicated above each lane (+) of the representative agarose gel image. An asterisk (*) indicates that the HeLa cell nuclear extract was heat treated at 95 °C for 10 min prior to adding it to the reaction. No RT, no RT control; H2O, water PCR controls. DNA size markers (in bp) are shown to the left of the gel image. The predicted position of the 305-bp U6/L1 RT-PCR product is noted on the left side of the gel image (white arrow, red font). (B) Summary of results of ligation reactions using HeLa cell extracts. Column 1, RNAs used in the reaction; column 2, number of RT-PCR products characterized for each reaction condition; column 3, number of RT-PCR products that correspond to the full-length ligation product; column 4, number of RT-PCR products that contain a variably 5′-truncated L1 sequence; column 5, number of putative RT-PCR artifact products. (C) Structures of U6/L1 chimeric RNAs containing 5′-variably truncated L1 sequences. A schematic of the L1 fragment used as a template for the in vitro transcription reaction is represented at the top. The horizontal black lines indicate the approximate length of the L1 sequence conjoined to the U6 poly(T) tract. U6/L1 chimeric RNAs were isolated from 19 independent experiments. (D) Depletion of RtcB protein expression in HeLa cell nuclear extracts. Western blot images depicting RtcB expression (green arrow) in HeLa nuclear extracts. Extract sources are indicated above each lane (HeLa indicates untransfected HeLa extracts). Each lane represents an independent biological replicate. The Western blot experiment was done twice. Nucleolin (NCL) (red arrow) was used as a loading control. Approximate molecular weights are indicated to the left of the gel image. (E) Depletion of RtcB affects U6/L1 ligation efficiency in HeLa extracts. The x axis indicates the experimental condition. The y axis indicates the normalized U6/L1 ligation efficiency. Ligation efficiencies were normalized to untransfected HeLa-JVM extracts, which are set to 1. The ligation efficiency value represents the average of 6 independent RT-qPCR experiments. Error bars indicate SDs. Two-tailed t tests were used to determine significance. An asterisk (*) indicates P value < 0.05; n.s., not significant.
Similar to experiments using purified RtcB, the predicted 305-bp U6/L1 cDNA product was detected in reactions containing U6 > P and OH-L1 (Fig. 3A, lane 3) but was not visible in negative control reactions that either lack or contain heat-treated HeLa cell nuclear extracts (Fig. 3A, lanes 1 and 2). Sequencing of RT-PCR products revealed that 47 of 106 (∼44%) products consisted of the 3′ end of a copy of a U6 snRNA cDNA ending in 4 thymidine nucleotides conjoined precisely to the 5′-OH of the full-length OH-L1 RNA (Fig. 3B). By comparison, 38 of 106 (∼36%) products consisted of the 3′ end of a copy of a U6 snRNA cDNA ending in 4 thymidine nucleotides conjoined to a 5′-truncated OH-L1 (Fig. 3B). The prevalence of U6/L1 chimeras conjoined to 5′-truncated L1 sequences was higher in reactions conducted with HeLa cell nuclear extracts when compared to in vitro reactions conducted with purified RtcB (36% vs. 7%), suggesting that L1 RNA may be cleaved by a ribonuclease activity that generates 5′-OH ends in the HeLa cell nuclear extracts. As described above, we also recovered 21 of 106 (∼20%) putative artifactual RT-PCR products (64) (SI Appendix, Fig. S3B).
Incubation of the U6-OH and OH-L1 substrates in HeLa cell nuclear extracts did not yield the predicted 305-bp U6/L1 cDNA product (Fig. 3A, lane 4). DNA sequence analyses revealed that 38 of 43 (∼88%) products likely represent artifacts generated during RT-PCR (64) (SI Appendix, Fig. S3B). Four out of 38 (∼9%) products contained the 3′ end of a U6 snRNA cDNA ending in 5 thymidine nucleotides conjoined precisely to the 5′-OH of the full-length OH-L1 RNA substrate. The synthetic U6 > P RNA ends in 4 uridine ribonucleotides whereas the mature forms of human U6 snRNA typically end in 5 uridine ribonucleotides (45); thus, endogenous U6 snRNA may have been ligated to the OH-L1 RNA in these reactions. Finally, 1 product contained the 3′ end of a copy of a U6 snRNA cDNA ending in 4 thymidine nucleotides followed by 5′-truncated L1 (Fig. 3B). Thus, a U6 2′,3′-cyclic phosphate is required for efficient U6/L1 ligation in HeLa cell nuclear extracts.
To test whether a 5′-OH was required for U6/L1 ligation, we incubated U6 > P and P-L1 RNAs in HeLa cell nuclear extracts. We did not detect the predicted 305-bp U6/L1 cDNA product (Fig. 3A, lane 5). Sequence analyses did not reveal the presence of the predicted U6/L1 chimeric cDNA; however, 16 of 23 (∼70%) of the cDNA products consisted of a copy of the 3′ end of a U6 snRNA cDNA ending in 4 thymidine nucleotides followed by a variably 5′-truncated P-L1 sequence (Fig. 3B). These U6/L1 junctions occurred throughout L1 RNA, and there was not an overt sequence motif within L1 RNA that facilitated U6/L1 formation (Fig. 3C and SI Appendix, Fig. S3C and Table S3). These data are consistent with results from reactions where U6 > P and OH-L1 were incubated with HeLa cell nuclear extracts, suggesting that L1 RNA may be processed by a ribonuclease activity in the HeLa cell nuclear extracts either prior to or during the ligation reaction. The remaining 7 of 23 (∼30%) cDNA products were characterized as putative RT-PCR artifacts (64) (SI Appendix, Fig. S3B). Thus, efficient U6/L1 ligation in HeLa cell nuclear extracts requires U6 2′,3′-cyclic phosphate and 5′-OH L1 substrates.
Depletion of RtcB from HeLa Cells Affects U6/L1 Ligation Efficiency.
Ligation reactions using HeLa nuclear extracts suggested that RtcB might ligate U6/L1 RNA in HeLa cells. To test this hypothesis, we utilized CRISPR/Cas9 gene editing to generate 2 HeLa cell lines (RtcB2.1 and RtcB2.2) exhibiting reduced RtcB protein expression (SI Appendix, Methods). Sanger sequencing revealed that the RtcB alleles in RtcB2.1 and RtcB2.2 contain genomic edits that are predicted to result in either frame-shift mutations or in-frame deletions in the RtcB amino acid sequence (SI Appendix, Fig. S3D). Western blots confirmed that steady state RtcB protein levels were reduced by ∼80% and ∼73% for the RtcB2.1 and RtcB2.2 clonal cell lines, respectively (Fig. 3D). We were unable to isolate a HeLa cell clone with a complete knockout of RtcB protein expression, likely because RtcB is an essential gene (68) and the total loss of RtcB protein expression would prevent cell growth and/or viability (69, 70).
To examine U6/L1 RNA ligation efficiency in RtcB-depleted HeLa nuclear extracts, we incubated the extracts with synthetic U6 RNA ending in a 2′,3′-cyclic phosphate and an L1 RNA that contained a 5′ hydroxyl. Quantitative reverse transcriptase PCR (RT-qPCR) revealed that U6/L1 ligation efficiency was reduced ∼4- to 5-fold in the RtcB2.1 and RtcB2.2 clonal cell lines, but that neither RtcB protein levels nor U6/L1 ligation efficiency was affected in a clonal cell line containing a negative control sgRNA that targeted GFP (Fig. 3 D and E).
To determine whether ectopic RtcB expression could complement the defect in the RtcB2.1 and RtcB2.2 cell lines, we transfected them with an RtcB cDNA expression plasmid. Western blots revealed that ectopic RtcB expression led to an ∼1.4-fold and ∼1.5-fold increase in RtcB protein expression in the RtcB2.1 and RtcB2.2 clonal cell lines when compared to HeLa-JVM cells (Fig. 3D). RT-qPCR experiments revealed that ectopic RtcB expression partially rescued ligation efficiency by ∼2-fold in the RtcB2.1 and RtcB2.2 extracts (Fig. 3E). These data indicate that endogenous RtcB contributes to U6/L1 RNA ligation in HeLa cell extracts.
U6 snRNA and GFP Are Ligated in HeLa Cell Nuclear Extracts.
The HGR contains chimeric pseudogenes that consist of a full-length copy of U6 conjoined to cDNAs derived from other cellular mRNAs (58), suggesting that U6 RNA can be ligated to non-L1 RNAs. To test this hypothesis, we generated a 310-nucleotide RNA fragment that corresponds to the 3′-end of a humanized Renilla GFP cDNA that contains a 5′-OH (OH-GFP) (Methods). We incubated U6 > P and OH-GFP RNA in HeLa cell nuclear extracts and then used RT-PCR to detect cDNA products (SI Appendix, Fig. S3 E and F and Methods). Incubation of the U6 > P and OH-GFP RNA substrates with HeLa cell nuclear extracts yielded the predicted 232-bp cDNA product (SI Appendix, Fig. S3G, lane 3). Sequencing of the cDNA products revealed that 9 of 23 (39%) consisted of a copy of the 3′ end of a U6 snRNA ending in 4 thymidine nucleotides conjoined precisely to the 5′-OH of the full-length GFP RNA fragment whereas 6 of 23 (26%) contained a copy of the 3′ end of a U6 snRNA ending in 4 thymidine nucleotides followed by a 5′-truncated GFP RNA fragment (SI Appendix, Fig. S3 G and H). The remaining 8 of 23 (∼35%) cDNA products contained either a 3′-truncated U6 snRNA sequence conjoined to a 5′-truncated GFP, U6 snRNA followed by a partial HDVr sequence conjoined to a 5′-truncated GFP, and/or untemplated nucleotides at the U6/GFP junction. These sequences sometimes exhibited 1 to 3 nucleotide microhomologies at the U6/L1 junction and likely represent artifacts generated during RT-PCR (similar in structure to SI Appendix, Fig. S2C, except GFP replaces L1) (64). These data suggest that chimeric RNA formation is not exclusive to L1 RNA.
Endogenous U6/L1 RNA Is Part of the Transcriptome in Human Cells.
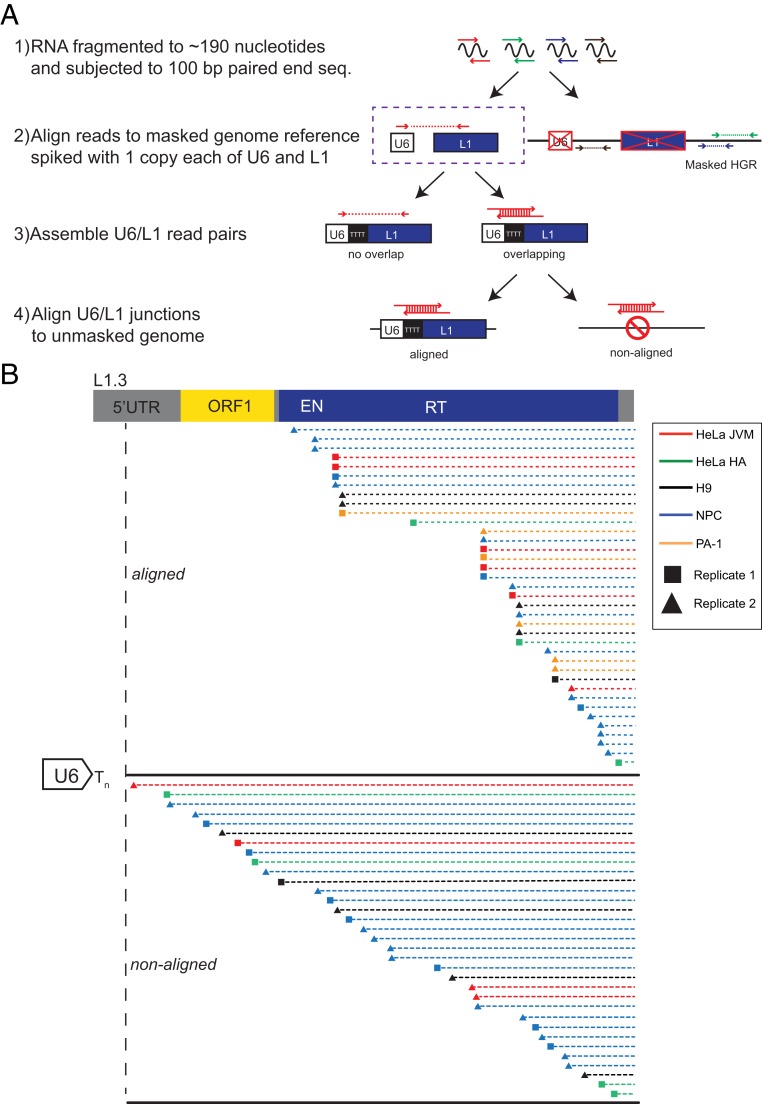
Data from transfection-based experiments and in vitro ligation experiments suggested that U6 snRNA could be ligated to L1 RNA in vivo; thus, we sought to determine whether U6/L1 chimeric RNAs were part of the transcriptome in human cells. We searched for U6/L1 junction reads in 100-base pair paired-end RNA sequencing (RNA-seq) data generated from 2 independent HeLa cell lines (HeLa-JVM and HeLa-HA), a human embryonic carcinoma cell line (PA-1), a human embryonic stem cell line (H9-hESCs), and H9-derived neural progenitor cells (NPCs) (Fig. 4A and SI Appendix, Methods). Each of these cell lines can accommodate the retrotransposition of engineered human L1s (22, 71–74). We identified 398 U6/L1 chimeric RNA read-pairs out of ∼1.1 × 109 RNA sequencing reads across the 5 cell lines. After removing duplicate PCR reads, we then merged overlapping reads to identify 64 intact U6/L1 junction sequences.
Fig. 4.
RNA-seq detection of endogenous U6/L1 chimeric RNAs in human cell lines. (A) Rationale of the RNA-seq experiments. Step 1: Ribosome-depleted RNA was fragmented to ∼190 nucleotides and subjected to 100-bp paired-end DNA sequencing. Step 2: RNA-seq read pairs (arrows) were aligned to a repeat masked version human reference genome (HGR/build Grch38), which contained “spiked-in” copies of a single U6 (white rectangle) and single L1.3 (blue rectangle) sequence. RNA-seq reads that did not map to U6 or L1 were discarded from subsequent analyses. Step 3: Overlapping U6/L1 read pairs were merged to determine the U6/L1 junction sequences. U6/L1 read pairs that contained a gap (i.e., “no overlap”) were discarded from subsequent analyses. Step 4: Overlapping U6/L1 junctions were aligned to the unmasked HGR. If the U6/L1 junction mapped to the HGR with >90% accuracy, it was designated as an “aligned” read. U6/L1 junctions that failed to map to the HGR with at least 90% accuracy were designated as “non-aligned” reads. (B) Structures of RNA-seq U6/L1 junctions. A schematic of a full-length RC-L1 is indicated at the top. The general structure of a U6/L1 chimeric junction sequence consists of the 3′ end of a U6 snRNA cDNA sequence ending in ∼4 to 8 thymidine nucleotides (left side of figure; white arrow ending in Tn) conjoined to a variably 5′−truncated L1 sequence. Two independent RNA-seq libraries were generated from HeLa-JVM, H9, NPC, and PA-1 cells, respectively (squares and triangles, respectively). One RNA-seq library was generated from HeLa-HA cells. Red horizontal lines, HeLa-JVM; green horizontal lines, HeLa-HA; yellow horizontal lines, PA-1; black horizontal lines, H9; blue horizontal lines, human NPCs. Each horizontal dashed line represents a single U6/L1 junction RNA-seq merged sequence read. The triangle or square at the left end of the horizontal dashed lines indicates the approximate location of the U6/L1 junction point relative to L1.3. The top set of dashed lines represent 16 U6/L1 junction sequences that mapped (“aligned”) to the HGR. The bottom set of dashed lines represent 33 U6/L1 junction sequences that did not map (“non-aligned”) to the HGR. These 33 U6/L1 junctions contained a copy of U6 conjoined to an L1 present in the same transcriptional orientation. The remaining 4 U6/L1 chimeras that did not map to the HGR (SI Appendix, Table S6) contained a copy of U6 conjoined to an L1 present in the opposite transcriptional orientation.
Hand annotation of the 64 U6/L1 junctions revealed that 53 (∼83%) consisted of the 3′ end of U6 snRNA cDNA ending in ∼4 to 8 thymidine nucleotides conjoined to a variably 5′-truncated L1 sequence (Fig. 4B and SI Appendix, Tables S4–S6). Of note, 4 of these 53 U6/L1 chimeras consisted of U6 ending in 5 to 7 thymidines conjoined to an L1 sequence in an antisense orientation, suggesting that U6 can become conjoined to both sense and antisense L1 RNAs (SI Appendix, Table S6). As above, there was not a specific sequence within L1 that appeared to facilitate U6/L1 chimera formation (Fig. 4B and SI Appendix, Fig. S4 A and B). The remaining 11 of 64 (∼17%) U6/L1 sequences contained a 3′-truncated U6 snRNA conjoined to a 5′-truncated L1 and were excluded from further analysis as they were structurally similar to template-switching artifacts generated during cDNA synthesis described above (64) (SI Appendix, Table S6). Thus, 53 bona fide unique U6/L1 chimeras were subjected to further analysis.
Most U6/L1 Chimeric RNA Sequences Do Not Align to the Genome.
The low proportion of U6/L1 chimera RNAs in our dataset suggested that they might represent a rare subset of RNAs in human cells. To determine whether the U6/L1 chimeras detected in RNA-seq experiments were derived from the transcription of an existing genomic U6/L1 or represented unique chimeric RNAs, the 53 U6/L1 chimeric RNAs were used as probes in BLAT searches of the HGR (Fig. 4A and SI Appendix, Methods). Sixteen out of 53 (∼30%) U6/L1 junctions were present in the HGR, suggesting that they could have resulted from the transcription of extant U6/L1 pseudogenes. Seven out of the 16 putative transcribed U6/L1 chimeric RNAs were detected in multiple cell lines (Fig. 4B and SI Appendix, Table S6), and 7 were supported by multiple reads from the same cell line (Fig. 4B and SI Appendix, Table S6). The 16 genomic U6/L1 chimeric pseudogenes that served as putative transcription templates that gave rise to chimeric U6/L1 RNAs exhibited L1 structural hallmarks (SI Appendix, Table S7). They consisted of a full-length U6 snRNA sequence ending in 5 to 7 thymidine nucleotides conjoined to a variably 5′-truncated L1, were flanked by 6- to 19-bp target site duplications, and inserted into an L1 EN consensus cleavage sequence. By comparison, 37 out of 53 (∼70%) U6/L1 junction sequences did not align to the HGR and were unique to a single cell line (Fig. 4B). Thirty-one of 37 junctions were supported by a single merged read pair, 5 of 37 junctions were supported by 2 merged reads, and 1 junction was supported by 3 merged reads (Fig. 4B and SI Appendix, Table S6 and Methods).
Human-specific L1 insertions can be polymorphic with respect to presence/absence in the human population (75); thus, it is conceivable that some of the cell lines used to generate RNA-seq data could contain a genomic U6/L1 chimeric pseudogene that is absent from the HGR. To examine this possibility, we used the 53 U6/L1 junctions as probes to query HeLa genome sequencing data available in the database of Genotypes and Phenotypes (dbGaP accession number phs000640.v1.p1) (76–78). Controls revealed that the 16 U6/L1 junction sequences that aligned to the HGR were also present in the HeLa genome data (Fig. 4B and see Methods). By comparison, the 37 non-aligned U6/L1 junction sequences were absent from HeLa genome data.
To further validate the uniqueness of the 37 U6/L1 junction sequences, we aligned the 53 U6/L1 junction sequences to 23 high-coverage individual genomes representing 23 distinct human geographic populations from the 1000 Genomes Project dataset (SI Appendix, Table S8 and Methods) (79). The 16 U6/L1 junction sequences that were present in the HGR and HeLa cell genomic datasets also were present in each of the 23 of high coverage 1000 Genomes Project individual genomes; 2 genomes (NA20845 and HG03742) contained an SNP in the U6 portion of the junction sequences (SI Appendix, Table S6). In contrast, the 37 non-aligned U6/L1 junction sequences were absent from the high coverage 1000 Genomes Project individual genomes. Thus, these data suggest that the 37 U6/L1 junctions detected in our RNA-seq experiments do not correspond to existing U6/L1 genomic pseudogenes and that different cell types may contain a unique cohort of chimeric RNAs that are generated by posttranscriptional RNA ligation events.
Discussion
RNA Ligation Generates Chimeric U6/L1 RNA.
Here, we demonstrated that U6/L1 chimeric RNAs are generated in transfected HeLa cells independently of L1 retrotransposition (Fig. 1 C and D). In vitro ligation assays demonstrated that purified RtcB can ligate U6 snRNA substrates ending in a 2′,3′-cyclic phosphate to L1 RNAs containing a 5′-OH (Fig. 2 C and D) and that replacement of either the U6 2′,3′-cyclic phosphate with a 3′-OH or the L1 5′-OH with a 5′ triphosphate dramatically reduced the formation of RtcB-mediated ligation products. Additional assays revealed that HeLa cell nuclear extracts can mediate ligation of a U6 snRNA substrate ending in a 2′,3′-cyclic phosphate to an L1 RNA containing a 5′-OH (Fig. 3A), depleting RtcB from HeLa extracts decreases U6/L1 ligation efficiency (Fig. 3E), and U6 RNA could be ligated to non-L1 RNAs (SI Appendix, Fig. S3G). Finally, RNA-seq experiments revealed that endogenous U6/L1 RNA chimeras are a component of the transcriptome of multiple human cell types (Fig. 4B). Together, these data provide a mechanistic explanation for how RtcB can mediate the ligation of U6 snRNA to a diverse cohort of cellular RNAs containing 5′-OH ends. It remains possible that another RNA ligation activity present in HeLa cells could also contribute to U6/L1 chimera formation. For example, previous studies have shown that a yeast-like tRNA ligase activity can join 2′,3′-cyclic phosphate and 5′-OH containing RNAs in vertebrate cells; however, this ligase remains to be identified (80).
RNA Ligation in HeLa Cell Extracts.
U6/L1 ligation reactions using purified bacterial RtcB required an oligonucleotide splint, presumably to keep the U6 snRNA 2′,3′-cyclic phosphate and L1 5′-OH ends in close proximity to one another. In contrast, U6/L1 ligation reactions using HeLa cell nuclear extracts did not require an oligonucleotide splint. The human form of RtcB is 505 amino acids in length, and protein sequence alignments using the Universal Protein Resource (UniProt) align tool (81) showed that human RtcB is highly conserved (>90%) among other vertebrate forms of RtcB (SI Appendix, Fig. S3I). However, human RtcB only shares ∼25% amino acid identity with RtcB from Escherichia coli (408 amino acids in length) (SI Appendix, Fig. S3I), suggesting that changes to the RtcB amino acid sequence over evolutionary time could have affected RtcB enzymatic activity and/or function.
It is also possible that cellular proteins within HeLa nuclear extracts may help facilitate U6/L1 RNA ligation. Previous studies have revealed that L1 RNPs are associated with numerous RNA binding proteins and cellular RNAs, including U6 snRNA (82–87). Moreover, RtcB has been detected in GFP-tagged L1 ORF1p cytoplasmic granules (83), cytoplasmic stress granules (88), and other protein/RNA complexes within the cell (60, 89, 90) whereas U6 snRNA associates with cellular RNA binding proteins, including La and the Lsm2-8 ring (38, 91, 92). Of note, the Lsm2-8 ring is ejected from U6 during splicing, and U6 snRNA must be recycled, which could provide other cellular proteins access to the 3′ end of U6 snRNA. Thus, it is plausible that protein–protein or protein–RNA interactions within HeLa cell nuclear extracts may help bring U6 snRNA and L1 RNA into close proximity to facilitate ligation.
The L1 RNA May Be Processed by Endoribonuclease Activity in HeLa Cells.
In contrast to experiments conducted with recombinant bacterial RtcB, almost half of the U6/L1 ligation products characterized using HeLa cell nuclear extracts consisted of the 3′ end of U6 snRNA cDNA ending in 4 thymidine nucleotides conjoined to a 5′-truncated L1 (Fig. 3 A and B and SI Appendix, Table S3). This result was evident in reactions with L1 RNA substrates containing either a 5′-OH or a 5′ triphosphate. Given that efficient ligation reactions were dependent upon U6 snRNA ending in a 2′,3′-cyclic phosphate and L1 RNA with a 5′-OH, these data raise the possibility that L1 RNAs may be processed by an endogenous endoribonucleolytic activity within HeLa cell nuclear extracts prior to their ligation to U6 snRNA. A number of cellular endoribonucleases generate RNA fragments with 5′-OH ends (93), including the tRNA splicing endonuclease (66), RNase L (94, 95), angiogenin (96, 97), and inositol-requiring enzyme 1 (IRE1) (98). RNase L, for example, can target viral as well as host cellular RNAs (94, 95, 99) and inhibits L1 retrotransposition by targeting L1 RNA (100). Recent reports have also provided evidence that RtcB associates with IRE1, an endoribonuclease involved in the UPR (69, 98). It is possible that other classes of RNA endoribonucleases and/or exoribonucleases that generate RNA 5′-phosphate ends also could process the L1 RNA. However, in these instances, the 5′-phosphate end would need to be hydrolyzed by a 5′-phosphatase to generate a 5′-OH end suitable for ligation by RtcB. Thus, the most parsimonious explanation of the data is that the L1 RNA is processed by an as-yet-unknown endoribonucleolytic activity within HeLa cell nuclear extracts to generate a truncated L1 RNA containing a 5′-OH.
Chimeric U6/L1 RNAs Are Present in Human Cells.
RNA-seq experiments demonstrated that U6/L1 chimeric RNAs are a component of the transcriptome in human cancer cell lines, hESCs, and human NPCs (Fig. 4B). The majority (∼70%) of U6/L1 chimeric RNA-seq reads failed to align to the HGR, HeLa, or 23 high-coverage genomes in the 1000 Genomes Project dataset (Fig. 4B), suggesting that they were generated de novo by a posttranscriptional mechanism that results in the formation of a unique U6/L1 chimeric RNA molecule. Consistent with this conclusion, the majority of the “non-aligned” junctions were supported by a single RNA-seq read and are unique to a single cell line. In contrast, ∼30% of the U6/L1 chimeric RNAs aligned to the HGR, HeLa, and 23 high-coverage genomes in the 1000 Genomes Project dataset (Fig. 4B), indicating that they are generated from existing U6/L1 pseudogenes. Of note, 13 of these U6/L1 pseudogenes are embedded within the introns of annotated RefSeq genes (SI Appendix, Table S7), with 8 oriented in the same transcriptional orientation as the gene, suggesting that U6/L1 chimeric RNAs could be expressed as part of an RNA polymerase II pre-mRNA transcript. Vertebrate U6 snRNA is transcribed by RNA polymerase III and relies on upstream promoter elements to drive its transcription (38, 40). Thus, unless U6/L1 chimeric pseudogenes fortuitously inserted downstream of a promoter that could augment RNA polymerase III transcription, it remains unlikely that U6/L1 chimeric RNAs are transcribed as discrete transcription units.
A Model of U6/L1 Pseudogene Formation.
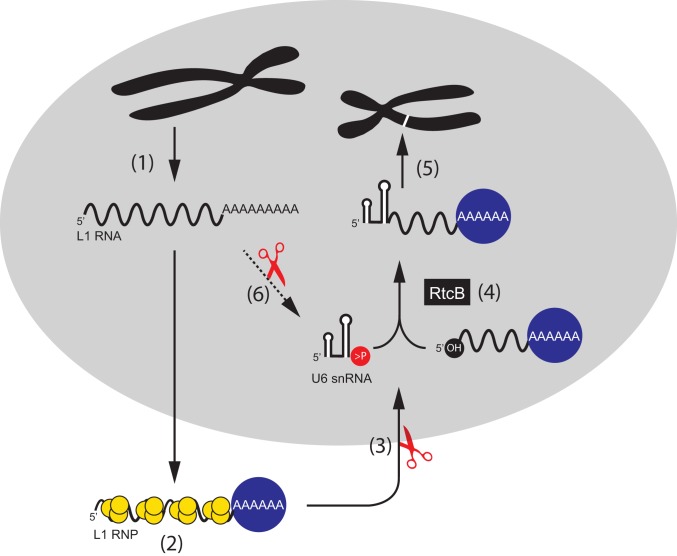
U6 snRNA is enriched in the nucleus (101) whereas RtcB is present in both the nucleus and cytoplasm (69, 102). Our data, in conjunction with previous knowledge of the L1 retrotransposition cycle, lead us to propose a mechanism for how U6/L1 chimeric pseudogenes are generated in human cells (Fig. 5). We posit that a ribonucleoprotein complex that minimally contains U6 snRNA, RtcB, and an undefined endoribonuclease can cleave the L1 RNA present in wild-type L1 RNPs to generate a 5′-OH end, which, in turn, allows L1 RNA to be ligated to the U6 snRNA 2′,3′-cyclic phosphate. As L1 ORF2p preferentially associates with the L1 poly(A) tail (23, 31), the resultant chimeric U6/L1 RNAs likely retrotranspose in cis. The majority of chimeric pseudogenes identified in the HGR consist of the full-length U6 snRNA sequence ending in a poly(T) tract conjoined to a variably 5′-truncated L1 sequence (53, 55, 57, 58); thus, this mechanism provides a plausible explanation for the generation of most U6/L1 chimeric pseudogenes. U6atac snRNA also ends in a 2′,3′-cyclic phosphate (103); thus, the above model also could explain how U6atac/L1 chimeric pseudogenes are generated in human cells (55, 57).
Fig. 5.
A model of U6/L1 RNA pseudogene formation. Following transcription, L1 RNA (black wavy line) is exported to the cytoplasm (step 1). L1 RNA is translated, and the L1 proteins, ORF1p (yellow circles) and ORF2p (blue circle) bind to their encoding L1 RNA to form an L1 ribonucleoprotein particle (RNP) (step 2). After RNP formation, the L1 RNA is cleaved by an unidentified endoribonuclease to generate 5′-truncated L1 RNA with a 5′-OH (step 3). RtcB, or a related ligase, ligates U6 snRNA to 5′-truncated L1 RNA (step 4). The resultant chimeric U6/L1 RNA is inserted into genomic DNA by TPRT (in cis), which results in the formation of a U6/L1 chimeric pseudogene (white band on black chromosome) (step 5). It is possible that L1 RNA could also be processed by nuclease activity (red scissors) and ligated to U6 while still in the nucleus (step 6). In this scenario, it is possible the chimeric U6/L1 RNA could undergo retrotransposition by trans-complementation.
Our data further demonstrate that U6 snRNA can become conjoined to retrotransposition-defective L1 and other cellular RNAs. However, because the retrotransposition of these RNAs would need to coopt the L1-encoded proteins in trans (31), they probably occur less frequently than U6/L1 chimeric pseudogenes that are formed in cis. Our model does not rule out the possibility that some snRNA/L1 chimeric pseudogenes arise during TPRT by a template-switching mechanism. For example, a small number of retro-pseudogene sequences consist of other small RNA species (e.g., U1, U3, U5, and 5s rRNA), which tend to be 3′-truncated and are conjoined to either 5′-truncated L1s or other cellular RNAs (53, 55, 57, 58). As these small RNAs are not normally modified at their 3′ ends to contain a 2′,3′-cyclic phosphate, it remains possible that the resultant chimeric products are formed by an RtcB-independent mechanism, or the 3′ truncation of the small RNAs is created by an endoribonuclease that generates a 2′,3′-cyclic phosphate or 3′-monophosphate end, either of which may serve as a substrate for ligation by RtcB to a 5′-OH (66, 67).
Conclusions.
We provide mechanistic evidence for how U6/L1 chimeric pseudogenes are formed in the human genome. Our data suggest that, in addition to their roles in mRNA and tRNA splicing, U6 snRNA and RtcB are involved in the formation of chimeric cellular RNAs, thereby contributing to RNA transcriptome diversity. The presence of a 2′,3′-cyclic phosphate on human U6 snRNA stabilizes it and prevents it from degradation (38). We speculate that RtcB-mediated ligation of U6 to L1 or other cellular RNAs containing a 5′-OH would eliminate the U6 2′,3′-cyclic phosphate, which could, in principle, facilitate U6 snRNA turnover.
Methods
Oligonucleotide Sequences.
Complete methods on cell culture, RT-PCR, RNA ligation assays, CRISPR gene editing, quantitative real-time PCR, and RNA-seq can be found in SI Appendix, Methods. A list of oligonucleotides used in this study is provided in SI Appendix, Table S9. The University of Michigan Pluripotent Stem Cell Research Oversight (HPSCRO) Committee has approved work with hESCs in the Moran lab (Record no. 1004/1023).
Supplementary Material
Acknowledgments
We thank Dr. Mitsuhiro Nakamura and Mr. Trenton Frisbie for critical reading of this manuscript and helpful discussions; and Dr. Diane Flasch and the University of Michigan DNA Sequencing Core Facility for generating the RNA-seq datasets. Henrietta Lacks and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951 have made significant contributions to scientific progress and advances in human health. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. J.V.M. was supported by National Institutes of Health (NIH) Grants GM060518 and U01MH106892 and funds from the Howard Hughes Medical Institute. R.E.M. was supported by NIH Grant HG007068. S.S. was supported by NIH Grant R35-GM126945.
Footnotes
Conflict of interest statement: J.V.M. is an inventor on patent US6150160, is a paid consultant for Gilead Sciences and a privately held company founded by Flagship Pioneering, and is on the American Society of Human Genetics Board of Directors.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805404116/-/DCSupplemental.
References
- 1.Lander E. S., et al. ; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). Erratum in: Nature411, 720 (2001) and Nature412, 565 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Smit A. F., The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6, 743–748 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Grimaldi G., Skowronski J., Singer M. F., Defining the beginning and end of KpnI family segments. EMBO J. 3, 1753–1759 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott A. F., et al. , Origin of the human L1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1, 113–125 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostertag E. M., Kazazian H. H. Jr, Twin priming: A proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 11, 2059–2065 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouha B., et al. , Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassaman D. M., et al. , Many human L1 elements are capable of retrotransposition. Nat. Genet. 16, 37–43 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Beck C. R., et al. , LINE-1 retrotransposition activity in human genomes. Cell 141, 1159–1170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dombroski B. A., Mathias S. L., Nanthakumar E., Scott A. F., Kazazian H. H. Jr, Isolation of an active human transposable element. Science 254, 1805–1808 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Martin S. L., Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11, 4804–4807 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes S. E., Singer M. F., Swergold G. D., Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267, 19765–19768 (1992). [PubMed] [Google Scholar]
- 12.Hohjoh H., Singer M. F., Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16, 6034–6043 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohjoh H., Singer M. F., Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15, 630–639 (1996). [PMC free article] [PubMed] [Google Scholar]
- 14.Khazina E., Weichenrieder O., Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl. Acad. Sci. U.S.A. 106, 731–736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin S. L., Bushman F. D., Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21, 467–475 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ergün S., et al. , Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 279, 27753–27763 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Doucet A. J., et al. , Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 6, e1001150 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodier J. L., Mandal P. K., Zhang L., Kazazian H. H. Jr, Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum. Mol. Genet. 19, 1712–1725 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Q., Moran J. V., Kazazian H. H. Jr, Boeke J. D., Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916 (1996). [DOI] [PubMed] [Google Scholar]
- 20.Mathias S. L., Scott A. F., Kazazian H. H. Jr, Boeke J. D., Gabriel A., Reverse transcriptase encoded by a human transposable element. Science 254, 1808–1810 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Dombroski B. A., et al. , An in vivo assay for the reverse transcriptase of human retrotransposon L1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 4485–4492 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran J. V., et al. , High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Doucet A. J., Wilusz J. E., Miyoshi T., Liu Y., Moran J. V., A 3′ poly(A) tract is required for LINE-1 retrotransposition. Mol. Cell 60, 728–741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swergold G. D., Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10, 6718–6729 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Athanikar J. N., Badge R. M., Moran J. V., A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32, 3846–3855 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becker K. G., Swergold G. D., Ozato K., Thayer R. E., Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum. Mol. Genet. 2, 1697–1702 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Leibold D. M., et al. , Translation of LINE-1 DNA elements in vitro and in human cells. Proc. Natl. Acad. Sci. U.S.A. 87, 6990–6994 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMillan J. P., Singer M. F., Translation of the human LINE-1 element, L1Hs. Proc. Natl. Acad. Sci. U.S.A. 90, 11533–11537 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alisch R. S., Garcia-Perez J. L., Muotri A. R., Gage F. H., Moran J. V., Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 20, 210–224 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esnault C., Maestre J., Heidmann T., Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24, 363–367 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Wei W., et al. , Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21, 1429–1439 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulpa D. A., Moran J. V., Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 13, 655–660 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Kulpa D. A., Moran J. V., Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14, 3237–3248 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H., Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 72, 595–605 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Cost G. J., Feng Q., Jacquier A., Boeke J. D., Human L1 element target-primed reverse transcription in vitro. EMBO J. 21, 5899–5910 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matera A. G., Wang Z., A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy R., Busch H., “Small nuclear RNAs: RNA sequences, structure, and modifications” in Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles, Birnstiel M. L., Ed. (Springer Berlin Heidelberg, Berlin, Heidelberg, 1988), pp. 1–37. [Google Scholar]
- 38.Didychuk A. L., Butcher S. E., Brow D. A., The life of U6 small nuclear RNA, from cradle to grave. RNA 24, 437–460 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brow D. A., Guthrie C., Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334, 213–218 (1988). [DOI] [PubMed] [Google Scholar]
- 40.Kunkel G. R., Maser R. L., Calvet J. P., Pederson T., U6 small nuclear RNA is transcribed by RNA polymerase III. Proc. Natl. Acad. Sci. U.S.A. 83, 8575–8579 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fica S. M., et al. , RNA catalyses nuclear pre-mRNA splicing. Nature 503, 229–234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp P. A., “Five easy pieces”. Science 254, 663 (1991). [DOI] [PubMed] [Google Scholar]
- 43.Madhani H. D., Guthrie C., A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71, 803–817 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Keating K. S., Toor N., Perlman P. S., Pyle A. M., A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16, 1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lund E., Dahlberg J. E., Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA’s. Science 255, 327–330 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Shchepachev V., Wischnewski H., Missiaglia E., Soneson C., Azzalin C. M., Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′-to-5′ RNA exonuclease processing U6 small nuclear RNA. Cell Rep. 2, 855–865 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Mroczek S., et al. , C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes Dev. 26, 1911–1925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volpi L., et al. , Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am. J. Hum. Genet. 86, 72–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dewannieux M., Esnault C., Heidmann T., LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Ostertag E. M., Goodier J. L., Zhang Y., Kazazian H. H. Jr, SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 73, 1444–1451 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raiz J., et al. , The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40, 1666–1683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hancks D. C., Goodier J. L., Mandal P. K., Cheung L. E., Kazazian H. H. Jr, Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 20, 3386–3400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buzdin A., et al. , A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics 80, 402–406 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Weber M. J., Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2, e205 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Perez J. L., Doucet A. J., Bucheton A., Moran J. V., Gilbert N., Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17, 602–611 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilbert N., Lutz S., Morrish T. A., Moran J. V., Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25, 7780–7795 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doucet A. J., Droc G., Siol O., Audoux J., Gilbert N., U6 snRNA pseudogenes: Markers of retrotransposition dynamics in mammals. Mol. Biol. Evol. 32, 1815–1832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buzdin A., et al. , The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 31, 4385–4390 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanaka N., Shuman S., RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 286, 7727–7731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Popow J., et al. , HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Englert M., Sheppard K., Aslanian A., Yates J. R. 3rd, Söll D., Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahl V., Keller H., Schmidt S., Weichenrieder O., Retrotransposition and crystal structure of an Alu RNP in the ribosome-stalling conformation. Mol. Cell 60, 715–727 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Kent W. J., BLAT–The BLAST-like alignment tool. Genome Res. 12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houseley J., Tollervey D., Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One 5, e12271 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peebles C. L., Gegenheimer P., Abelson J., Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell 32, 525–536 (1983). [DOI] [PubMed] [Google Scholar]
- 66.Chakravarty A. K., Shuman S., The sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res. 40, 8558–8567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka N., Chakravarty A. K., Maughan B., Shuman S., Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 286, 43134–43143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blomen V. A., et al. , Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Lu Y., Liang F. X., Wang X., A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol. Cell 55, 758–770 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Unlu I., Lu Y., Wang X., The cyclic phosphodiesterase CNP and RNA cyclase RtcA fine-tune noncanonical XBP1 splicing during ER stress. J. Biol. Chem. 293, 19365–19376 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Perez J. L., et al. , Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466, 769–773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coufal N. G., et al. , L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia-Perez J. L., et al. , LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 16, 1569–1577 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Macia A., et al. , Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 27, 335–348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beck C. R., Garcia-Perez J. L., Badge R. M., Moran J. V., LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 12, 187–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mailman M. D., et al. , The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 39, 1181–1186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Landry J. J., et al. , The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3, 1213–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adey A., et al. , The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 500, 207–211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Auton A., et al. ; 1000 Genomes Project Consortium , A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zillmann M., Gorovsky M. A., Phizicky E. M., Conserved mechanism of tRNA splicing in eukaryotes. Mol. Cell. Biol. 11, 5410–5416 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.UniProt Consortium T., UniProt: The universal protein knowledgebase. Nucleic Acids Res. 46, 2699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moldovan J. B., Moran J. V., The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet. 11, e1005121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goodier J. L., Cheung L. E., Kazazian H. H. Jr, Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. 41, 7401–7419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goodier J. L., Zhang L., Vetter M. R., Kazazian H. H. Jr, LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 27, 6469–6483 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goodier J. L., Ostertag E. M., Engleka K. A., Seleme M. C., Kazazian H. H. Jr, A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 13, 1041–1048 (2004). [DOI] [PubMed] [Google Scholar]
- 86.Taylor M. S., et al. , Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155, 1034–1048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mandal P. K., Ewing A. D., Hancks D. C., Kazazian H. H. Jr, Enrichment of processed pseudogene transcripts in L1-ribonucleoprotein particles. Hum. Mol. Genet. 22, 3730–3748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Markmiller S, et al. , Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Popow J., Jurkin J., Schleiffer A., Martinez J., Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature 511, 104–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kanai Y., Dohmae N., Hirokawa N., Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Pannone B. K., Xue D., Wolin S. L., A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 17, 7442–7453 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verdone L., Galardi S., Page D., Beggs J. D., Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr. Biol. 14, 1487–1491 (2004). [DOI] [PubMed] [Google Scholar]
- 93.Li W. M., Barnes T., Lee C. H., Endoribonucleases–Enzymes gaining spotlight in mRNA metabolism. FEBS J. 277, 627–641 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Wreschner D. H., McCauley J. W., Skehel J. J., Kerr I. M., Interferon action–sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289, 414–417 (1981). [DOI] [PubMed] [Google Scholar]
- 95.Cooper D. A., Jha B. K., Silverman R. H., Hesselberth J. R., Barton D. J., Ribonuclease L and metal-ion-independent endoribonuclease cleavage sites in host and viral RNAs. Nucleic Acids Res. 42, 5202–5216 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fett J. W., et al. , Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 24, 5480–5486 (1985). [DOI] [PubMed] [Google Scholar]
- 97.Lyons S. M., Fay M. M., Akiyama Y., Anderson P. J., Ivanov P., RNA biology of angiogenin: Current state and perspectives. RNA Biol. 14, 171–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gonzalez T. N., Sidrauski C., Dörfler S., Walter P., Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 18, 3119–3132 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Silverman R. H., Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J. Virol. 81, 12720–12729 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang A., et al. , RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 42, 3803–3820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boelens W. C., Palacios I., Mattaj I. W., Nuclear retention of RNA as a mechanism for localization. RNA 1, 273–283 (1995). [PMC free article] [PubMed] [Google Scholar]
- 102.Kosmaczewski S. G., et al. , RNA ligation in neurons by RtcB inhibits axon regeneration. Proc. Natl. Acad. Sci. U.S.A. 112, 8451–8456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shchepachev V., Wischnewski H., Soneson C., Arnold A. W., Azzalin C. M., Human Mpn1 promotes post-transcriptional processing and stability of U6atac. FEBS Lett. 589, 2417–2423 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.