Significance
Innate immune memory (i.e., immune priming) is found in many invertebrates. In some cases, immune priming provides protection against infection only when the same bacteria are used for priming and challenge; that is, priming can be specific. However, we still know little about the conditions favoring the evolution of immunological specificity. We present evidence that immune priming and its specificity can rapidly evolve in an insect through experimental selection by repeated bacterial exposure. Our populations evolved treatment-specific differences in expression profiles of immune, metabolic, and transcription-regulatory genes, pointing to similar mechanisms acting in vertebrate trained immunity. Hence, immune memory combines deeply rooted resemblances across systems with enormous evolutionary plasticity.
Keywords: immune priming, innate immunity, immune memory, immunological specificity, trained immunity
Abstract
Memory and specificity are hallmarks of the adaptive immune system. Contrary to prior belief, innate immune systems can also provide forms of immune memory, such as immune priming in invertebrates and trained immunity in vertebrates. Immune priming can even be specific but differs remarkably in cellular and molecular functionality from the well-studied adaptive immune system of vertebrates. To date, it is unknown whether and how the level of specificity in immune priming can adapt during evolution in response to natural selection. We tested the evolution of priming specificity in an invertebrate model, the beetle Tribolium castaneum. Using controlled evolution experiments, we selected beetles for either specific or unspecific immune priming toward the bacteria Pseudomonas fluorescens, Lactococcus lactis, and 4 strains of the entomopathogen Bacillus thuringiensis. After 14 generations of host selection, specificity of priming was not universally higher in the lines selected for specificity, but rather depended on the bacterium used for priming and challenge. The insect pathogen B. thuringiensis induced the strongest priming effect. Differences between the evolved populations were mirrored in the transcriptomic response, revealing involvement of immune, metabolic, and transcription-modifying genes. Finally, we demonstrate that the induction strength of a set of differentially expressed immune genes predicts the survival probability of the evolved lines upon infection. We conclude that high specificity of immune priming can evolve rapidly for certain bacteria, most likely due to changes in the regulation of immune genes.
Specific immune memory is considered the hallmark of the vertebrate adaptive immune system (1). It describes the ability of the immune system to store and recall information of previously encountered pathogens to mount a fast and specific immune response during secondary exposure to the same or similar pathogens. However, over the past few years, evidence has rapidly accumulated indicating that plants and invertebrates also have forms of immune memory, and that it can even be generated by the vertebrate innate immune system (2–4). The consequences of these observations for our current concept of immune memory are hotly debated (5–7), and one of the controversial questions is whether all these phenomena fulfill the requirements of a true memory effect (8). Specificity, defined as the ability to discriminate among different antigens or pathogens, is an important aspect in this controversy, as it varies across different taxa. For example, immune priming, a form of immune memory in invertebrates, is often rather unspecific (9, 10), but has occasionally been demonstrated to enable discrimination between different pathogen species, strains, or even genotypes (11–16). By contrast, trained immunity, a form of memory in the innate immune system of vertebrates, seems to provide only broad protection with rather low specificity (2).
Specificity of the inducible immune system provides a response tailored toward the particular type of pathogen encountered and might help avoid autoimmunity (17, 18). Such specificity needs a very elaborate, and thus likely costly, system that is simultaneously specific and also covers most of the theoretically possible antigenic space (19). The evolutionary benefit of specificity in immune memory depends on the likelihood with which the same type of pathogen is encountered repeatedly during the lifespan, a parameter that will vary across environments and time (20, 21). Given the strong and fluctuating selection pressure of pathogens on host fitness (22, 23), we hypothesize that immunological specificity should be a trait that itself is able to adapt to the characteristics of the pathogenic environment. However, to the best of our knowledge, no attempt has yet been made to assess the extent to which specificity can evolve within short periods.
Insect immune priming provides an informative model for addressing this hypothesis, because many insects are often easy to handle in the laboratory, allowing for selection experiments under controlled conditions and for elucidation of molecular underpinnings of experimentally evolved traits (24–26). Recent transcriptome analyses of primed individuals suggest that mechanisms underlying the priming effect depend on the particular host-pathogen system used and on the routes of infection (14–16). For example, oral priming and challenge of Tribolium castaneum larvae with the entomopathogenic bacteria Bacillus thuringiensis tenebrionis induced the expression of immune genes with reported activity against the same bacterium, while transgenerational priming of adult beetles via septic wounding with a related B. thruringiensis strain induced changes in the amino acid metabolism and transcriptional control of their offspring (14, 15). Importantly, priming was found to vary substantially among host populations even for the same host-pathogen system (27), suggesting that part of the ample phenotypic variability should be based on standing genetic variation that evolution could act upon.
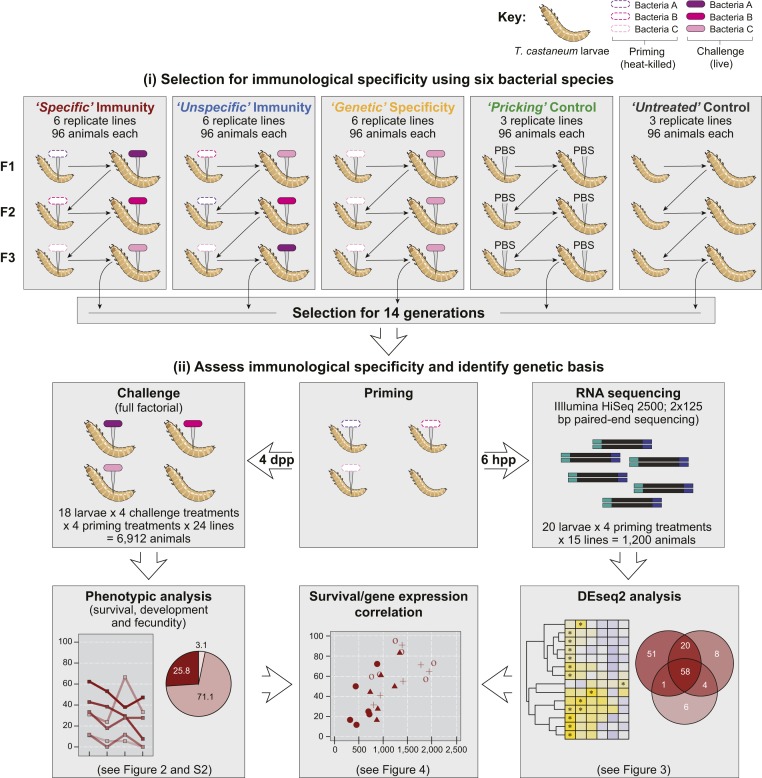
Here we conducted an experimental evolution study to investigate whether specificity in immune priming can rapidly evolve using data from more than 48,000 animals over a period of 3 y. For the septic priming and challenge procedure, we used larvae of the red flour beetle T. castaneum as the host and confronted them with alternating combinations of 6 bacteria: gram-negative Pseudomonas fluorescens, gram-positive Lactococcus lactis, and 4 strains of gram-positive B. thuringiensis (Fig. 1 and SI Appendix). The contrasting selection treatments consisted of either exposing individual larvae to the same type of bacterium for both priming and challenge (specific selection treatment) or to different ones (unspecific selection treatment). We thereby varied the degree to which specificity of the primed response confers fitness benefits to the host and thus expected selection for vs. against the ability of the host to raise a specific primed response. In both cases, different bacteria were chosen for subsequent generations so as to avoid adaptation to one particular bacterium. In another selection treatment, we evolved lines for such a genetically encoded rather than primed specificity (denoted as genetic), making use of priming and challenge with always the same bacteria within and across generations. Moreover, we kept phosphate-buffered saline-pricked (pricking) and naïve beetles (untreated) as control lines. After 14 host generations, immunological specificity was tested in terms of survival of infection following homologous vs. heterologous priming. We also tested for potential fitness costs of evolved differences in priming specificity in terms of development and fecundity, and studied gene expression after priming, using an RNA sequencing approach. Our results demonstrate that gene expression upon priming differs significantly between the specific and unspecific selection treatments, and that the strength of induction of several immune genes upon priming correlates with the likelihood of survival after homologous challenge.
Fig. 1.
Experimental design of experimental evolution treatments and subsequent phenotypic and transcriptomic analyses. Three selection treatments were selected for specific immunity, unspecific immunity, or genetic specificity using 6 bacteria species or strains: L. lactis (Ll), P. fluorescens (Pf), B. thuringiensis tenebrionis (Btt), B. thuringiensis (Bt1), B. thuringiensis yunnanensis (Bt2), and B. thuringiensis 407 (Bt407). Two additional evolution treatments were used to control for the effect of wounding (pricking control) and laboratory conditions (untreated control). After 14 generations of evolution, several phenotypic assays and a transcriptomic analysis were performed to assess the degree of immune priming and its specificity and genetic basis in each treatment.
Results
Experimental Evolution Increased Immunological Specificity for the Entomopathogen B. thuringiensis.
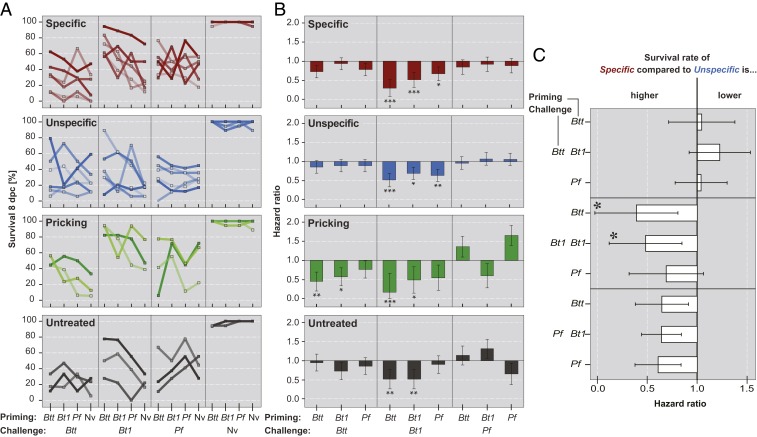
Our selection protocol was aimed at either increasing or decreasing immunological specificity (Fig. 1). After 7 and 14 generations of experimental evolution, we relaxed selection for 2 generations (to reduce epigenetic effects) and then tested the F2 offspring for immune priming, using a full factorial priming challenge design with 3 different bacteria (Fig. 1). No significant effects of the selection treatments were found after 7 generations (raw data in Dataset S1; statistical analysis results in Dataset S2). However, experimental evolution for 14 generations resulted in phenotypic differences. All lines showed a significant priming response when primed with either of the 2 strains of B. thuringiensis (Btt or Bt1) and subsequently challenged with the most pathogenic bacterium Bt1 (Fig. 2 A and B, Cox proportional hazard of primed vs. naive treatments; Dataset S3a). This Bt-specific priming benefit seemed more pronounced in the lines selected for increased specificity (Fig. 2B, Cox proportional hazard of primed vs. naive treatments for the specific selection treatment; Dataset S3a). Thus, we directly compared the specific and unspecific selection treatments and found significantly improved survival of Bt1 challenge for beetles from the specific selection treatment when primed with either strain of Bt but not when primed with P. fluorescens (Pf) (Fig. 2C, Cox proportional hazard of specific lines vs. unspecific lines for each priming/challenge treatment combination; Dataset S3b). This shows that selection for specificity increased the bacterial species-specific (but not the strain-specific) priming response.
Fig. 2.
Survival of evolved populations at 8 d after priming and challenge. (A) Survival rates by selection treatment and challenge bacteria. Each line corresponds to 1 replicate line. (B) Hazard ratios of primed treatments compared with naive controls by selection treatment and challenge bacteria. Each column corresponds to the median hazard ratio of 6 replicate lines for the specific and unspecific selection treatments and 3 replicate lines for the pricking and untreated control treatments, with SEs. (C) Hazard ratios of the specific selection treatment compared with the unspecific selection treatment by priming and challenge bacteria combination.
It is noteworthy that the pricking selection treatment, which served as a control for wounding responses, survived challenge with both Btt and Bt1 better after priming with either of the 2 Bt strains (Fig. 2B, Cox proportional hazard of primed vs. naive treatments for the pricking selection treatment; Dataset S3a).
The genetic beetle lines selected for nonplastic genetic specificity responded to selection only weakly in terms of survival of infection (SI Appendix, Fig. S1 and Dataset S3c), evolving resistance for only 1 of the bacteria, Bt1. However, the genetic lines showed significantly faster larval development than all other lines (SI Appendix, Fig. S2A, cumulative link mixed model, priming × challenge × selection, P = 0.0181; Dataset S3d). We did not observe any differences in fecundity or development among the selection treatments (Datasets S3e and f and S4).
Evolution of Divergent Gene Expression Profiles in Response to Priming.
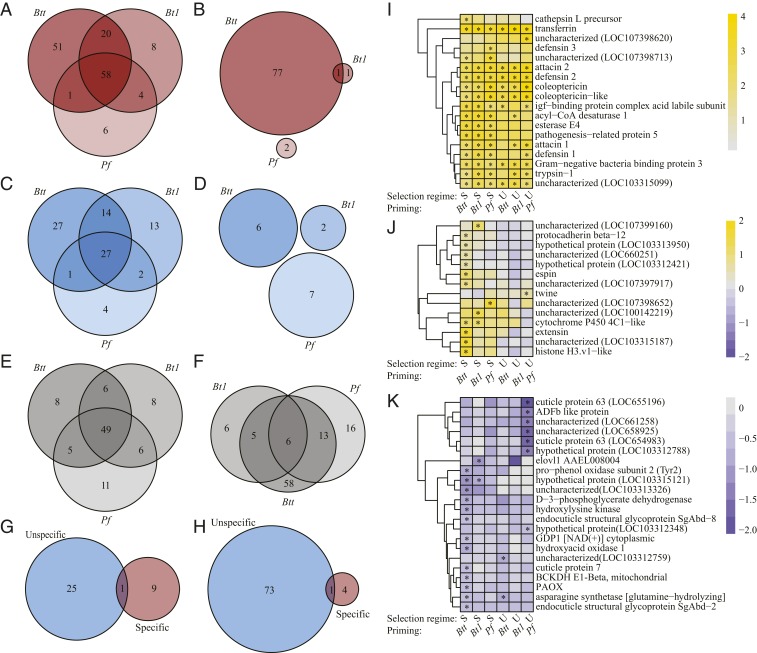
We next examined the degree to which the evolved beetles differed in their gene expression profiles after priming. We compared transcriptomes among the specific, unspecific, and untreated selection treatments, each at 6 h after priming with Btt, Bt1, and Pf, and related them to the corresponding unprimed control animals (Fig. 3 and Dataset S5; gene annotation in Dataset S6 and Gene Ontology terms in Dataset S7) (28).
Fig. 3.
DEGs at 6 h after priming by selection treatment. (A–F) The number of DEGs at 6 h after priming compared to naive (i.e., unprimed) animals. (A) Specific treatment, up-regulated. (B) Specific treatment, down-regulated. (C) Unspecific treatment, up-regulated. (D) Unspecific treatment, down-regulated. (E) Untreated treatment, up-regulated. (F) Untreated treatment, down-regulated. (G and H) DEGs of naive (i.e., unprimed) animals of the specific and unspecific selection treatments compared with the untreated control treatment. (G) Up-regulated. (H) Down-regulated. (I–K) Heatmaps for selected genes that were significantly differentially regulated for at least 1 of the 6 selection/priming treatment combinations, indicated by asterisks. (I) Heatmap of the 10 most up-regulated genes for each treatment combination. (J) Heatmap of genes showing contrasting expression patterns among the treatment combinations. (K) Heatmap of the 10 most down-regulated genes for each treatment combination. For I–K, DEGs that are shared by treatments are shown only once.
Across all selection treatments, priming with any of the included bacteria caused the up-regulation of a core set of immune-related genes (Fig. 3 A, C, E, and I), such as the iron scavenger transferrin. Far more genes were differentially up- or down-regulated upon priming in beetles from the specific selection treatment than with the unspecific selection treatment (Fig. 3 A–D). This pattern was particularly pronounced after priming with Btt, when large numbers of genes were uniquely up-regulated (n = 51) or down-regulated (n = 77) (Fig. 3 A and B). Several of the most strongly up-regulated genes were significant only in beetles from the specific selection treatment, such as thaumatin (pathogenesis-related protein 5) (Fig. 3I). By contrast, down-regulated genes were often involved in metabolism and cuticle processes (Fig. 3I). Several genes even showed contrasting directions of regulation between beetles from the specific and unspecific selection treatments, such as a histone H3-like gene with reported functions in epigenetic regulation of immunity in vertebrates (Fig. 3J) (29).
Given these strong differences in priming-responsive gene expression profiles, we also asked whether there are any differences between the selection treatments already in the nonprimed state. A lower number of differentially regulated genes (compared with the untreated selection treatment) in the specific treatment indicated that the unspecific lines already differed from the controls in the naive, nonprimed state (Fig. 3 G and H and Dataset S6). The immune genes PPO1, 2, and 3 were all down-regulated in the unspecific selection treatment, while the antimicrobial peptide (AMP) Attacin 1 was up-regulated in the specific selection treatment (Dataset S6). We also identified 2 genes involved in epigenetic control of transcription as up-regulated in the specific selection treatment: mediator of RNA polymerase II transcription subunit 15 and exosome complex exonuclease RRP6-like.
Likelihood of Survival after Priming/Challenge Treatment Correlates with the Expression Level of Immune Genes after Priming.
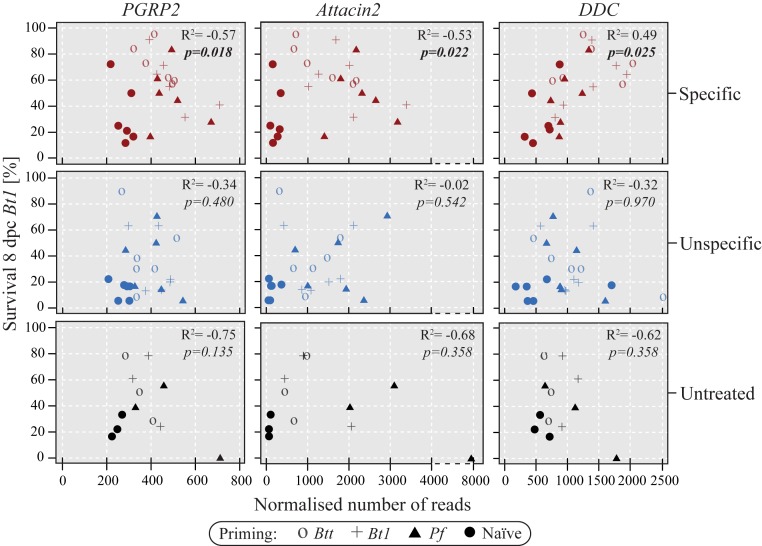
The phenotypic analysis of survival after priming and challenge, along with the analysis of gene expression patterns upon priming, uncovered differences between the selection treatments, suggesting a link between transcriptional responses and survival. Thus, we combined the survival and gene expression data to test whether transcriptional activation of specific genes underpin the survival differences among selection treatments and replicate lines. For this analysis, we focused on survival after Bt1 challenge, because the strongest survival differences were observed for this bacterium (Fig. 2C). Fig. 4 shows some examples of the observed correlations. The immune gene peptidoglycan receptor 2 (PGRP 2) was induced on priming, but counterintuitively, those specific lines that showed the highest expression had relatively lower survival, as indicated by the negative correlation (Fig. 4; Kendall rank correlation coefficient, P = 0.018). A similar pattern was observed for several AMPs, such as Attacin 2 (Fig. 4; Kendall rank correlation coefficient, P = 0.022). By contrast, those specific selection lines with high expression of dopa decarboxylase, a key enzyme in the melanization pathway of insects, had the best survival (Fig. 4, Kendall rank correlation coefficient, P = 0.025). In contrast to the specific selection treatment, we did not find any such correlations in the unspecific or untreated treatments (Fig. 4).
Fig. 4.
Correlation of survival rates and absolute expression levels for immune DEGs. Absolute transcript levels after normalization with DESeq2 were correlated with survival rates after Bt1 challenge for immune DEGs in all primed groups. Correlations are shown separately for the specific (red) and unspecific (blue) selection treatments and the untreated (gray) control treatment from left to right per gene. n = 6 for specific and unspecific; n = 3 for untreated.
Discussion
Our study reveals that selection for immunological specificity over a rather small number of 14 generations already results in strongly differing transcriptional responses upon immune priming. These differences correspond to survival benefits during a subsequent infection. This demonstrates a general evolutionary responsiveness of a phenotypically plastic system that provides immune memory. Moreover, the evolutionary changes appear to be targeted at a limited set of pathogens; we observed that selection aiming at a generally higher degree of specificity yielded a more pronounced primed immune response for one bacterial species, the entomopathogen B. thuringiensis.
B. thuringiensis is a gram-positive bacterial pathogen of insects and nematodes. Some strains infect T. castaneum (30), which in turn can activate an immune priming response conferring some degree of specificity upon both oral and septic exposure (31, 32). In the present study, we found specificity of priming on the level of bacterial species but not on strains as has been previously reported (31, 32), most likely due to the use of distinct strains of B. thuringiensis. Evolution of increased priming responses were restricted to B. thuringiensis but did not extend to the other tested bacterial species. This suggests that priming specificity and its evolvability might be related to the likelihood of infection or coevolution with a certain pathogen (33–35). It might well be that the lower receptor diversity of the invertebrate immune system compared with vertebrate adaptive immunity is responsible for the limitation of specificity toward a certain set of pathogens. The extent to which immune systems with somatic diversification of receptors (36, 37), such as the adaptive immune system of vertebrates, show more or less limited evolution of immunological specificity remains to be studied.
An interesting observation is that the pricking control lines showed increased general survival when primed and challenged with either Btt or Bt1, even though they were not selected with any bacteria during the selection process. A number of studies have demonstrated how pathogen-associated molecular patterns (PAMPs; e.g., epitopes with bacterial origin, such as lipopolysaccharides) and danger-associated molecular patterns (DAMPs; e.g., actin) (38) have strongly overlapping signaling pathways in insects (39, 40). Furthermore, danger signaling has been indicated to play a vital role in trained immune responses mediated by long-term functional reprogramming in cells of the innate immune system in vertebrates (41). Based on this, we hypothesize that the increased survival phenotype observed in the pricking control lines might be due to a DAMP-mediated evolved priming response. Given the low number of replicates in this control line (n = 3) and the high within-replicate variation (Fig. 2A), this should be repeated in separate selection lines with proper replication and controls for wounding.
The changes in priming specificity did not lead to any apparent trade-offs in adult short-term fecundity, which have been reported for primed immune responses against pathogens in other insect species (42, 43). This could be a consequence of our selection protocol, as we selected for larval phenotypes under ad libitum rearing conditions, or our fecundity readouts over a short time frame without specific testing for early and late life fecundity. However, we observed that the genetic lines, which were selected for nonplastic resistance (or tolerance) rather than for priming ability, developed faster than any other selection treatment, which may suggest that the plastic priming ability could be developmentally costly. Alternatively, fast development could also be an adaptation to escape an unfavorable, pathogenic environment (44).
The transcriptomic signature of priming in the evolved beetles supported the observed enhanced specificity toward B. thuringiensis and suggests that immune priming consists of divergent sets of genes that confer general priming and bacteria-specific responses, respectively. A core set of genes was induced by priming with any of the 3 bacteria used (Fig. 3I) and resembles patterns of gene expression previously reported during infection of T. castaneum with B. thuringiensis (14, 45, 46). For example, the iron sequestration factor transferrin was strongly up-regulated upon priming, resembling the acute-phase response of a mosquito cell line infected with Escherichia coli to create a low free-iron environment (47).
The divergent transcriptome signatures for the specific vs. unspecific selection treatments (Fig. 3 and Dataset S6) suggest that the microevolution of specificity relies on changes in metabolism, which is often a deciding factor in the outcome of host-pathogen interactions (48, 49), and immunity. Within the specific selection treatment, the strong transcriptional response upon priming with Btt included gram-positive responsive genes, such as a Toll-3-like receptor and Persephone (14, 45, 46, 50, 51), indicative of Toll pathway activation. By contrast, down-regulated genes contain metabolism-associated genes, such as hexokinase type 2 and sedoheptulokinase, which have previously been implied in shifting the energy metabolism of immune cells in response to immune activation (52, 53). Trained immunity in vertebrates is similarly based on changes in the energy metabolism of immune cells (54). Several genes previously reported to be involved in the epigenetic reprogramming of immune cells during trained immunity were also up-regulated in our evolved populations (Fig. 3J and Dataset S6), pointing to an evolutionarily conserved mechanism of innate immune memory. Our data thereby support similar results obtained by Tate et al. (15) in T. castaneum upon transgenerational immune priming with Bt. In addition, the finding that a histone H3 gene is down-regulated in the unspecific treatment but up-regulated in the specific treatment supports recent findings of correlation between IFN memory in vertebrate trained immunity with histone H3.3 and H3K36me3 chromatin marks (29). More generally, partially similar trends in gene expression changes were observed for within-generational, transgenerational, and evolved differences in innate immune memory. This supports the view that gene expression patterns, potentially mediated via epigenetic processes, could be a first step toward evolved differences that might lead to gene sequence evolution in the long term (55).
We further identified significant correlations between gene expression and survival rates of infection after priming (Fig. 4). Lines from the specific selection treatment with highest survival of Bt1 infection showed the strongest expression of dopa decarboxylase (ddc), a gene involved in the phenoloxidase (PO) response (56). The role of the PO response in priming and immune defense against bacterial pathogens is well known in T. castaneum and other insect species (57–59). Nodulation and phagocytosis of E. coli in the medfly Ceratitis capitata have been shown to be dependent on dopa decarboxylase activity (60). Several AMPs were induced in response to priming with any of the bacteria (Fig. 3I and Dataset S6), but expression of Attacin 2 was lower in lines that survived best. This suggests a more fine-tuned response in lines derived from the specific selection treatment that involves a shift toward the PO response (61, 62). Recent studies describe priming-induced shifts in the transcriptional basis of the immune response in T. castaneum (14) and the snail Biomphalaria glabrata (63), as well as transcriptional underpinnings of genotype-by-genotype specificity in the immune response of Bombus terrestris colonies infected with their natural gut parasite Crithidia bombi (64). Thus, specificity might be facilitated through adaptational changes in transcription to orchestrate a more fine-tuned and distinct response upon subsequent encounters. Such adaptational responses could be achieved through, for example, changes in oligomerization of transcription factors, as has been shown for other physiological systems that form memory (65).
In conclusion, we demonstrate that the specificity of immune priming in an invertebrate species can be changed by experimental evolution within a few host generations. Similarities of the evolved differences with the vertebrate trained immune response suggest that some features of acquired immune responses might be the result of convergent evolution or even share a common origin. Indeed, a recent review of trained immunity discussed striking parallels of animal, plant, and even bacterial acquired immune systems (66). Thus, future studies might use a comparative approach (67). Finally, given the existence of acquired immunity in multiple arthropod disease vectors, the combination of insights gained from molecular immunology and evolutionary ecology approaches could lead to improved strategies for vector control (68).
Materials and Methods
All methods are described in more detail in SI Appendix. T. castaneum (Cro1 population) were wild collected in Slavonski Brod, Croatia in 2010 and allowed to adapt to laboratory conditions for at least 12 generations (approximately 12 mo) before the experiments. We used approximately 10,000 2- to 3-wk-old adult beetles as an ancestral parental generation to produce animals for 3 different selection treatments—specific immunity (specific), unspecific immunity (unspecific), and genetic specificity (genetic)—and 2 control treatments: pricking control (pricking) and untreated control (untreated). Each selection treatment was replicated 6 times, and each control treatment was replicated 3 times. The specific selection treatment was used to select for the ability to raise a specific immune response upon homologous priming; therefore, this line was primed and challenged with the same bacteria within generations but with different bacteria across generations. The unspecific selection treatment was used to select for unspecific immunity in the sense of a broad-range innate immune response. To achieve this, we primed and challenged with different bacteria within and across generations. We included the genetic selection treatment to test for the evolution of resistance against the bacteria used in the selection procedure. All bacteria challenge doses were adjusted to LD20. To control for any effects of repeated wounding, the pricking treatment was aseptically primed and challenged using sterile PBS (Calbiochem). Finally, the untreated treatment was reared at densities like the wounded and infected selection treatments to control for the effects of population size on the response to selection (Fig. 1).
We performed a full reciprocal priming and challenge postselection experiment with all selection treatments and replicates. We monitored the survival, developmental speed, and short-term fecundity of surviving adults. We used an injection method to prime and challenge the larvae to expose them to controlled numbers of bacteria. The injections were performed with a Nanoject II Auto-Nanoliter Injector (Drummond) equipped with 2-step pulled, cut, and back-filled glass capillaries. Each larva was either injected with 18.4 nL of a bacteria suspension or left untreated for priming and challenge, resulting in a dose of 18,400 heat-inactivated bacteria for all priming groups and a LD50 inducing dose of live bacteria for all challenge groups. (Specific bacteria concentrations and doses are provided in SI Appendix, Table S1). Censored survival data, ordinal developmental data, and fecundity count data were analyzed in R. Details of the statistical analyses are provided in SI Appendix.
To identify the genetic bases for the responses to selection after 14 generations, we performed a transcriptomic analysis with primed individuals of the same populations used for the phenotypic readout, excluding the genetic and pricking selection treatments. In brief, for each replicate, 20 larvae were pooled at 6 h after priming. The libraries for Illumina sequencing were prepared using the TruSeq RNA Sample V2 Kit (Illumina) following the manufacturer’s protocol and sequenced with the TruSeq SBS Kit V4 on 2 lanes of the Illumina HiSeq 2500, yielding 2 × 125-bp paired reads per sample. RNA-seq data were analyzed using a custom pipeline, as described in SI Appendix. We performed a differential gene expression analysis and generated heatmaps using the “DESeq2” package in R. The annotation of reads was performed on the basis of version 5.2 of the T. castaneum genome. The sequencing results have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (accession no. GSE133892).
Finally, the absolute expression values for the differentially expressed genes (DEGs) identified in the DESeq2 analysis were correlated with the corresponding data for survival rates for all primed treatment groups. For each gene, Kendall’s test was performed to determine statistical significance, followed by correction for multiple testing.
Supplementary Material
Acknowledgments
We thank Barbara Hasert for support with the experiments; Megan Kutzer and Sophie A. O. Armitage for support with data analysis; Hendrik Eggert and Barbara Milutinović for helpful discussions; Mark Miller for help with the figure designs; Kristina U. Wensing, Nora K. E. Schulz, Anne Kaminski, Madeleine Hamley, Jennifer Schauer, and Jana Dobelmann for support in the selection experiments; and Britt-Sabrina Petersen for library preparation for RNA-seq. We also thank 2 reviewers for comments that greatly improved the manuscript. The project was funded by the Deutsche Forschungsgemeinschaft to J.K. (DFG, KU 1929/4-2), H.S. (SCHU 1415/9-2), and P.R. (RO 2994/3-2) within the priority program SPP 1399, “Host–Parasite Coevolution”. W.Y. was also supported by the International Max Planck Research School for Evolutionary Biology at Kiel University. K.F. acknowledges support by the Münster Graduate School of Evolution (MGSE).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequencing results have been deposited in the National Center for Biotechnology Information’s (NCBI) Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. GSE133892).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904828116/-/DCSupplemental.
References
- 1.Murphy K. P., Janeway C. A., Mowat A., Janeway’s Immunobiology (Garland Science, New York, ed. 8, 2012). [Google Scholar]
- 2.Netea M. G., Quintin J., van der Meer J. W. M., Trained immunity: A memory for innate host defense. Cell Host Microbe 9, 355–361 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Reimer-Michalski E.-M., Conrath U., Innate immune memory in plants. Semin. Immunol. 28, 319–327 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Milutinović B., Kurtz J., Immune memory in invertebrates. Semin. Immunol. 28, 328–342 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kurtz J., Specific memory within innate immune systems. Trends Immunol. 26, 186–192 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Rimer J., Cohen I. R., Friedman N., Do all creatures possess an acquired immune system of some sort? Bioessays 36, 273–281 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Müller V., de Boer R. J., Bonhoeffer S., Szathmáry E., An evolutionary perspective on the systems of adaptive immunity. Biol. Rev. Camb. Philos. Soc. 93, 505–528 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Pradeu T., Du Pasquier L., Immunological memory: What’s in a name? Immunol. Rev. 283, 7–20 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Moret Y., Siva-Jothy M. T., Adaptive innate immunity? Responsive-mode prophylaxis in the mealworm beetle, Tenebrio molitor. Proc. Biol. Sci. 270, 2475–2480 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Ami F., Orlic C., Regoes R. R., Disentangling unspecific and specific transgenerational immune priming components in host-parasite interactions. bioRxiv 429498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milutinović B., Peuß R., Ferro K., Kurtz J., Immune priming in arthropods: An update focusing on the red flour beetle. Zoology (Jena) 119, 254–261 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Kurtz J., Franz K., Innate defence: Evidence for memory in invertebrate immunity. Nature 425, 37–38 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Little T. J., O’Connor B., Colegrave N., Watt K., Read A. F., Maternal transfer of strain-specific immunity in an invertebrate. Curr. Biol. 13, 489–492 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Greenwood J. M., et al. , Oral immune priming with Bacillus thuringiensis induces a shift in the gene expression of Tribolium castaneum larvae. BMC Genomics 18, 329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tate A. T., Andolfatto P., Demuth J. P., Graham A. L., The within-host dynamics of infection in trans-generationally primed flour beetles. Mol. Ecol. 26, 3794–3807 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barribeau S. M., Schmid-Hempel P., Sadd B. M., Royal decree: Gene expression in trans-generationally immune primed bumblebee workers mimics a primary immune response. PLoS One 11, e0159635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank S. A., Immunology and Evolution of Infectious Disease (Princeton Univ Press, Princeton, NJ, ed. 1, 2002). [PubMed] [Google Scholar]
- 18.Schmid-Hempel P., Ebert D., On the evolutionary ecology of specific immune defence. Trends Ecol. Evol. 18, 27–32 (2003). [Google Scholar]
- 19.Borghans J. A. M., Noest A. J., De Boer R. J., How specific should immunological memory be? J. Immunol. 163, 569–575 (1999). [PubMed] [Google Scholar]
- 20.Zander C. D., Four-year monitoring of parasite communities in gobiid fishes of the southwestern Baltic, II: Infracommunity. Parasitol. Res. 93, 17–29 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Gonçalves R. A., Oliveira M. S. B., Neves L. R., Tavares-Dias M., Seasonal pattern in parasite infracommunities of Hoplerythrinus unitaeniatus and Hoplias malabaricus (Actinopterygii: Erythrinidae) from the Brazilian Amazon. Acta Parasitol. 61, 119–129 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Koskella B., Lin D. M., Buckling A., Thompson J. N., The costs of evolving resistance in heterogeneous parasite environments. Proc. R. Soc. B Biol. Sci. 279, 1896–1903 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tellier A., Brown J. K., Spatial heterogeneity, frequency-dependent selection and polymorphism in host-parasite interactions. BMC Evol. Biol. 11, 319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerstes N. A. G., Martin O. Y., Insect host-parasite coevolution in the light of experimental evolution. Insect Sci. 21, 401–414 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Koch E. L., Guillaume F., Combining transcriptomic and fitness data reveals additive and mostly adaptive plastic responses of gene expression to multiple stress in Tribolium castaneum. bioRxiv 442145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan I., Prakash A., Agashe D., Experimental evolution of insect immune memory versus pathogen resistance. Proc. R. Soc. B Biol. Sci. 284, 20171583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan I., Prakash A., Agashe D., Divergent immune priming responses across flour beetle life stages and populations. Ecol. Evol. 6, 7847–7855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferro K., et al. , Tribolium castaneum experimental evolution. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE133892. Deposited 5 July 2019.
- 29.Kamada R., et al. , Interferon stimulation creates chromatin marks and establishes transcriptional memory. Proc. Natl. Acad. Sci. U.S.A. 115, E9162–E9171 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milutinović B., Höfling C., Futo M., Scharsack J. P., Kurtz J., Infection of Tribolium castaneum with Bacillus thuringiensis: Quantification of bacterial replication within cadavers, transmission via cannibalism, and inhibition of spore germination. Appl. Environ. Microbiol. 81, 8135–8144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth O., Sadd B. M., Schmid-Hempel P., Kurtz J., Strain-specific priming of resistance in the red flour beetle, Tribolium castaneum. Proc. Biol. Sci. 276, 145–151 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Futo M., Sell M. P., Kutzer M. A. M., Kurtz J., Specificity of oral immune priming in the red flour beetle Tribolium castaneum. Biol. Lett. 13, 20170632 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woolhouse M. E. J., Webster J. P., Domingo E., Charlesworth B., Levin B. R., Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32, 569–577 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Schulenburg H., Kurtz J., Moret Y., Siva-Jothy M. T., Introduction. Ecological immunology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 3–14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Best A., Tidbury H., White A., Boots M., The evolutionary dynamics of within-generation immune priming in invertebrate hosts. J. R. Soc. Interface 10, 20120887 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pancer Z., et al. , Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 430, 174–180 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Schultz J. H., Bu L., Adema C. M., Comparative immunological study of the snail Physella acuta (Hygrophila, Pulmonata) reveals shared and unique aspects of gastropod immunobiology. Mol. Immunol. 101, 108–119 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Srinivasan N., et al. , Actin is an evolutionarily-conserved damage-associated molecular pattern that signals tissue injury in Drosophila melanogaster. Elife 5, 1–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heil M., Land W. G., Danger signals: Damaged-self recognition across the tree of life. Front. Plant Sci. 5, 578 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-García M., Recio-Tótoro B., Claudio-Piedras F., Lanz-Mendoza H., Injury and immune response: Applying the danger theory to mosquitoes. Front. Plant Sci. 5, 451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crișan T. O., Netea M. G., Joosten L. A. B., Innate immune memory: Implications for host responses to damage-associated molecular patterns. Eur. J. Immunol. 46, 817–828 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Contreras-Garduño J., Rodríguez M. C., Rodríguez M. H., Alvarado-Delgado A., Lanz-Mendoza H., Cost of immune priming within generations: Trade-off between infection and reproduction. Microbes Infect. 16, 261–267 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Bartlett L. J., Wilfert L., Boots M., A genotypic trade-off between constitutive resistance to viral infection and host growth rate. Evolution 72, 2749–2757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth O., Kurtz J., The stimulation of immune defence accelerates development in the red flour beetle (Tribolium castaneum). J. Evol. Biol. 21, 1703–1710 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Tate A. T., Graham A. L., Dissecting the contributions of time and microbe density to variation in immune gene expression. Proc. R. Soc. B Biol. Sci. 284, 20170727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behrens S., et al. , Infection routes matter in population-specific responses of the red flour beetle to the entomopathogen Bacillus thuringiensis. BMC Genomics 15, 445 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshiga T., Hernandez V. P., Fallon A. M., Law J. H., Mosquito transferrin, an acute-phase protein that is up-regulated upon infection. Proc. Natl. Acad. Sci. U.S.A. 94, 12337–12342 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kutzer M. A. M., Armitage S. A. O., The effect of diet and time after bacterial infection on fecundity, resistance, and tolerance in Drosophila melanogaster. Ecol. Evol. 6, 4229–4242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kangassalo K., et al. , Intra- and trans-generational effects of larval diet on susceptibility to an entomopathogenic fungus, Beauveria bassiana, in the greater wax moth, Galleria mellonella. J. Evol. Biol. 28, 1453–1464 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Ashok Y., Drosophila toll pathway: The new model. Sci. Signal. 2, jc1 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Valanne S., Wang J.-H., Rämet M., The Drosophila Toll signaling pathway. J. Immunol. 186, 649–656 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Kelly B., O’Neill L. A. J., Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagy C., Haschemi A., Sedoheptulose kinase regulates cellular carbohydrate metabolism by sedoheptulose 7-phosphate supply. Biochem. Soc. Trans. 41, 674–680 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Cheng S. -C., et al. , mTOR- and HIF-1 -mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jablonka E., The evolutionary implications of epigenetic inheritance. Interface Focus 7, 20160135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Binggeli O., Neyen C., Poidevin M., Lemaitre B., Prophenoloxidase activation is required for survival to microbial infections in Drosophila. PLoS Pathog. 10, e1004067 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dhinaut J., Chogne M., Moret Y., Immune priming specificity within and across generations reveals the range of pathogens affecting evolution of immunity in an insect. J. Anim. Ecol. 87, 315–526 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Roth O., et al. , Paternally derived immune priming for offspring in the red flour beetle, Tribolium castaneum. J. Anim. Ecol. 79, 403–413 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Moret Y., Schmid-Hempel P., Immune defence in bumble-bee offspring. Nature 414, 506–507 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Sideri M., Tsakas S., Markoutsa E., Lampropoulou M., Marmaras V. J., Innate immunity in insects: Surface-associated dopa decarboxylase-dependent pathways regulate phagocytosis, nodulation and melanization in medfly haemocytes. Immunology 123, 528–537 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chambers M. C., Lightfield K. L., Schneider D. S., How the fly balances its ability to combat different pathogens. PLoS Pathog. 8, e1002970 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tschirren B., Richner H., Parasites shape the optimal investment in immunity. Proc. Biol. Sci. 273, 1773–1777 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pinaud S., et al. , A shift from cellular to humoral responses contributes to innate immune memory in the vector snail Biomphalaria glabrata. PLoS Pathog. 12, e1005361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barribeau S. M., Sadd B. M., du Plessis L., Schmid-Hempel P., Gene expression differences underlying genotype-by-genotype specificity in a host-parasite system. Proc. Natl. Acad. Sci. U.S.A. 111, 3496–3501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khan M. R., et al. , Amyloidogenic oligomerization transforms Drosophila Orb2 from a translation repressor to an activator. Cell 163, 1468–1483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gourbal B., et al. , Innate immune memory: An evolutionary perspective. Immunol. Rev. 283, 21–40 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Loker E. S., Macroevolutionary immunology: A role for immunity in the diversification of animal life. Front. Immunol. 3, 25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shaw D. K., et al. , Vector immunity and evolutionary ecology: The harmonious dissonance. Trends Immunol. 39, 862–873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.