A central postulate in chromatin biology is that nucleosomes are inherited through replication, and evidence for the recycling of nucleosomes from ahead of the replication fork to behind goes back more than 40 y (1, 2). Early electron microscopic observations of chromatin fibers revealed that nucleosomes form directly behind the replication fork (3), confirmed by later kinetic studies (4). However, it has remained uncertain as to whether histones from a nucleosome ahead of the fork return to the same position on a daughter strand after the fork has passed through. This is a critical question to resolve, because any dispersion of histones behind the fork disperses histone features such as posttranslational modifications that have been causally implicated in the propagation of gene expression states (5). The restoration of nucleosome positions may also be important for transcriptional regulation, given that nucleosomes act as barriers to transcriptional elongation but are disrupted when RNA polymerase passes through (6). Thus both replication and transcription can potentially disperse nucleosomes. To address this uncertainty, Schlissel and Rine (7) devise an elegant strategy to permanently mark histones within a 4-nucleosome region of the budding yeast genome, which allows them to precisely determine whether or not those nucleosomes shift positions after replication fork passage. By engineering the marked region within the repressible and inducible GAL10 gene, this system also allows them to separate the effects of replication fork passage and transcription on nucleosome positioning.
Biochemical studies have examined the process of nucleosome redeposition postreplication, but the question of positional memory has not been resolved. Unwinding of a nucleosome in vitro by the action of a helicase and a DNA polymerase resulted in transfer of the histone core to the leading-strand DNA duplex (8). As the leading strand is replicated before the lagging strand in vivo, a similar passive capture process may underlie the asymmetric segregation of old nucleosomes to the leading strand in Drosophila male germline stem cells and during testes development (9). Also, loss of histone chaperones responsible for actively mediating nucleosome redeposition to achieve nearly equal frequencies on leading and lagging strands resulted in a leading-strand bias (10, 11). These studies suggest that the default redeposition mechanism is passive transfer of the histone core to the leading strand nearby, consistent with positional memory. However, only random dispersion of nucleosomes was observed postreplication using a standard eukaryotic in vitro replication system. Some degree of positional retention was seen when Xenopus extracts were used, although not enough to maintain a nucleosome position for more than a cell cycle or so (12).
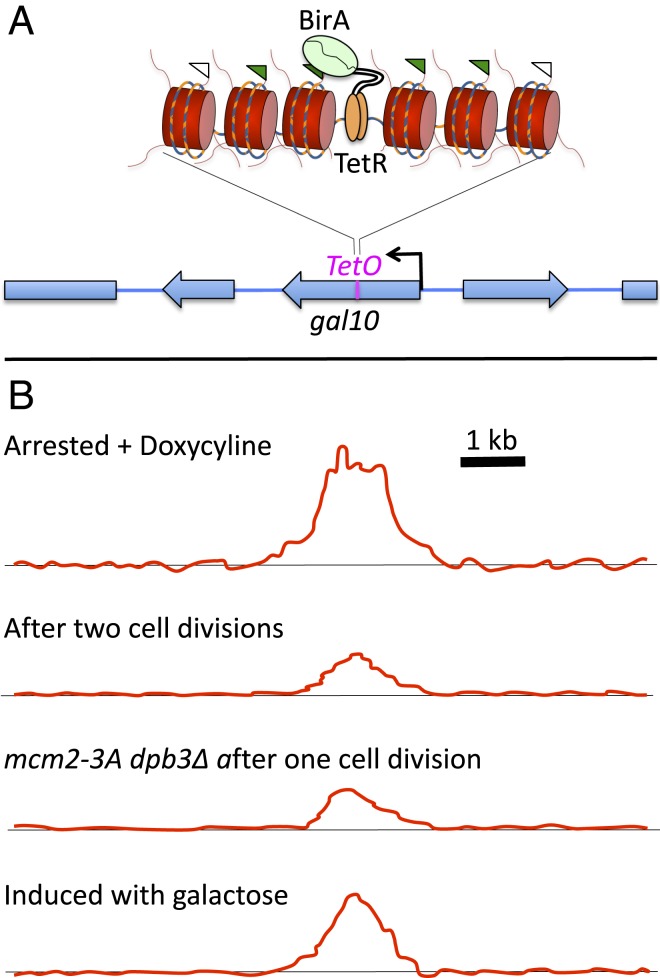
The reporter insertion into GAL10 constructed by Schlissel and Rine (7) consisted of the 19-bp Escherichia coli tetracycline operator sequence (TetO) for binding the tetracycline repressor (TetR) protein (Fig. 1A). To mark nucleosomes around the TetO site, the authors replaced each of the 2 yeast histone H3 coding sequences with an H3 sequence fused at its C-terminal end to a 15-amino acid “AviTag” substrate for biotinylation by the E. coli BirA biotin ligase. When a chimeric protein consisting of TetR fused to BirA was expressed, the binding of TetR−BirA to TetO resulted in biotinylation of the 4 nucleosomes closest to TetO. Although the yeast engineered to locally biotinylate nucleosomes within GAL10 were not affected in growth, gene expression, or silencing, it proved challenging to detect the cluster of biotinylated nucleosomes above background. Chromatin immunoprecipitation was unable to detect the TetR−BirA fusion protein bound to its site using its V5 epitope tag, and streptavidin pulldown of biotinylated H3 yielded only a very weak signal around TetO above background. To reduce the background signal, Schlissel and Rine used a hypomorphic BirA allele, reasoning that lowering the rate of biotinylation would reduce the background proportionally, while the targeted sites would still be saturated. A much greater reduction in background was obtained by splitting the BirA protein into 2 halves that could fold together to form an active enzyme, such that only when TetR−BirA formed a heterodimer with both halves of BirA would biotinylation occur. By attenuating biotinylation activity in this way, the authors observed consistently strong labeling above the genomic background at TetO, and, at a much lower level, a single site found to have a partial TetO sequence. By synchronizing yeast cells under conditions that repress GAL10, they could follow events over the course of successive cell cycles to determine whether the 4 marked nucleosomes returned to same positions or were dispersed or lost. Doxycycline was added to prevent further TetR−BirA binding, and, after 2 rounds of replication, the shape of the biotinylation profile remained unchanged, indicating that no nucleosome dispersal had occurred (Fig. 1B). Thus, nucleosomes returned to exactly where they were despite being severely disrupted by replication fork passage.
Fig. 1.
Testing positional memory. (A) Binding of TetR to its site (TetO) within the GAL10 gene tethers an attenuated version of E. coli biotin ligase (split BirA) to the site. AviTagged histone H3 (flags) in ∼4 adjacent nucleosomes becomes biotinylated (green flags). (B) After 2 rounds of cell division, the shape of the biotinylation profile does not change, although the signal becomes diluted. Mutations in the MCM2 and DPB3 histone chaperones required for equal reassembly of nucleosomes between leading and lagging strand do not change the shape of the profile, although they further weaken the signal. High-level transcription through GAL10 induced by growth in galactose weakens the signal but has no effect on its shape. Thus, nucleosomes remember their position through replication and transcription.
The question remained as to whether positional memory occurs by default or is an actively regulated process. Replication-coupled nucleosome reassembly is actively regulated by the histone chaperone activities of the MCM2 subunit of the replicative helicase on the leading strand (10) and the DPB3 histone chaperone on the lagging strand (11). Indeed, mutating either chaperone resulted in partial loss of marked nucleosomes, even though no local change in position of the 4 marked nucleosomes that were inherited through cell division was detected (Fig. 1B) (7). Similarly, growth of yeast cells under conditions that induced gal10 to express at a high level resulted in partial loss of marked nucleosomes, as expected for transcription-coupled nucleosome turnover, but, again, no change was observed in the position of those nucleosomes that reassembled (Fig. 1B). Although previous work had suggested that nucleosomes move in a 3′-to-5′ direction with RNA polymerase passage (13), this interpretation was based on averaging over the spans of thousands of genes, and variations in repositioning or in the degree of nucleosome turnover over gene bodies might have resulted in a net effect not detected near the middle of a single gene. Perhaps the surrounding nucleosomes in a tightly packed array within the gene body constrain nucleosomes to return to the same position following disruption, either when redeposition occurs behind the replication fork or when nucleosomes are redeposited in the wake of RNA polymerase.
The marking strategy developed by Schlissel and Rine (7) should be applicable in any eukaryotic biological context, and so the extent to which their findings generalize to multicellular eukaryotes will likely be a subject of future publications, for example, to test for local retention of transcription factors and propagation of histone marks. A challenge to the presumption that nucleosomes transmit epigenetic information during development comes from work in Drosophila, where the current paradigm of developmental silencing originated (14). Key components of the Polycomb Repressive Complexes (PRCs) 1 and 2 were first identified genetically in flies as members of the so-called Polycomb Group (15), where trimethylation of histone H3K27 by PRC2 is the feature that many studies have shown is inherited through replication (5). However, it is not necessary to assume that a nucleosome marked by H3K27me3 must return to the same position behind the fork to explain epigenetic memory, because the presence of this mark in flies is regulated in cis by short Polycomb Response Elements (PREs), which are bound by multiple sequence-specific DNA-binding proteins (16). In contrast, the H3K27me3 mark is present in broad domains that encompass PREs (17), containing perhaps hundreds of nucleosomes, and so dispersion of nucleosomes during replication within these broad domains is consistent with epigenetic inheritance being mediated by transcription factor rebinding, not by nucleosomes per se. So, although inheritance of H3K27me3-marked nucleosomes through replication is a central feature of developmental silencing, we need not assume that faithful inheritance is inconsistent with some degree of nucleosome dispersal behind the replication fork. However, the work by Schlissel and Rine indicates that precise positional inheritance can occur, and thus might be used in some cases of epigenetic inheritance. In the case of Polycomb silencing in mammals, there is, as yet, no known counterpart of Drosophila PREs, in which epigenetic memory maps genetically to discrete regulatory sites, and so positional memory remains an attractive explanation for the long-term maintenance of developmental silencing (5).
More generally, there are profound differences in replication-coupled nucleosome assembly between yeast and animals at regulatory elements, which caution against generalizing results from yeast to multicellular eukaryotes. For example, in yeast, nucleosome-depleted regions remain depleted immediately behind the replication fork (18), whereas, in both Drosophila and mammals, nucleosomes are redeposited seemingly at random over regulatory elements and require more than an hour to be cleared out to restore nucleosome-depleted regions (19, 20). The occlusion of regulatory elements by nucleosomes behind the replication fork may enforce specificity of transcription factor rebinding in the context of multicellular development, whereas being “open for business” may be the norm where selection for rapid growth would favor unimpeded transcription factor binding over specificity in a small genome. Likewise, positional memory may be an adaptation that promotes efficient retention of nucleosomes, which, in turn, would better protect DNA from damage during disruptions caused by replication and transcription.
Footnotes
The authors declare no conflict of interest.
See companion article on page 20605.
References
- 1.Leffak I. M., Grainger R., Weintraub H., Conservative assembly and segregation of nucleosomal histones. Cell 12, 837–845 (1977). [DOI] [PubMed] [Google Scholar]
- 2.Annunziato A. T., Assembling chromatin: The long and winding road. Biochim. Biophys. Acta 1819, 196–210 (2013). [DOI] [PubMed] [Google Scholar]
- 3.McKnight S. L., Miller O. L. Jr, Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12, 795–804 (1977). [DOI] [PubMed] [Google Scholar]
- 4.Gruss C., Wu J., Koller T., Sogo J. M., Disruption of the nucleosomes at the replication fork. EMBO J. 12, 4533–4545 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinberg D., Vales L. D., Chromatin domains rich in inheritance. Science 361, 33–34 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Weber C. M., Ramachandran S., Henikoff S., Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol. Cell 53, 819–830 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Schlissel G., Rine J., The nucleosome core particle remembers its position through DNA replication and RNA transcription. Proc. Natl. Acad. Sci. U.S.A. 116, 20605–20611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan L. D., Forties R. A., Patel S. S., Wang M. D., DNA looping mediates nucleosome transfer. Nat. Commun. 7, 13337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wooten M., et al. , Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat. Struct. Mol. Biol. 26, 732–743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petryk N., et al. , MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Yu C., et al. , A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madamba E. V., Berthet E. B., Francis N. J., Inheritance of histones H3 and H4 during DNA replication in vitro. Cell Reports 21, 1361–1374 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Radman-Livaja M., et al. , Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 9, e1001075 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pirrotta V., Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 13, 314–318 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Simon J. A., Kingston R. E., Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Orsi G. A., et al. , High-resolution mapping defines the cooperative architecture of Polycomb response elements. Genome Res. 24, 809–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmad K., Spens A. E., Separate Polycomb Response Elements control chromatin state and activation of the vestigial gene. PLoS Genet. 15, e1007877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fennessy R. T., Owen-Hughes T., Establishment of a promoter-based chromatin architecture on recently replicated DNA can accommodate variable inter-nucleosome spacing. Nucleic Acids Res. 44, 7189–7203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart-Morgan K. R., Reverón-Gómez N., Groth A., Transcription restart establishes chromatin accessibility after DNA replication. Mol. Cell 75, 408–414 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Ramachandran S., Henikoff S., Transcriptional regulators compete with nucleosomes post-replication. Cell 165, 580–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]