Significance
Batrachochytrium dendrobatidis [Bd] is one of the most devastating wildlife pathogens ever documented. Most surveys for Bd report only the presence/absence of the pathogen. However, Bd has distinct genetic lineages that vary in geographic extent and virulence, thus reporting Bd presence alone is not particularly informative. Our study uses a custom method for genotyping degraded Bd DNA samples, such as those nondestructively collected from live animal or museum specimen skin swabs, and presents the discovery of a divergent lineage of Bd—BdASIA3. This study advances our understanding of the evolutionary origins of Bd, highlights areas of the world where Bd lineages are coming into contact, and opens the door to affordable, rapid genetic monitoring of this pathogen.
Keywords: Batrachochytrium dendrobatidis, amphibian, conservation, genetic monitoring
Abstract
Biodiversity loss is one major outcome of human-mediated ecosystem disturbance. One way that humans have triggered wildlife declines is by transporting disease-causing agents to remote areas of the world. Amphibians have been hit particularly hard by disease due in part to a globally distributed pathogenic chytrid fungus (Batrachochytrium dendrobatidis [Bd]). Prior research has revealed important insights into the biology and distribution of Bd; however, there are still many outstanding questions in this system. Although we know that there are multiple divergent lineages of Bd that differ in pathogenicity, we know little about how these lineages are distributed around the world and where lineages may be coming into contact. Here, we implement a custom genotyping method for a global set of Bd samples. This method is optimized to amplify and sequence degraded DNA from noninvasive skin swab samples. We describe a divergent lineage of Bd, which we call BdASIA3, that appears to be widespread in Southeast Asia. This lineage co-occurs with the global panzootic lineage (BdGPL) in multiple localities. Additionally, we shed light on the global distribution of BdGPL and highlight the expanded range of another lineage, BdCAPE. Finally, we argue that more monitoring needs to take place where Bd lineages are coming into contact and where we know little about Bd lineage diversity. Monitoring need not use expensive or difficult field techniques but can use archived swab samples to further explore the history—and predict the future impacts—of this devastating pathogen.
Emerging infectious diseases are increasingly recognized as a threat to both human and wildlife health (1–3). One reason emerging infectious diseases are on the rise is the facilitated spread of pathogen propagules via globalized trade. With the aid of modern shipping, pathogens have been introduced to naïve remote areas (4). These new introductions can have grave consequences, in some cases causing mass mortality in wildlife populations (e.g., refs. 4 and 5). Understanding the pathways for disease spread is critical to predicting and addressing disease outbreaks (1).
Amphibians have been hit particularly hard by emerging infectious disease in the last century. Hundreds of amphibian species have been impacted by the pathogenic chytrid fungus Batrachochytrium dendrobatidis [Bd] (6, 7). Bd invades the keratinized tissue of amphibians (skin in adults, mouthparts in tadpoles), disrupting critical skin functions such as the regulation of osmotic pressure and electrolyte balance (8). The resulting disease—chytridiomycosis—can be deadly to susceptible amphibians once the pathogen reaches a critical infection load (9). However, some host species can maintain Bd infection without disease development (10, 11). These tolerant species can be important reservoir vectors of Bd. For example, the American bullfrog (Rana catesbeiana) is a highly traded, Bd-tolerant species that has been implicated in spreading Bd throughout the Western United States, Brazil, and Korea (12–14).
Molecular studies have played an important role in illuminating the evolutionary history of Bd, patterns of spatial and temporal spread, and pathogen genetic diversity (e.g., refs. 15–17). There are currently 4 documented major Bd lineages based on the most recent whole-genome phylogeny (14). First the “global panzootic lineage,” BdGPL, is globally distributed and associated with most mass mortalities in wild amphibian populations (17). Second, BdCAPE was first described from an isolate collected in Cape Province, South Africa, and has since been found in Cameroon, Mallorca, and the United Kingdom (14, 17). Third, BdASIA1 was recently described from 8 isolates collected in South Korea but also includes the previously-named BdCH lineage collected in Switzerland (14). Fourth, BdBrazil/ASIA2 was first described from samples collected in Brazil (18) and renamed to include the clade previously known as BdKorea after whole-genome sequencing revealed their close relationship (14). The observed phylogenetic relationship among the 4 currently described Bd lineages suggests that the earliest diverging lineage is BdASIA1, and the most recent is BdGPL (14).
In addition to the 4 major lineages, some hybridization between lineages has been reported. Schloegel et al. (18) documented the first evidence of sexual recombination in Bd, a finding later supported by whole-genome sequencing (15). Alarmingly, one of the recombinant lineages studied (an F1 hybrid of BdGPL and BdBrazil) was found to be more virulent than either parental lineage when tested against a native Brazilian host (19). Thus, the spread and recombination of different Bd lineages can lead to unpredictable and potentially dangerous outcomes. Therefore, documenting the spatial distribution of Bd genotypes is a high priority for amphibian conservation.
Recent genetic and genomic studies have provided unprecedented insight into the evolutionary history of Bd. However, a comprehensive understanding of historical and contemporary patterns of Bd diversity and spread has been limited by the lack of robust Bd genotype data in many areas of the world. One key limitation in using molecular tools to study Bd has been the need for pure cultures, which yield high-quality and -quantity DNA and have been required for whole-genome sequencing. However, the process of isolating and maintaining live Bd cultures is time-consuming and challenging, particularly in remote areas. In contrast, skin-swab samples are plentiful because they are easy to collect and are part of a standardized protocol to detect the presence of Bd via qPCR (20). Swab samples provide ample DNA for sensitive PCR techniques but often do not have enough high-quality DNA for whole-genome sequencing. Some studies have attempted to address this problem by sequencing small, hypervariable loci present in high copy number—specifically the ribosomal intragenic spacer (ITS-1) region. However, phylogenetic inferences made from this region produce relationships that are highly discordant with those inferred from high-coverage, whole-genome sequencing (14). These challenges have resulted in a large gap in knowledge between our robust understanding of Bd presence and prevalence in many parts of the world and our patchy knowledge of genetic variation and lineage distributions.
Thus, many critical questions remain in the Bd–amphibian system. First, are there additional undiscovered Bd lineages present in wild amphibian populations? Second, what is the current distribution of known Bd lineages in unsampled or undersampled areas of the world? Third, where are divergent Bd lineages coming into contact? Answering these questions would provide a truly global understanding of the threat that Bd poses to amphibians around the world and identify geographic centers of high conservation urgency. Here, we employ a microfluidic PCR genotyping method targeting almost 200 loci across the Bd genome from swab samples (21). With this technique, we can now leverage a global library of amphibian skin swabs that have never been genotyped. Our analysis provides a deeper understanding of the diversity and distribution of Bd globally and highlights cryptic variation in this pathogen around the world.
Results
Global Bd Diversity.
We used our swab genotyping assay to assign 222 samples from 24 different countries to major Bd clades (Dataset S1). The dataset includes 189 field-collected swabs, 18 museum swabs, and 15 pure Bd isolates collected between 1984 and 2017 (Dataset S1 and SI Appendix, Fig. S2). The samples represent all continents where Bd occurs and were chosen to target areas of the world where genotype data are lacking and explore localities where lineages may be coming into contact. We first describe our findings at the global scale, integrating our dataset with 47 previously published Bd whole genomes (14, 15, 17), some of which we resequenced using our method (SI Appendix, Table S1). Fig. 1A shows the most current and complete global survey of Bd lineage distributions. Our global phylogeny (Fig. 1B) recapitulates the structure of a recent whole-genome phylogeny (14), with the addition of a divergent Bd clade found only in Asia that we name BdASIA3. Below, we highlight results from each of 4 regions of the world. The regional results are summarized in Fig. 2, where we show a separate phylogeny for each region of the world.
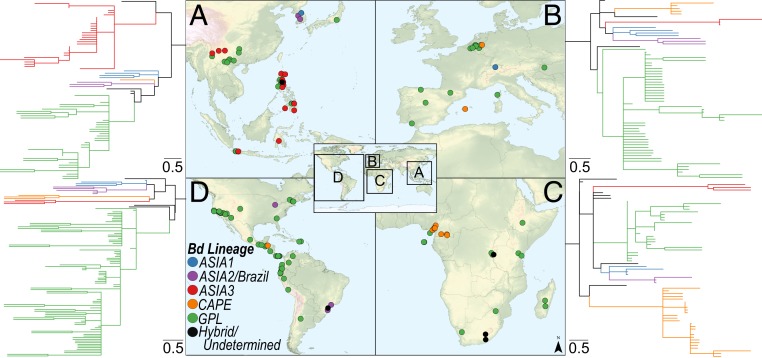
Fig. 1.
(A) Global map of Bd genotypes. Points within 100 m are dispersed to decrease overlap and demonstrate sampling effort; therefore, map point locations are approximate. Colors indicate major Bd lineage, circles are newly genotyped samples (n = 222), and squares are previously published Bd genotype data (n = 334; data from refs. 14–17 and 27). (B) Best scoring unrooted maximum-likelihood tree estimated from 172 concatenated nuclear loci (23,651 bp) and 100 bootstrap replicates performed in RAxML. Branches on phylogeny are colored by major Bd lineage. This tree includes newly sequenced samples with at least 84 loci (n = 131) and whole-genome data (n = 47). Nodes with bootstrap support <50 have been collapsed and nodes >70 bootstrap support are labeled. Phylogeny with tip labels is available in SI Appendix, Fig. S1.
Fig. 2.
Maps and regional phylogenies showing Bd sampling locations and lineages in Asia (n = 78) (A), Europe (n = 66) (B), Africa (n = 66) (C), and the Americas (n = 108) (D). Points and clades are colored as in Fig. 1. Sample sets include representatives of each major clade in addition to all newly genotyped samples collected in that region. Overlapping points on the map are offset by 1° longitude for display purposes. Phylogenies are species tree consensus topologies calculated in Astral (v.5.6.2) from maximum-likelihood gene trees, individually estimated in RAxML for each locus. Full-size versions of the phylogenies with tip and node labels are available in SI Appendix, Figs. S3–S6.
Asia.
Our most significant finding in Asia is a unique and divergent Bd lineage that we name BdASIA3 (Fig. 2A). This lineage is clearly differentiated in the phylogenetic analyses and appears to be widespread in the Philippines, Indonesia, and parts of China. BdASIA3 co-occurs with BdGPL in all 3 countries. In the Philippines, 56% (19/34) of samples harbored the BdASIA3 lineage and 41% (14/34) of samples had the BdGPL lineage. In Java, Indonesia, 62% (8/13) of samples were in the BdASIA3 lineage and 38% (5/13) were BdGPL. In China, 43% (3/7) of samples were BdASIA3 and 57% (4/7) were BdGPL. Thus, this previously undescribed Bd lineage appears to be relatively common in samples collected from various parts of Asia.
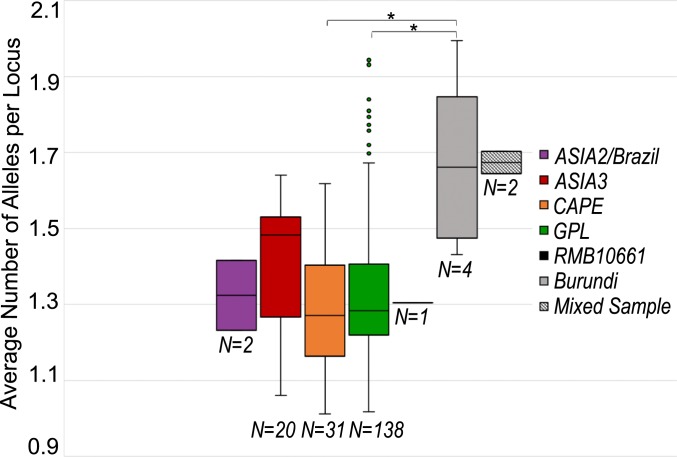
One additional sample from the Philippines had a unique genetic signature and could not be confidently assigned to a known Bd lineage (RMB10661). To assess whether this sample represents a mixed infection or a hybrid between 2 lineages, we plotted the average number of alleles per locus (Fig. 3). RMB10661 has a similar degree of heterozygosity as the average for each of the major lineages, so it does not appear to be a hybrid or mixed sample. In addition, this sample was sister to the BdASIA3 clade in the phylogeny and has unique haplotypes at some loci. Therefore, this sample appears to be distinct from currently named lineages and possibly represents another undescribed, early branching lineage.
Fig. 3.
Average number of alleles for each major Bd lineage and ambiguous samples sequenced via the Fluidigm Access Array method. The mixed sample represents an experimental mixture of BdGPL and BdBrazil/ASIA2 isolates. *A significant difference between the Burundi samples and the BdGPL/BdCAPE lineages (Mann–Whitney U test: P < 0.01).
Europe.
In Europe, we report the presence of 3 major lineages: BdGPL, BdCAPE, and BdASIA1 (Fig. 2B), reinforcing the key finding that multiple divergent Bd lineages are now commonly found at the regional scale. Of our genotyped samples from Europe, 90% (38/42) belong to BdGPL and 10% (4/42) belong to the BdCAPE lineage. The presence of BdASIA1 in Europe was documented in a prior study (18). Remarkably, we found that 4 swabs collected from bullfrogs (R. catesbeiana) in The Netherlands carried the BdCAPE genotype. This expands the known range of BdCAPE in Europe.
Africa.
In Africa, we found that the BdCAPE lineage is ubiquitous in Cameroon, while BdGPL dominates nearby parts of West Africa and previously uncharacterized parts of Central Africa (Fig. 2C). All 25 Bd samples collected from Cameroon are members of the BdCAPE lineage, indicating BdCAPE is the dominant, and perhaps exclusive, Bd lineage in Cameroon. Furthermore, we found additional support for previous studies documenting the presence of BdGPL in Madagascar (22) and provide a report of BdGPL in Burundi and Kenya. In Burundi, 43% (3/7) of samples were in the BdGPL lineage and 57% (4/7) of samples were of an undetermined lineage. To further understand why these ambiguous samples did not group with a major lineage, we plotted the average number of alleles sequenced per locus (Fig. 3). We found that the ambiguous samples from Burundi had a significantly higher average allele per locus than BdCAPE and BdGPL samples (Mann–Whitney U test: P < 0.01). These samples were most similar in average number of alleles per locus to an experimental mixture of 2 divergent Bd isolates and so may be instances of coinfection or hybridization.
Americas.
BdGPL is the dominant lineage in the Americas (excluding Brazil, where both BdGPL and BdASIA2/Brazil are found). However, we report BdCAPE—a lineage that previous studies have found only in Africa and Europe—in Latin America (Fig. 2D). We found that 11% (2/19) of Bd samples collected from Cusuco National Park in Honduras in 2014 were BdCAPE, whereas 89% (17/19) of samples were BdGPL. BdCAPE may be newly introduced (or detected) in the Americas and occurs in very close proximity to BdGPL in Honduras. All other genotyped samples from the Americas were members of the BdGPL clade.
Discussion
Are There Undiscovered Bd Lineages in Wild Populations?
Our discovery of a divergent lineage of Bd endemic to Asia (BdASIA3) supports the hypothesis that Bd originated in Asia and highlights our contention that substantial gaps remain in our understanding of the global genetic diversity in Bd. Recent whole-genome studies have proposed an Asian origin for Bd, citing the genetic signatures of long-term endemism in the BdASIA1 lineage and noting the high lineage diversity in Southeast Asia (14). Interestingly, our global phylogeny (Fig. 1A) shows that BdASIA3 is now the earliest diverging named Bd lineage. In addition, BdASIA3 has the longest interior branch lengths of any described lineage, indicating that it may have persisted in isolation and/or that closely related lineages have not yet been found or have gone extinct. Furthermore, there are additional well-supported nodes within the BdASIA3 clade, indicating some within-clade genetic structure. This phylogenetic pattern is consistent with constant population-size dynamics for this lineage (23) and supports the hypothesis that BdASIA3 is an endemic Southeast Asian lineage. In contrast, the BdGPL clade shows long external branch lengths, indicating periods of exponential growth—a pattern consistent with the documented global spread of this lineage. It is likely that additional Bd lineages remain to be discovered, which may further alter our understanding of Bd’s evolutionary history, including the time and place of its origin.
Another line of evidence suggesting that our current understanding of Bd genetic diversity is incomplete comes from samples that could not be confidently assigned to a known major Bd clade. For example, one sample (RMB10661 collected from the relatively pristine forests of Luzon Island in the Philippines) was collected in an area where both BdGPL and BdASIA3 are present (Fig. 2A) and was phylogenetically estimated to be sister to the BdASIA3 clade (Fig. 1B and SI Appendix, Fig. S3). Our analyses indicate that this sample is not a mixed infection—nor a hybrid—of 2 different Bd lineages. Thus, RMB10661 may represent genetic diversity that is not yet present in our current library of Bd genotypes. In fact, this sample may come from yet another undescribed, early diverging Asian Bd lineage. However, we refrain from naming this lineage given that there is only one representative sample. It is possible that additional cryptic Bd diversity remains undocumented in isolated, unstudied amphibian populations around the world.
What Is the Current Distribution of Bd Lineages in Previously Understudied Parts of the World?
Our study expands the understanding of Bd lineage distributions in many parts of the world where Bd diversity was previously uncharacterized. While we are not the first to report BdCAPE in Cameroon (14), we increased the sample size for Cameroon Bd genotypes (Fig. 2B). The ubiquity of BdCAPE in Cameroon is unique—we do not currently know of any other country occupied only by this lineage. Another study reported the presence of BdGPL and other unidentified lineages in Cameroon (24) but used the ribosomal ITS region to genotype Bd, which is not phylogenetically informative (14). Our findings point to either a long relationship of BdCAPE in Cameroon or a recent complete sweep. A previous study that did not include genotype data reported Bd in Cameroon dating back to 1933 (25). Indeed, BdCAPE may have originated in this area and spread to other parts of Africa, Europe, and now Central America, or it may have recently invaded and spread in Cameroon as well. This highlights an important point: Bd lineages are often named for the areas where they were first discovered (i.e., BdCAPE was first discovered in Cape Province, South Africa; ref. 17), but these names may become misleading as we discover more about the history and distribution of each lineage. Some lineage names have been changed or combined as more sequence data become available (such as the joining of BdKorea and BdBrazil into BdASIA2/Brazil; ref. 14). We raise this point to recognize that lineage names can sometimes introduce biases that may arise from historical attachments to original lineage designations and to suggest that alternative lineage naming schemes (i.e., numeric) may be worth considering in this system.
In East Africa, our data reveal an interesting pattern in the newly sequenced region of Burundi. Here, we also encountered samples we could not assign to a major Bd clade. However, unlike the ambiguous sample from Asia, the unassigned Burundi samples had an average number of alleles per locus that was similar to levels of allelic diversity found in experimental mixtures of 2 divergent Bd isolates (Fig. 3). Thus, these ambiguous samples may be a coinfection (on single hosts) of different Bd lineages, or a possible hybrid—as they lie between the BdCAPE and BdGPL clades in the Africa phylogeny (SI Appendix, Fig. S5). However, they do not appear closely related to previously published BdGPL/BdCAPE hybrids (14). Thus, additional work will be needed to differentiate between coinfection versus hybridization and to test whether these samples represent a separate hybridization event between BdGPL and BdCAPE. Our ongoing work includes sequencing more samples from this region to test these alternative hypotheses (for example, by comparing mitochondrial and nuclear loci and analyzing patterns of linkage disequilibrium between loci).
Where Are Divergent Bd Lineages Coming into Contact?
As more data become available, they reveal that divergent Bd lineages are overlapping across fine spatial scales. Our study documents multiple instances where 2 different lineages coexist at the same time and place (e.g., sampled meters apart). For example, we find both BdCAPE and BdGPL in Honduras. While previous studies have documented Bd in this area and attributed amphibian declines to the pathogen (26), none have reported BdCAPE in the Americas. Our finding of BdCAPE outside of its previously reported range is alarming for a number of reasons. First, we know that Bd is capable of hybridizing across lineages, as has been documented in multiple parts of the world (14, 16, 18). Second, hybrid lineages can sometimes be more virulent than parental lineages (19). Third, although some amphibian species may have developed resistance and/or tolerance to a particular Bd lineage, it remains unclear how they might respond to the introduction of a new lineage or exposure to a hybrid lineage (27). Finally, although some amphibian host communities are beginning to recover from Bd outbreaks (e.g., refs. 27 and 28), many populations are persisting only in small numbers, making them especially vulnerable to new disease outbreaks.
We also found co-occurrence of divergent Bd lineages in parts of Asia. For example, we found BdGPL and BdASIA3 at almost every sampling locality in the Philippines. Our data indicate that these lineages have been coexisting in this region for at least 7 y. The earliest samples (from Mindanao Island in 2005) and more recent samples (from the same island in 2012) had both BdGPL and BdASIA3 present (Dataset S1). Previous studies found Bd to be widespread in the Philippines, but the genotype of these samples was unknown (29). Our findings are consistent with either a slow spread of BdGPL through the Philippines or a longer, more stable coexistence of divergent lineages. In West Java, Indonesia, we found similar evidence of lineage co-occurrence in high montane amphibian communities. However, we do not yet have time-series samples from this area and so cannot make inferences concerning the timing of arrival of different lineages.
In Europe and Asia, we see additional examples of Bd lineages co-occurring at small spatial scales, this time in populations of invasive bullfrogs (R. catesbeiana). In The Netherlands, some samples collected from R. catesbeiana had BdCAPE and others had BdGPL, despite being collected in the same year in close geographic proximity. In the Yunnan province of China, the single R. catesbeiana sampled was infected with BdGPL, while the native species from the same locality carried BdASIA3. These findings support other recent studies suggesting that invasive R. catesbeiana are contributing to the spread of Bd around the world (14, 18). R. catesbeiana are consumed as food by humans globally and are one of the most commonly traded amphibian species. Commercial farms that raise R. catesbeiana may create disease spillover in regions with high amphibian-species richness, including Brazil and Asia (30). Thus, our study provides additional evidence that bullfrog trade should be a major concern as it creates potential pathways for short- and long-distance Bd dispersal (14).
Expanded Threats for Amphibian Conservation.
Our dataset expands our understanding of how Bd lineages are distributed around the world; however, there remain unexplored frontiers in this system. First, there are many parts of the world where we know Bd exists, but it remains unclear which lineages are present. For example, Bd in Asia is widespread but exists at very low prevalence and often at low infection intensities (29). Moreover, one recent study found that the traditional qPCR assay for Bd (20) may not accurately quantify endemic Asian Bd lineages because of variation at the ribosomal RNA ITS primer-binding sites (31). This not only could lead to underreporting the presence of Asian Bd in wild populations but also could generate a sampling bias for studies like ours that select samples for genotyping based on positive qPCR results. If we exclude samples because they ostensibly have too little Bd DNA, it could skew our results in favor of reporting more BdGPL genotypes. Therefore, our current estimates of Bd diversity may still be grossly underestimated, and there may be additional endemic Bd lineages that remain undiscovered in Asia and other parts of the world. Additional Bd genotyping in undersampled areas will be critical for fully understanding the evolutionary relationships between Bd and amphibian hosts.
Second, we have yet to fully explore temporal variation in Bd genotypes to understand the timing of lineage arrival, turnover, and spread. Swabbing museum specimens to record the historic presence/absence of Bd over the last century has produced a rich library of DNA samples for which our genotyping method is ideal (e.g., refs. 32–34). Our current dataset includes 18 successfully genotyped museum swabs collected from around the world (Dataset S1 and SI Appendix, Fig. S2), the oldest from a specimen collected in 1984 in Peru. By genotyping museum swabs, we can test hypotheses for factors driving Bd-related declines. Understanding the dynamics of historical amphibian declines is key for predicting future risk.
Third, our data indicate that Bd lineages are continually spreading and are co-occurring in close proximity. Given that novel Bd lineages and hybrid lineages could be a threat to naïve populations (19), it is increasingly important that we continue to monitor Bd presence, prevalence, genetic diversity, and host health. In addition to monitoring, best practices for limiting Bd spread must be communicated not only to scientists but also to the public traveling to remote areas and commercial farms. Furthermore, steps should be taken to mitigate cross-continental lineage spread such as restrictions on amphibian imports and exports and mandatory testing and treatment protocols. These precautions not only could prevent new Bd outbreaks but also could help curb the spread of many other plant and wildlife diseases.
Conclusions
Our study provides a broader understanding of the cryptic variation in one of the deadliest wildlife pathogens ever documented. We can now better track pathways of disease spread in this system and link specific pathogen lineages to outcomes in wild populations. Our genotyping method, optimized for low-quality DNA samples, can be further implemented across different sample types (e.g., museum specimen swabs, environmental DNA samples) to further understand the ecology and evolution of Bd and to inform management and mitigation strategies. Although Bd has a global distribution, individual lineages that vary in pathogenicity still occur in geographically limited ranges. Thus, as Bd genotypes continue to expand their range, we need to consider broader actions that may be necessary to halt Bd lineage spread and secondary contact that could have grave consequences for amphibian hosts.
Materials and Methods
The full description of methods can be found in SI Appendix, SI Methods. Briefly, we genotyped 222 Bd samples using a custom amplicon sequencing assay (21) targeting 191 regions of the Bd genome. We generated consensus sequences for each sample at each locus. We then integrated our data with previously published whole-genome data and produced both global and regional phylogenies. To create the global phylogeny, we concatenated loci for each sample with <50% missing data and used RAxML (v.8.2.11; ref. 35) to iterate over 100 bootstrap samples. We created the regional phylogenies using a gene-tree to species-tree approach. First, we generated gene trees for each loci using RAxML. Second, we used Astral (v.5.6.2; ref. 36) to estimate an unrooted species tree given the set of input gene trees from each regional sample group. Finally, to estimate heterozygosity in sample groups, we calculated the average number of alleles by summing the number of unique sequence variants for each locus, per sample, and dividing by the number of loci sequenced for that sample.
Supplementary Material
Acknowledgments
We thank Obed Hernández-Gómez and Thomas Jenkinson for assisting with laboratory work and providing comments on the manuscript and Max Lambert and Andrew Rothstein for comments on the manuscript. This work was supported by NSF Grant IOS 1354241 (to E.B.R.), NSF Grant DEB 1557190 (to C.J.B., R.A.K., and E.B.R.), NSF Grant DEB 1551488 (to J.V., C.L.R.-Z., and E.B.R.), and the NSF Graduate Research Fellowship Program (A.Q.B.). Sequencing done at the University of Idaho Institute of Bioinformatics and Evolutionary Studies Genomics Resources Core is supported by NIH Centers of Biomedical Research Excellence Phase III Grant P30GM103324. Sample collection was supported by grants from the NSF (DEB-1202609 [to D.C.B.]; DEB-0743491, 0334952, 1418895, and 1654388 [to R.M.B.]; and Belmont Forum 1633948 [to V.T.V.]), the Czech Science Foundation and Institute of Parasitology (Grants P506/10/2330 and RVO 60077344 [to M.J.]), the National Natural Science Foundation of China (Grant 31702008 [to Z.Y.]), and the Yunnan Applied Basic Research Project (Grant 2018FD047 [to Z.Y.]).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (Bioproject PRJNA555719).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908289116/-/DCSupplemental.
References
- 1.Jones K. E., et al. , Global trends in emerging infectious diseases. Nature 451, 990–993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daszak P., Cunningham A. A., Hyatt A. D., Emerging infectious diseases of wildlife–Threats to biodiversity and human health. Science 287, 443–449 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Fisher M. C., Garner T. W. J., The relationship between the emergence of Batrachochytrium dendrobatidis, the international trade in amphibians and introduced amphibian species. Fungal Biol. Rev. 21, 2–9 (2007). [Google Scholar]
- 4.Foley J., Clifford D., Castle K., Cryan P., Ostfeld R. S., Investigating and managing the rapid emergence of white-nose syndrome, a novel, fatal, infectious disease of hibernating bats. Conserv. Biol. 25, 223–231 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Skerratt L. F., et al. , Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth 4, 125–134 (2007). [Google Scholar]
- 6.Wake D. B., Vredenburg V. T., Colloquium paper: Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl. Acad. Sci. U.S.A. 105 (suppl. 1), 11466–11473 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Longcore J. E., Pessier A. P., Nichols D. K., Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to Amphibians. Mycologia 91, 219–227 (1999). [Google Scholar]
- 8.Voyles J., et al. , Pathogenesis of chytridiomycosis, a cause of catastrophic amphibian declines. Science 326, 582–585 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Vredenburg V. T., Knapp R. A., Tunstall T. S., Briggs C. J., Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc. Natl. Acad. Sci. U.S.A. 107, 9689–9694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs C. J., Knapp R. A., Vredenburg V. T., Enzootic and epizootic dynamics of the chytrid fungal pathogen of amphibians. Proc. Natl. Acad. Sci. U.S.A. 107, 9695–9700 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeder N. M. M., Pessier A. P., Vredenburg V. T., A reservoir species for the emerging amphibian pathogen Batrachochytrium dendrobatidis thrives in a landscape decimated by disease. PLoS One 7, e33567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap T. A., Koo M. S., Ambrose R. F., Vredenburg V. T., Introduced bullfrog facilitates pathogen invasion in the western United States. PLoS One 13, e0188384 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schloegel L. M., et al. , The North American bullfrog as a reservoir for the spread of Batrachochytrium dendrobatidis in Brazil. Anim. Conserv. 13, 53–61 (2010). [Google Scholar]
- 14.O’Hanlon S. J., et al. , Recent Asian origin of chytrid fungi causing global amphibian declines. Science 360, 621–627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenblum E. B., et al. , Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc. Natl. Acad. Sci. U.S.A. 110, 9385–9390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkinson T. S., et al. , Amphibian-killing chytrid in Brazil comprises both locally endemic and globally expanding populations. Mol. Ecol. 25, 2978–2996 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Farrer R. A., et al. , Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc. Natl. Acad. Sci. U.S.A. 108, 18732–18736 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schloegel L. M., et al. , Novel, panzootic and hybrid genotypes of amphibian chytridiomycosis associated with the bullfrog trade. Mol. Ecol. 21, 5162–5177 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Greenspan S. E., et al. , Hybrids of amphibian chytrid show high virulence in native hosts. Sci. Rep. 8, 9600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle D. G., Boyle D. B., Olsen V., Morgan J. A., Hyatt A. D., Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis. Aquat. Organ. 60, 141–148 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Byrne A. Q., et al. , Unlocking the story in the swab: A new genotyping assay for the amphibian chytrid fungus Batrachochytrium dendrobatidis. Mol. Ecol. Resour. 17, 1283–1292 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Bletz M. C., et al. , Widespread presence of the pathogenic fungus Batrachochytrium dendrobatidis in wild amphibian communities in Madagascar. Sci. Rep. 5, 8633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grenfell B. T., et al. , Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303, 327–332 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Miller C. A., et al. , Distribution modeling and lineage diversity of the chytrid fungus Batrachochytrium dendrobatidis (Bd) in a central African amphibian hotspot. PLoS One 13, e0199288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soto‐Azat C., Clarke B. T., Poynton J. C., Cunningham A. A., Widespread historical presence of Batrachochytrium dendrobatidis in African pipid frogs. Divers. Distrib. 16, 126–131 (2010). [Google Scholar]
- 26.Kolby J. E., Padgett-Flohr G. E., Field R., Amphibian chytrid fungus Batrachochytrium dendrobatidis in Cusuco National Park, Honduras. Dis. Aquat. Organ. 92, 245–251 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Voyles J., et al. , Shifts in disease dynamics in a tropical amphibian assemblage are not due to pathogen attenuation. Science 359, 1517–1519 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Knapp R. A., et al. , Large-scale recovery of an endangered amphibian despite ongoing exposure to multiple stressors. Proc. Natl. Acad. Sci. U.S.A. 113, 11889–11894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swei A., et al. , Is chytridiomycosis an emerging infectious disease in Asia? PLoS One 6, e23179 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schloegel L. M., et al. , Magnitude of the US trade in amphibians and presence of Batrachochytrium dendrobatidis and ranavirus infection in imported North American bullfrogs (Rana catesbeiana). Biol. Conserv. 142, 1420–1426 (2009). [Google Scholar]
- 31.Mutnale M. C., et al. , Enzootic frog pathogen Batrachochytrium dendrobatidis in Asian tropics reveals high ITS haplotype diversity and low prevalence. Sci. Rep. 8, 10125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng T. L., Rovito S. M., Wake D. B., Vredenburg V. T., Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc. Natl. Acad. Sci. U.S.A. 108, 9502–9507 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez D., Becker C. G., Pupin N. C., Haddad C. F. B., Zamudio K. R., Long-term endemism of two highly divergent lineages of the amphibian-killing fungus in the Atlantic Forest of Brazil. Mol. Ecol. 23, 774–787 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Burrowes P. A., De la Riva I., Unraveling the historical prevalence of the invasive chytrid fungus in the Bolivian Andes: Implications in recent amphibian declines. Biol. Invasions 19, 1781–1794 (2017). [Google Scholar]
- 35.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C., Rabiee M., Sayyari E., Mirarab S., ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19 (suppl. 6), 153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.