Significance
The unicellular apicomplexan parasite, Plasmodium falciparum, causes malaria in humans that results in >400,000 deaths per year. The parasite must produce force and motion to progress through its lifecycle and cause disease. The core of the macromolecular complex that powers invasion and motile stages consists of a unique single-headed myosin and a divergent actin (PfAct1). We used total internal reflection fluorescence microscopy to visualize polymerization of individual PfAct1 filaments in real time in the absence of stabilizing agents. The unique dynamic properties of both ends of the polar filament, and the high actin concentration needed for it to polymerize, provide a molecular explanation for why these filaments have been notoriously hard to visualize both in vitro and in the parasite.
Keywords: Plasmodium, malaria, actin polymerization, treadmilling, Toxoplasma
Abstract
Gliding motility and host cell invasion by the apicomplexan parasite Plasmodium falciparum (Pf), the causative agent of malaria, is powered by a macromolecular complex called the glideosome that lies between the parasite plasma membrane and the inner membrane complex. The glideosome core consists of a single-headed class XIV myosin PfMyoA and a divergent actin PfAct1. Here we use total internal reflection fluorescence microscopy to visualize growth of individual unstabilized PfAct1 filaments as a function of time, an approach not previously used with this actin isoform. Although PfAct1 was thought to be incapable of forming long filaments, filaments grew as long as 30 µm. Polymerization occurs via a nucleation–elongation mechanism, but with an ∼4 µM critical concentration, an order-of-magnitude higher than for skeletal actin. Protomers disassembled from both the barbed and pointed ends of the actin filament with similar fast kinetics of 10 to 15 subunits/s. Rapid treadmilling, where the barbed end of the filament grows and the pointed end shrinks while maintaining an approximately constant filament length, was visualized near the critical concentration. Once ATP has been hydrolyzed to ADP, the filament becomes very unstable, resulting in total dissolution in <40 min. Dynamics at the filament ends are suppressed in the presence of inorganic phosphate or more efficiently by BeFX. A chimeric PfAct1 with a mammalian actin D-loop forms a more stable filament. These unusual dynamic properties distinguish PfAct1 from more canonical actins, and likely contribute to the difficultly in visualizing PfAct1 filaments in the parasite.
The unicellular apicomplexan parasite that causes malaria in humans, Plasmodium falciparum (Pf), results in more than 400,000 deaths per year (1). This obligate intracellular parasite has a complex life cycle that involves both human and mosquito hosts, with motile and nonmotile stages in each. Once an infected Anopheles mosquito bites a human, the highly motile (∼2 µm/s) sporozoites from the mosquito salivary glands progress to dermal capillaries in the human, where they are transported to the liver to invade hepatocytes. The parasite replicates in the hepatocyte and ultimately releases many thousands of nonmotile merozoites that invade the red blood cell in a force-requiring process, where they develop and cause the symptoms of malaria. Motion and force production by the parasite are thus necessary elements for disease progression. The malaria parasite moves via a substrate-dependent form of gliding motility that does not involve cell shape changes. The core of the macromolecular complex responsible for force and motion production, called the glideosome, is composed of PfMyoA, a class XIV single-headed tailless myosin with 2 associated light chains, ELC and MTIP (myosin tail interacting protein) and a divergent actin, P. falciparum actin 1 isoform (PfAct1) (reviewed in ref. 2) (Fig. 1A). PfMyoA generates force by an atypical mechanism as a result of sequence adaptations in this myosin, and phosphorylation of an N-terminal extension of the heavy chain regulates force output (3). The motor is anchored to the inner membrane complex by an interaction between the N-terminal extension of the MTIP light chain and glideosome-associated proteins (GAP40/GAP45/GAP50). The actin filaments are anchored to the parasite plasma membrane via transmembrane adhesins that bind receptors on the host cell plasma membrane. Despite numerous lines of evidence supporting the belief that the motility of Plasmodium spp. and its ability to invade host cells relies on an actomyosin system (reviewed in ref. 2), it has been notoriously hard to directly visualize either the myosin motor or actin filaments in the parasite, even using cryoelectron microscopy (cryo-EM) techniques (4). More recently, conditional knockouts established that both PfMyoA (3) and PfAct1 (5) are critical for red blood cell invasion.
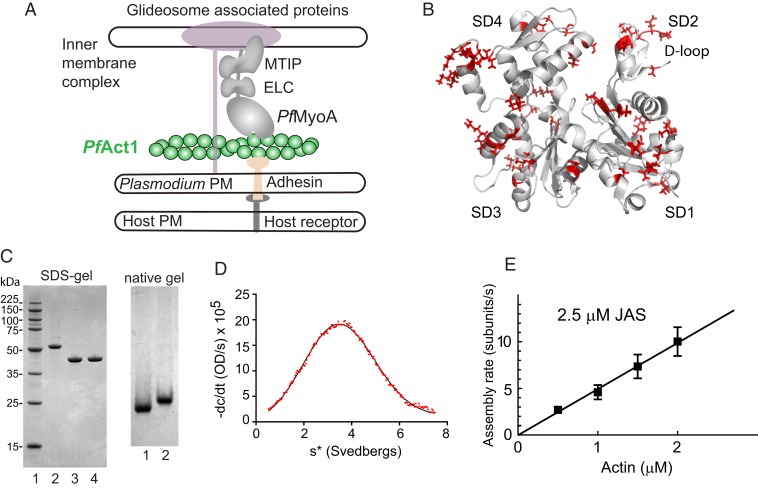
Fig. 1.
Schematic of PfAct1 in the glideosome, the crystal structure of PfAct1, characterization of expressed PfAct1, and polymerization in the presence of JAS. (A) Schematic of a model of the Plasmodium spp. glideosome (2), the macromolecular complex responsible for gliding motility. The core consists of PfMyoA, a single-headed class XIV myosin with 2 light chains (ELC and MTIP), and a divergent actin, PfAct1. The N terminus of MTIP anchors the myosin motor to the inner membrane complex via glideosome-associated proteins. PfAct1 filaments are anchored to the Plasmodium plasma membrane (PM) via adhesins that bind host receptors from the host plasma membrane. (B) Cartoon of PfAct1 crystal structure (PDB ID code 4CBU), with the 65 residues differing from those found in human β-cytoplasmic actin illustrated with red side chains. The location of the D-loop in subdomain 2 is indicated. SD, subdomain. (C, Left) SDS-gel of (lane 1) molecular mass markers; (lane 2) PfAct1-thymosin following HIS-column; (lane 3) PfAct1-thymosin following chymotryptic cleavage; (lane 4) purified PfAct1 after ion-exchange chromatography. (Right) Native gels of (lane 1), purified PfAct1; (lane 2) skeletal muscle actin. (D) Homogeneity of purified PfAct1 demonstrated by analytical ultracentrifugation. PfAct1 sedimented at 3.2 ± 0.01S (0.2 M ammonium acetate, 5 mM Tris, 0.2 mM Na2ATP, 0.2 mM CaCl2, 0.5 mM DTT, pH 7.5, 20 °C, 0.9 mg/mL PfAct1). s*, sedimentation coefficient. (E) Polymerization rate as a function of actin concentration in the presence of 2.5 µM JAS. See Movie S1 and Table 1 for polymerization rate constants.
In addition to PfAct1, which is ubiquitously expressed in all Plasmodium life cycles, a second isoform, called PfAct2, is present only in gametocytes and mosquito stages. Although actin is one of the most highly conserved proteins, PfAct1 shares only 81% sequence identity with human skeletal actin, and PfAct2 even less (76%). The 2 Plasmodium isoforms are also only 80% identical with each other, the lowest sequence identity seen between actins in 1 species (6). The overall fold of the crystal structures of the 2 Plasmodium actins not surprisingly closely resembles that of canonical actins (Fig. 1B), but sequence differences in the D-loop in subdomain 2 were shown to influence filament formation (7).
Here we focus on PfAct1 because of its importance for host cell invasion. In the absence of stabilizing agents, no long filaments of PfAct1 have been visualized to date in in vitro studies. Only short filaments <200 nm were observed, whether the actin was purified from merozoites (8, 9), expressed and purified from yeast (10), or expressed and purified from insect cells (7). These observations led to the idea that filaments formed from PfAct1 are short and highly dynamic. In Plasmodium, a superresolution microscopy study revealed a structural F-actin cytoskeleton in the nonmotile gametocytes that was mainly composed of the PfAct1 isoform and a colocalizing formin (11). Very recently, actin-binding chromobodies were used to visualize in vivo filamentous actin dynamics in Plasmodium for the first time (12). Our study used the same approach to visualize PfAct1 filament growth in vitro.
Addition of the cyclic peptide jasplakinolide (JAS) promoted polymerization of long PfAct1 filaments (7, 9, 13). Atomic force microscopy (9) and a low-resolution cryo-EM study (7) both showed that the JAS-stabilized PfAct1 filament has an ∼10% longer pitch than skeletal actin filaments (∼41 vs. 38 nm, respectively) due to an ∼1° change in the rotational angle between subunits. A more recent near-atomic cryo-EM reconstruction (∼3.8 Å average resolution) of JAS-stabilized PfAct1 filaments confirmed these differences and furthermore showed a few minor but important differences at inter- and intrastrand contacts that were inferred to account for the instability of the PfAct1 filament (13). Merozoites treated with JAS failed to invade erythrocytes (14), implying that the ability of the filament to not only assemble but to disassemble is important for its biological function.
Actin filaments are polar, with a fast-growing barbed end and a slower-growing pointed end. Two differing theories have been proposed for how apicomplexan actin polymerizes. One study using the sole actin ortholog from the apicomplexan Toxoplasma gondii (TgAct1), which is 93% identical to PfAct1, concluded that TgAct1 assembles via a unique isodesmic polymerization mechanism in which all steps occur with the same rate constant, resulting in no lag for nucleation and no critical concentration, meaning that there is no actin concentration below which assembly does not occur (15). A subsequent paper disagreed with this finding, and concluded that PfAct1 polymerization does follow the canonical nucleation–elongation mechanism, and has a critical concentration in the range of ∼0.1 µM (16), similar to that observed for skeletal actin (17). Both studies used indirect techniques to follow polymerization (i.e., centrifugation, light scattering, pyrene fluorescence).
Here we use total internal reflection fluorescence (TIRF) microscopy to quantify growth of individual PfAct1 filaments as a function of time, using actin–chromobody Emerald to visualize the filaments. We show that PfAct1 actin is capable of forming transient long (∼30 µm) and dynamic filaments in the absence of the stabilizing agent JAS. We show that assembly follows a canonical nucleation–elongation mechanism, but with a critical concentration of ∼4 µM, a value that is an order-of-magnitude higher than observed for skeletal actin (reviewed in ref. 18). Moreover, both the barbed and pointed ends of the PfAct1 filament exhibit fast depolymerization rates (∼10 to 15 s−1). Near the critical concentration, rapid actin filament treadmilling was visualized. Our data support the previously proposed idea that dynamic actin filaments are essential for gliding motility (19).
Results
Expressed PfAct1 Polymerizes with a Low Critical Concentration in the Presence of JAS.
PfAct1 was expressed as an actin–β-thymosin–HIS tag fusion construct using the baculovirus/Sf9 insect cell-expression system (20). Following purification on a nickel-affinity column, the C-terminal β-thymosin/HIS-tag was removed by chymotryptic cleavage after the last amino acid of actin (Phe375), leaving no nonnative amino acids in the final product (Fig. 1C). The purified actin migrated as a single band on native gels (Fig. 1C), and showed a single homogenous peak that migrated at 3.2S by sedimentation velocity in the analytical ultracentrifuge (Fig. 1D), both indicating that expressed PfAct1 forms a homogeneous monomer.
Polymerization was followed in real time by TIRF microscopy using actin–chromobody Emerald to visualize the growing PfAct1 filaments. This approach has the advantage of requiring no modification of the actin, which has the potential to alter actin function (21). Actin–chromobodies as in vivo F-actin sensors in apicomplexan parasites have been recently validated in T. gondii (22) and in Plasmodium (12). We also show here that the rate constants for skeletal muscle actin polymerization in the presence of actin–chromobody agree with literature values obtained by visualizing actin labeled at Cys-374 with the fluorescent dye Oregon green (17) (Table 1 and SI Appendix, Fig. S1).
Table 1.
PfAct1 polymerization rate constants compared with smooth and skeletal muscle actin
| Actin | Assembly rate constant (subunits/µM·s) | Disassembly rate constant (subunits/s) | Critical concentration (µM) |
| PfAct1 + JAS* | 4.9 | 0.03 | 0.005 |
| PfAct1† | 3.8 ± 1.0 | 14.8 ± 0.4 | 4.1 ± 1.0 |
| PfAct1 + BeFx‡ | 2.6 ± 0.7 | 11.2 ± 3.7 | 4.2 ± 0.3 |
| PfAct1-ADP§ | 0.18 | 9.8 | 55 |
| PfAct1 (human D-loop)¶ | 9.2 ± 3.1 | 7.4 ± 0.5 | 0.86 ± 0.33 |
| Smooth muscle actin (ACTA2)# | 15.9 ± 3.4 | 0.7 ± 0.6 | 0.05 ± 0.04 |
| Skeletal muscle actin (ACTA1)‖ | 7.4 ± 0.5 | 0.8 ± 0.8 | 0.13 ± 0.17 |
| Skeletal muscle actin (ACTA1) visualized with actin-chromobody** | 8.2 | 2.1 | 0.25 |
Rate constants were obtained from the plot of assembly rate versus actin concentration, with the slope defining the assembly rate constant, the y-intercept the disassembly rate constant, and the x-intercept the critical concentration. Error bars ± SD. Rates of PfAct1 assembly were obtained during the first 6 min of polymerization for actin-ATP, or the first 2 to 3 min for actin-ADP. Filaments that grew at the barbed end and showed no shrinkage at the pointed end were analyzed. PfAct1 polymerization buffer: 10 mM imidazole, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 2.5 mM MgATP,10 mM DTT, 0.25% methylcellulose, 0.13 mg/mL glucose oxidase, 50 μg/mL catalase, and 3 mg/mL glucose. Data with PfACt1 were obtained at 37 °C; ACTA1 data are at 25 °C.
Data from 1 experiment with 1 PfAct1 preparation.
Data from 4 experiments with 3 PfAct1 preparations.
Data from 2 experiments with 2 PfAct1 preparations.
Bound nucleotide in PfAct1 was converted to ADP prior to polymerization (Materials and Methods). Fit to data from 2 independent experiments combined, 1 PfAct1 preparation.
Data from 2 experiments with 1 PfAct1 preparation.
Data from ref. 23.
Data from ref. 17, 25 °C.
Data obtained here using actin-chromobody, at 25 °C for comparison with published values. 1 experiment, 1 skeletal actin preparation (SI Appendix, Fig. S1).
We first investigated polymerization in the presence of the stabilizing compound JAS, which was used to obtain a near atomic structure of the PfAct1 filament (13). The rate of polymerization as a function of actin concentration in the presence of 2.5 µM JAS defined an assembly rate constant (slope) of 4.9 subunits/µM·s, a disassembly rate constant (y-intercept) of 0.03 subunits/s, and a critical concentration (x-intercept) of 5 nM (Fig. 1E, Table 1, and Movie S1). The low critical concentration is consistent with JAS-enhancing filament stability via hydrophobic interactions (13). These results verify that our expressed, purified PfAct1 is polymerization competent. Rate constants for PfAct1 assembly in the presence of JAS versus those for unstabilized expressed smooth muscle α-actin (23) or tissue-purified skeletal muscle actin (17) are shown in Table 1. Common features are low critical concentrations and slow disassembly rate constants.
Unstabilized PfAct1 Shows Unusual Dynamic Polymerization Behavior.
In the absence of JAS, no polymerization was observed at the same actin concentrations (<2 µM) used when filaments formed in the presence of JAS (Movie S2). When the monomeric PfAct1 concentration was increased to ∼7 µM, however, robust polymerization was observed, with filament lengths reaching several tens of microns (Fig. 2A and Movie S3). This result shows that PfAct1 can form filaments in the absence of stabilizing agents that are as long as those polymerized from mammalian actin.
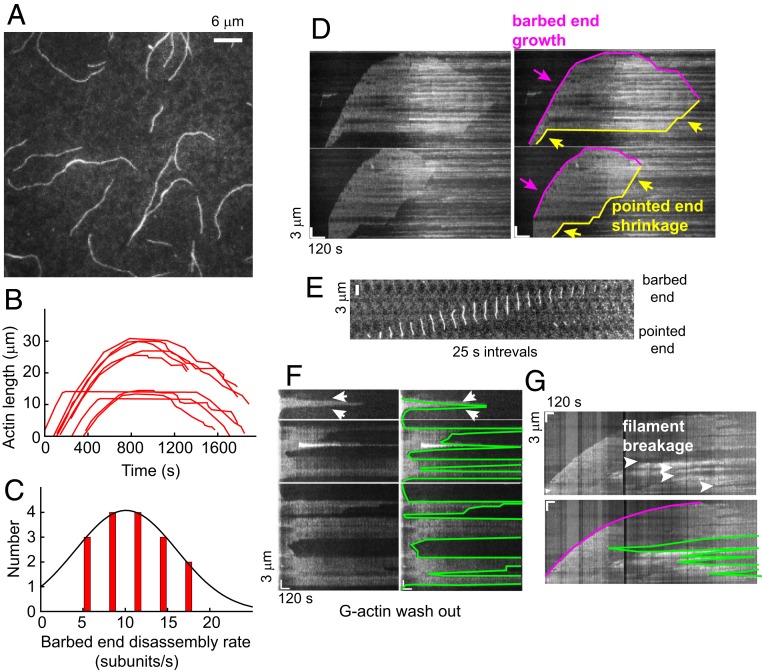
Fig. 2.
Dynamics of unstabilized PfAct1 filaments. (A) The 8 µM PfAct1 forms >10-µm-long filaments after 10 min in the absence of JAS. Filaments were visualized by TIRF microscopy using actin–chromobody Emerald (Movies S2–S4). (B) Time course of growth and shrinkage of PfAct1 barbed ends in 9 individual actin filaments. (C) Histogram of PfAct1 barbed-end disassembly rate during the depolymerization stage. The solid curve is a Gaussian fit to the data (11 ± 4 subunits/s). (D) Raw kymographs (Left), along with a skeletonized version of each (Right), illustrating growth of the barbed end, followed by a period of relatively constant length and then shrinkage at longer time (magenta). The pointed end (yellow) showed periods of shortening (sloped lines) interspersed with pauses (horizontal lines) where the length did not change. (E) Time-lapse (25-s intervals) of a filament in which pointed end depolymerization caught up with the growing barbed end resulting in filament dissolution. (F) Raw kymographs (Left), along with a skeletonized version of each (Right), showing actin filament breakage and depolymerization, sometimes from both ends (arrows), following removal of free G-actin monomers from the solution at the start of the kymograph (Left). (G) Raw kymograph (Upper) and a skeletonized version (Lower, magenta tracks the barbed end) illustrating multiple cleavage events along a filament and shrinkage of the cleaved filaments from both ends with time. Polymerization buffer: 10 mM imidazole, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 2.5 mM MgATP,10 mM DTT, 0.25% methylcellulose, 0.13 mg/mL glucose oxidase, 50 μg/mL catalase, and 3 mg/mL glucose.
The PfAct1 filament showed complex growth behavior compared with that of mammalian actin, which grows and then stays at a constant length. The barbed end of the PfAct1 actin filament initially grew linearly, but then growth slowed and finally stalled. With time, the barbed end started to shrink, as illustrated by the length profiles of 9 representative individual filaments as a function of time (Fig. 2B). The rate of shrinkage was not always constant, with 1 or more pauses occasionally observed. This polymerization profile is distinct from the classic growth phase followed by steady-state behavior observed for canonical actins. The average rate of barbed-end disassembly was 11 ± 4 subunits/s, which likely reflects dissociation of actin in various nucleotide states from the filament, counterbalanced by the association of decreasing free PfAct1-ATP in solution (Fig. 2C).
The behavior at the pointed end was also unusual. Some pointed ends started to shrink as soon as the filaments formed, while others remained intact for many minutes before they started to depolymerize (Fig. 2D, yellow arrows indicate pointed-end shrinkage). Depolymerization from the pointed end occasionally caught up with the growing barbed end before the barbed end began to shrink (Fig. 2E). The combination of disassembly from both ends resulted in the total disintegration of the filaments after ∼30 min. At that point, the flow cell surface was covered with fluorescent dots, which are probably small actin filament fragments. At our resolution, the pointed end does not seem to grow over tens of minutes even at the highest concentration of G-actin we tested (∼13 µM), suggesting that polymerization at the pointed end has a very high critical concentration.
When the flow cell contents were exchanged with buffer devoid of PfAct1 after 10 min of polymerization, the filaments started to depolymerize within seconds. Filaments often shrank from both ends (Fig. 2F), suggesting that both the barbed and pointed ends participate. The rate of disassembly was variable, ranging from 1 to 12 subunits/s (average ∼7 subunits/s), probably reflecting different nucleotide states of the actin protomers at the filaments ends. Some filaments broke in the middle following the G-actin washout, which could also be observed at late stages of a normal polymerization reaction (>30 min) (Fig. 2G), suggesting that they may be caused by the same process. These observations highlight the fragility of the PfAct1 filament.
PfAct1 Filament Instability Is Due to a Very Fast Barbed-End Dissociation Rate.
Growth at the PfAct1 barbed end was approximately linear during the initial phase (∼6 min) of polymerization (Fig. 2B), which allowed us to quantify the rate of subunit addition as a function of increasing PfAct1 concentration (Fig. 3A). Only filaments without shrinking pointed ends were analyzed. The relationship is a standard second-order reaction similar to that observed for skeletal muscle actin (17, 24), suggesting that PfAct1 shares the same general mechanism of nucleation and elongation as canonical actins. The average assembly rate constant (slope) is 3.8 ± 1.0 subunits/µM·s, the disassembly rate constant (y-intercept) is 14.8 ± 0.4 subunits/s, and the critical concentration (x-intercept) is 4.1 ± 1.0 µM (Table 1). This disassociation rate is the fastest reported for a native actin, underlying the extremely dynamic behavior of PfAct1 filaments compared with canonical actins. Compared with values obtained for PfAct1 in the presence of JAS, the assembly rate constant for unstabilized filaments was almost unchanged, but the disassembly rate increased by 2 orders-of-magnitude, suggesting that JAS stabilizes the filament by suppressing the fast dissociation of protomers from filament ends.
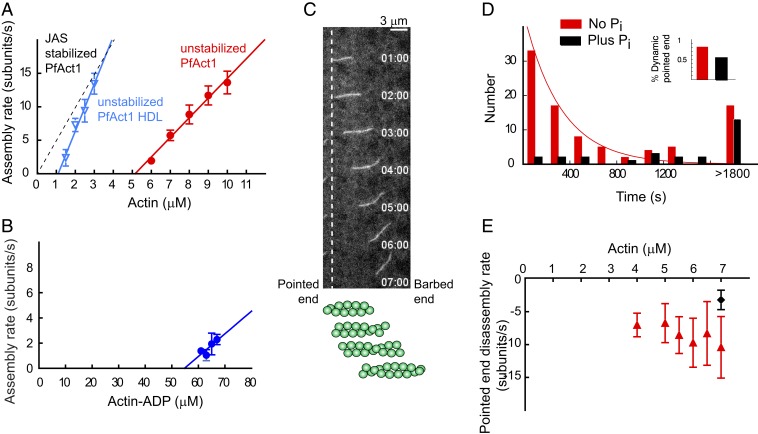
Fig. 3.
Assembly parameters of unstabilized PfAct1 filaments and the stabilizing effect of inorganic phosphate on pointed-end dynamics. (A) Polymerization rate at the barbed end as a function of actin concentration (red circles). Note the extremely high critical concentration of 5.0 µM actin (x-intercept), with an assembly rate constant of 3.0 subunits/µM·s (slope) and a disassembly rate constant of 15.0 subunits/s (y-intercept). The distinctly different polymerization curve in the presence of JAS (dashed black line) is shown for comparison. Compared with PfAct1, a mutant containing the D-loop from human actin (PfAct1-HDL; blue inverted triangles) decreased the critical concentration to 1.1 µM, with an assembly rate constant of 7.7 subunits/µM·s and a disassembly rate constant of 7.8 subunits/s. See Table 1 for average assembly and disassembly rate constants and critical concentration from multiple experiments. (B) Polymerization rate of actin-ADP at the barbed end as a function of actin-ADP concentration. See Materials and Methods for preparation of actin-ADP. The critical concentration (x-intercept) of ∼55 µM is 10-fold higher than for actin-ATP. See Table 1 for polymerization rate constants. (C) A treadmilling filament of PfAct1 near the critical concentration. Polymerization occurs at the barbed end simultaneously with depolymerization at the pointed end (vertical dashed line), without much change in overall length. The schematic below the data illustrates the flux of protomers. See also Movies S3 and S4. (D) The distribution of starting times at which the pointed end begins to depolymerize, fitted to an exponential with a decay constant of 0.003 s−1 (red bars). The last bin consists of filaments that remain intact after 30 min. Data from 5.5 to 7 µM G-actin were pooled; SI Appendix, Fig. S3 shows data at each actin concentration. Addition of 12.5 mM Pi (black bars) suppresses pointed-end depolymerization. Data from 2 experiments with 2 PfAct1 preparations (Movie S5). Inset shows that the percent of dynamic ends decreased from 81 to 53% in the presence of phosphate. (E) The pointed-end depolymerization rate is independent of actin concentration (7 to 10 s−1), and is reduced to approximately half (3.2 ± 1.5 s−1) in the presence of 12.5 mM Pi (black diamond) by favoring formation of bound ADP.Pi. Polymerization buffer: 10 mM imidazole, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 2.5 mM MgATP,10 mM DTT, 0.25% methylcellulose, 0.13 mg/mL glucose oxidase, 50 μg/mL catalase, and 3 mg/mL glucose.
The D-Loop Is Responsible for Much of the Instability.
Because the D-loop in subdomain 2 of actin is involved in contacts important for polymerization, we determined the polymerization properties of PfAct1 with a chimeric actin that contained the D-loop sequence found in all isoforms of canonical human actins (PfAct1-HDL; human D-loop). TIRF polymerization assays showed that the PfAct1-HDL chimera had an ∼5-fold lower critical concentration than PfAct1, suggesting that this region of the molecule influences the overall stability of the PfAct1 filament, although not to the extent seen in the presence of JAS (Fig. 3A). The decreased critical concentration is due to both a faster assembly rate and a slower disassembly rate than wild-type PfAct1 (Table 1).
Polymerization of PfAct1-ADP.
With the expectation that the critical concentration for barbed-end growth of PfAct1-ADP would be at least 10-fold higher than that of PfAct1-ATP, we carried out experiments with ∼60 µM PfAct1-ADP. Only short filaments, less than 3-µm long, slowly formed (SI Appendix, Fig. S2). Fits to the plot of assembly rate (initial 2 to 3 min) versus actin-ADP concentration gave an assembly rate constant of ∼0.18 subunits/µM·s, a disassembly rate constant of ∼9.8 subunits/s, and a critical concentration of ∼55 µM for the barbed end (Fig. 3B and Table 1). The fitting was based on limited data due to the difficulty of achieving actin concentrations >70 µM. Pointed-end growth was not detected, nor was fast depolymerization from the pointed end observed. The growth of PfAct1-ADP filaments stalled and started to shrink from the barbed end after 4 to 5 min, suggesting that disassembled protomers may be in a conformational state that is not compatible with rapid repolymerization.
Pointed-End Dynamics.
At actin concentrations near the critical concentration, rapid (∼10 subunits/s) actin filament treadmilling was observed: That is, simultaneous actin growth from the barbed end and depolymerization from the pointed end, with little change in net length with time. By TIRF microscopy, the images look like the actin filament is moving as a function of time, similar to what is observed in in vitro motility assays when actin movement is driven by a myosin motor (Fig. 3C and Movies S3 and S4).
Under closer scrutiny, depolymerization from the pointed end was not uniform for all filaments; some filaments started to shrink from the pointed end as soon as the experiment started, while the pointed end of other filaments remained intact even after thousands of seconds. To understand the mechanism of pointed end depolymerization, the number of filaments whose pointed ends started to shorten was plotted as a function of time. The results showed 2 distinct populations. The time distribution of 1 population was exponentially distributed, with a decay constant of 0.003 s−1 (half-life of 290 s) (Fig. 3D, red bars, and SI Appendix, Fig. S3), similar to the 350-s half-life directly measured for the rate of γ-phosphate dissociation from mammalian actin (25). The second population of filaments, ∼19% of the total, had pointed ends that remained intact even after >1,800 s.
To further test that pointed-end dynamics are related to ATP hydrolysis and subsequent release of Pi from the actin protomers in the filaments, the same experiment was carried out in the presence of 12.5 mM Pi (Movie S5). Phosphate decreased the percent of the total population with dynamic pointed ends from 81 to 53% (Fig. 3 D, Inset), and delayed the distribution of pointed end shortening starting times (Fig. 3D, black bars), by favoring bound ADP.Pi. The percentage of stable filaments that do not shrink from the pointed end increased with G-actin concentration (SI Appendix, Fig. S3), consistent with addition of G-actin–ATP to the pointed end suppressing pointed-end dynamics. Dynamic pointed ends thus have ADP-protomers, while stable pointed ends are capped with ADP.Pi/ATP protomers.
The rate of disassembly from the pointed end in the absence of phosphate was plotted as a function of G-actin concentration (Fig. 3E). The measured depolymerization rate from the pointed end was very fast (7 to 10 s−1), and essentially constant over 4 to 7 µM actin, as expected because this process does not involve monomer addition. That the rapidly dissociating species from the pointed end is actin-ADP was further supported by the slowing of this rate to 3.2 ± 1.5 s−1 in the presence of 12.5 mM phosphate (Fig. 3E, black symbol).
Beryllium Fluoride Inhibits Pointed-End Depolymerization.
We postulated that more efficiently locking the protomer in the PfAct1 filaments in an ADP.Pi-like state may further slow pointed-end depolymerization. Polymerization was carried out in the presence of 2.5 mM beryllium fluoride (BeFx), which binds to ADP with much higher affinity than Pi (26). Addition of BeFx completely inhibited pointed-end disassembly (Fig. 4A), with all pointed ends staying intact more than 30 min, compared with the 53% dynamic pointed ends remaining in the presence of 12.5 mM Pi (Fig. 3 D, Inset). This observation supports our hypothesis that ADP-protomers are responsible for the fast pointed-end depolymerization.
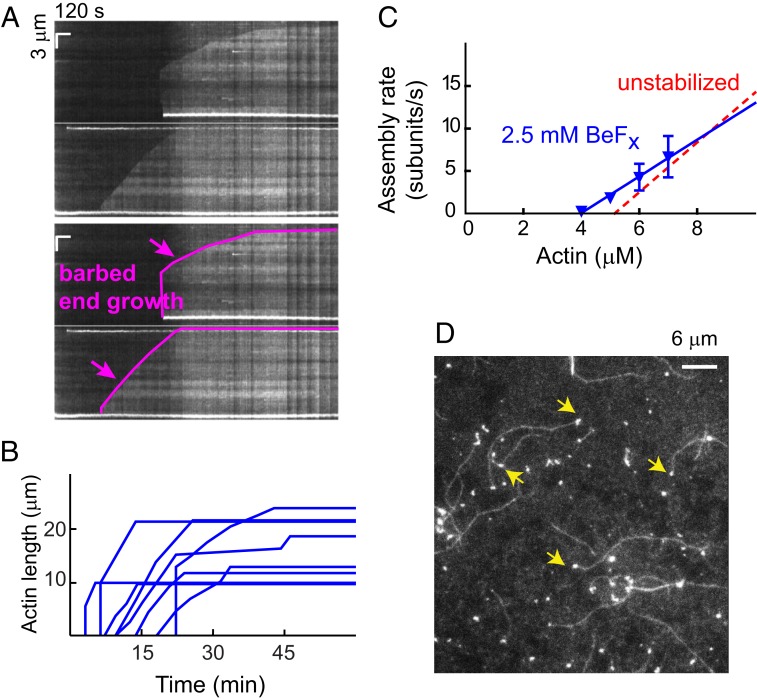
Fig. 4.
BeFx suppresses pointed-end dynamics. (A) Raw kymographs (Upper), and a skeletonized version of each (Lower), illustrating growth of the barbed end (magenta) and the absence of pointed-end dynamics, similar to canonical actins (Movie S6). (B) Time course of growth of 8 PfAct1 filaments in the presence of BeFx shows growth followed by a steady-state phase. (C) Polymerization rate as a function of actin concentration in the presence of BeFx (blue inverted triangles). BeFx slowed both the assembly and disassembly rates, thus maintaining a critical concentration similar to that observed in its absence (red dashed line). The assembly rate constant (slope) was 2.2 subunits/µM·s, the disassembly rate constant (y-intercept) 8.6 subunits/s, and the critical concentration (x-intercept) 4.0 µM. See Table 1 for average polymerization parameters from multiple experiments. (D) Image showing that in the presence of BeFx a large percentage of PfAct1 filaments have a bright, short oligomer at the pointed end (yellow arrows). A number of short oligomers are also seen on the surface. Polymerization buffer: 10 mM imidazole, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 2.5 mM MgATP,10 mM DTT, 0.25% methylcellulose, 0.13 mg/mL glucose oxidase, 50 μg/mL catalase, and 3 mg/mL glucose.
The shrinkage from the barbed end that we previously observed after steady state was also abolished. In the presence of BeFx, the PfAct1 filaments behaved more like canonical actin with a polymerization phase and a steady-state phase, suggesting that BeFx also reduced the barbed-end depolymerization rate (Fig. 4B). The apparent assembly rate was plotted as a function of G-actin concentration in the presence of BeFx (Fig. 4C). Fitting gave an assembly rate constant of 2.2 subunits/s·µM (slope), a disassembly rate constant of 8.6 subunits/s (y-intercept), and a critical concentration of 4.0 µM. Both rate constants are slowed to approximately 70% that observed in the absence of BeFx, leaving the critical concentration unchanged at ∼4 µM (Table 1).
Many bright, short pieces of actin appeared on the surface as soon as the experiment began. The filaments that ultimately grew showed a delay of a few minutes before they appeared on the surface and started to grow. A large percentage of growing filaments originated from these shards (Fig. 4D and Movie S6), suggesting that they act as actin nuclei prior to the elongation phase. These oligomers may also protect the pointed end from depolymerization.
Discussion
Our study reveals a number of unique and previously unreported dynamic aspects of PfAct1 assembly and disassembly. These observations were enabled by direct visualization of the growth of single PfAct1 filaments in real time from unlabeled monomers by TIRF microscopy using actin–chromobody, a single-chain anti-actin antibody fused to the fluorescent protein Emerald. Actin–chromobodies have been recently validated as F-actin sensors in apicomplexan parasites in vivo (12, 22). Prior studies that have not observed these dynamics relied on light scattering, centrifugation, and changes in fluorescence of pyrene-labeled monomers to follow polymerization (15, 16).
PfAct1 Assembles via a Nucleation–Elongation Mechanism with a Very High Critical Concentration.
Long apicomplexan actin filaments have never been seen in prior studies, and thus it was concluded that formation of short filaments (<200 nm) is an intrinsic property of PfAct1, and that short, unstable filaments are necessary for gliding motility (19). This idea was supported by studies showing that treatment of Plasmodium with the actin-stabilizing agent JAS adversely affected growth, invasion, and the actin cytoskeleton (14). Here we show that PfAct1 is capable of forming filaments that are tens of microns long, enough to span the whole organism, whether as a motile spindle-shaped sporozoite (10- to 15-μm long), or as the nonmotile merozoite (∼1- to 2-μm long). It is possible that actin-binding proteins maintain filaments at a short length in vivo, but this is not an intrinsic property of PfAct1. It is also possible that long filaments were not seen in vivo due to our observation that filaments disassemble at longer times. Another consideration is that the filaments readily break and appear to be fragile, and thus specimen preparation may have underestimated the actual filament length (8).
The concentration of actin in the apicomplexan T. gondii was reported to be ∼8 to 10 µM (27), with most of it in the monomeric form (28). Assuming similar values for Plasmodium, despite the extremely high critical concentration of 4 µM that we measured, the cell should have sufficient actin to form filaments by a nucleation–elongation mechanism. Moreover, the space between the Plasmodium plasma membrane and the inner membrane complex where the glideosome is located is restricted in size (∼20 nm), which could increase the local actin concentration. The regulation of assembly in vivo is, however, likely more complex. Based on our characterization of PfAct1 alone, we are now well-situated to further investigate the interaction of actin with the limited repertoire of actin-binding proteins found in Plasmodium, which include 2 formins, 1 profilin, 2 ADFs, 1 cyclase associated protein (CAP/srv2), 1 heterodimeric αβ-capping protein, and the actin filament cross-linking protein coronin (reviewed in ref. 29).
Although a recent study agreed with our conclusion that PfAct1 polymerizes via a nucleation–elongation mechanism, they measured a low critical concentration of ∼100 nM from pyrene fluorescence measurements (16), far lower than the 4 µM we measure from plotting observed filament growth as a function of actin concentration. Their experiment was performed using actin that was polymerized overnight, a time scale over which we showed long filaments would not persist. We speculate that the low critical concentration they measured may relate to residual small oligomers that are spontaneously formed in the absence of ammonium acetate, as they first described in ref. 7. In general, using methods less direct than visualization to quantify assembly complicates the interpretation of results obtained with this dynamic actin filament. Our data do not agree with the isodesmic polymerization mechanism proposed to occur with T. gondii TgAct1 (15), which may result from the indirect techniques used to follow assembly, but differences between PfAct1 and TgAct1 cannot be ruled out.
Once stabilized by JAS, the PfAct1 filament loses its dynamic polymerization properties and behaves similar to skeletal actin. A recent near-atomic resolution structure of JAS-stabilized PfAct1 filaments showed that while small but important differences were seen at inter- and intrastrand contacts, the general architecture is similar to mammalian actin filaments (13). Our results would predict that the native, unstabilized PfAct1 filament, with its significantly different kinetics from the JAS-stabilized filament, will show more differences in inter- and intrastrand contacts compared with canonical actin filaments.
In vitro motility experiments showed that the class XIV myosin motor PfMyoA moved JAS-stabilized PfAct1 actin filaments at the same speed as JAS-stabilized skeletal actin filaments (20). One possibility is that the PfMyoA motor primarily interacts with regions of actin that are conserved between PfAct1 and skeletal muscle actin. Alternatively, the inclusion of JAS may have masked differences that occur when PfMyoA interacts with unstabilized PfAct1. The latter will be difficult to investigate experimentally because of the treadmilling seen with actin alone.
Nucleotide-Dependent Dynamics.
The rates of subunit disassembly from both ends of the PfAct1 filament are fast (Fig. 5A). Direct observation of barbed-end depolymerization showed a rate of 11 ± 4 subunits/s, and the y-intercept of the plot of barbed-end growth rate versus actin concentration gave ∼15 subunits/s. Pointed-end disassembly was observed to be 7 to 10 subunits/s. Rapid filament treadmilling at ∼10 subunits/s was observed near the critical concentration. Although treadmilling was previously observed in skeletal muscle actin (17, 30), the rate of pointed-end shrinkage was 2 orders-of-magnitude slower (pointed-end shrinkage of ∼0.1 to 0.4 subunits/s) than that observed here with PfAct1.
Fig. 5.
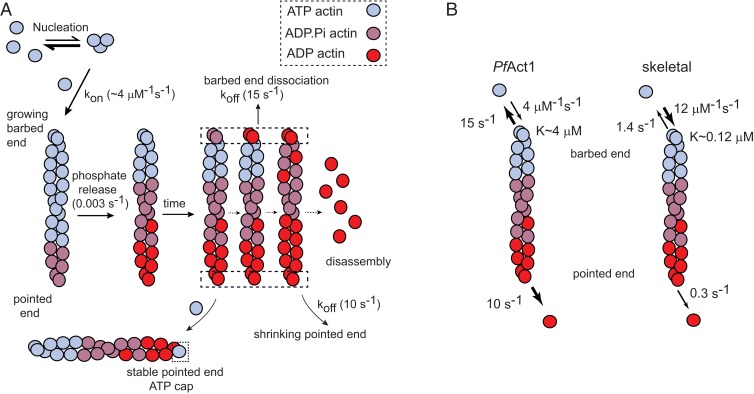
Schematic of PfAct1 filament dynamics. (A) Like canonical actins, PfAct1 follows a nucleation–elongation mechanism, but has a very high critical concentration (∼4 µM) due to both a lower assembly rate and a faster disassembly rate than canonical actins. The rates of protomer disassembly from both the barbed and pointed ends are fast (10 to 15 s−1). Treadmilling can be observed near the critical concentration when the rate of elongation becomes similar to the pointed-end disassembly rate. In the absence of added inorganic phosphate or BeFx, the filament composed of ADP protomers completely depolymerizes at long times. Inorganic phosphate, or to a greater extent BeFx, suppresses pointed end dynamics by capping the pointed end with an ATP-like protomers. (B) A comparison of the kinetics of PfAct1 versus skeletal actin filaments. Skeletal rates were taken from ref. 37. Compared with skeletal actin filaments, PfAct1 filaments have a higher critical concentration, a slower assembly rate at the growing barbed end, and faster disassembly kinetics at both ends. The off rates at the barbed and pointed ends are more similar for PfAct1 than for skeletal actin.
The observation that unstabilized filaments stop growing and start to shrink and cleave with increasing time suggests that once ATP is hydrolyzed into ADP in the actin protomer and the γ-phosphate is released with a half-life of ∼300 s (25), filament stability is decreased and the rate of polymerization slowed, resulting in filament dissolution in less than 40 min (Fig. 5A). The critical concentration for PfAct1-ADP monomers is extremely high, ∼55 µM. These dynamics are suppressed by either Pi or more effectively by BeFx (ADP.BeFX is an ATP analog), suggesting that a filament whose end is capped with an ATP/ADP.Pi/ADP.BeFX protomer is stable (Fig. 5A).
Role of the D-Loop.
When the entire PfAct1 D-loop was replaced with the canonical D-loop from human actins, the chimera formed long filaments with an average length of 1.6 µm, as observed by negative-stain EM, in contrast to short, irregular structures of ∼100 nm that were seen with the wild-type PfAct1 (7). Consistent with this observation, we showed here that this chimeric actin had a 4-fold lower critical concentration than wild-type PfAct1, implicating this loop as a primary determinant of filament stability. The sequence alignment of the 2 loops (protein residues 39 to 60), with significant differences highlighted in bold is: 39KNPGIMVGMEEKDAFVGDEAQT60 PfAct1 and 39RHQGVMVGMGQKDSYVGDEAQS60 mammalian actin.
Although a cryo-EM reconstruction of unstabilized PfAct1 filaments should definitively show how interactions between adjacent protomers of PfAct1 filaments differ from that of skeletal muscle actin, one can speculate based on the sequence differences that the change of polar Q41 at the base of the loop in canonical actins to proline in PfAct1 may force the D-loop to take a different orientation. In addition, high-resolution cryo-EM of skeletal actin filaments showed that Q49 in subunit “a” of canonical actins forms a π–cation interaction with conserved Y169 in subdomain 3 of subunit “a-2” (31) that will be disrupted by the E49 substitution in PfAct1, which would likely weaken the interaction between neighboring actin protomers. A single-point mutation, N41H, was also shown to be a key determinant for PfAct1 monomers to incorporate into mammalian actin filaments in a skin cell line, suggesting that this residue is important for an incoming monomer to dock onto a barbed end (32).
Comparison with Skeletal Actin and Mical-Oxidized Skeletal Actin.
The difference in kinetics between skeletal muscle and PfAct1 filaments are striking and explain why the PfAct1 filament is so unstable relative to skeletal actin (Fig. 5B). The small difference in the fast off rates at both the barbed and pointed ends of PfAct1 contrast with slower rates and a 5-fold difference in rates at the 2 ends of skeletal actin. In addition, the assembly rate at the barbed end is ∼3-fold slower for PfAct1 compared with skeletal muscle actin.
While native PfAct1 filaments are intrinsically dynamic because of their kinetic properties, a recently identified posttranslational modification to skeletal muscle actin causes its dynamics to more closely resemble that of PfAct1. Oxidation of M44 and M47 in the D-loop of skeletal actin by Mical redox enzymes results in either a 10-fold higher depolymerization rate than unmodified actin (∼2.6 vs. 0.2 s−1) or in catastrophic actin disassembly at ∼84 s−1 (33). The structural basis for the fast disassembly is novel interactions involving the oxidized D-loop residues that differ from those seen in the native filament and that are incompatible with a stable filament (31, 33). Our polymerization data with the D-loop chimera (PfAct1-HDL) is also consistent with the interaction of the D-loop with the adjacent actin protomer being critical for filament stability.
There are several other parallels between unmodified PfAct1 filaments and Mical-oxidized skeletal F-actin. Both show assembly rates that are ∼3-fold slower than skeletal actin. Mical-oxidized skeletal actin-ADP did not form filaments even at 18 µM actin, implying that the Cc > 18 µM. Here we show that PfAct1-ADP has the very high critical concentration of ∼55 µM. Addition of phosphate, or more efficiently the addition of BeFx, to either actin slowed the rapid disassembly, likely due to the stabilizing effect of capping filament ends by ADP.Pi/ADP.BeFx monomers.
Perspectives.
It is interesting to note that the polymerization/depolymerization kinetics for PfAct1 are much closer to that reported for the prokaryotic actin homolog ParM, such as a critical concentration in the micromolar range and fast disassembly rates from both ends, compared with canonical actins (34). Our findings may thus help to elucidate the evolution of actin filaments among different species. What are the biological implications of having such a dynamic actin filament in the Plasmodium parasite? Invasion of host cells by the Plasmodium parasite takes place in less than a minute, and thus a stable filament would not be required, in contrast to muscle actins that need to remain assembled. It was previously proposed that controlled polymerization of actin filaments in T. gondii dictates the timing, duration, and directionality of gliding motility (35). Heavy-chain phosphorylation of PfMyoA enhances the speed at which it moves actin 2-fold at the expense of lower ensemble force (3), but a more robust “on–off” switch has not been identified for PfMyoA activity. Our findings are thus consistent with the idea that the availability of filamentous actin may contribute to regulating actomyosin interactions in this system. In addition, this study emphasizes the importance of reinvestigating the functional interaction of the limited repertoire of Plasmodium actin-binding proteins with unstabilized PfAct1.
Materials and Methods
Plasmodium Actin Expression and Purification.
Infected Sf9 cells (2 × 109) were harvested 3 d after baculovirus infection and lysed by sonication in 50 mL of 10 mM Hepes, pH 8.0, 0.3 M NaCl, 0.25 mM CaCl2, 0.5 mM Na2ATP, 0.5 mM DTT containing protease inhibitors (0.5 mM 4-benzenesulfonyl fluoride hydrochloride, 5 µg/mL leupeptin, and 0.5 mM tosyl-l-lysyl-chloromethane hydrochloride), clarified, and immediately bound to a nickel-affinity column (HIS Select Nickel Affinity Gel, Sigma). Nonspecifically bound protein was eluted with 2 column volumes of 10 mM imidazole, 10 mM Hepes, pH 8.0, 0.3 M NaCl, 0.25 mM CaCl2, 0.25 mM Na2ATP, 0.5 mM DTT,1 µg/mL leupeptin. Actin was eluted with wash buffer containing 200 mM imidazole. Pooled fractions were dialyzed versus G-buffer (5 mM Tris, pH 8.2 at 4 °C, 0.2 mM CaCl2, 0.2 mM Na2ATP, 0.5 mM DTT, 1 μg/mL leupeptin). Samples were either flash-frozen for storage at −80 °C and subsequent removal of the tag, or digested following dialysis. The thymosin and HIS-tag were cleaved off by digestion with a 1:20 to 1:30 weight ratio of chymotrypsin:actin for 15 min at room temperature. Actin was separated from the thymosin/His-tag using a 1 mL TSKgel SuperQ-5PW column (Tosoh) with a 22-mL gradient of 0 to 0.3 M NaCl in G-buffer, followed by a step to 0.5 M NaCl. Peak fractions were pooled, concentrated, and dialyzed against G-buffer containing 0.2 M ammonium acetate, pH 8 (4 °C). The inclusion of ammonium acetate to maintain PfAct1 in a monomeric state was discovered by Kumpula et al. (16). Fractions were either flash-frozen for storage at −80 °C and subsequent gel filtration, or purified by gel filtration following dialysis. PfAct1 G-actin (3 to 5 mg) was applied to a Superdex 10/300 GL column (Pharmacia) equilibrated with G-buffer plus 0.2 M ammonium acetate, pH 8 at 4 °C. Fractions eluting at the position of monomeric actin were pooled, concentrated with an Amicon Ultra-4 centrifugal filter (Regenerated Cellulose 10,000 NMWL, EMD Millipore) to 4 to 5 mg/mL, and either used immediately or flash-frozen for storage at −80 °C. Protein concentration was determined using the Bradford protein assay (Bio-Rad) with BSA as standard.
A mutant PfAct1 in which the native D-loop was replaced with the D-loop from mammalian actins (7) was also cloned and expressed. The native residues of PfAct1 between Pro38 and Lys61 (39KNPGIMVGMEEKDAFVGDEAQT60) were replaced with the corresponding amino acids found in all isoforms of canonical human actins (RHQGVMVGMGQKDSYVGDEAQS).
Actin–Chromobody Expression and Purification.
Actin–chromobody expression and purification procedures were as described in ref. 20.
Analytical Ultracentrifugation.
Sedimentation velocity runs were performed at 20 °C in an Optima XL-I analytical ultracentrifuge (Beckman Coulter) using an An60Ti rotor at 40,000 rpm. The sedimentation coefficient, s*, was determined by curve fitting to 1 species using the dc/dt program (36).
Preparation of PfAct1 for Polymerization.
Monomeric PfAct1 in G-buffer plus 0.2 to 0.3 M ammonium acetate, pH 8 at 4 °C, either directly following gel filtration or thawed from a frozen aliquot (−80 °C), was centrifuged at 393,000 × g for 30 min. The ammonium acetate was removed using a Zeba spin-desalting column (0.5 mL, 7K MWCO, Thermo Fisher Scientific 89882) equilibrated in G-buffer, according to the manufacturer’s protocol. Actin concentration was determined with the Bradford protein assay (Bio-Rad) with BSA as standard.
PfAct1 with MgADP at the active site was prepared by first converting actin-CaATP (0.2 M ammonium acetate, 5 mM Tris pH 8.0 [4 °C], 0.2 mM CaCl2, 0. 2 mM NaATP, 0.5 mM DTT) to MgATP-actin by incubating with a 1.2-fold molar excess of MgCl2 and 250 µM EGTA for 10 min at 4 °C. Mg-ATP actin was then incubated with 1 mM glucose and 5 U hexokinase (Sigma H6380) for 10 min at room temperature, followed by addition of 1 mM ADP (Sigma A2754).
Visualization of Actin Polymerization by TIRF Microscopy.
PfAct1 polymerization was visualized with 0.5 µM actin–chromobody Emerald (ChromoTek) at 37 °C as described in ref. 20. The plasmid was a gift from Markus Meissner, University of Glasgow, United Kingdom, and Aoife Heaslip, University of Connecticut, Storrs, CT. Briefly, the flow cell was blocked with 5 mg/mL BSA in G-buffer for 2 min and rinsed once with 20 µL of 10 mM imidazole, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.25% methylcellulose. Control experiments (SI Appendix, Fig. S4) showed that the same assembly rates were observed whether the flow cell was blocked with only BSA, or when both BSA and NEM-myosin (an ATP-insensitive modified myosin that binds actin filaments and keeps them in the TIRF field) were used. Twenty microliters of G-actin (4 to 12 μM, also containing 0.5 µM actin chromobody) in G-buffer was mixed with 20 μL of 2× polymerization buffer to initiate polymerization. The final polymerization buffer contains 10 mM imidazole, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 2.5 mM MgATP,10 mM DTT, 0.25% methylcellulose, 0.13 mg/mL glucose oxidase, 50 μg/mL catalase, and 3 mg/mL glucose. The mixture was then flowed into the flow cell (2 times, 20 μL each time). Excess solution was removed, and the flow cell was sealed with nail polish. Depending on the experiment as detailed in the text, the buffer also contained either 2.5 µM JAS (J7473, Thermo Fisher Scientific), 12.5 mM sodium phosphate, or 2.5 mM BeF2 (CAS 7787-49-7, Santa Cruz Biotechnology). Polymerization was measured at 37 °C.
TIRF microscopy was carried out on a Nikon ECLIPSE Ti microscope, run by the Nikon NIS Elements software package, and equipped with through-objective type TIRF and a temperature-control unit that enclosed the flow cell. The samples were excited with the TIRF field of a 488-nm laser line, and emission was observed with a 525/50 filter. The fluorescence image was observed with a 100× objective and recorded on an Andor EMCCD camera (Andor Technology) at various frame rates (1 frame per second or 0.2 frame per second) for 6 min to an hour with automatic focus correction. The final resolution is 0.1066 μm per pixel. Length increases in actin were converted into subunits using 370 subunits per 1 µm actin.
Fluorescence Data Processing.
Image drift was first corrected in ImageJ with a modified “manual drift correction” plug-in. Barbed end polymerization and depolymerization rates were obtained as described previously (23). The time at which the pointed end began to shrink was determined manually. Pauses in the depolymerization events were excluded from rate determinations. Kymographs of actin growth and shrinkage were generated in ImageJ using the “multiplekymograph” plug-in.
Supplementary Material
Acknowledgments
We thank Elena Krementsova for performing analytical ultracentrifugation. This work was funded by NIH Grant AI 132378 (to K.M.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906600116/-/DCSupplemental.
References
- 1.WHO , World Malaria Report 2018 (World Health Organization, 2018). [Google Scholar]
- 2.Tardieux I., Baum J., Reassessing the mechanics of parasite motility and host-cell invasion. J. Cell Biol. 214, 507–515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robert-Paganin J., et al. , Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism. Nat. Commun. 10, 3286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kudryashev M., Lepper S., Baumeister W., Cyrklaff M., Frischknecht F., Geometric constrains for detecting short actin filaments by cryogenic electron tomography. PMC Biophys. 3, 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S., Lemgruber L., Tay C. L., Baum J., Meissner M., Multiple essential functions of Plasmodium falciparum actin-1 during malaria blood-stage development. BMC Biol. 15, 70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wesseling J. G., Smits M. A., Schoenmakers J. G., Extremely diverged actin proteins in Plasmodium falciparum. Mol. Biochem. Parasitol. 30, 143–153 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Vahokoski J., et al. , Structural differences explain diverse functions of Plasmodium actins. PLoS Pathog. 10, e1004091 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz S., et al. , Malaria parasite actin filaments are very short. J. Mol. Biol. 349, 113–125 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Schmitz S., et al. , Malaria parasite actin polymerization and filament structure. J. Biol. Chem. 285, 36577–36585 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schüler H., Mueller A. K., Matuschewski K., Unusual properties of Plasmodium falciparum actin: New insights into microfilament dynamics of apicomplexan parasites. FEBS Lett. 579, 655–660 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Hliscs M., et al. , Organization and function of an actin cytoskeleton in Plasmodium falciparum gametocytes. Cell. Microbiol. 17, 207–225 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Stortz J. F., et al. , Formin-2 drives polymerisation of actin filaments enabling segregation of apicoplasts and cytokinesis in Plasmodium falciparum. eLife 8, e49030 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pospich S., et al. , Near-atomic structure of jasplakinolide-stabilized malaria parasite F-actin reveals the structural basis of filament instability. Proc. Natl. Acad. Sci. U.S.A. 114, 10636–10641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuno Y., et al. , Effect of jasplakinolide on the growth, invasion, and actin cytoskeleton of Plasmodium falciparum. Parasitol. Res. 88, 844–848 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Skillman K. M., et al. , The unusual dynamics of parasite actin result from isodesmic polymerization. Nat. Commun. 4, 2285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumpula E. P., et al. , Apicomplexan actin polymerization depends on nucleation. Sci. Rep. 7, 12137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn J. R., Pollard T. D., Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard T. D., Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8, a018226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skillman K. M., et al. , Evolutionarily divergent, unstable filamentous actin is essential for gliding motility in apicomplexan parasites. PLoS Pathog. 7, e1002280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bookwalter C. S., et al. , Reconstitution of the core of the malaria parasite glideosome with recombinant Plasmodium class XIV myosin A and Plasmodium actin. J. Biol. Chem. 292, 19290–19303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujiwara I., Zweifel M. E., Courtemanche N., Pollard T. D., Latrunculin a accelerates actin filament depolymerization in addition to sequestering actin monomers. Curr. Biol. 28, 3183–3192.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Periz J., et al. , Toxoplasma gondii F-actin forms an extensive filamentous network required for material exchange and parasite maturation. eLife 6, e24119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu H., Fagnant P. M., Bookwalter C. S., Joel P., Trybus K. M., Vascular disease-causing mutation R258C in ACTA2 disrupts actin dynamics and interaction with myosin. Proc. Natl. Acad. Sci. U.S.A. 112, E4168–E4177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pollard T. D., Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlier M. F., Pantaloni D., Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry 25, 7789–7792 (1986). [DOI] [PubMed] [Google Scholar]
- 26.Combeau C., Carlier M. F., Characterization of the aluminum and beryllium fluoride species bound to F-actin and microtubules at the site of the gamma-phosphate of the nucleotide. J. Biol. Chem. 264, 19017–19021 (1989). [PubMed] [Google Scholar]
- 27.Sahoo N., Beatty W., Heuser J., Sept D., Sibley L. D., Unusual kinetic and structural properties control rapid assembly and turnover of actin in the parasite Toxoplasma gondii. Mol. Biol. Cell 17, 895–906 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobrowolski J. M., Niesman I. R., Sibley L. D., Actin in the parasite Toxoplasma gondii is encoded by a single copy gene, ACT1 and exists primarily in a globular form. Cell Motil. Cytoskeleton 37, 253–262 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Kumpula E. P., Kursula I., Towards a molecular understanding of the apicomplexan actin motor: On a road to novel targets for malaria remedies? Acta Crystallogr. F Struct. Biol. Commun. 71, 500–513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujiwara I., Takahashi S., Tadakuma H., Funatsu T., Ishiwata S., Microscopic analysis of polymerization dynamics with individual actin filaments. Nat. Cell Biol. 4, 666–673 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Chou S. Z., Pollard T. D., Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U.S.A. 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas R. G., et al. , Inter-subunit interactions drive divergent dynamics in mammalian and Plasmodium actin filaments. PLoS Biol. 16, e2005345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grintsevich E. E., et al. , Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat. Commun. 8, 2183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garner E. C., Campbell C. S., Mullins R. D., Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021–1025 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Wetzel D. M., Håkansson S., Hu K., Roos D., Sibley L. D., Actin filament polymerization regulates gliding motility by apicomplexan parasites. Mol. Biol. Cell 14, 396–406 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philo J. S., A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal. Biochem. 279, 151–163 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara I., Vavylonis D., Pollard T. D., Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 8827–8832 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.