Significance
SOX2, one of the pluripotency-associated stem cell factors, is aberrantly amplified in many types of human cancer and could be an attractive target for cancer therapy. We report here that MLN4924, a small-molecule inhibitor of neddylation, down-regulates SOX2 expression via inactivating FBXW2 E3 ligase to cause accumulation of MSX2, a transcription repressor known to repress SOX2 expression. We therefore established an FBXW2–MSX2–SOX2 axis and found it regulates cancer stem cell property and drug resistance. Specifically, FBXW2 or SOX2 promotes tumor sphere formation and tamoxifen resistance, which is blocked by MSX2. Our study revealed an anticancer mechanism of MLN4924 by which MLN4924 inactivates the FBXW2–MSX2–SOX2 axis to suppress stem cell property and sensitize cancer cells to chemotherapy.
Keywords: SOX2, MSX2, FBXW2, ubiquitylation, degradation
Abstract
SOX2 is a key transcription factor that plays critical roles in maintaining stem cell property and conferring drug resistance. However, the underlying mechanisms by which SOX2 level is precisely regulated remain elusive. Here we report that MLN4924, also known as pevonedistat, a small-molecule inhibitor of neddylation currently in phase II clinical trials, down-regulates SOX2 expression via causing accumulation of MSX2, a known transcription repressor of SOX2 expression. Mechanistic characterization revealed that MSX2 is a substrate of FBXW2 E3 ligase. FBXW2 binds to MSX2 and promotes MSX2 ubiquitylation and degradation. Likewise, FBXW2 overexpression shortens the protein half-life of MSX2, whereas FBXW2 knockdown extends it. We further identified hypoxia as a stress condition that induces VRK2 kinase to facilitate MSX2–FBXW2 binding and FBXW2-mediated MSX2 ubiquitylation and degradation, leading to SOX2 induction via derepression. Biologically, expression of FBXW2 or SOX2 promotes tumor sphere formation, which is blocked by MSX2 expression. By down-regulating SOX2 through inactivation of FBXW2 E3 ligase, MLN4924 sensitizes breast cancer cells to tamoxifen in both in vitro and in vivo cancer cell models. Thus, a negative cascade of the FBXW2–MSX2–SOX2 axis was established, which regulates stem cell property and drug resistance. Finally, an inverse correlation of expression was found between FBXW2 and MSX2 in lung and breast cancer tissues. Collectively, our study revealed an anticancer mechanism of MLN4924. By inactivating FBXW2, MLN4924 caused MSX2 accumulation to repress SOX2 expression, leading to suppression of stem cell property and sensitization of breast cancer cells to tamoxifen.
A small subpopulation of cancer cells within a tumor mass possesses characteristics associated with stem cells, thus designated cancer stem cells (CSCs). CSCs exhibit the ability of self-renewal and multilineage differentiation, which is responsible for tumor initiation, progression, metastasis, drug resistance, and tumor recurrence (1). Despite initial excellent tumor responses to conventional or targeted chemotherapies, tumor relapse is a common event that leads to treatment failure, since chemotherapies usually target the fast-proliferating group of differentiated cancer cells, while sparing the tumor-initiating CSCs (2, 3). Therefore, CSCs are considered major culprits for chemoresistance and tumor recurrence, which call for novel therapy to selectively target cancer stem cells (4).
Sry-related high-mobility box 2 (SOX2), one of the pluripotency-associated stem cell factors, plays essential roles in the maintenance of stem cell property and determination of cell fate, thereby regulating developmental processes (5). SOX2 was found to be aberrantly expressed in many types of cancer, including carcinomas in the lung, breast, colon, ovary, prostate, and others (6). Importantly, SOX2 expression is positively correlated with cancer cell stemness and poor patient outcome, suggesting its important roles in CSC generation and biology (7–9). Furthermore, SOX2-dependent activation of Wnt signaling in cancer stem/progenitor cells was reported to be responsible for the development of tamoxifen resistance in breast cancer cells (10). Thus, SOX2 could be an attractive therapeutic target for eliminating CSCs and overcoming chemoresistance. A recent report showed that SOX2 is transcriptionally down-regulated by the muscle segment homeobox 2 (MSX2) transcription repressor (11), suggesting a new approach to target SOX2.
Cullin-RING ligase (CRL) is the largest E3 ubiquitin ligase, consisting of multiple components with 8 cullin family members (cullin-1, -2, -3, -4A, -4B, -5, -7, and -9) (12). CRL1, also known as SCF (SKP1-cullin-1-F-box protein) E3 ubiquitin ligase, is the founding member of CRL E3 ligases, consisting of the scaffold protein cullin-1, adaptor protein SKP1, RING component RBX1 or RBX2, and substrate receptor component F-box protein (13). The F-box protein is a substrate recognition subunit and determines the substrate specificity of the SCF complex (14), whereas the activity of CRL requires cullin neddylation, a process catalyzed by NEDD8-activating enzyme (NAE, E1), NEDD-8–conjugating enzyme (E2), and NEDD-8 ligase (E3). Given that some CRL components and neddylation enzymes were abnormally activated in many types of human cancer, CRL E3 and neddylation enzymes were validated as attractive anticancer targets (12, 15).
MLN4924/pevonedistat is a small-molecule inhibitor of NAE. By inhibiting the entire process of neddylation modification (16), MLN4924 inactivates all members of the CRL ligase family (17), to cause accumulation of many CRL substrates which regulate a number of important biological processes, including cell-cycle progression, DNA replication, signal transduction, development, and tumorigenesis. In preclinical settings, MLN4924 has been shown to suppress growth and survival of a variety of cancer cell lines, and sensitizes cancer cells to chemoradiation by inducing growth arrest, apoptosis, autophagy, and senescence via the mechanism mainly associated with accumulation of tumor suppressor substrates (17, 18). Due to its impressive anticancer activity, MLN4924 was advanced to phase I/II clinical trials both alone and in combination with chemo drugs (17, 19). Interestingly, we have previously shown that MLN4924 regulates tumor sphere formation with a stimulating effect at low doses but inhibitory effect at high doses. While the stimulating effect appears to involve c-MYC accumulation via blocking its degradation, and continued activation of EGFR via inducing EGFR dimerization (20), the mechanism for its inhibitory effect is unknown.
In this study, we found that MLN4924 down-regulates SOX2 expression in a dose-dependent manner by inducing accumulation of MSX2, a known transcription repressor of SOX2 (11). Mechanistic studies revealed that MSX2 is a new substrate of SCFFBXW2 E3 ubiquitin ligase. Under hypoxia, VRK2 kinase was induced to facilitate FBXW2–MSX2 binding and subsequent MSX2 ubiquitylation. By inactivating SCFFBXW2 E3, MLN4924 caused MSX2 accumulation to repress SOX2 expression, leading to inhibition of tumor sphere formation and sensitization of breast cancer to tamoxifen. In lung and breast cancer tissues, we found an inverse correlation in the levels of FBXW2 and MSX2, and that high SOX2 levels predict poor survival of lung cancer patients. Collectively, our study established a previously unknown signaling cascade of the FBXW2–MSX2–SOX2 axis that regulates cancer cell stemness and chemoresistance. Our study also revealed an anticancer mechanism of MLN4924 via repressing SOX2 through targeting the FBXW2–MSX2 axis.
Results
MLN4924 Down-Regulates SOX2 through MSX2.
MLN4924 is a small-molecule inhibitor of NEDD8-activating enzyme that inhibits CRLs by deneddylation, thus causing accumulation of many CRL substrates. However, whether and how MLN4924 affects gene transcription were previously unknown. To this end, we performed RNA-seq analysis in lung cancer A549 cells after treatment with MLN4924. One of the down-regulated genes, among the hundreds of candidates, was SOX2 (SI Appendix, Fig. S1A). For more details, check the GEO database https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134190 (21). Given the significance of SOX2 in cancer biology, we focused on this lead and found that consistent with the RNA-seq data, SOX2 expression, as measured by RT-PCR, was down-regulated in both time- and dose-dependent manners after MLN4924 treatment (Fig. 1A and SI Appendix, Fig. S1B). The dose-dependent reduction of SOX2 by MLN4924 was also detected at the protein level in multiple lung cancer cell lines (Fig. 1B). Thus, MLN4924 suppresses SOX2 transcription to reduce its mRNA levels, and then protein levels.
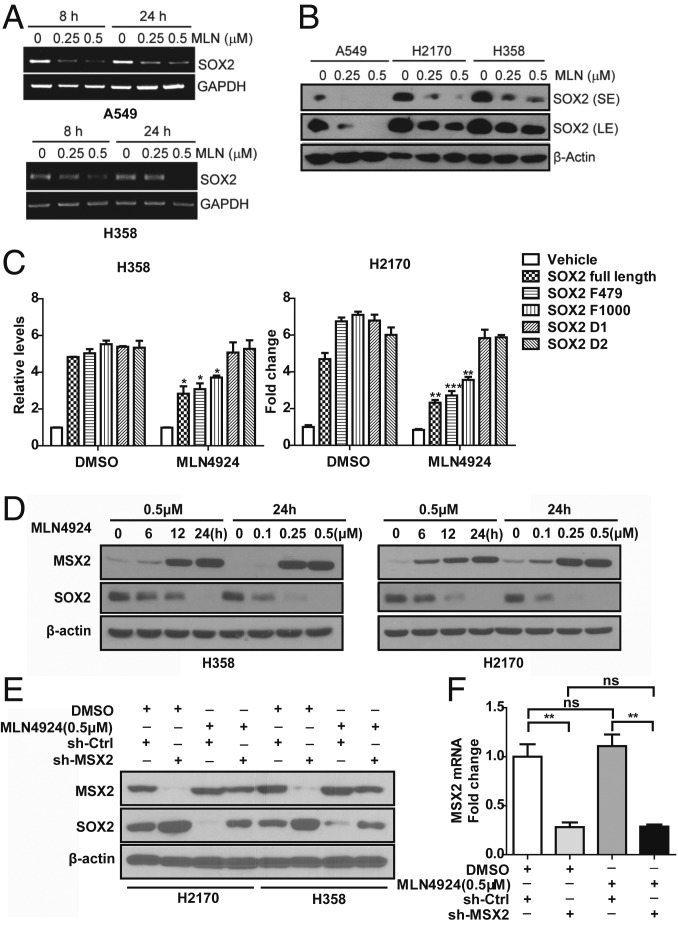
Fig. 1.
MLN4924 down-regulates SOX2 through MSX2. (A) MLN4924 treatment decreases SOX2 mRNA expression in both time- and dose-dependent manners. Human lung cancer A549 and H358 cells were treated with the indicated concentrations of MLN4924 for 8 and 24 h, followed by RT-PCR analysis. (B) MLN4924 reduces the protein levels of SOX2 in a dose-dependent manner. Cells were treated with the indicated doses of MLN4924 for 24 h, followed by immunoblotting (IB) with the indicated antibodies (Abs). LE, long exposure; SE, short exposure. (C) MLN4924-induced SOX2 reduction depends on 2 MSX2 binding sites in the SOX2 promoter. H358 and H2170 cells were transfected with the indicated plasmids and then treated with MLN4924 (0.5 μM) for 24 h, followed by dual-luciferase reporter assay. (D) MLN4924 increases MSX2 but decreases SOX2 protein levels in both time- and dose-dependent manners. Cells were treated with various concentrations of MLN4924 for the indicated time periods, followed by IB with the indicated Abs. (E) MSX2 depletion rescues SOX2 reduction by MLN4924. Cells infected with lentivirus expressing small-hairpin RNA (shRNA) targeting MSX2 or scrambled control shRNA were treated with MLN4924 (0.5 μM) for 24 h, followed by IB with the indicated Abs. (F) MLN4924 treatment has no effect on MSX2 mRNA expression. Cells infected with lentivirus expressing shRNA targeting MSX2 or scrambled control shRNA were treated with MLN4924 (0.5 μM) for 24 h, followed by qRT-PCR analysis. Shown are mean ± SEM of 3 independent biological experiments. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
A recent study has shown that SOX2 is a transcriptional downstream target of MSX2, a transcription repressor, acting via 2 MSX2 binding sites found in the SOX2 gene promoter (SI Appendix, Fig. S1C) (11). We therefore used a luciferase-based reporter assay to determine whether these 2 sites are responsible for MLN4924-induced SOX2 reduction. Indeed, luciferase reporter activity, driven by the SOX2 promoter, was significantly inhibited upon MLN4924 treatment, and this inhibitory effect was completely abrogated when either of the MSX2 binding sites was deleted (Fig. 1C), suggesting that MLN4924 suppresses SOX2 transcription via modulating MSX2.
Given MLN4924 inhibits cullin neddylation, thus inactivating CRLs to cause accumulation of CRL substrates, we hypothesized that MSX2 could be one of the CRL substrates whose accumulation blocks SOX2 transcription. Indeed, MLN4924 caused a dose- and time-dependent accumulation of MSX2 with consequential reduction of SOX2 protein in lung cancer cells (Fig. 1D). Importantly, MLN4924 suppression of SOX2 was MSX2-dependent, since MSX2 depletion completely rescued MLN4924 effects (Fig. 1E). We confirmed that MLN4924 treatment did not affect MSX2 mRNA levels (Fig. 1F), suggesting that MSX2 is a bona fide substrate of CRLs. To exclude possible off-target effects, we used a small-interfering RNA (siRNA) oligonucleotide targeting MSX2 at its 3′ UTRs and showed that siMSX2-3′ UTR indeed knocked down MSX2 and caused SOX2 increase, which can be completely rescued by transfection of MSX2 cDNA (encoding the ORF only) resistant to siMSX2-3′ UTR (SI Appendix, Fig. S1D).
FBXW2 Binds to MSX2 and Regulates MSX2 Levels.
We next determined whether MSX2 is a substrate of SCF, which would bind to one of the substrate receptor F-box proteins. An immunoprecipitation (IP)-based pull-down screening with 8 F-box proteins identified FBXW2 as an F-box protein specifically binding to endogenous MSX2 upon transfection (Fig. 2A). We have recently defined the consensus-binding motif (TSXXXS) required for FBXW2 to bind to its substrates, such as SKP2 (22) and β-catenin (23). Examination of the MSX2 protein sequence indeed identified such a motif, which is evolutionarily conserved (43SSLPFS48) (SI Appendix, Fig. S2A). Further IP-based pull-down assays using ectopically expressed hemagglutinin (HA)-FBXW2 confirmed an in vivo interaction between FBXW2 and MSX2 (Fig. 2 B and C). Importantly, the FBXW2–MSX2 binding is binding motif-dependent, since an MSX2 mutant with mutations on 3 serine residues to alanines (MSX2-3A) on the motif (SSLPFS to AALPFA) completely abrogated its FBXW2 binding (Fig. 2D).
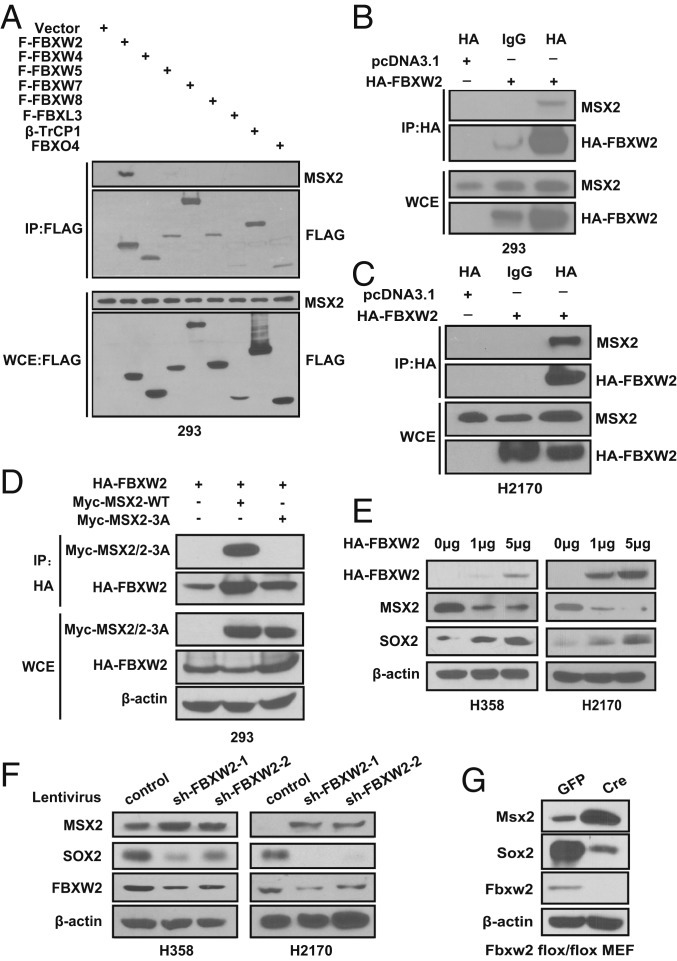
Fig. 2.
FBXW2 binds to MSX2 and negatively regulates MSX2 levels. (A) FBXW2 specifically binds to MSX2. HEK293 cells were transfected with the indicated plasmids, followed by IP with FLAG beads and IB with the indicated Abs. WCE, whole-cell extract. (B and C) FBXW2 binds to endogenous MSX2. Cell lysates from HEK293 (B) or H2170 (C) cells transfected with the indicated plasmids were pulled down with HA Ab or normal rat IgG, followed by IB with the indicated Abs. (D) FBXW2–MSX2 binding is dependent on the 3 serine residues on the degron motif. HEK293 cells were transfected with the indicated plasmids, followed by IP with FBXW2 Ab and IB with the indicated Abs. Note that Myc-MSX2/2-3A is for detection of both Myc-MSX2 and the Myc-MSX2-3A mutant. (E) FBXW2 overexpression causes a dose-dependent decrease of MSX2 levels and consequent increase of SOX2 levels. Cells were transfected with the indicated amounts of HA-FBXW2, followed by IB with the indicated Abs. (F) FBXW2 knockdown causes accumulation of MSX2 levels and consequent decrease of SOX2 levels. H358 or H2170 cells were infected with lentivirus expressing shRNA targeting FBXW2 or scrambled control shRNA, followed by IB with the indicated Abs. (G) Fbxw2 depletion causes accumulation of Msx2 levels and consequent reduction of Sox2 levels. Primary MEFs generated from E13.5 embryos with Fbxw2fl/fl were infected with Ad-GFP or Ad-Cre for 72 h, followed by IB with the indicated Abs.
We further confirmed the relationship between FBXW2 and MSX2 as E3 ligase vs. substrate. In both H358 and H2170 lung cancer cells, FBXW2 transfection caused a dose-dependent decrease of MSX2 levels and a consequent increase of SOX2 levels (Fig. 2E). Likewise, FBXW2 knockdown caused an accumulation of MSX2 levels and a consequent decrease of SOX2 levels (Fig. 2F). Again, to exclude possible off-target effects, we used an siRNA oligonucleotide targeting FBXW2 at its 3′ UTRs and showed that siFBXW2-3′ UTR knocked down FBXW2 and caused MSX2 accumulation, which can be rescued by transfection of FBXW2 cDNA (encoding the ORF only) resistant to siFBXW2-3′ UTR (SI Appendix, Fig. S2B). Finally, we generated an Fbxw2 conditional knockout mouse model and used Fbxw2fl/fl mouse embryonic fibroblasts (MEFs) derived from the embryos to determine the E3 ligase and substrate relationship in normal cells. Indeed, Fbxw2 depletion via Ad-Cre infection caused a remarkable accumulation of Msx2 and consequent reduction of Sox2 (Fig. 2G). Similar results were obtained in several MEFs derived from the littermate embryos with 3 genotypes of Fbxw2, showing the Fbxw2Δ/Δ MEFs have Fbxw2 depleted with very high Msx2 levels and no Sox2 expression (SI Appendix, Fig. S2 C and D). Collectively, MSX2 appears to be a bona fide substrate of SCFFBXW2 E3 ligase.
FBXW2 Shortens MSX2 Protein Half-Life and Promotes MSX2 Ubiquitylation.
To determine if FBXW2 regulates the protein half-life of MSX2, we first used cycloheximide to block new protein synthesis and found that MSX2 protein half-life was between 8 and 12 h and reduction of MSX2 level can be completely rescued by the proteasome inhibitor MG132 (SI Appendix, Fig. S3A). We then determined whether FBXW2 shortened MSX2 protein half-life. Remarkably, FBXW2 knockdown extended MSX2 half-life, whereas FBXW2 expression shortened it (Fig. 3 A and B and SI Appendix, Fig. S3 B and C). Moreover, although FBXW2 expression shortened the protein half-life of ectopically expressed wild-type MSX2, it had no effect on the MSX2-3A mutant (Fig. 3C and SI Appendix, Fig. S3D), which failed to bind to FBXW2 (Fig. 2D). These results indicate that the stability of MSX2 is negatively regulated by FBXW2 in a manner dependent on the FBXW2-binding motif.
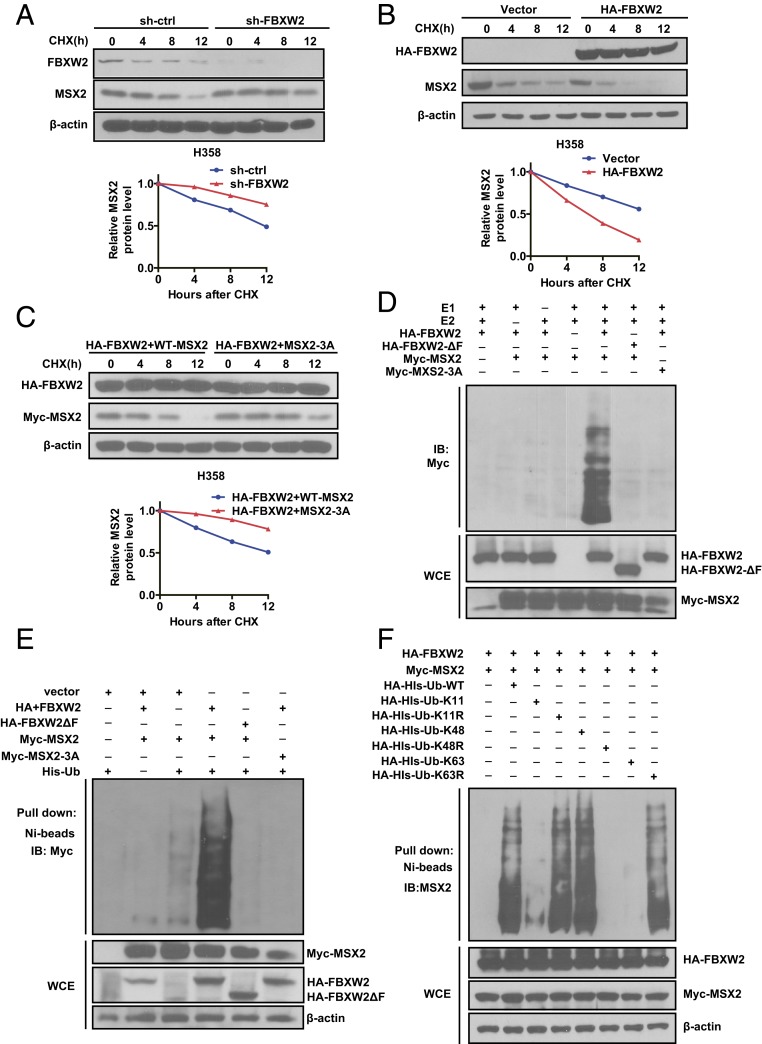
Fig. 3.
FBXW2 shortens MSX2 protein half-life and promotes MSX2 ubiquitylation. (A and B) FBXW2 manipulation alters the protein half-life of MSX2. H358 cells were infected with lentivirus expressing shRNA targeting FBXW2 or scrambled control for 48 h (A) or transfected with HA-FBXW2 or mock vector for 48 h (B). Cells were then treated with cycloheximide (CHX) for the indicated time periods and harvested for IB (Top). The band density was quantified using ImageJ software, and the decay curves are shown (Bottom). (C) Overexpression of FBXW2 shortens the protein half-life of MSX2-WT, but has no effects on MSX2-3A: H358 cells were transfected with HA-FBXW2, along with MSX2-WT or MSX2-3A for 48 h. Cells were then treated with CHX for the indicated time periods and harvested for IB (Top). The band density was quantified using ImageJ software, and the decay curves are shown (Bottom). (D) FBXW2, but not its ΔF mutant, promotes polyubiquitylation of MSX2-WT, but has no effects on MSX2-3A in vitro: FBXW2 and FBXW2-ΔF were purified using HA beads from HEK293 cells transfected with HA-FBXW2 and HA-FBXW2-ΔF, respectively. MSX2 and MSX2-3A were pulled down by IP with Myc beads from HEK293 cells transfected with Myc-MSX2 and Myc-MSX2-3A, respectively. Purified FBXW2 or FBXW2-ΔF (E3s), and purified MSX2 or MSX2-3A (substrate), were added into a reaction mixture containing ATP, ubiquitin, E1, and E2, followed by IB using anti-Myc Ab. (E) FBXW2, but not its ΔF mutant, promotes polyubiquitylation of MSX2-WT, but has no effect on MSX2-3A in vivo: HEK293 cells were transfected with the indicated plasmids, followed by pull-down using Ni-NTA beads (Top) or direct IB with the indicated Abs (Bottom). (F) FBXW2 promotes MSX2 ubiquitylation via K48 linkage: HEK293 cells were transfected with the indicated plasmids, lysed under denaturing conditions with 6 M guanidine solution, followed by pull-down using Ni-NTA beads (Top) or direct IB with the indicated Abs (Bottom). Data shown are mean ± SEM of 3 independent experiments.
To provide the biochemical basis of FBXW2 shortening MSX2 protein half-life, we performed both in vitro and in vivo ubiquitylation assays. We found that 1) wild-type FBXW2, but not its ΔF mutant, which fails to recruit other SCF components, promoted MSX2 polyubiquitylation, and 2) FBXW2 promoted polyubiquitylation of wild-type MSX2 but not its 3A mutant (Fig. 3 D and E). Finally, we used various ubiquitin mutants and defined that FBXW2 promoted MSX2 polyubiquitylation through the K48 linkage (Fig. 3F). Taken together, our combined results showed that MSX2 is a substrate of the SCFFBXW2 E3 ligase, which ubiquitylates it and targets it for proteasome degradation.
Hypoxia-Inducible VRK2 Is Required for MSX2–FBXW2 Binding and FBXW2-Mediated MSX2 Ubiquitylation.
A typical substrate of SCF E3 ligase is, in general, phosphorylated on Ser or Thr residues at the consensus-binding motif prior to being recognized by an F-box protein. To define the upstream kinase mediating MSX2 phosphorylation on the serine residues, we used computer-aided algorithms (GSP 3.0; gps.biocuckoo.org) and identified CK1/VRK2 (vaccinia-related kinase 2) and GRK2 (G protein-coupled receptor kinase) as the top candidate kinases (SI Appendix, Table S1). We then used approaches of siRNA knockdown or small-molecule inhibitor treatment to inactivate VRK2 or GRK2, respectively, followed by determining their effect on MSX2 levels. We found that VRK2 inactivation by VRK2 knockdown or treatment with the small-molecule inhibitor IC-261 largely blocked MSX2 degradation, whereas GRK2 knockdown had no effect (Fig. 4 A and B and SI Appendix, Fig. S4 A and B). Consistently, VRK2 knockdown and/or IC-261 treatment reduced FBXW2 binding to endogenous MSX2, leading to abrogation of FBXW2-mediated MSX2 polyubiquitination (Fig. 4 C and D). Collectively, VRK2 appears to be a modifying kinase to phosphorylate MSX2 that facilitates its FBXW2 binding for subsequent ubiquitylation.
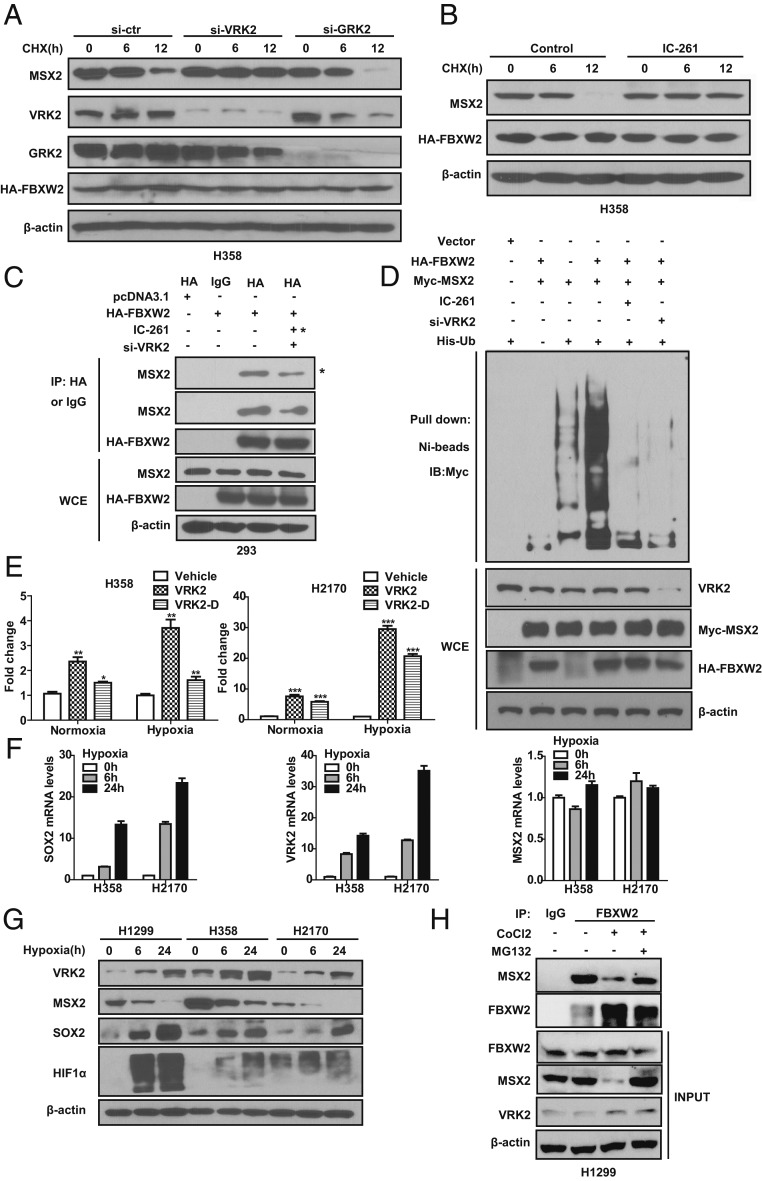
Fig. 4.
VRK2 is a hypoxia-inducible protein that facilitates MSX2–FBXW2 binding and MSX2 ubiquitylation and degradation. (A) Silencing of VRK2, but not GRK2, blocks MSX2 degradation. H358 cells were transfected with siRNA oligos targeting VRK2, GRK2, or scrambled control siRNA for 24 h, and then transfected with HA-FBXW2 for 48 h. Cells were treated with CHX for the indicated time periods, followed by IB with the indicated Abs. (B) VRK2 inactivation by IC-261 abrogates MSX2 degradation. H358 cells transfected with HA-FBXW2 were left untreated or treated with CK1 inhibitor IC-261 (10 μM), along with CHX for the indicated time periods, followed by IB with the indicated Abs. (C) VRK2 knockdown or inactivation reduces FBXW2–MSX2 binding. HEK293 cells transfected with the indicated siRNA oligos and indicated plasmids were left untreated or treated with IC-261 (10 μM) for 24 h. Cells were harvested for IP with HA Ab and IB with the indicated Abs. Note that the far right sample in the Top IP panel was treated with IC-261 (indicated by asterisks), whereas the far right sample in the second IP panel was transfected with si-VRK2. (D) VRK2 knockdown or inactivation abrogates FBXW2-mediated MSX2 polyubiquitylation. HEK293 cells were transfected with siRNA targeting VRK2 or scrambled control siRNA for 24 h, and then transfected with the indicated plasmids for 48 h. Cells were left untreated or treated with IC-261 (10 μM) for 24 h, followed by treatment with MG132 for the last 6 h before being harvesting for pull-down using Ni-NTA beads and IB with the indicated Abs. (E) Hypoxia activates VRK2 promoter activity in a manner dependent of the HIF1α binding site. H358 and H2170 cells transfected with the indicated constructs were subjected to normoxia or hypoxia conditions for 24 h, followed by dual-luciferase reporter assay. (F) Hypoxia induces SOX2 mRNA expression but has no effect on MSX2 mRNA levels. Cells were grown in a hypoxia chamber for the indicated time periods, followed by qRT-PCR analysis. (G) Hypoxia increases the levels of VRK2 and SOX2 but decreases the levels of MSX2 in a time-dependent manner. Cells were grown in a hypoxia chamber for the indicated time periods, followed by IB with the indicated Abs. (H) Endogenous FBXW2–MSX2 binding. H1299 cells were left untreated or treated with CoCl2 or CoCl2 in combination with MG132 as indicated. Cells lysates were prepared and subjected to IP and IB with the indicated Abs. Data are shown as mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
We next defined a physiological condition that would activate VRK2 kinase to trigger MSX2 degradation. It has been previously reported that hypoxia induced the expression of several embryonic stem cell markers, including SOX2 (24). Since we demonstrated that the FBXW2–MSX2 axis regulates SOX2 levels, we therefore determined whether VRK2 kinase is subjected to hypoxia regulation. A computer search identified a putative HIF1 binding site (ACGTGC) in the promoter of the VRK2 gene (SI Appendix, Fig. S4C). A luciferase-based reporter assay showed that hypoxia significantly activated VRK2 promoter activity in a manner largely dependent of the HIF1 binding site (Fig. 4E). We further found that SOX2 mRNA was subjected to hypoxia induction in a time-dependent manner, whereas MSX2 mRNA was resistant to hypoxia induction (Fig. 4F), indicating that SOX2 is mainly regulated at the transcription level whereas MSX2 is at the posttranslational level. More importantly, we found a highly coordinated regulation among VRK2, MSX2, and SOX2 by hypoxia. That is, hypoxia induced by either hypoxia chamber or CoCl2 increased HIF1α levels, followed by a time-dependent VRK2 increase, MSX2 decrease, and SOX2 increase (Fig. 4G and SI Appendix, Fig. S4D). In 2 lines of renal cell carcinoma 786-O and RCC4 cells with VHL deletion and constitutive HIF1α expression, VRK2 was no longer subjected to hypoxia induction, nor to the changes of MSX2 and SOX2 levels (SI Appendix, Fig. S4E), further indicating a HIF1-dependent event. Finally, we measured endogenous binding of FBXW2–MSX2 under normoxia and chemical hypoxia conditions and found that the 2 proteins indeed bind to each other under physiological conditions. The binding was reduced upon CoCl2 treatment, likely due to CoCl2-mediated VRK2 induction, which facilitates the FBXW2–MSX2 binding for subsequent ubiquitylation and degradation, since reduced binding can be fully rescued by MG132 treatment (Fig. 4H). Collectively, it appears that hypoxia induces VRK2 to phosphorylate MSX2, which facilitates the FBXW2–MSX2 binding for subsequent MSX2 ubiquitylation and degradation, leading to SOX2 induction as a result of MSX2 derepression.
The FBXW2–MSX2–SOX2 Axis Regulates Tumor Sphere Formation.
SOX2 is a key transcription factor in maintaining pluripotent properties of stem cells. It also plays critical roles in maintaining stemness, one of the characteristics of cancer-initiating cells, and conferring resistance to chemotherapy. We therefore determined whether the negative cascade of the FBXW2–MSX2–SOX2 axis would regulate stemness and chemoresistance of cancer cells.
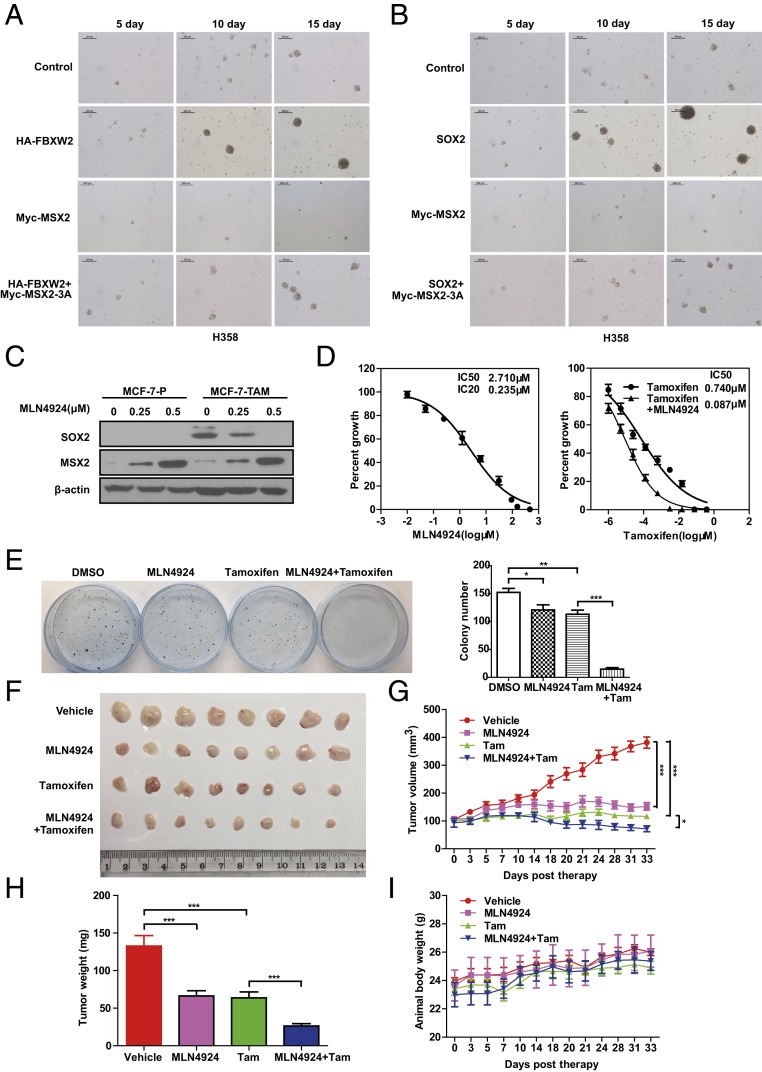
To determine stem cell-like property, we first used a tumor sphere formation assay in a serum-free suspension culture, along with the standardized sphere score (SSS) method we developed previously (25) to quantify the sphere volume. The results showed that in H358 cells, transfection of FBXW2 or MSX2 significantly promoted or inhibited sphere formation, respectively. Interestingly, FBXW2-mediated enhancement of sphere formation was completely blocked by simultaneous transfection of an FBXW2-resistant MSX2-3A mutant (Fig. 5A and SI Appendix, Fig. S5A) (P < 0.0001), indicating an MSX2-dependent event.
Fig. 5.
FBXW2–MSX2–SOX2 axis regulates tumor sphere formation and tamoxifen resistance. (A and B) Manipulation of the FBXW2–MSX2–SOX2 axis alters the ability of tumor sphere formation. H358 cells stably expressing the indicated plasmids after G418 selection at a concentration of 1 mg/mL for 2 wk were plated into ultralow attachment (ULA) plates for tumor sphere formation for up to 15 d. Representative pictures were taken. (C) MLN4924 treatment reduces SOX2 levels but increases MSX2 levels in tamoxifen-resistant cells. Cells were treated with the indicated concentrations of MLN4924 for 24 h, followed by IB with the indicated Abs. MCF-7-P, MCF-7 parental cells; MCF-7-TAM, tamoxifen-resistant MCF-7 cells. (D) MLN4924 treatment sensitizes resistant cells to tamoxifen. MCF-7-TAM cells were treated with various concentrations of MLN4924 for 72 h followed by ATPlite assay (Left). MCF-7-TAM cells were treated with various concentrations of tamoxifen alone, or in combination with an IC20 concentration of MLN4924 for 72 h, followed by ATPlite assay (Right). (E) The combined treatment of MLN4924 and tamoxifen significantly inhibits colony formation of tamoxifen-resistant cells. MCF-7-TAM cells were treated with MLN4924 or tamoxifen alone, or in combination at respective IC20 concentrations for 72 h, followed by clonogenic survival assay. Representative pictures were taken (Left) and colonies were counted and are shown as mean ± SEM of 3 independent experiments (Right). *P < 0.05, **P < 0.01, ***P < 0.001. (F–I) The combined treatment of MLN4924 and tamoxifen significantly suppresses in vivo tumor growth: A total of 1.75 × 106 MCF-7-TAM cells were injected subcutaneously into both flank sides of nude mice. When the tumors reached a volume of ∼100 mm3, the mice were randomized and treated with HPBCD (vehicle control), MLN4924, or tamoxifen alone, or in combination, as indicated. Tumor tissues were photographed (F) and weighed (H) at 33 d. The tumor growth as monitored up to 33 d and growth curves were plotted (G). The body weight of the mice was also measured and plotted (I). Shown are mean ± SEM (n = 8 for each group). The significance of the data was determined by 1-way ANOVA test. *P < 0.05, ***P < 0.001.
To determine whether induced alterations in sphere formation by FBXW2 or MSX2 manipulation are indeed mediated by SOX2, we measured endogenous SOX2 levels in the spheres of H358 cells stably expressing FBXW2 or MSX2 alone or in combination. The results showed that transfection of FBXW2 or MSX2 alone increases or decreases SOX2 levels, respectively, whereas FBXW2-mediated SOX2 increase was blocked at least in part by simultaneous transfection of an FBXW2-resistant MSX2-3A mutant (SI Appendix, Fig. S5B), suggesting a causal role of SOX2 in the process.
We then directly examined the role of the MSX2–SOX2 interaction in sphere formation and found that, as expected, SOX2 promotes sphere formation, whereas MSX2 suppressed sphere formation. Significantly, the SOX2-promoting effect was completely blocked by the MSX2-3A mutant (Fig. 5B and SI Appendix, Fig. S5C). Similar results were observed in H2170 cells, although to a lesser extent (SI Appendix, Fig. S5 D and E), suggesting a general phenomenon.
Taken together, our results clearly demonstrate that tumor sphere formation is subjected to regulation by the FBXW2–MSX2–SOX2 axis with a promoting effect by FBXW2 and SOX2 and an inhibiting effect by MSX2. Mechanistically, FBXW2-induced tumor sphere formation is mediated by depletion of MSX2 to derepress SOX2, whereas MSX2 suppresses the sphere formation by down-regulating SOX2.
The FBXW2–MSX2–SOX2 Axis Regulates Tamoxifen Resistance.
Tamoxifen-resistant breast cancer MCF-7-TAM cells were reported to express high levels of SOX2, and elevated SOX2 levels correlate with poor prognosis and development of recurrence in breast cancer patients (10). We next determined whether MLN4924 treatment would down-regulate SOX2 and induce tamoxifen sensitization in this cancer cell model. Indeed, in MCF-7-TAM cells, MLN4924 treatment significantly caused MSX2 accumulation and SOX2 reduction in a dose-dependent manner (Fig. 5C). We then determined the IC20 or IC50 values of MLN4924 in MCF-7-TAM cells, and the IC50 value of tamoxifen alone or in combination with MLN4924 at IC20 concentration. Remarkably, MLN4924 increased tamoxifen sensitivity by 8.5-fold (a reduction of IC50 value from 0.74 to 0.087 μM; Fig. 5D). A clonogenic survival assay also showed a significant enhancement of tamoxifen toxicity by MLN4924 when both were used at respective IC20 concentrations, reaching 90% of suppression (Fig. 5E).
Finally, we used an in vivo xenograft tumor model to evaluate the combined anticancer effect of tamoxifen and MLN4924 by inoculating subcutaneously the MCF-7-TAM cells into the flank sides of nude mice, followed by treatment with vehicle control, MLN4924, or tamoxifen alone or in combination. When used in nontoxic concentrations, MLN4924 or tamoxifen alone significantly suppressed in vivo tumor growth, respectively. Greater suppression was seen in combination, which is statistically significant (Fig. 5 F–I). Thus, MLN4924 sensitized MCF-7-TAM cells to tamoxifen as tested in both in vitro cell-culture and in vivo xenograft models. Taken together, the anticancer activity of MLN4924 can also be mediated by down-regulating SOX2 via targeting the FBXW2–MSX2 axis to suppress stem cell property and overcome drug resistance.
Negative Correlation of FBXW2–MSX2 Levels in Human and Mouse Tumor Tissues.
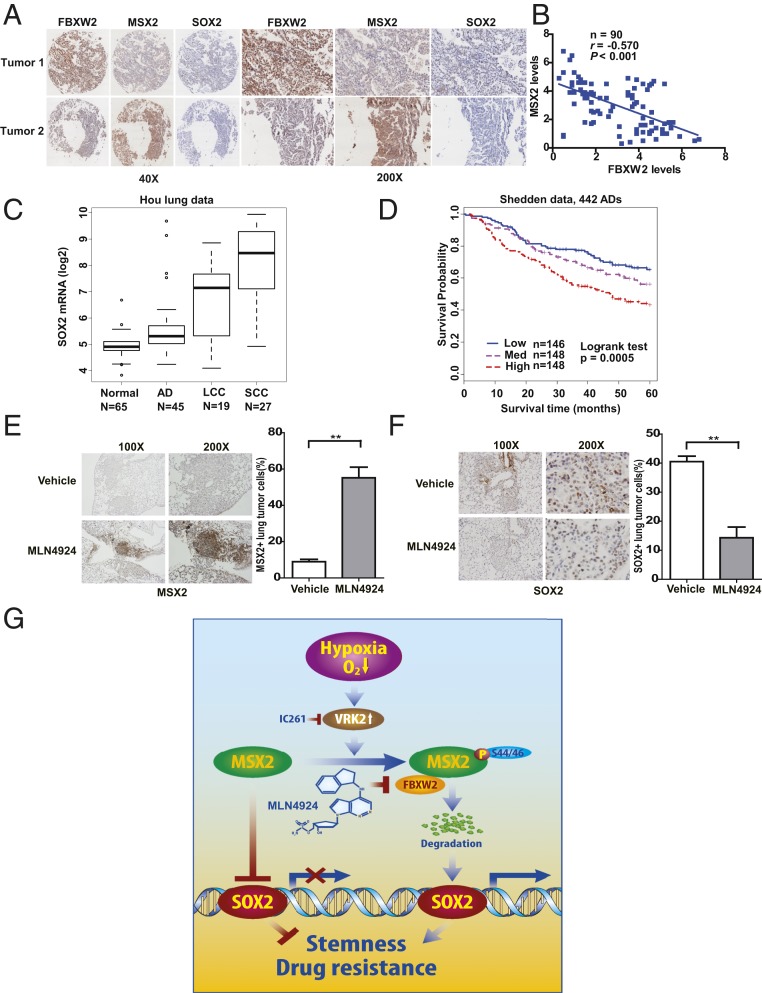
To elucidate the clinical relevance of our findings, we performed immunohistochemistry (IHC) staining to compare the levels of FBXW2, MSX2, and SOX2 in lung cancer tissues and potential correlation among them, as well as with patient survival. Examination of a set of lung cancer tissue microarrays with 90 samples revealed that SOX2 staining was overall weak in most of the samples, which precluded a correlation study. Comparison of the levels between FBXW2 and MSX2 with relatively stronger staining revealed an inverse correlation, which is statistically significant (Fig. 6 A and B). We also analyzed 2 published Affymetrix microarray datasets (26, 27) for the relationship between SOX2 expression and patient survival, and found that SOX2 mRNA levels were higher in all 3 types of lung cancer tissues than in normal tissues, and patients with high SOX2 mRNA levels had a statistically significant poorer survival (Fig. 6 C and D).
Fig. 6.
Negative correlation of FBXW2–MSX2–SOX2 expression in lung cancer tissues and the association with patient survival. (A and B) The expression of FBXW2, MSX2, and SOX2 in human lung cancer tissues: Human lung cancer tissue microarrays containing 90 human lung cancer specimens were stained with the indicated antibodies. Representative staining pictures of tumors are shown (A). Quantification of the staining intensity was performed by semiquantitative scoring with all 90 tumors (r = −0.570, P < 0.001) (B). (C and D) SOX2 mRNA levels are up-regulated in human lung cancer tissues (C) and its high expression is related to poor patient survival in lung adenocarcinomas (D). AD, adenocarcinoma; LCC, large cell carcinoma; SCC, squamous cell carcinoma. Survival fractions were plotted using the Kaplan–Meier method, and comparison of survival fractions was performed by the log-rank test. The Student’s t test was used for comparing marker expression between lung tumor and normal lung tissues. Boxplots were used to show the difference between tumor and normal. (E and F) High expression of MSX2 and low expression of SOX2 in lung tumor tissues from KrasG12D mice treated with MLN4924. KrasG12D mice were treated with MLN4924 for 4 wk, as described previously (28); the lung tissues were harvested and fixed and the sections were subjected to IHC staining with anti-MSX2 (E, Left) and anti-SOX2 Abs (F, Left). The staining was quantified and plotted (E and F, Right). Error bars represent the SEM. **P < 0.01 (n = 4). (G) Model for the FBXW2–MSX2–SOX2 axis in regulation of stem cell property and drug resistance of breast cancer cells (see text for details).
We further performed IHC staining to compare the levels of FBXW2, MSX2, and SOX2 in estrogen receptor (ER)-positive breast cancer tissue microarrays to elucidate the clinical relevance. Examination of a set of breast cancer tissue microarrays with 95 samples revealed that SOX2 staining was again overall weak in most of the samples, which precluded a correlation study. Comparison of the staining intensity between FBXW2 and MSX2 revealed again an inverse correlation, which is statistically significant (SI Appendix, Fig. S6 A and B). Interestingly, we found that FBXW2 levels are higher in ER+ tumor tissues derived from patients with recurrence, whereas MSX2 levels are lower in these samples (SI Appendix, Fig. S6 C and D). Although we do not have information on tamoxifen treatment of these patients, the data do suggest a potential association of the FBXW2–MSX2 axis with drug resistance.
Finally, we determined the expression levels and potential correlation between MSX2 and SOX2 in vivo, using a KrasG12D-triggered mouse lung tumor model, in which MLN4924 treatment significantly suppressed tumor formation (28). The results showed that in tumors with MLN4924 treatment, MSX2 expression was high whereas SOX2 expression was low (Fig. 6 E and F), demonstrating that observations made in in vitro cell-culture settings can be extended to in vivo models and thus are pathologically relevant. These association studies also suggested that the FBXW2–MSX2–SOX2 axis could be operating in the development of human lung cancer.
Discussion
MLN4924, also known as pevonedistat, is a small-molecule inhibitor of NEDD8-activating enzyme, which inhibits the entire neddylation pathway. As the first-in-class neddylation inhibitor, MLN4924 is currently in phase I/II clinical trials for anticancer application as a single agent or in combination with chemotherapeutic drugs (17, 18). MLN4924 has shown impressive anticancer activity by inactivating cullin-RING ligases to cause accumulation of tumor suppressor substrates (12). Whether MLN4924 regulates RNA expression by modulating the levels of transcription factors/repressors remains elusive. In this study, we found through RNA-seq profiling that MLN4924 significantly decreases SOX2 mRNA level. The follow-up mechanistic studies revealed that MLN4924 inhibits FBXW2 E3 ligase to cause MSX2 accumulation, which in turn transcriptionally represses SOX2, leading to suppression of stem cell property and sensitization of breast cancer cells to tamoxifen.
Given SOX2 plays a critical role in embryonic stem cell pluripotency and maintenance, cell-fate determination, and differentiation (29), SOX2 levels are precisely regulated at both transcriptional and posttranslational levels. At the transcriptional level, SOX2 is repressed by MSX2 via binding to its promoter (11), or by p21 via binding to its enhancer (30). An opposite regulation of SOX2 by cell-cycle effectors E2f3a and E2f3b was reported by targeting the SOX2 locus (31). On the other hand, SOX2 is induced by TGFβ via SOX4 (32), or by SIRT1 via epigenetic modification (33). At the posttranslational level, SOX2 was reported to be ubiquitylated and destabilized by CUL4ADET1-COP1 E3 ligase (34) and by ubiquitin-conjugating enzyme UBE2S (35), whereas methylated SOX2 was ubiquitylated and destabilized by HECT domain-containing WWP2 E3 ligase (36) and by the L3MBTL3-CRL4DCAF5 ubiquitin ligase complex (37).
MSX2 is a regulator of human pluripotent stem cell differentiation involved in the development of the skull vault, hair follicle, tooth, and mammary gland (38, 39). It was recently reported that MSX2 transcriptionally represses SOX2 expression (11). How MSX2 stability is regulated and by which E3 ligase and whether MLN4924 down-regulation of SOX2 is mediated by MSX2 are, however, totally unknown. Here we demonstrated that FBXW2 is an E3 ligase that ubiquitylates MSX2 for proteasome degradation and, by inactivation of FBXW2 E3, MLN4924 causes MSX2 accumulation to repress SOX2 expression. Specifically, we found that FBXW2 binds to MSX2 in a manner dependent on an evolutionarily conserved FBXW2 consensus-binding motif on MSX2 (43SSLPFS48). Ectopic expression or siRNA knockdown of FBXW2 decreases or increases MSX2 levels by shortening or extending MSX2 protein half-life, respectively. FBXW2 promotes MSX2 ubiquitylation and proteasome degradation, which are facilitated by VRK2 kinases. Collectively, MSX2 joins the list of GCM1, SKP2, and β-catenin as a physiological substrate of the SCFFBXW2 E3 ubiquitin ligase.
VRK2 was first identified in 1997 as a putative serine/threonine kinase with structural similarity to vaccinia virus B1R kinase (40), and was recently characterized as a candidate gene for psychiatric and neurological disorders (41). Involvement of VRK2 in human cancer is much less studied, with few reports on its interactions with p53 (42), Bcl-xL (43), and Akt (44) and its inhibition of MAPK (45). How VRK2 expression is regulated under physiological or stressed conditions is completely unknown. We recently found that VRK2 level is low in arrested cells but increases when cells enter the cell cycle (22). Here, we found that VRK2 is subjected to hypoxia up-regulation via a HIF1-binding cis-element in the promoter. VRK2 induction reduces MSX2 levels, leading to the increase of SOX2 level as a result of derepression. Our study also showed that SOX2 is subjected to hypoxia induction as well. One previous study showed that in prostate cancer, the levels of HIF1α and SOX2 were positively correlated and hypoxia induced SOX2 expression in a HIF1α-dependent manner, but as yet the mechanism remains elusive (24). We showed here that hypoxia induction of SOX2 expression is indirect and rather complicated, with a mechanism involving the VRK2–FBXW2–MSX2 axis through the removal of the SOX2 negative regulator MSX2.
After establishing the FBXW2–MSX2–SOX2 axis, we examined the potential biological significance of their negative cascade regulation. We first used a tumor sphere formation assay to measure the stem cell property of lung cancer cells after manipulation of each component. Under our assay condition, ectopic expression of SOX2 or MSX2 either triggers stemness to promote sphere formation or inhibits it, respectively. These results are expected, given the stemness-promoting role of SOX2 and the SOX2 transcription repression role of MSX2. However, it was quite surprising that ectopic FBXW2 expression significantly stimulated sphere formation.
We have recently shown that FBXW2 inhibits growth of lung cancer cells by promoting ubiquitylation and degradation of oncogenic SKP2, thus acting as a tumor suppressor (22). Most recently, we found that FBXW2 promotes ubiquitylation and degradation of oncogenic β-catenin to suppress migration and invasion of lung cancer cells, while also acting as a tumor suppressor (23). Here we showed that by promoting ubiquitylation and degradation of MSX2 to derepress SOX2, FBXW2 actually stimulated sphere formation, acting as an oncogene. The fact that the FBXW2 effect can be completely abrogated by simultaneous expression of the degradation-resistant MSX2 mutant indicates a causal role of MSX2 in mediating FBXW2 activity. Thus, in serum-starved suspension culture, FBXW2 appears to act as an oncogene to promote tumor sphere formation. Thus, it appears that FBXW2 is a context-dependent tumor suppressor or oncogene, analogous to SAG/RBX2, which acts as an oncogene in the lung (28) and prostate (46) but as a tumor suppressor in the skin (47) during mouse tumorigenesis triggered by Kras activation or Pten deletion.
It was previously reported that in MCF7 breast cancer cells, tamoxifen resistance was causally related to SOX2 overexpression (10). Given MLN4924 down-regulates SOX2, we tested our hypothesis that MLN4924 would overcome tamoxifen resistance. Indeed, by eliminating SOX2 via increasing MSX2, MLN4924 sensitized resistant breast cancer cells to tamoxifen, as measured by in vitro proliferation and survival assays in a cell-culture setting and an in vivo xenograft tumor model in mice. Given the promoting role SOX2 played in cancer progression, cancer stem cell maintenance, and drug resistance, SOX2 was validated as an attractive cancer target (48). However, SOX2 is a transcription factor, which makes direct targeting difficult. Thus, MLN4924 appears to be a promising SOX2-targeting agent via SOX2 depletion at the transcription level by inactivation of the FBXW2–MSX2 axis to abrogate cancer stem cells and to overcome drug resistance, 2 major causes of cancer therapeutic failure (49–51).
Finally, we confirmed the negative cascade of the FBXW2–MSX2–SOX2 axis can be extended to tumor tissues. An inverse correlation between the levels of FBXW2 and MSX2 was found in lung and breast cancer TMA, whereas negative correlation between the levels of MSX2 and SOX2 was evident in mouse lung tissues triggered by KrasG12D, followed by MLN4924 treatment. Thus, the FBXW2–MSX2–SOX2 axis may operate during the development of lung cancer.
In summary, our study reveals an interesting biochemical interplay among FBXW2, MSX2, and SOX2 with biological significance. Upon hypoxia, VRK2 is induced to facilitate FBXW2–MSX2 binding for subsequent MSX2 ubiquitylation and degradation. By inactivating FBXW2, MLN4924 causes MSX2 accumulation to transcriptionally repress SOX2, leading to suppression of tumor sphere formation and sensitization of tamoxifen resistance (Fig. 6G). Our study therefore establishes a negative cascade of the FBXW2–MSX2–SOX2 axis which regulates cancer cell stemness and drug resistance, and reveals a mechanism of MLN4924 anticancer action.
Materials and Methods
Cell Culture and the Generation of the Fbxw2fl/fl Model.
Human embryonic kidney 293 (HEK293) cells and lung cancer H1299 and A549 cells were obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (vol/vol) FBS (Invitrogen). Lung cancer H2170 and H358 cells were cultured in RPMI medium 1640 with 10% FBS. Breast cancer MCF-7 parental (MCF-7-P) cells were cultured in DMEM supplemented with 10% FBS, whereas tamoxifen-resistant MCF-7-TAM cells were cultured in phenol red-free DMEM supplemented with 5% charcoal-stripped serum as described (52, 53).
The Fbxw2fl/fl mouse model was generated at the UM Transgenic Core by injecting Fbxw2-targeted ES cell clones (purchased from the European Mouse Mutant Cell Repository; https://www.eummcr.org/) into blastocysts to generate heterozygous Fbxw2fl-neo/+ lines with germline transmission. FLP mice were used to remove the neomycin cassette, and intercrossing of Fbxw2fl/+ to generate Fbxw2fl/fl. The Fbxw2∆/∆ mice were generated via crossing Fbxw2fl/fl mice with EIIa-Cre transgenic mice and were viable. Mouse embryonic fibroblasts were generated from embryonic day (E)13.5 embryos of these mice as described (44) and cultured in DMEM with 15% FBS, 2 mM l-glutamine, and 0.1 mM MEM nonessential amino acids. Fbxw2fl/fl MEFs were infected with Ad-Cre to remove exon 4 to deplete Fbxw2, along with Ad-GFP control. All cell lines were routinely examined to ensure they were free of mycoplasma contamination.
In Vivo Ubiquitylation Assay.
Cells were cotransfected with FBXW2, His-HA-Ub, and MSX2 or MSX2-3A, along with mock vector or FBXW2-∆F controls. To define ubiquitin linkage in the MSX2 polyubiquitylation chain, 3 ubiquitin mutants, K11R, K48R and K63R, were used, along with a wild-type control in the transfection. Cells were then lysed in 6 M guanidine denaturing solution as described (54). Polyubiquitinated MSX2 was pulled down by Ni-NTA beads (QIAGEN) and detected by immunoblotting using anti-MSX2 or anti-Myc antibody.
In Vitro Ubiquitylation Assay.
HA-FBXW2 or HA-FBXW2-∆F was respectively pulled down from HEK293 cells transfected with either plasmid using HA beads (Sigma) and then eluted with HA peptide (Sigma), serving as the source of E3. Myc-tagged MSX2 or MSX2-3A was pulled down by Myc beads (Sigma) from HEK293 cells transfected with the corresponding construct, serving as the substrate. The reaction was carried out at 30 °C for 1 h in 30 μL reaction buffer (40 mM Tris⋅HCl, pH 7.5, 2 mM DTT, 5 mM MgCl2) in the presence of purified substrate, E1, E2, the above E3s, ATP, and ubiquitin. The reaction products were then resolved by SDS/PAGE and detected by immunoblotting with anti–c-Myc antibody.
Sphere Formation Assay.
For the tumor sphere assay, cells were counted and plated on 24-well ultralow attachment plates (Corning) at a density of 1 cell per microliter in conditioned medium. Cells were then grown in serum-free DMEM/F12 medium supplemented with 0.4% BSA, 5 mM Hepes, 1× penicillin-streptomycin (Gibco), 20 ng/mL EGF (Life Technologies), and 10 ng/mL bFGF (Sigma), unless otherwise indicated. SSSs are documented, as previously described (25), every 5 d for up to 15 d.
In Vivo Antitumor Study.
Five- to 6-wk-old BALB/c athymic nude mice (nu/nu, female) were used according to a protocol approved by the University of Michigan Committee for Use and Care of Animals. A total of 1.75 × 106 MCF-7 tamoxifen-resistant cells (MCF-7-TAM) were mixed 1:1 with Matrigel (BD Biosciences) in a total volume of 0.2 mL and were then subcutaneously injected into both flanks of mice. When the tumors reached a volume of ∼100 mm3, the mice were then randomized into 4 groups (4 mice per group). MLN4924 (30 mg/kg, s.c.) was given once a day, 5 d a week, for 33 d; tamoxifen (20 mg/kg, i.p.) was administered once every 3 d for 33 d. Mice in the drug control group received 10% 2-hydroxypropyl-β-cyclodextrin (HPBCD) as the vehicle control. The growth of tumors was measured at the indicated time points and average tumor volumes were calculated according to the equation, volume = (length × width × width)/2.
Primary Tumor-Derived Gene Expression Datasets.
Two published Affymetrix microarray datasets (26, 27) were used in patient survival or tumor vs. normal comparison analyses for SOX2 expression. The CEL files of microarray data were normalized using the robust multiarray average method and log2-transformed data were used as described (28).
Statistical Analysis.
Overall survival was the outcome for Shedden et al.’s dataset, and it was censored at 5 y (27). Survival was plotted using the Kaplan–Meier method, and comparison of survival was performed by log-rank test. The Student’s t test was used for comparing gene expression in lung tumor and normal lung tissues. Boxplots were used to show the difference between tumor and normal tissue. The data were expressed as mean ± SEM, and were subjected to Student’s t test, Mann–Whitney U test, or 1-way ANOVA test. All statistical analyses were 2-sided, and different cutoff values, *P < 0.05, **P < 0.01, and ***P < 0.001, were considered significant. Statistical significance was determined as P < 0.05. The SPSS 16.0 package was used for the statistical analyses.
Supplementary Material
Acknowledgments
This work was supported in part by the National Key R&D Program of China (2016YFA0501800) (to Y.S.); National Cancer Institute Grant CA156744 (to Y.S.); National Natural Science Foundation of China Grant 81772636 (to Y.Y.); and in part by the Breast Cancer Research Foundation (BCRF-18-145), National Cancer Institute grants, Specialized Programs of Research Excellence Grant P50CA186784, and Cancer Center Grant P30CA125123 (to R.S.) for the establishment of MCF-7-TAM cells.
Footnotes
Conflict of interest statement: R.S. has received research funding from AstraZeneca, GlaxoSmithKline, Gilead Sciences, and PUMA Biotechnology and is a consulting/advisory committee member for Macrogenics and Eli Lilly. These funds, however, had no impact on the content of this study.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE134190).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905973116/-/DCSupplemental.
References
- 1.Ajani J. A., Song S., Hochster H. S., Steinberg I. B., Cancer stem cells: The promise and the potential. Semin. Oncol. 42 (suppl. 1), S3–S17 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Oskarsson T., Batlle E., Massagué J., Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell 14, 306–321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck B., Blanpain C., Unravelling cancer stem cell potential. Nat. Rev. Cancer 13, 727–738 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Dean M., Fojo T., Bates S., Tumour stem cells and drug resistance. Nat. Rev. Cancer 5, 275–284 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Sarkar A., Hochedlinger K., The Sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu K., et al. , The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell. Signal. 25, 1264–1271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bass A. J., et al. , SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 41, 1238–1242 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leis O., et al. , Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 31, 1354–1365 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Zhang J., Chang D. Y., Mercado-Uribe I., Liu J., Sex-determining region Y-box 2 expression predicts poor prognosis in human ovarian carcinoma. Hum. Pathol. 43, 1405–1412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piva M., et al. , Sox2 promotes tamoxifen resistance in breast cancer cells. EMBO Mol. Med. 6, 66–79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q., et al. , MSX2 mediates entry of human pluripotent stem cells into mesendoderm by simultaneously suppressing SOX2 and activating NODAL signaling. Cell Res. 25, 1314–1332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y., Sun Y., Cullin-RING ligases as attractive anti-cancer targets. Curr. Pharm. Des. 19, 3215–3225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardozo T., Pagano M., The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Jin J., et al. , Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia L., Sun Y., SCF E3 ubiquitin ligases as anticancer targets. Curr. Cancer Drug Targets 11, 347–356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soucy T. A., et al. , An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Zhou L., Zhang W., Sun Y., Jia L., Protein neddylation and its alterations in human cancers for targeted therapy. Cell. Signal. 44, 92–102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y., Morgan M. A., Sun Y., Targeting neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox Signal. 21, 2383–2400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nawrocki S. T., Griffin P., Kelly K. R., Carew J. S., MLN4924: A novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin. Investig. Drugs 21, 1563–1573 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Zhou X., et al. , Blockage of neddylation modification stimulates tumor sphere formation in vitro and stem cell differentiation and wound healing in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, E2935–E2944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie C., Sun Y., Regulation of stem cell property and drug resistance of cancer cells by targeting transcriptional machinery via inhibition of neddylation. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134190. Deposited 12 July 2019.
- 22.Xu J., et al. , The β-TrCP-FBXW2-SKP2 axis regulates lung cancer cell growth with FBXW2 acting as a tumour suppressor. Nat. Commun. 8, 14002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang F., et al. , FBXW2 suppresses migration and invasion of lung cancer cells via promoting β-catenin ubiquitylation and degradation. Nat. Commun. 10, 1382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae K. M., Dai Y., Vieweg J., Siemann D. W., Hypoxia regulates SOX2 expression to promote prostate cancer cell invasion and sphere formation. Am. J. Cancer Res. 6, 1078–1088 (2016). [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X., Wang G., Sun Y., A reliable parameter to standardize the scoring of stem cell spheres. PLoS One 10, e0127348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou J., et al. , Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One 5, e10312 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shedden K., et al. ; Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma , Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat. Med. 14, 822–827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., et al. , Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J. Clin. Invest. 124, 835–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng R., Wen J., Overview of the roles of Sox2 in stem cell and development. Biol. Chem. 396, 883–891 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Marqués-Torrejón M. A., et al. , Cyclin-dependent kinase inhibitor p21 controls adult neural stem cell expansion by regulating Sox2 gene expression. Cell Stem Cell 12, 88–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Julian L. M., et al. , Opposing regulation of Sox2 by cell-cycle effectors E2f3a and E2f3b in neural stem cells. Cell Stem Cell 12, 440–452 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Ikushima H., et al. , Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell 5, 504–514 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Liu L., et al. , SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells. Hepatology 64, 814–827 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Cui C. P., et al. , Dynamic ubiquitylation of Sox2 regulates proteostasis and governs neural progenitor cell differentiation. Nat. Commun. 9, 4648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., et al. , Ube2s regulates Sox2 stability and mouse ES cell maintenance. Cell Death Differ. 23, 393–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang L., et al. , A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 55, 537–551 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Zhang C., et al. , Proteolysis of methylated SOX2 protein is regulated by L3MBTL3 and CRL4DCAF5 ubiquitin ligase. J. Biol. Chem. 294, 476–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Liu J., Hudson M., Kim S., Hatch N. E., FGF2 promotes Msx2 stimulated PC-1 expression via Frs2/MAPK signaling. J. Cell. Biochem. 111, 1346–1358 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Satokata I., et al. , Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat. Genet. 24, 391–395 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Nezu J., Oku A., Jones M. H., Shimane M., Identification of two novel human putative serine/threonine kinases, VRK1 and VRK2, with structural similarity to vaccinia virus B1R kinase. Genomics 45, 327–331 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Li M., Yue W., VRK2, a candidate gene for psychiatric and neurological disorders. Mol. Neuropsychiatry 4, 119–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanco S., Klimcakova L., Vega F. M., Lazo P. A., The subcellular localization of vaccinia-related kinase-2 (VRK2) isoforms determines their different effect on p53 stability in tumour cell lines. FEBS J. 273, 2487–2504 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Monsalve D. M., et al. , Human VRK2 modulates apoptosis by interaction with Bcl-xL and regulation of BAX gene expression. Cell Death Dis. 4, e513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirata N., et al. , Functional characterization of lysosomal interaction of Akt with VRK2. Oncogene 37, 5367–5386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernández I. F., Blanco S., Lozano J., Lazo P. A., VRK2 inhibits mitogen-activated protein kinase signaling and inversely correlates with ErbB2 in human breast cancer. Mol. Cell. Biol. 30, 4687–4697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan M., Xu J., Siddiqui J., Feng F., Sun Y., Depletion of SAG/RBX2 E3 ubiquitin ligase suppresses prostate tumorigenesis via inactivation of the PI3K/AKT/mTOR axis. Mol. Cancer 15, 81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie C. M., et al. , Erbin is a novel substrate of the Sag-βTrCP E3 ligase that regulates KrasG12D-induced skin tumorigenesis. J. Cell Biol. 209, 721–737 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hüser L., Novak D., Umansky V., Altevogt P., Utikal J., Targeting SOX2 in anticancer therapy. Expert Opin. Ther. Targets 22, 983–991 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Dawood S., Austin L., Cristofanilli M., Cancer stem cells: Implications for cancer therapy. Oncology (Williston Park) 28, 1101–1107, 1110 (2014). [PubMed] [Google Scholar]
- 50.Prieto-Vila M., Takahashi R. U., Usuba W., Kohama I., Ochiya T., Drug resistance driven by cancer stem cells and their niche. Int. J. Mol. Sci. 18, E2574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu G., et al. , Overcoming treatment resistance in cancer: Current understanding and tactics. Cancer Lett. 387, 69–76 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Nardone A., et al. , The oral selective oestrogen receptor degrader (SERD) AZD9496 is comparable to fulvestrant in antagonising ER and circumventing endocrine resistance. Br. J. Cancer 120, 331–339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fu X., et al. , FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc. Natl. Acad. Sci. U.S.A. 113, E6600–E6609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu Q., Tan M., Sun Y., SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 67, 3616–3625 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.