Significance
Many parasites escape the host immune system by undergoing antigenic variation, a process in which surface antigens are regularly shed and replaced by new ones. Trypanosoma brucei employs multiple sophisticated molecular mechanisms to ensure the expression of a homogeneous VSG coat. We generated a mutant parasite that expresses multiple distinct VSGs and studied the consequences of having a multi-VSG coat during an infection. We showed that expression of multiple VSGs makes the parasites more vulnerable to the immune response, which can now control the trypanosomes from the onset of the infection, allowing most mice to survive. In the future, trypanosome infections may be treated using drugs that generate parasites with multi-VSG coats.
Keywords: Trypanosoma brucei, variant surface glycoprotein, monoallelic expression, adaptive immune response, TDP1
Abstract
Trypanosoma brucei parasites successfully evade the host immune system by periodically switching the dense coat of variant surface glycoprotein (VSG) at the cell surface. Each parasite expresses VSGs in a monoallelic fashion that is tightly regulated. The consequences of exposing multiple VSGs during an infection, in terms of antibody response and disease severity, remain unknown. In this study, we overexpressed a high-mobility group box protein, TDP1, which was sufficient to open the chromatin of silent VSG expression sites, to disrupt VSG monoallelic expression, and to generate viable and healthy parasites with a mixed VSG coat. Mice infected with these parasites mounted a multi-VSG antibody response, which rapidly reduced parasitemia. Consequently, we observed prolonged survival in which nearly 90% of the mice survived a 30-d period of infection with undetectable parasitemia. Immunodeficient RAG2 knock-out mice were unable to control infection with TDP1-overexpressing parasites, showing that the adaptive immune response is critical to reducing disease severity. This study shows that simultaneous exposure of multiple VSGs is highly detrimental to the parasite, even at the very early stages of infection, suggesting that drugs that disrupt VSG monoallelic expression could be used to treat trypanosomiasis.
High-mobility group box (HMGB) proteins are characterized by their small size, rapid mobility, and by containing at least one high-mobility group box domain, responsible for DNA binding (1, 2). HMGB proteins influence transcription by 3 different mechanisms: They facilitate nucleosome sliding by chromatin remodeling proteins (3, 4), they serve as transient chaperones for stable binding of transcription factors (5), or they participate in transcription blockage as part of a complex that inhibits the assembly of a preinitiation complex on promoters (6). HMGB proteins also participate in the regulation of ribosomal DNA (rDNA) genes, which are transcribed by RNA polymerase I (Pol I). Upstream binding factor (UBF) in mammals and Hmo1 in yeast are 2 HMGB proteins important for the chromatin structure and transcription of rDNA loci (7, 8). Both UBF and Hmo1 bind throughout actively transcribed rDNA (9, 10) while only UBF binds to the rDNA promoter as part of the preinitiation complex (7). In yeast, a similar role is fulfilled at the promoter by upstream-associated factor (UAF) (11). Nonetheless, UBF and Hmo1 are important to establish and maintain an open chromatin conformation at Pol I loci (12).
Trypanosoma brucei is a unicellular parasite that causes African trypanosomiasis, generally a deadly disease if untreated. T. brucei is able to evade the host immune response, which is dependent on B cells to control peaks of parasitemia, although it remains unclear which immunoglobulins (Igs) are involved in this process (13, 14). To avoid being eliminated by the immune system, trypanosomes use antigenic variation, a process that consists in periodically switching the major cell-surface protein, the variant surface glycoprotein (VSG). Although the T. brucei genome encodes more than 2,000 VSG genes and pseudogenes (15), only one VSG is transcribed at any time in bloodstream forms (BSFs), from 1 of the ∼15 subtelomeric bloodstream expression sites (BESs) that are present in the genome (16). Monoallelic VSG expression, together with VSG switching, is the hallmark of antigenic variation.
During the life cycle of T. brucei, parasites shift between a mammalian host and the tsetse (Glossina) vector (17). To survive in such different hosts, parasites undergo major changes in gene expression, which include replacement of VSGs by procyclins in procyclic stages (18), and culminate in reexpression of VSGs in the metacyclic forms, which reinfect a mammalian host (19). Metacyclic VSG genes are transcribed from subtelomeric loci called metacyclic expression sites (MESs). MESs are transcribed monocistronically (16, 20, 21), in contrast to BESs, which are polycistronic units that also encode a variable number of expression site-associated genes (ESAGs). On the other hand, procyclin genes are located in the internal region of chromosomes distributed among 2 loci (22). Strikingly, both VSG- and procyclin-coding genes are transcribed by Pol I (23, 24).
In bloodstream forms, the chromatin structures of transcribed and silent BESs are significantly different. The actively transcribed BES is depleted of nucleosomes (open state) while silent BESs are enriched in regularly spaced nucleosomes (closed state) (25, 26). Procyclin loci possess a more open chromatin structure than silent BESs, which is consistent with the fact that procyclin genes are transcribed in BSFs, but at a lower rate than in procyclic forms (27). Posttranscriptional repression mechanisms also contribute to preventing expression of procyclin proteins (28). MESs are not (or are very poorly) transcribed in BSF and have a closed chromatin structure (29).
Besides the multisubunit class I transcription factor A (CITFA), which binds only to active BES promoters and is necessary to initiate BES transcription (30), TDP1 is the only core structural component described so far in the active BES (31). TDP1, one of the few HMGB genes in T. brucei, encodes the only identified nuclear HMGB protein, which is highly enriched in regions transcribed by Pol I, namely the active BES and rDNA genes (31). TDP1 is regarded as a Pol I transcription facilitator, and its depletion results in a striking growth arrest, increased histone abundance, with concomitant chromatin repression at actively transcribed Pol I loci, and reduction of VSG transcription. We have previously shown that TDP1 is important in maintaining the open chromatin structure of the active BES after inducing transcriptional silencing (32). This functional role allows the probing of silent BESs before commitment to switching to a new BES, or returning to the initial BESs, demonstrating the importance of TDP1 in antigenic variation.
In the current study, we characterized the phenotype of a TDP1 overexpression mutant in VSG gene regulation. We found that TDP1 overexpression decondensed the chromatin of silent loci transcribed by Pol I, leading to a major disruption of VSG monoallelic expression, without significantly affecting Pol II transcription. During a mouse infection, these mutant parasites were less proficient at escaping the host immune system, which now generated antibodies against several VSGs, prolonging survival of the host. Together with our previous results, these observations highlight the importance of TDP1 as a key factor in antigenic variation where its levels need to be tightly regulated.
Results
TDP1 Overexpression Is Well-Tolerated In Vitro.
HMGB proteins UBF and Hmo1 are structural chromatin components with an essential role in Pol I elongation (8, 33). Overexpression of UBF1, one of the proteins that compose the UBF complex, increases rDNA transcription (34). On the other hand, overexpression of Hmo1 causes yeast cells to enter vegetative growth (35). The T. brucei HMGB protein TDP1 is a core component of chromatin at Pol I loci. In this study, we assessed whether TDP1 overexpression interferes with antigenic variation in T. brucei.
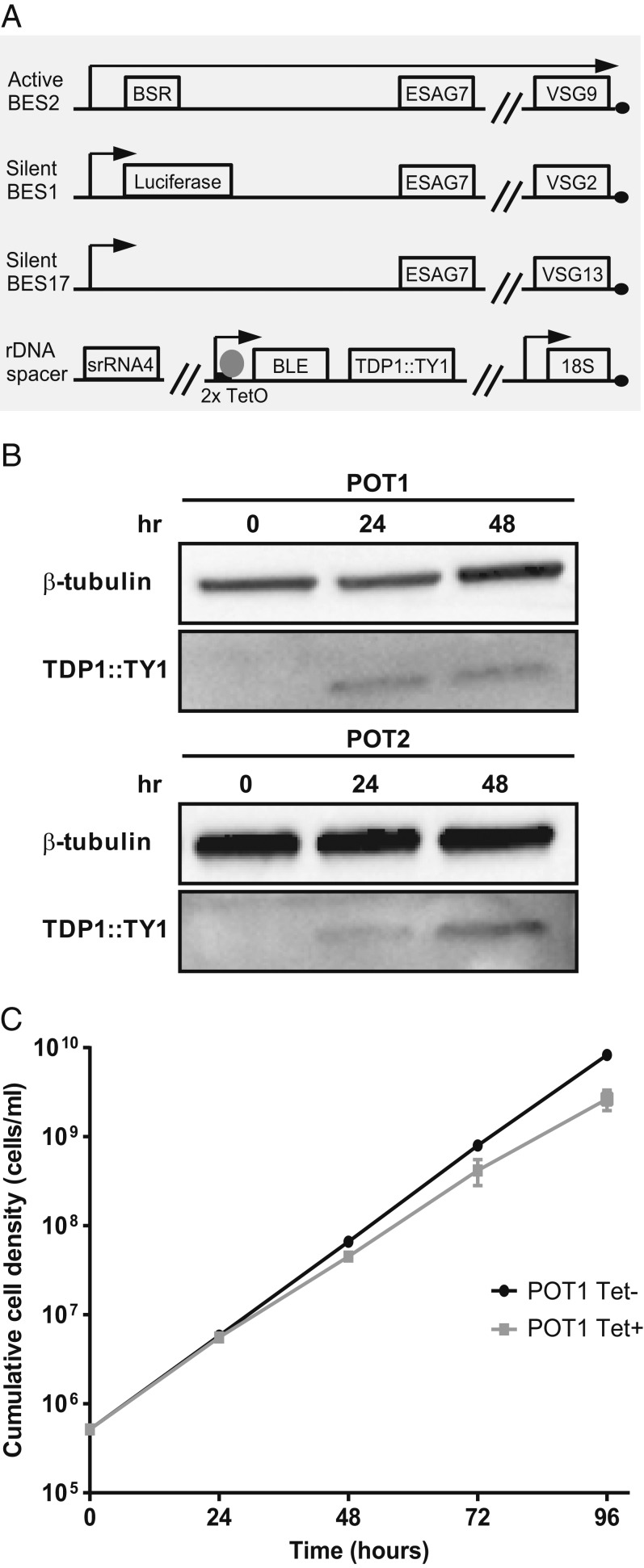
A tetracycline-inducible system was first generated to conditionally overexpress a C-terminal TY1 epitope-tagged TDP1 gene from the rDNA spacer locus in the PL1S cell line (Fig. 1A) (36). PL1S expresses VSG9 from BES2 and contains a luciferase reporter gene in the promoter region of silent BES1. Two independent clones from this PL1S overexpressing TDP1 cell line were named POT1 and POT2. Overexpression was first confirmed by Western blotting at 24 and 48 h (Fig. 1B). Expression of the TY1-tagged allele was undetectable before induction, indicating a tight control of the inducible expression in both clones. After inducing expression, clear bands of the expected size could be detected in both clones. However, at 24 h, intensity was higher for POT1 while, at 48 h, it was higher for POT2, indicating that overexpression was successful for each clone, but to different extents. Both clones grew normally within the first 24 h of overexpression, after which a small reduction in growth was detected (Fig. 1C and SI Appendix, Fig. S1A), but lethality was never observed at later time points. In accordance with the minor growth rate changes, the cell cycle profile of POT2 clone was normal after 24 h of TDP1 overexpression but showed small but significant changes at 48 h (SI Appendix, Fig. S1B). It is possible that this small growth defect may be due to the expression of multiple VSGs since this defect has also been observed in histone H1-depleted parasites (29) and VEX1 overexpressors (37). We conclude that, unlike Hmo1 in yeast, TDP1 overexpression causes only mild growth defects.
Fig. 1.
Inducible overexpression of TDP1 in bloodstream forms. (A) The cell line POT is a derivative of PL1S (36), in which a tetracycline-dependent overexpression construct for TDP1::TY1 was integrated in an rDNA spacer locus. (B) Western blotting analysis of TDP1 protein after 24 and 48 h of overexpression in 2 POT clones. Time point 0 h indicates noninduced condition and shows no TDP1::TY1 protein. Each lane corresponds to lysates from 1 × 106 cells. (C) Growth curve of POT1 before (Tet−, black curve) or after (Tet+, gray curve) induction of overexpression. Three independent experiments were analyzed and are represented as mean ± SEM.
Deposition of TDP1 Specifically Facilitates Pol I Transcription.
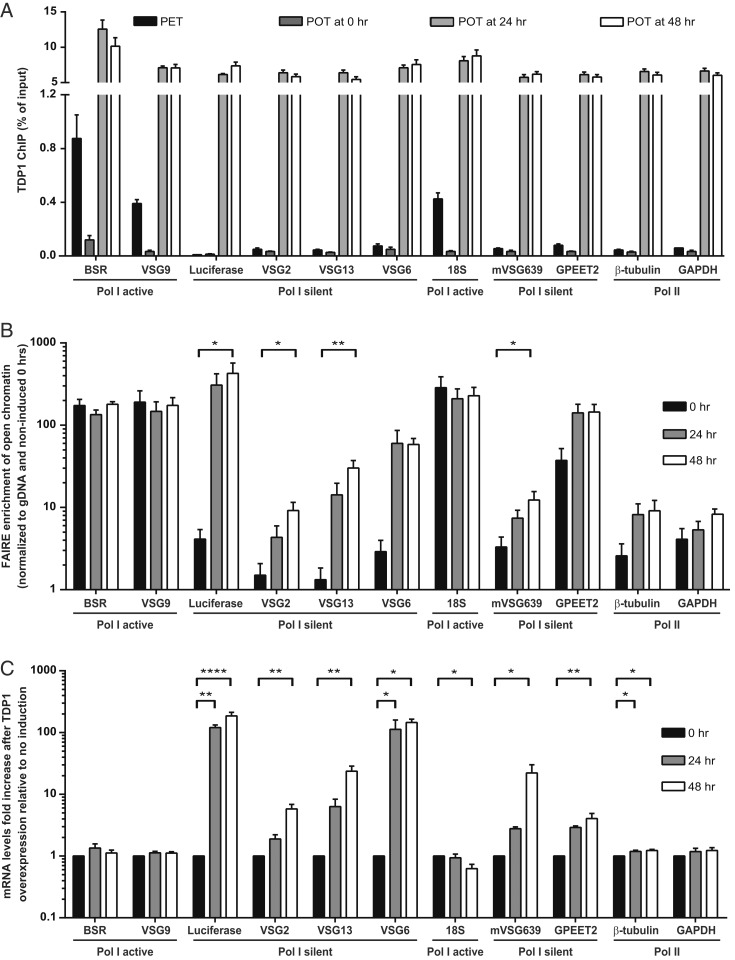
T. brucei TDP1 keeps chromatin of active BESs open in the presence (31) and absence (32) of transcription. Given that TDP1 is a chromatin component of the actively transcribed BES, we hypothesized that overexpression of TDP1 would lead to its loading in sites typically devoid of TDP1. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) using an antibody that recognizes the TY1 tag present in the induced TDP1 allele (Fig. 2A). To quantify the enrichment of TDP1 relative to wild-type levels, we generated an isogenic cell line, PET (PL1S with endogenous TDP1 tag), which TY1 tags an endogenous TDP1 allele. As previously reported (31, 32), ChIP revealed that the endogenous TDP1 is present mainly in the actively transcribed Pol I loci BSR (blasticidin-S resistance gene), VSG9, and 18S rDNA genes, with very low levels detected at Pol II and Pol III transcribed genes (Fig. 2A). Upon TDP1 overexpression, the levels of TDP1 dramatically increased in all tested loci, including Pol II transcribed genes (β-tubulin and GAPDH), Pol I silent loci (silent BESs, GPEET2, and mVSG639), and the actively transcribed BESs (BSR and VSG9). The efficiency of chromatin immunoprecipitation was very similar for all loci, suggesting that TDP1 overexpression reduces its specificity.
Fig. 2.
TDP1 overexpression opens the chromatin structure of many loci, but only Pol I silent genes become significantly up-regulated. (A) TDP1 ChIP at 0, 24, and 48 h after TDP1 overexpression induction in POT1 and in PET cell line, a control cell line isogenic to POT cell line in which an endogenous allele of TDP1 is TY1-tagged instead of containing a TDP1 overexpression construct. Immunoprecipitated DNA was compared to the total input material. Four independent experiments were analyzed and are represented as mean with SEM. (B) Chromatin conformation was measured by FAIRE at 24 and 48 h after overexpression induction in POT1. DNA isolated by FAIRE was quantified by qPCR and normalized to genomic DNA (gDNA) copy number and ampicillin gene from DNA spike. Four independent experiments were analyzed and are represented as mean with SEM. (C) Fold-increase of mRNA levels after 24 h of overexpression induction in POT1, measured by qPCR and normalized to noninduced cells and VSG9 gene. Three to 4 independent experiments were performed and are represented as mean ± SEM. (B and C) Statistical significance was determined by 1-way ANOVA with Bonferroni posttest comparison for each gene. *P value < 0.05; **P value < 0.01; ****P value < 0.0001.
To measure the effect of increased TDP1 in chromatin conformation, we employed formaldehyde-assisted isolation of regulatory elements (FAIRE), a method that preferentially extracts chromosomal DNA that is poorly bound to histones: i.e., genomic loci present in an open chromatin conformation (25, 29, 32). Chromatin was prepared 24 and 48 h after TDP1 overexpression in POT1 (Fig. 2B). At both time points, the chromatin of silent BESs was clearly more open relative to the noninduced condition (Luciferase, VSG2, VSG13, VSG6). At 48 h, the promoter region of silent BESs, marked by Luciferase, opened ∼100-fold whereas the corresponding subtelomeric VSG (VSG2) opened only 9-fold. This indicates that chromatin of silent BESs is not fully open/decondensed and that there is a position-dependent effect, as previously reported in other mutants (38). Interestingly, although the levels of TDP1 were similar in the silent VSG2 and VSG13, the chromatin of the latter became 22-fold more open, indicating that TDP1 is not the sole determinant of chromatin structure at silent BESs.
Albeit less dramatically, the chromatin of other Pol I loci also became more open. Chromatin at the silent MES (mVSG639) became 4-fold more open whereas the procyclin loci (GPEET2) became 5-fold more open (Fig. 2B). Although we observed increased binding of TDP1 in genes from the active BESs (BSR and VSG9) and rDNA (18S), the chromatin conformation remained unchanged, probably because their chromatin is already fully open in the noninduced condition. Finally, we also detected a slight but statistically insignificant increase in chromatin accessibility of 2 genes transcribed by Pol II: β-tubulin (4-fold) and GAPDH (3-fold). The combined results show that, despite the overall increase in TDP1 association with chromatin, the effects on different gene loci were varied and dependent on both the starting levels of chromatin condensation and the RNA polymerase transcribing that locus.
Next, we asked whether an increase in chromatin accessibility in silent BESs would lead to higher levels of BES transcription. To answer this question, transcript levels of several genes were measured 24 and 48 h after inducing TDP1 overexpression (Fig. 2C). Transcript levels of genes in silent BESs were increased although the increase was higher in the promoter region (Luciferase, ∼70- to 200-fold increase) relative to the telomeric region (VSG2 and VSG13, 3- to 5-fold and 20- to 30-fold increase, respectively). These results are highly consistent with the chromatin accessibility measured by FAIRE; those genes in which chromatin opened more were also more up-regulated. Transcripts of life cycle-regulated MESs (mVSG639, 5- to 16-fold increase) and procyclin (GPEET2, 3- to 4-fold increase) genes were also slightly up-regulated, indicating that all Pol I low-transcribed genes were derepressed. Conversely, active BESs (BSR and VSG9) and rDNA (18S) genomic regions maintained their transcription status, which is consistent with no changes in chromatin conformation (Fig. 2B). Interestingly, transcript levels of Pol II highly transcribed genes (i.e., β-tubulin and GAPDH) and lowly transcribed genes (inhibitor of serine peptidase gene [ISP] and procyclin associated gene 3 [PAG3]) were not affected by TDP1 overexpression, even though the chromatin of these loci was opened to a similar degree to the procyclin (GPEET2) and mVSG639 (Fig. 2 B and C and SI Appendix, Fig. S2). These results show that TDP1 is sufficient to increase transcript levels of several types of Pol I-dependent loci, but not of Pol II loci.
Overall, we observed that TDP1 overexpression opened the chromatin structure of all tested genes whose chromatin is organized in regularly spaced nucleosomes, but its effect is more pronounced in the chromatin of silent BESs. Consistently, decondensation of chromatin was associated with an increase of transcript levels of Pol I loci but not of Pol II, which agrees with the role of TDP1 as a Pol I transcription facilitator.
TDP1 Overexpression Disrupts VSG Monoallelic Expression.
VSG monoallelic expression is a hallmark of antigenic variation. Although derepression of silent BESs has been previously observed in several T. brucei mutants (39, 40), the expression of several VSGs at the cell surface has only been assessed and detected in a few cases (36, 37, 41–44). Hence, next, we tested if the transcriptional up-regulation in silent BESs caused by TDP1 overexpression could be detected at the protein level.
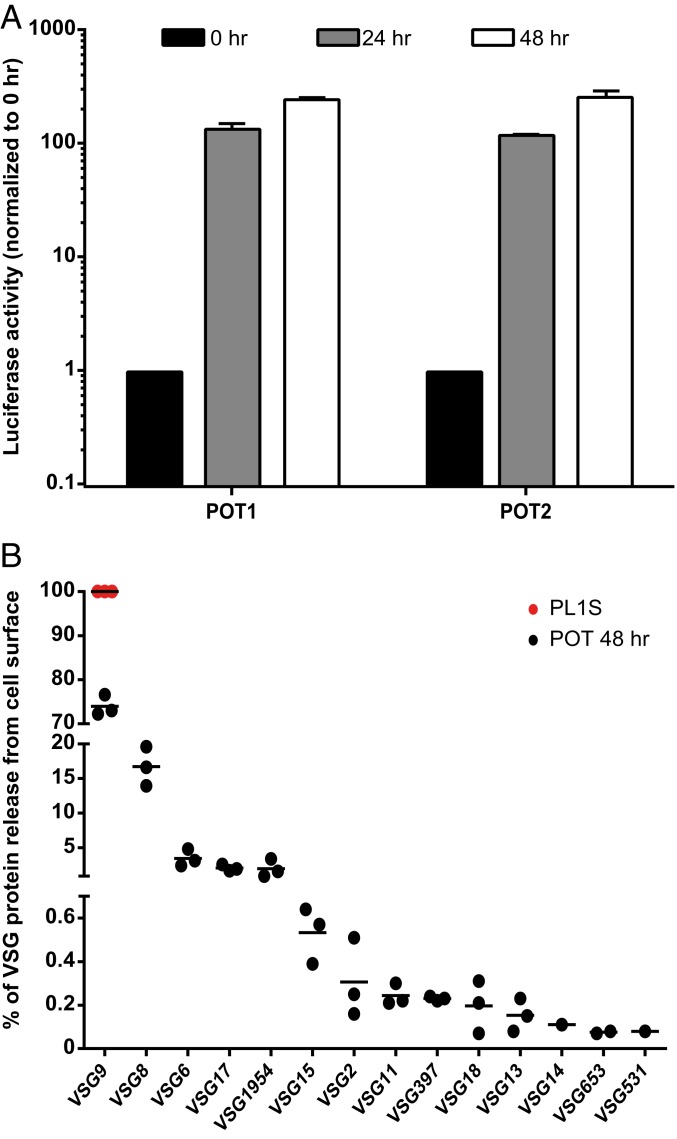
First, we measured the activity of the luciferase reporter, which is located downstream of the promoter of the silent BES1. Given that the mRNA levels of this gene were highly increased (Fig. 2C), we expected its luminescence activity to be concomitantly higher (Fig. 3A). Indeed, after 48 h of TDP1 overexpression, the luciferase activity was higher in both clones and proportional to the increase at the mRNA levels. Luciferase activity increased almost 250-fold, which is much higher than found in previous studies (maximum of 40-fold in TbRAP1 knockdown cells and maximum of 10-fold in histone H1 knockdown cells) (29, 36). Although this increase is very significant, it is still below the levels of luciferase activity when its gene is localized in the active BES (typically 1,000- to 4,000-fold higher) (36), indicating that TDP1 is necessary but not sufficient for full activation of silent BESs.
Fig. 3.
TDP1 overexpression derepresses previously silent VSGs, which become exposed at the cell surface. (A) Luciferase activity fold-increase of TDP1 overexpressed cells relative to noninduced cells after 24 and 48 h of induction for POT1 and POT2. Five to 6 independent experiments were analyzed for each time point and are represented as mean ± SEM. (B) Quantification of VSG proteome at the cell surface after 48 h of TDP1 overexpression in POT2. The PL1S parental cell line was used as a noninduced control. Three independent experiments were analyzed.
To test the presence of derepressed VSG protein, we used quantitative proteomics (37) (Fig. 3B). Cell-surface VSG was obtained by activating the parasite endogenous phospholipase C, which cleaves the dimyristoylglycerol lipid component of the VSG GPI anchor and releases the soluble VSG onto the supernatant (45), from which it was purified and analyzed by mass spectrometry (MS). As expected, the control cell line PL1S only presented VSG9 at the cell surface (100%). However, 48 h after TDP1 overexpression, VSG9 relative abundance decreased to 74%. The rest of the VSG coat was composed of 13 BES- and MES-associated VSGs, especially VSG8, VSG6, VSG17, and metacyclic VSG1954. As observed at the RNA level, the 3 independent MS experiments consistently showed that some BESs are more easily up-regulated than others or confer greater fitness in culture. Furthermore, of the 13 derepressed VSGs, 10 were detected in all 3 experiments, and the 3 remaining (VSG14, VSG653, and VSG531) were the least abundant VSGs detected in this assay. This differential VSG deregulation has also been previously observed upon VEX1 depletion and overexpression, albeit with a different VSG expression pattern (37).
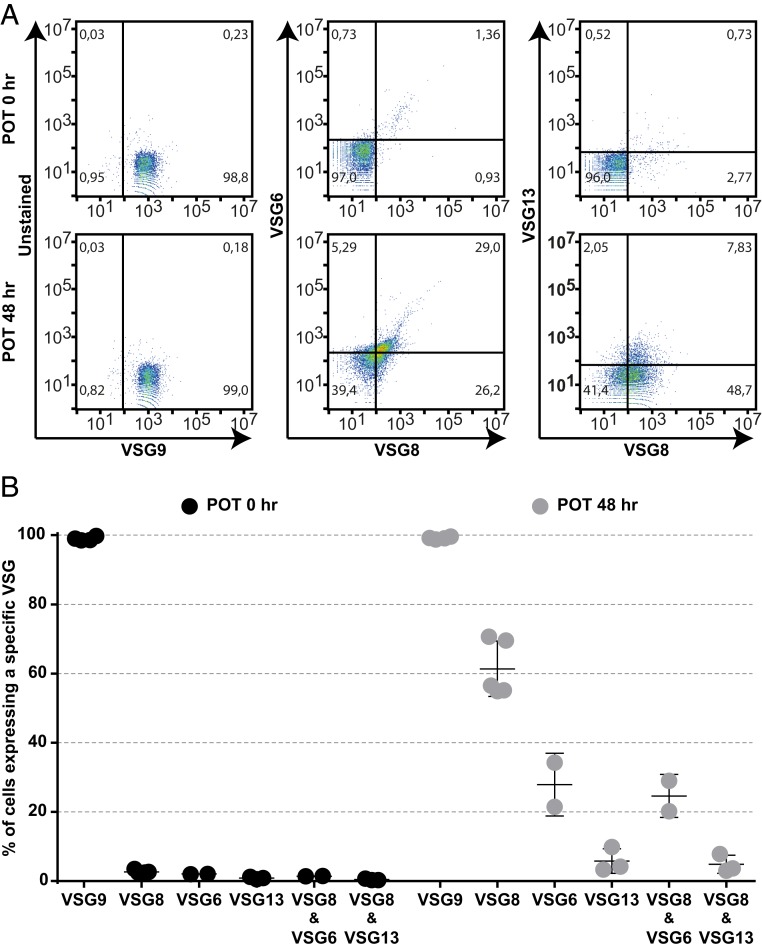
To test if VSG derepression leads to monoallelic disruption at a single-cell level, we performed flow cytometry analysis of TDP1-expressing parasites using antibodies against VSG9 and VSG8 (raised in mouse) and VSG6 and VSG13 (raised in rabbit) (Fig. 4). As expected, ∼100% of the noninduced cells were recognized by an anti-VSG9 serum, but not by antibodies against the silent VSG8, VSG6, and VSG13. Most cells continued to be recognized by VSG9 serum (99%) 48 h after TDP1 overexpression, which is expected because this remains the actively transcribed VSG. Importantly, TDP1 overexpressors were also positive for all 3 silent VSGs, with 61%, 28%, and 6% of the cells expressing VSG8, VSG6, and VSG13, respectively, at the surface. Moreover, 23% to 29% of cells are simultaneously positive in the double-staining with VSG6 and VSG8 sera while 8% of TDP1 overexpressors are simultaneously positive with VSG13 and VSG8 sera. The higher percentage of cells expressing VSG6/VSG8 compared to VSG13/VSG8 is consistent with the proteomic analysis, in which we found that VSG6 is more abundant than VSG13 (3.5% and 0.2% of the total surface VSG content, respectively) (Fig. 3B). Overall, this analysis shows that, while ∼100% of TDP1 overexpressors express the same active VSG (VSG9), at least 2 combinations of additional VSGs are present at the parasite surface (VSG13 and VSG8, or VSG6 and VSG8). Thus, we show at the single cell level that VSG monoallelic expression is clearly disrupted.
Fig. 4.
TDP1 overexpression compromises VSG monoallelic expression in individual cells. (A) Expression of VSG9, VSG6, VSG8 and VSG13 in POT2 before (POT 0 h) or after (POT 48 h) induction of TDP1 overexpression. Representative experiment out of 2 to 3 independent experiments. Sera against VSG9, VSG8 were produced in mice, while sera against VSG6, VSG13 were produced in rabbit. (B) Quantification of percentage of parasite population that express VSG9 (the active VSG) and VSG6, VSG8 and VSG13 (silent or derepressed VSGs).
TDP1 Overexpression Leads to a Multi-VSG Antibody Immune Response.
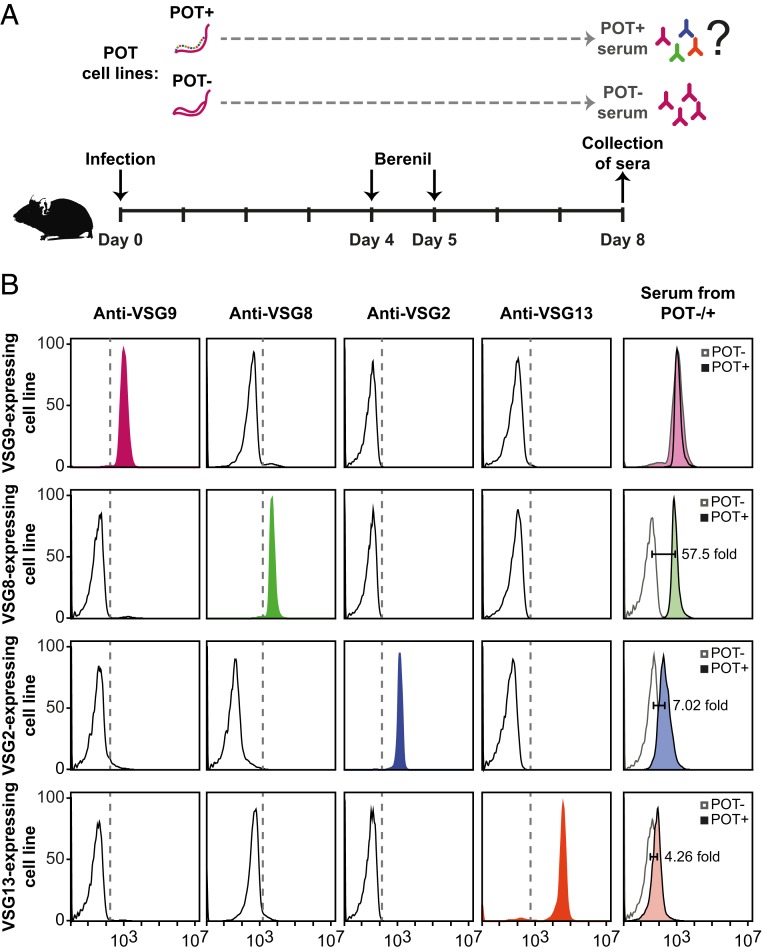
Next, we asked whether the mouse immune system would be sensitive to the loss of monoallelic expression in TDP1-overexpressing parasites. By flow cytometry, we tested whether the antisera of mice infected with POT2 parasites recognized multiple VSGs or only the actively expressed VSG9. Mice were infected with POT2 parasites in which TDP1 overexpression was induced (with doxycycline) or not (without doxycycline). Parasitemia was confirmed at 4 and 5 d postinfection, and mice were then treated with Berenil. Four days of infection is sufficient to activate the immune system to produce immunoglobulins while Berenil treatment ensures that mice survive for 8 d producing high titers of immunoglobulins (Fig. 5A). To test VSG specificity, the 2 POT sera (with or without induction of TDP1 overexpression) were individually incubated with 4 different parasite cell lines that express different VSGs (VSG9, VSG8, VSG2, and VSG13). First, we confirmed that each parasite cell line homogenously expressed the expected VSG and was not recognized by another anti-VSG antibody (Fig. 5B). As expected, each parasite cell line was recognized by its specific antibody, but not by any of the other 3. Next, we tested whether serum from the noninduced POT2 infection could recognize parasites from the cell lines that singly express VSG9, VSG8, VSG2, or VSG13. As expected, the serum recognized the actively transcribed VSG (VSG9), but none of the other VSGs (VSG8, VSG2, or VSG13). In contrast, sera obtained from mice infected with induced POT2 parasites recognized not only VSG9, but also the remaining 3 VSGs tested (Fig. 5B). Among the previously silent VSGs (VSG8, VSG2, and VSG13), the staining intensity was proportional to the abundance of these VSGs in the proteome (Fig. 3B) although it could also be influenced by other factors, including antibody affinity and avidity. Nevertheless, the observation that POT2+ antisera did not strongly recognize VSG13-expressing parasites is not surprising as VSG13 is only slightly derepressed in TDP1-expressing parasites (Figs. 3B and 4B).
Fig. 5.
Loss of VSG monoallelic expression triggers a multi-VSG antibody response. (A) Diagram of the pipeline used to assess the antibody response in mice infected with parasites expressing multiple VSGs. Experimental mice were infected with POT2 clones (with or without TDP1 overexpression). Mouse survival was prolonged by eliminating parasites on days 4 and 5 with Berenil, and sera were collected at day 8 postinfection. (B) FACS analysis to confirm VSG specificity of 4 cell lines expressing VSG9 (pink), VSG8 (green), VSG2 (blue), and VSG13 (orange). Each cell line was stained with 4 sera against the same panel of VSGs. In the last column, the 4 cell lines were used as a substrate to test by FACS if the sera from POT−- or POT+-infected mice recognized the 4 VSGs. The fold change between staining intensity of the induced vs. the noninduced POT sera is indicated. Representative experiment of 2 independent experiments.
Overall, we conclude that disruption of VSG monoallelic expression in TDP1-overexpressing parasites elicits a different immune response from wild-type parasites. Instead of generating immunoglobulins against a single VSG, mice generate a multi-VSG antibody immune response.
The Adaptive Immune Response Is Required for Reduced Parasite Burden and Longer Host Survival.
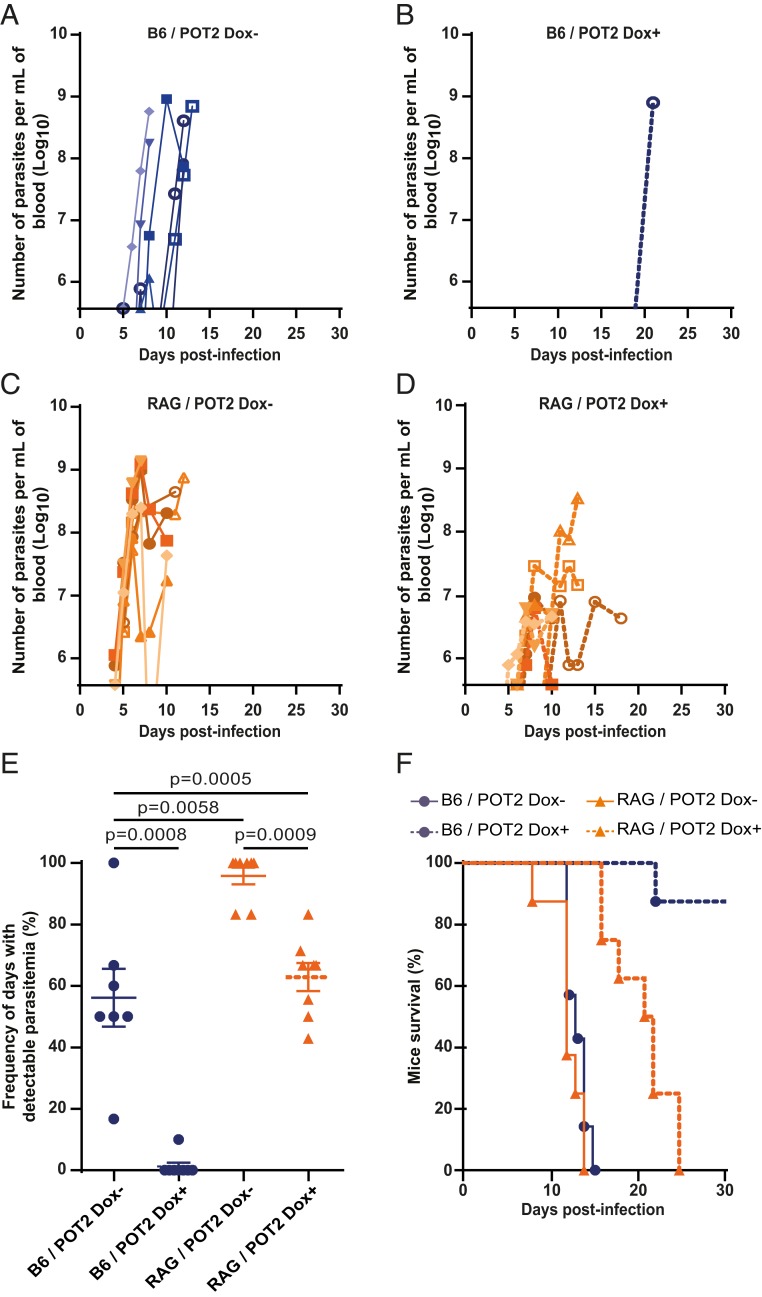
To evaluate the impact of a multi-VSG antibody response in the development of an infection in vivo, we induced TDP1 overexpression for 24 h in vitro and inoculated 20 parasites in wild-type C57BL/6J mice. Doxycycline was administered throughout the infection to maintain TDP1 overexpression. Infection with noninduced POT2 parasites led to a rapid increase in parasitemia in the first days, reaching 108 to 109 parasites per milliliter (Fig. 6 A and E). Remarkably, upon TDP1 overexpression, no parasitemia could be detected at any day in 7 out of 8 mice (Fig. 6 B and E). These 7 animals showed no signs of disease until they were killed, suggesting that they may have resolved the infection (Fig. 6F). The same tendency was observed in infections with the POT1 clone, but to a lesser extent (SI Appendix, Fig. S3). TDP1 overexpression in POT1 caused lower parasitemia in the first week than in noninduced clones, and, beyond the first peak, parasitemia was detectable in ∼50% of the days scored (SI Appendix, Fig. S3 A and B). Overall, in both clones, TDP1 overexpression resulted in lower parasitemia and a prolonged mouse survival, with 88% of mice infected with POT2 and 40% of mice infected with POT1 surviving a 25-d infection (Fig. 6F and SI Appendix, Fig. S3F).
Fig. 6.
TDP1 overexpression reduces disease severity via the interaction with the adaptive immune response. (A–D) Curves of parasitemia upon infection with POT2 parasites in C57BL/6J (A and B) or RAG2−/− mice (C and D) with or without induction of TDP1 overexpression with doxycycline (Dox) (A and C; B and D, respectively). Parasitemia was monitored daily from day 4 until day 10 postinfection for mice 1 to 5, and from day 4 until morbidity signs for mice 6 to 8. In A–D, each animal is represented with a unique symbol. Infections in wild-type animals are shown in blue, infections in RAG−/− mice are shown in orange. (E) Frequency of the number of days where parasitemia was detected relative to infection length for POT2 clones. Note that parasitemia started to be detected in C57BL/6J mice at day 5 postinfection and in RAG2−/− mice at day 4 postinfection. Statistical significance was determined by a 2-tailed Mann–Whitney U test. (F) Survival curves of mice infected with POT2 clones. Statistical significance was determined by a Log-rank test obtaining the following P values: B6/POT2 Tet− vs. Tet+, <0.0001; RAG/POT2 Tet− vs. Tet+, <0.0001; B6/POT2 Tet− vs. RAG/POT2 Tet−, not significant; B6/POT2 Tet+ vs. RAG/POT2 Tet+, 0.0004.
In a normal infection, the control of parasitemia and host survival strongly depend on the immune response. Given that infection with TDP1-overexpressing parasites results in a multi-VSG antibody immune response (Fig. 5B), we asked how the lack of an adaptive immune response would affect the infection course. For that, we repeated the infections using RAG2 knock-out (RAG2−/−) mice, and we followed parasitemia and host survival. RAG2−/− mice fail to produce mature B and T lymphocytes due to loss of initiation of V(D)J recombination (46), but they retain an innate immune response. Infection of RAG2−/− mice with noninduced POT2 showed an earlier detection of parasitemia relative to an infection in wild-type mice, suggesting that lymphocytes exert a slight negative impact in the growth of noninduced POT2 clone (Fig. 6 A and C). Importantly, infection of RAG2−/− mice with TDP1-overexpressing parasites led to profound changes in the parasitemia curves relative to wild-type mice: For POT2, only a single mouse showed detectable parasitemia in wild-type mice where all RAG2−/− mice presented parasitemia during the course of the infection (Fig. 6 B and D). In infections with induced POT1 clone, we observed the same tendency: In RAG2−/− mice, parasitemia appeared earlier and reached higher levels sooner than in wild-type mice (SI Appendix, Fig. S3 B and D). These results clearly indicate that the adaptive immune response is responsible for the significant suppression of infection with TDP1-induced POT clones.
Interestingly, when we compared the parasitemia curves of RAG2−/− mice infected with noninduced and induced POT clones, we observed that the parasitemia patterns were not identical: TDP1 overexpression led to only 63% of days with detectable parasitemia while this was close to 100% when TDP1 was not overexpressed (Fig. 6E). These differences may be due to the slight growth defect associated with multiple-VSG expression (Fig. 1C) or to a component of the innate immune system that modestly contributes to the control of mutant parasites. These differences were consequently reflected in mice survival: 21 d or 12 to 13 d when TDP1 is overexpressed or not, respectively (Fig. 6F).
The different parasitemia patterns in the 4 infection conditions translated into differences in host survival. Upon doxycycline-induced TDP1 expression in POT2 clone, wild-type mice survived longer than the 30 d of experiment while RAG2−/− mice died at 21-d postinfection. In contrast, in noninduced conditions, both wild-type and RAG2−/− mice showed similar survival times (12 to 13 d) (Fig. 6F). The same trend was observed with POT1 clone, albeit the infection phenotype was milder (SI Appendix, Fig. S3F).
The infections in mice lacking an adaptive immune response clearly show that TDP1 overexpression alone has a slight detrimental impact on infection: parasitemia does not rise as sharply in the presence as in the absence of doxycycline (Fig. 6 C and D). Nevertheless, our results clearly show that the most dramatic phenotype is detected due to an interaction between the mutant parasite and the adaptive immune response (Fig. 6B). The fact that wild-type mice infected with TDP1-overexpressing parasites mount a multi-VSG antibody response appears to enable a more effective control of the infection, leading to very low (or undetectable) parasitemia and better disease prognosis.
Discussion
TDP1 is the only identified nuclear HMGB protein in T. brucei, and it seems to play a pivotal role in antigenic variation. This protein has been previously shown to be a Pol I transcription facilitator (31), and, in the absence of transcription, it is crucial to stabilize chromatin in an open conformation, similar to the yeast ortholog Hmo1 (47). Here, we show that overexpression of TDP1 induced 3 different molecular phenotypes.
First, upon TDP1 overexpression, the chromatin conformation of Pol II-transcribed regions became slightly more open, but this did not result in an increase in the transcript levels of those genes. Yeast Hmo1 also binds to Pol II-transcribed loci (48, 49), but, in contrast to T. brucei, this binding contributes to Pol II transcription initiation (50, 51) and leads to a major growth defect (35). In T. brucei, TDP1 is insufficient to increase transcription of Pol II-transcribed loci. In fact, this is not so surprising, given the polycistronic nature of transcription in trypanosomes. Previous studies have also found that chromatin alterations that have an effect on Pol I transcript levels have no significant impact on Pol II genes (29, 37).
Second, upon TDP1 overexpression, the chromatin conformation and transcription rates of the active BES and rDNA loci were unaltered, which is presumably because these loci have fully open chromatin by default. This is consistent with the previous observation that depletion of histone H1 leads to chromatin opening of several Pol I loci, but not 18S (29), suggesting that all, or almost all, rDNA genes in T. brucei are always in an open chromatin conformation.
Finally, upon TDP1 overexpression, we observed that the chromatin of BESs, MESs, and procyclin loci acquired a more open conformation and that the transcript levels of these loci increased. Why silent BESs are never fully derepressed in TDP1 overexpression POT clones remains unknown, but this has been observed in all mutants described so far to have a phenotype in VSG expression loci (36–38, 41–44). It is possible that the levels of TDP1 were not high enough to displace all histones from silent BESs or alternatively that additional levels of regulation or cross-talk between BESs prevent a silent BES from being transcribed at the same level as the active BES. There are previous reports that trypanosomes may be able to count the number of VSG RNA molecules so overall VSG levels remain constant upon expression of a second VSG (52, 53). Overall, these results indicate that TDP1 shares several functional features with UBF (structural component and transcriptional facilitator of Pol I loci in mammals) and Hmo1 (structural component of both Pol I and Pol II loci in yeast). However, it is not a bona fide ortholog of either of them because UBF does not bind Pol II-transcribed loci and Hmo1 is not involved in Pol I transcription initiation (this function is fulfilled by UAF).
TDP1 overexpression increases the diversity of VSGs detectable at the cell surface of this mutant. A similar VSG diversity has been previously observed in VEX1 mutant although the proportion of silent VSGs at the cell surface upon TDP1 overexpression is higher than upon overexpression of VEX1 (28% vs. 9%) (37). Interestingly, both studies revealed the same group of VSGs becoming preferentially up-regulated, suggesting a preference for certain VSGs in vitro. The molecular basis for this preference is unknown, but it may be related to the chromatin environment of their genes. The detection of very high levels of VSGs at the cell surface in POT clones is consistent with the luciferase assay. Luciferase activity increased 250-fold upon TDP1 overexpression while, in other mutants (histone H1 and RAP1 knockdowns), this increase was around 10- to 40-fold (29, 36). Overall, our results indicate that TDP1 has a strong influence on VSG gene expression.
Expression of only one VSG at the cell surface of T. brucei is thought to be crucial to focus the repertoire of antibodies generated by the host immune system at a given time, hence favoring a long-term persistent infection. Although each peak of parasitemia contains parasites monoallelically expressing dozens of VSG variants, the large repertoire of VSGs in the genome, together with the ability to generate mosaic VSGs, allow the maintenance of a chronic infection (54). The monomorphic Lister 427 strain used in this study avoids the formation of the nondividing stumpy form (55) and, in mice, typically leads to death of the host in the first or second peak of parasitemia (29). However, under TDP1 overexpression, the survival of mice was remarkably prolonged.
When we compared the outcome of infections of TDP1-overexpressing parasites in wild-type and RAG2−/− mice, we observed that RAG2−/− mice succumbed to infection much faster than wild-type mice. These data show that the adaptive immune response is very important in the control of infection of TDP1-overexpressing parasites. This is consistent with current knowledge about the immune response against African trypanosomes, in which B cells and anti-VSG Ig responses are considered to be the first line of host defense (14, 56). We propose that simultaneous exposure to multiple VSGs triggers the activation of multiple B cells, resulting in a faster kinetics of Ig production, ultimately killing parasites and reducing parasitemia. This mechanism may also underly the prolonged survival and reduced parasitemia observed in mice infected with histone H1-depleted parasites (29). In this mutant, VSGs were also derepressed 4- to 10-fold at RNA level, and, although this was not tested, probably several VSGs were expressed at the cell surface. In the future, it will be interesting to check if parasites that express more than one VSG at the cell surface (29, 37, 57) generate the production of multiple types of immunoglobulins by the host and whether prolonged survival depends on a minimum number of different VSGs and on their levels or identities.
In the absence of an adaptive immune response, overexpression of TDP1 has a slight detrimental impact on infection. The reasons for this defect are not clear. Given that mutant parasites grow slower in vitro, they may also do so in vivo. Indeed, other mutants that express multiple VSGs at the surface show slight growth defects in vitro (29, 36, 37). This could be due to a saturation of the endocytic pathway that is overloaded with recycling VSG (58) and/or due to the presence at the surface of more than 1.5-fold the normal levels of VSG, a molecular crowding threshold estimated by Hartel et al. (59). An alternative explanation is that the innate immune response (still present in RAG2 knock-out mice) has a stronger effect in TDP1-overexpressing parasites than wild-type parasites. As a potential therapeutic approach, it would actually be desirable if a multi-VSG–expressing parasite were not only more efficiently eliminated by the adaptive immune response but also grew slower or were more susceptible to innate immune response.
Dubois et al. (60) had previously shown that an infection with another trypanosome subspecies (Trypanosoma brucei rhodesiense) simultaneously expressing 2 VSGs did not activate early T-independent B cell responses to produce VSG-specific antibodies, suggesting that transient expression of 2-VSG coats provided an immunological advantage for the parasite. The authors mentioned that, despite the reduced production of antibodies, mice survival was independent of parasites having single or double-VSG coats. The differences between this study and ours are numerous and could contribute to the apparent opposite phenotypes: 1) In our study, POT parasites expose VSG9 and minor quantities of several other VSGs while parasites in the Dubois et al. study expressed only 2 VSGs (VSG LouTat1 and VSG117); it is likely that the variety of epitopes and perhaps of sugars present in POT clones is higher than in double-VSG coats, affecting immune response; 2) infections were established with different T. brucei subspecies, which could trigger different immune responses; 3) we used a more virulent monomorphic strain, which cannot produce tsetse-transmissible forms, while Dubois et al. used a transmission-competent pleomorphic strain; 4) the number of parasites used to initiate the infection was very different (Dubois et al. used 100,000 parasites while we used 20); this likely has a significant impact in the activation of the immune response and favored the early parasite growth in their study.
Giardia lamblia, a causative agent of intestinal disease, also undergoes antigenic variation by switching its variant surface proteins (VSPs) (61). Knocking down the RNA-dependent RNA polymerase (RnRP) or Dicer in this parasite induces expression of multiple VSPs at the cell surface of trophozoites (62). Infection with these parasites generates a strong immunological response that prevents infection with subsequent Giardia challenges, showing that antigenic variation is essential for parasite survival in vivo (63). A major difference between the in vivo effects of the loss of monoallelic expression in Giardia and African trypanosomes is that the loss of RnRP or Dicer does not affect the outcome of the primary infection. In contrast, the TDP1-overexpressing mutant is highly susceptible to the adaptive immune response, and infection is often resolved. Thus, while disrupting antigenic variation in Giardia could be used as a vaccine strategy, in T. brucei, it could only provide a therapeutic strategy.
Overall, TDP1 overexpression disrupts VSG monoallelic expression as a consequence of its high loading in silent BESs and stimulation of transcription. Infection with parasites that overexpress TDP1 results in reduced parasitemia and prolonged host survival, an effect strongly dependent on the presence of an adaptive immune response. Our study reveals that the generation of a multi-VSG antibody response leads to a dramatic reduction in parasitemia that remains undetectable in most mice on most days, resulting in a better prognosis for the host. We conclude that TDP1 is pivotal for antigenic variation and its endogenous levels are critical for trypanosomes to evade the immune system and survive in the mammalian host. Moreover, our study raises the possibility for the development of new strategies to treat African trypanosomiasis, in which forced exposure to multiple VSGs at the surface of individual cells would make them more susceptible to the host’s immune system.
Materials and Methods
For details of T. brucei growth and manipulation, plasmids, RNA quantification, Western blotting, flow cytometry, FAIRE, ChIP, quantitative mass spectrometry, animal infections, and sera characterization, see SI Appendix, Materials and Methods. All T. brucei cell lines derive from strain Lister 427. PL1S and VSG13-expressing cell lines were described in refs. 36 and 42, respectively. VSG8- and VSG11-expressing cell lines were described in ref. 64. All mass spectrometry measurement files are deposited at the ProteomeXchange consortium via PRIDE (PXD014803). The animal facility and the experimental procedures complied with European Union regulations and were approved by the Instituto de Medicina Molecular Animal Care and Ethics Committee (AWB_2016_07_LF_Tropism).
Supplementary Material
Acknowledgments
We thank Lucy Glover (Institut Pasteur) for helping and sharing the protocol for the soluble-form VSG isolation and sharing the serum against VSG6. We also thank Douglas Lamont and Mike Ferguson (Fingerprints Proteomics and Mass Spectrometry Facility, School of Life Sciences, University of Dundee) for assistance with quantitative mass spectrometry; and Nina Papavasiliou for sharing VSG8-expressing cell lines, serum against VSG2, and fruitful discussion of the manuscript. Ruy Ribeiro helped with statistical analysis, and Afonso Bravo helped generating pFAB9. We thank Sara Silva Pereira for useful comments and George A. M. Cross for interrupting his retirement schedule to give us the honor of proofreading the manuscript. This work was supported in part by Fundação para a Ciência e Tecnologia Grants PTDC/SAU-MIC/113225/2009 and SFRH/BD/80718/2011 (to F.A.-B.) and PD/BD/105838/2014 (to M.S.-V.); a European Molecular Biology Organization Installation grant (Project 2151); and Howard Hughes Medical Institute International Early Career Scientist Program Grant 55007419. L.M.F. is an Investigator of the Fundação para a Ciência e Tecnologia. Publication of this work was funded by UID/BIM/50005/2019, project funded by Fundação par a Ciência e Tecnologia (FCT), Ministério da Ciência, Tecnologia e Ensino Superior (MCTES) through Fundos do Orçamento de Estado.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Mass spectrometry measurement files reported in this paper have been deposited in the ProteomeXchange Consortium, http://proteomecentral.proteomexchange.org, via the PRIDE partner repository (dataset identifier PXD014803).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905120116/-/DCSupplemental.
References
- 1.Goodwin G. H., Sanders C., Johns E. W., A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem. 38, 14–19 (1973). [DOI] [PubMed] [Google Scholar]
- 2.Stros M., Launholt D., Grasser K. D., The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 64, 2590–2606 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonaldi T., Längst G., Strohner R., Becker P. B., Bianchi M. E., The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 21, 6865–6873 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hepp M. I., Alarcon V., Dutta A., Workman J. L., Gutiérrez J. L., Nucleosome remodeling by the SWI/SNF complex is enhanced by yeast high mobility group box (HMGB) proteins. Biochim. Biophys. Acta 1839, 764–772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agresti A., Bianchi M. E., HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13, 170–178 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Ge H., Roeder R. G., The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem. 269, 17136–17140 (1994). [PubMed] [Google Scholar]
- 7.Clos J., Buttgereit D., Grummt I., A purified transcription factor (TIF-IB) binds to essential sequences of the mouse rDNA promoter. Proc. Natl. Acad. Sci. U.S.A. 83, 604–608 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gadal O., Labarre S., Boschiero C., Thuriaux P., Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21, 5498–5507 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Sullivan A. C., Sullivan G. J., McStay B., UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell. Biol. 22, 657–668 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merz K., et al. , Actively transcribed rRNA genes in S. cerevisiae are organized in a specialized chromatin associated with the high-mobility group protein Hmo1 and are largely devoid of histone molecules. Genes Dev. 22, 1190–1204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keys D. A., et al. , Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10, 887–903 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Hamperl S., et al. , Chromatin states at ribosomal DNA loci. Biochim. Biophys. Acta 1829, 405–417 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Campbell G. H., Esser K. M., Weinbaum F. I., Trypanosoma rhodesiense infection in B-cell-deficient mice. Infect. Immun. 18, 434–438 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magez S., et al. , The role of B-cells and IgM antibodies in parasitemia, anemia, and VSG switching in Trypanosoma brucei-infected mice. PLoS Pathog. 4, e1000122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross G. A., Kim H. S., Wickstead B., Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol. Biochem. Parasitol. 195, 59–73 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hertz-Fowler C., et al. , Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 3, e3527 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenn K., Matthews K. R., The cell biology of Trypanosoma brucei differentiation. Curr. Opin. Microbiol. 10, 539–546 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziegelbauer K., Quinten M., Schwarz H., Pearson T. W., Overath P., Synchronous differentiation of Trypanosoma brucei from bloodstream to procyclic forms in vitro. Eur. J. Biochem. 192, 373–378 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Vickerman K., On the surface coat and flagellar adhesion in trypanosomes. J. Cell Sci. 5, 163–193 (1969). [DOI] [PubMed] [Google Scholar]
- 20.Johnson P. J., Kooter J. M., Borst P., Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell 51, 273–281 (1987). [DOI] [PubMed] [Google Scholar]
- 21.Alarcon C. M., Son H. J., Hall T., Donelson J. E., A monocistronic transcript for a trypanosome variant surface glycoprotein. Mol. Cell. Biol. 14, 5579–5591 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roditi I., Clayton C., An unambiguous nomenclature for the major surface glycoproteins of the procyclic form of Trypanosoma brucei. Mol. Biochem. Parasitol. 103, 99–100 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Figueiredo L. M., Cross G. A., Janzen C. J., Epigenetic regulation in African trypanosomes: A new kid on the block. Nat. Rev. Microbiol. 7, 504–513 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Günzl A., et al. , RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2, 542–551 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figueiredo L. M., Cross G. A., Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot. Cell 9, 148–154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanne T. M., Rudenko G., Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot. Cell 9, 136–147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanhamme L., Berberof M., Le Ray D., Pays E., Stimuli of differentiation regulate RNA elongation in the transcription units for the major stage-specific antigens of Trypanosoma brucei. Nucleic Acids Res. 23, 1862–1869 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotz H. R., Hartmann C., Huober K., Hug M., Clayton C., Mechanisms of developmental regulation in Trypanosoma brucei: A polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 25, 3017–3026 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pena A. C., et al. , Trypanosoma brucei histone H1 inhibits RNA polymerase I transcription and is important for parasite fitness in vivo. Mol. Microbiol. 93, 645–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandenburg J., et al. , Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 26, 4856–4866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narayanan M. S., Rudenko G., TDP1 is an HMG chromatin protein facilitating RNA polymerase I transcription in African trypanosomes. Nucleic Acids Res. 41, 2981–2992 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aresta-Branco F., Pimenta S., Figueiredo L. M., A transcription-independent epigenetic mechanism is associated with antigenic switching in Trypanosoma brucei. Nucleic Acids Res. 44, 3131–3146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefanovsky V., Langlois F., Gagnon-Kugler T., Rothblum L. I., Moss T., Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 21, 629–639 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Hannan R. D., Stefanovsky V., Taylor L., Moss T., Rothblum L. I., Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: Implications for cardiac hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 93, 8750–8755 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshikawa K., et al. , Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28, 349–361 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Yang X., Figueiredo L. M., Espinal A., Okubo E., Li B., RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell 137, 99–109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover L., Hutchinson S., Alsford S., Horn D., VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 113, 7225–7230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Günzl A., Kirkham J. K., Nguyen T. N., Badjatia N., Park S. H., Mono-allelic VSG expression by RNA polymerase I in Trypanosoma brucei: Expression site control from both ends? Gene 556, 68–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cestari I., Stuart K., Transcriptional regulation of telomeric expression sites and antigenic variation in trypanosomes. Curr. Genomics 19, 119–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller L. S. M., et al. , Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature 563, 121–125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benmerzouga I., et al. , Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol. Microbiol. 87, 196–210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Figueiredo L. M., Janzen C. J., Cross G. A., A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 6, e161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DuBois K. N., et al. , NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 10, e1001287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cestari I., Stuart K., Inositol phosphate pathway controls transcription of telomeric expression sites in trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 112, E2803–E2812 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cross G. A., Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71, 393–417 (1975). [DOI] [PubMed] [Google Scholar]
- 46.Shinkai Y., et al. , RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68, 855–867 (1992). [DOI] [PubMed] [Google Scholar]
- 47.Wittner M., et al. , Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell 145, 543–554 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Hall D. B., Wade J. T., Struhl K., An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 3672–3679 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasahara K., et al. , Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 27, 6686–6705 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasahara K., Ki S., Aoyama K., Takahashi H., Kokubo T., Saccharomyces cerevisiae HMO1 interacts with TFIID and participates in start site selection by RNA polymerase II. Nucleic Acids Res. 36, 1343–1357 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kasahara K., Ohyama Y., Kokubo T., Hmo1 directs pre-initiation complex assembly to an appropriate site on its target gene promoters by masking a nucleosome-free region. Nucleic Acids Res. 39, 4136–4150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batram C., Jones N. G., Janzen C. J., Markert S. M., Engstler M., Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. eLife 3, e02324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridewood S., et al. , The role of genomic location and flanking 3'UTR in the generation of functional levels of variant surface glycoprotein in Trypanosoma brucei. Mol. Microbiol. 106, 614–634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mugnier M. R., Cross G. A., Papavasiliou F. N., The in vivo dynamics of antigenic variation in Trypanosoma brucei. Science 347, 1470–1473 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Breidbach T., Ngazoa E., Steverding D., Trypanosoma brucei: In vitro slender-to-stumpy differentiation of culture-adapted, monomorphic bloodstream forms. Exp. Parasitol. 101, 223–230 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Reinitz D. M., Mansfield J. M., T-cell-independent and T-cell-dependent B-cell responses to exposed variant surface glycoprotein epitopes in trypanosome-infected mice. Infect. Immun. 58, 2337–2342 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muñoz-Jordán J. L., Davies K. P., Cross G. A., Stable expression of mosaic coats of variant surface glycoproteins in Trypanosoma brucei. Science 272, 1795–1797 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Engstler M., et al. , Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J. Cell Sci. 117, 1105–1115 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Hartel A. J., et al. , N-glycosylation enables high lateral mobility of GPI-anchored proteins at a molecular crowding threshold. Nat. Commun. 7, 12870 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dubois M. E., Demick K. P., Mansfield J. M., Trypanosomes expressing a mosaic variant surface glycoprotein coat escape early detection by the immune system. Infect. Immun. 73, 2690–2697 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adam R. D., et al. , Antigenic variation of a cysteine-rich protein in Giardia lamblia. J. Exp. Med. 167, 109–118 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prucca C. G., et al. , Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 456, 750–754 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Rivero F. D., et al. , Disruption of antigenic variation is crucial for effective parasite vaccine. Nat. Med. 16, 551–557 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Kim H. S., Cross G. A., TOPO3alpha influences antigenic variation by monitoring expression-site-associated VSG switching in Trypanosoma brucei. PLoS Pathog. 6, e1000992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.