Significance
Diabetes and pancreatic cancer are devastating diseases of the pancreas. The identification of organ-specific adult progenitor/stem cells is important for understanding their origin and designing therapeutic interventions. We identified progenitor cells in the adult mouse pancreas characterized by the expression of the mitochondrial enzyme Aldh1b1. These cells give rise to all 3 cell types in the organ and single-cell gene expression analysis showed that they preferentially express Kras, oncogenic mutations of which account for more than 90% of the cases of human pancreatic cancer. Importantly, Aldh1b1 function is required for tumor development in a pancreatic cancer mouse model, suggesting that these cells play a key role in the disease and potentially constitute a therapeutic target for pancreatic cancer.
Keywords: adult stem and progenitor cells, aldehyde dehydrogenase, organoids, single-cell RNA sequencing, pancreatic ductal adenocarcinoma
Abstract
The presence of progenitor or stem cells in the adult pancreas and their potential involvement in homeostasis and cancer development remain unresolved issues. Here, we show that mouse centroacinar cells can be identified and isolated by virtue of the mitochondrial enzyme Aldh1b1 that they uniquely express. These cells are necessary and sufficient for the formation of self-renewing adult pancreatic organoids in an Aldh1b1-dependent manner. Aldh1b1-expressing centroacinar cells are largely quiescent, self-renew, and, as shown by genetic lineage tracing, contribute to all 3 pancreatic lineages in the adult organ under homeostatic conditions. Single-cell RNA sequencing analysis of these cells identified a progenitor cell population, established its molecular signature, and determined distinct differentiation pathways to early progenitors. A distinct feature of these progenitor cells is the preferential expression of small GTPases, including Kras, suggesting that they might be susceptible to Kras-driven oncogenic transformation. This finding and the overexpression of Aldh1b1 in human and mouse pancreatic cancers, driven by activated Kras, prompted us to examine the involvement of Aldh1b1 in oncogenesis. We demonstrated genetically that ablation of Aldh1b1 completely abrogates tumor development in a mouse model of KrasG12D-induced pancreatic cancer.
The adult pancreas can respond to injury or metabolic stimuli to repair damage and regulate cell numbers. Although evidence for the presence of progenitor cells has been obtained under conditions of injury, there is an ongoing debate whether this is a result of the inherent plasticity of terminally differentiated cells or due to the presence of unidentified rare adult pancreatic stem/progenitor cells (1, 2). A related controversy concerns the cell of origin of pancreatic ductal adenocarcinoma (PDAC), which is the most lethal among malignancies (5-y survival rate ∼6%). In more than 90% of the cases, PDAC is initiated by an oncogenic mutation of the GTPase Kras (Kras*) predominantly in codon 12 (e.g., KrasG12D). It is thought that the precancerous lesion originates either in a stem/progenitor cell or in an acinar cell that undergoes acinar-to-ductal metaplasia, perhaps reinforced by cellular injury (3–8).
Rare acinar cells can be reprogrammed in vivo into endocrine cells or induced to differentiate into endocrine cells after streptozotocin-mediated ablation of β cells or pancreatic duct ligation (9–12). Other experiments suggested the presence of progenitor cells in the ductal tree. Pancreatic duct ligation induced the appearance of endocrine progenitors near the ducts, but lineage tracing experiments did not identify their origin conclusively (13–16). Inactivation of the tumor-suppressor gene Fbw7 resulted in the conversion of some duct cells into β cells (17), while ductal cells have been isolated and expanded clonally in vitro as organoid-like structures from human and mouse adult pancreata (18, 19). The outcome of these experiments may be attributed to the plasticity of acinar and ductal cells or, alternatively, to the presence of rare stem/progenitor cells.
Terminal duct/centroacinar cells, hereafter referred to simply as CACs, are rare, small cells with minimal cytoplasm, numerous mitochondria, and long cytoplasmic extensions. They are contiguous with ductal cells at the end of the ductules forming a fenestrated lining on the luminal acinar surface (20, 21). In adult zebrafish, CACs generate new β cells following β-cell ablation or partial pancreatectomy (22). In the mouse, they have been prospectively isolated through their high aldhehyde dehydrogenase (Aldh) activity using the Aldefluor reagent that detects several Aldh isoforms (23, 24). These cells have been described as both progenitor-like and tumor-initiating cells (TICs) (22, 25–27) and it has been shown that they can generate in vitro pancreatospheres containing endocrine and exocrine cells (26). However, the large number of the Aldh superfamily genes encoding enzymes with diverse specificities (28) had so far precluded further analysis. Interestingly, the mitochondrial enzyme Aldh1b1 is expressed in all mouse embryonic pancreatic progenitors but in the adult organ is confined to rare elongated cells (29). The number of adult Aldh1b1+ cells is dramatically up-regulated upon pancreatic injury of either the acinar compartment, using cerulein-induced pancreatitis, or of the endocrine compartment, using streptozotocin-induced β-cell ablation (29). Importantly, the levels of ALDH1B1 in human pancreatic cancers, as assessed by immunohistochemistry, was found to be ∼12-fold higher than normal (30).
Results
Aldh1b1 Is a Specific Molecular Marker of Centroacinar Cells.
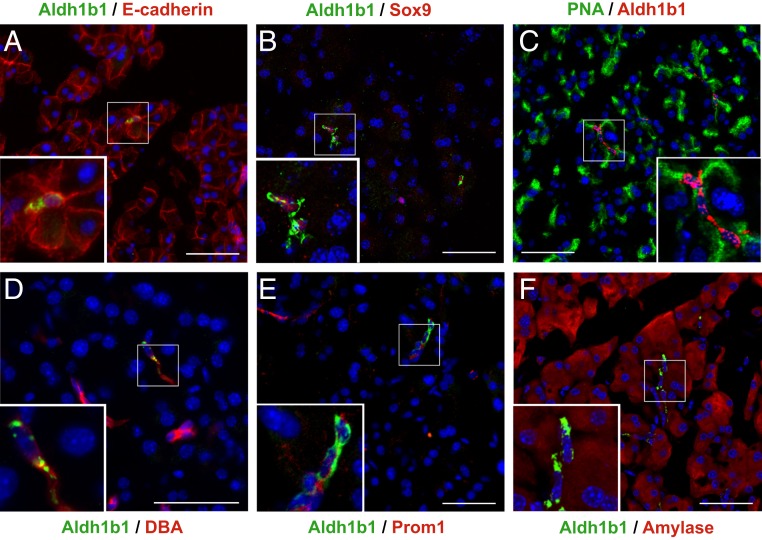
First, we analyzed in the pancreas of adult mice (8 to 10 mo old unless otherwise stated) the coexpression of Aldh1b1 with pancreas lineage and progenitor markers using a specific Aldh1b1 antibody (SI Appendix, Fig. S1 A and B). All these Aldh1b1+ cells are epithelial (Fig. 1A and SI Appendix, Fig. S1C) with invariably centroacinar location and morphology (Fig. 1 A–F). The majority of Aldh1b1+ cells were positive for the acinar surface marker PNA (71 ± 12%) and the embryonic pancreas or duct progenitor marker Sox9 (64 ± 11%) (Fig. 1 B and C and SI Appendix, Fig. S1C). Some Aldh1b1+ cells were also positive for the ductal marker DBA (17 ± 6%) and many (40 ± 14%) were weakly stained for the ductal epithelial progenitor marker Prom1 (Fig. 1 D and E and SI Appendix, Fig. S1C). Aldh1b1+ cells exhibiting weak amylase expression were also observed (Fig. 1F and SI Appendix, Fig. S1 D–F). For each marker examined, between 70 and 100 Aldh1b1+ cells per pancreas (number of pancreata examined n = 3) were scored. Adult Aldh1b1+ cells were never found in islets (at least 500 islets examined, n = 10) and were never positive for the progenitor and endocrine markers Pdx1 and Nkx6.1 (>1,000 cells, n = 6). There was no coexpression of Aldh1b1 with endothelial (PECAM), mesenchymal (Vimentin), or hematopoietic (CD45) markers (n = 3) (SI Appendix, Fig. S1 G–I). The coexpression of Aldh1b1 with pancreas progenitor markers, but also with acinar and ductal markers, is consistent with the position of CACs at the interface of acinar and ductal cells and indicated that Aldh1b1+ cells might include uncommitted stem/progenitor cells, as well as differentiating derivatives.
Fig. 1.
Coexpression of Aldh1b1 with differentiation and progenitor markers. (A–F) Immunofluorescence showed complete coexpression of Aldh1b1 with E-cadherin (A) and partial coexpression with Sox9 (B), PNA (C), DBA (D), Prom1 (E), and amylase (F). (Scale bars: 50 μm for main figures and 20 μm for insets.)
Aldh1b1-Expressing Cells Are Necessary and Sufficient to Generate Adult Mouse Pancreatic Organoids.
Adult pancreatic organoids were derived from terminal duct fragments (15), but the identity of the cells involved remains elusive. Aldh+ cells reside at the interface of terminal ducts with the acinar compartment (26) and they also express Aldh1b1, while maintaining features of both ductal and acinar identity.
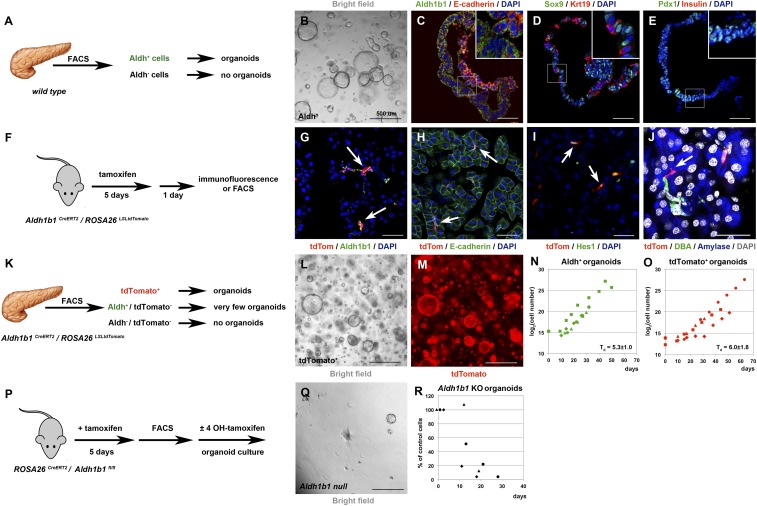
To examine whether Aldh+ cells of the adult mouse pancreas can give rise to organoids, we isolated Aldh+ cells by fluorescence-activated cell sorter (FACS) using the Aldefluor reagent (23) (SI Appendix, Fig. S2 A and B) and placed them in a 3D Matrigel-based culture (18, 31). This medium promotes expansion of progenitors and differs from the medium that allows only minimal expansion and formation of pancreatospheres containing endocrine and acinar cells (26). Aldh+ cells, which constituted 2.1 ± 0.4% (n = 12) of the cells in the adult pancreas, readily formed organoids at a seeding density of 500 cells per μL (Fig. 2 A and B). These organoids could be passaged at least 11 times over 60 d. In contrast, the fraction of Aldh− cells did not form organoids (n = 4). Similarly to duct-derived organoids (18), those derived from Aldh+ cells contained epithelial E-cadherin+ and Sox9+ cells and were predominantly of ductal (Krt19+) character. The organoids retained strong Aldh1b1 expression and contained Pdx1+ cells (Fig. 2 C–E and SI Appendix, Fig. S2D). There was no expression of endocrine progenitor (Nkx6.1, Ngn3), terminal endocrine (Ins), or acinar (Ptf1a and Amy) markers. We also evaluated the expression levels of progenitor and terminal differentiation genes at successive organoid passages by qPCR and compared them to expression levels in 14.5 dpc (days post coitum) embryonic pancreata. The expression levels of Aldh1b1, Sox9, Pdx1, and Nkx6.1 were relatively stable but lower in comparison to the corresponding embryonic values, with the notable exception of Sox9 (SI Appendix, Fig. S2C). There was no appreciable expression of terminal endocrine or acinar genes (Ins1, Ptf1a, and Amy).
Fig. 2.
Aldh1b1 function is necessary for adult pancreas organoid formation. (A–E) Among FACS-separated adult pancreatic cells (A) only Aldh+ cells form organoids (B) which maintain expression of Aldh1b1 and E-cadherin (C) while expressing Sox9, Krt19 (D), and Pdx1 but not insulin (E). (F–J) Tamoxifen administration to Aldh1b1CreERT2/ROSA26LSLtdTomato double homozygotes (F) labels efficiently Aldh1b1+ cells (arrows in G). All tdTomato+ cells are E-cadherin+ (H), many are Hes1+ (I), and some are clearly centroacinar (J). (K–O) Among FACS-fractionated adult pancreatic cells (K) only tdTomato+ cells generate organoids (L and M). Growth rates of Aldh+ (N) and tdTomato+ organoids (O) are very similar (distinct symbols indicate independent experiments). (P–R) Deletion of Aldh1b1 initiated in vivo by tamoxifen administration and completed in culture in the presence of 4-OHT after FACS isolation of Aldh+ cells (P) showed that Aldh1b1 null cells fail to generate self-renewing organoids (Q) while control cells grown in vitro in the absence of 4-OHT continue to grow (R) (distinct symbols indicate independent experiments). (Scale bars in B, L, M, and Q, 500 μm; C–E, G, and I, 50 μm; J, 25 μm.)
These data indicated that adult mouse pancreatic organoids are derived exclusively from Aldh+ cells. Considering that not all Aldh+ cells are necessarily Aldh1b1+, we assessed the contribution of Aldh1b1+ cells in organoid formation by generating an Aldh1b1 lineage-tracing mouse allele. To maintain Aldh1b1 expression levels, while ensuring specific expression of a tamoxifen-activated Cre recombinase (CreERT2), we inserted the CreERT2 transgene into the Aldh1b1 locus immediately before the stop codon and preceded by a sequence encoding the self-cleaving peptide P2A. The selection cassette was removed in vivo by Flp-mediated recombination. This strategy preserved all endogenous sequences, including the single intron of the gene, thus ensuring specific expression of CreERT2 (SI Appendix, Fig. S2E). Mice carrying the Aldh1b1CreERT2 allele were then crossed with mice carrying the ROSA26LSLtdTomato allele that encodes a Cre-inducible form of a stable red fluorescent green fluorescent protein variant. Five consecutive doses of tamoxifen to 8-wk-old Aldh1b1CreERT2/ROSA26LSLtdTomato double heterozygotes resulted in efficient labeling of 81.4 ± 2.8% of the Aldh1b1+ cells as assessed by immunofluorescence 1 d later (n = 3) (Fig. 2 F and G). Tamoxifen treatment did not affect the number of Aldh1b1+ cells. Immunofluorescence and image analysis showed that in 8-wk-old mice not treated with tamoxifen the percentage of Aldh1b1+ cells was 1.4 ± 0.5% (an average of 16 K cells per pancreas scored; n = 3), whereas the percentage of Aldh1b1+ cells treated with 5 consecutive doses of tamoxifen was 1.8 ± 0.6% (an average of 8.5 K cells per pancreas scored; n = 3). Similarly to Aldh1b1+ cells, all tdTomato+ cells were epithelial E-cadherin+ cells (478/479 cells scored; n = 3) (Fig. 2H) and their majority (59.5 ± 10%, n = 3) expressed Hes1, another centroacinar cell marker (14) (Fig. 2I). Our lineage tracer allowed the occasional unequivocal assignment of centroacinar position (Fig. 2J and SI Appendix, Fig. S2H). Similarly to Aldh1b1+ cells, TdTomato+ cells did not express CD45, a hematopoietic cell marker used for the isolation of resident macrophages (32) (448/450 cells scored; n = 3) (SI Appendix, Fig. S2F). Aldh1a1+ cells, although also centroacinar by location and morphology (SI Appendix, Fig. S2H), were clearly fewer and distinct from tdTomato+ cells (SI Appendix, Fig. S2G). A total of 608 tdTomato+ and 137 Aldh1a1+ cells were scored (n = 3) in the same cryosections and no coexpression was detected. This indicated that Aldh1b1+ cells are a subpopulation of centroacinar Aldh+ cells and distinct from Aldh1a1+ cells.
Aldh+/tdTomato− and tdTomato+ cells were FACS-isolated (Fig. 2K and SI Appendix, Fig. S2 N and O) and placed in organoid culture. TdTomato+ cells readily formed organoids at ≤400 cells per μL, which could be passaged at least 7 times, while retaining a similar doubling time (Td = 6.0 ± 1.8 d) to that of Aldh+ cells (Td = 5.3 ± 1.0 d) (Fig. 2 L–O). Time-lapse imaging suggested that single tdTomato+ cells have the capacity to form organoids (Movies S1 and S2). In contrast, Aldh+/tdTomato− cells very rarely formed organoids that could not be passaged, indicating that adult pancreas organoids are derived from Adlh1b1-expressing cells. Organoids derived from tdTomato+ cells were identical to Aldh+ organoids, as shown by immunofluorescence (n = 4) (compare Fig. 2 C–E with SI Appendix, Fig. S2 J–M). The expression levels of Aldh1b1, Sox9, Pdx1, and Nkx6.1 were also strikingly similar to Aldh+ organoids and stable at successive passages, while there was no appreciable expression of terminal endocrine or acinar genes (Ins1, Ptf1a, and Amy) (n = 3) (compare SI Appendix, Fig. S2C with SI Appendix, Fig. S2P). Aldh1b1 transcript levels were significantly lower in organoid cells than in embryonic pancreata. This could reflect an adaptation to culture conditions or a genuine difference in Aldh1b1 expression between embryonic and adult progenitors. Embryonic progenitors need to quickly down-regulate Aldh1b1 levels upon differentiation (33, 34). This may necessitate lower protein stability and, therefore, correspondingly higher transcript levels.
To evaluate whether a functional Aldh1b1 gene was necessary for adult pancreatic organoid formation, we generated a Cre conditionally inactivated Aldh1b1 allele (Aldh1b1fl) (SI Appendix, Fig. S2Q). These mice were intercrossed with ROSA26CreERT (2) mice to generate double homozygotes. To examine the consequences of Aldh1b1 deletion in organoid formation and to ensure timely deletion while in culture we initiated Aldh1b1 inactivation in vivo by tamoxifen administration to 8-wk-old Aldh1b1fl/l/ROSA26CreERT2/CreERT2 mice. This regime resulted in partial Aldh1b1 inactivation as seen by the presence of both intact and recombined alleles (SI Appendix, Fig. S2R). Pancreatic Aldh+ cells, comparable in numbers to those recovered from untreated animals, were FACS-isolated and placed in organoid culture either in the presence of 1 μM 4-hydroxytamoxifen (4-OHT) to complete Aldh1b1 inactivation, or in its absence (SI Appendix, Fig. S2P). Exposure to 4-OHT in culture effected the completion of Aldh1b1 deletion and this resulted in failure of these organoids to grow, in contrast to control organoids that have not been treated with 4-OHT (Fig. 2 Q and R and SI Appendix, Fig. S2R).
Altogether, these analyses revealed that Aldh1b1-expressing cells are a subset of centroacinar cells of the adult mouse pancreas, which are necessary and sufficient for the formation of organoids. Importantly, Aldh1b1 function was required for organoid self-renewal.
Aldh1b1-Expressing Cells Self-Renew and Contribute to All 3 Pancreatic Lineages in Homeostasis.
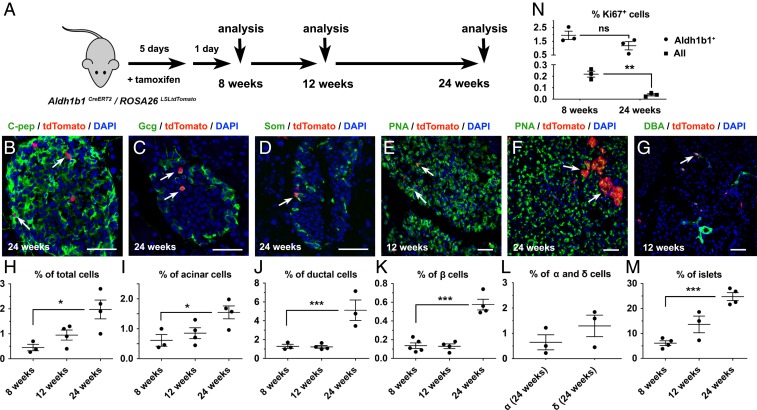
Next, we examined whether Aldh1b1-expressing cells contribute to all 3 lineages of the adult pancreas in vivo under homeostatic conditions. Aldh1b1CreERT2/ROSA26LSLtdTomato double-heterozygous mice were treated with 5 consecutive doses of tamoxifen and the descendants of pancreatic Aldh1b1-expressing cells, constitutively expressing tdTomato from the ROSA26 locus (tdTomato+ cells), were then analyzed at 3 time points: 1 d after the end of the labeling period (i.e., at the age of 8 wk) as well as at the ages of 12 and 24 wk (Fig. 3A). During this 16-wk period, tdTomato+ cells were found in all 3 pancreatic lineages at all 3 time points of analysis (Fig. 3 B–G).
Fig. 3.
The progeny of Aldh1b1-expressing cells contribute to all 3 pancreatic lineages. (A) Aldh1b1CreERT2/ROSA26LSLtdTomato double heterozygotes were treated with tamoxifen and analyzed 1 d later (8 wk) as well as at 12 and 24 wk of age. (B–G) tdTomato+ progeny became C-pep+ (C), Gcg+ (C), or Som+ (D) cells, acinar cells (E and F), or duct cells (G). (H–M) tdTomato+ cells (H), tdTomato+ acinar cells (I), tdTomato+ duct cells (J), tdTomato+ β cells (I), and the number of islets containing tdTomato+ β cells (M) increase over time. The contribution of tdTomato+ cells to α and δ cells was quantified at 24 wk (L). (N) Percentage of Aldh1b1+ or all pancreatic cells that were Ki67+ at 8 and 24 wk of age. Individual data points for each time point represent animals analyzed. (Scale bars: 50 μm.) In G–K the mean (wide horizontal bar) and the SEM (narrow horizontal bars) are shown. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
To analyze the contribution of the descendants of Aldh1b1-expressing cells in the adult pancreas under homeostatic conditions, we used flow cytometry to assess the numbers of descendants of Aldh1b1-expressing cells as tdTomato+, acinar (tdTomato+/PNA+), and duct (tdTomato+/DBA+) cells. In these experiments, an average of 100,000 single viable cells were analyzed for each marker and time point (n = 3 to 5 per marker and time point). This analysis showed a gradual increase of tdTomato+ cells but the numbers of tdTomato+/PNA+ and tdTomato+/DBA+ cells were significantly increased only at the 24-wk time point. This suggested that labeled cells first underwent expansion and then differentiation (Fig. 3 H–J and SI Appendix, Fig. S3 A–D). Labeled duct cells were found primarily in terminal ducts, but some were also incorporated in larger ducts (Fig. 3G and SI Appendix, Fig. S3 E and F). Consistent with the flow cytometry analysis, acinar tdTomato+ cells analyzed by immunofluorescence were most conspicuous at 24 wk (Fig. 3 E and F).
The contribution of the descendants of Aldh1b1-expressing cells to the endocrine lineage was assessed by immunofluorescence and image analysis at all 3 time points for β cells and at 24 wk for α and δ cells. On average 50 islets per animal, consisting of a total of 2,000 to 5,000 C-peptide+ cells, were imaged and analyzed (n = 3 to 5 per time point). Similarly to the acinar and duct lineages, the increase in β cells was only apparent at 24 wk (Fig. 3 K and L). Mature tdTomato+ β cells were also detected within the islets as Ucn3+ cells (SI Appendix, Fig. S3G). To confirm that labeled β cells are derived from progenitors residing outside the islets we scored the percentage of islets containing tdTomato+ β cells for all 3 time points of the analysis. We found that the number of islets containing tdTomato+ cells increased over time, consistent with a model according to which tdTomato+ β cells originate from progenitor cells outside the islets (Fig. 3M). The distribution of tdTomato+ β cells in islets over time was also consistent with such a model, resulting in an increase of the number of islets containing single tdTomato+ β cells as well as the increase islets containing more than 1 labeled β cell (SI Appendix, Fig. S3H).
As expected, differentiation was accompanied by loss of Aldh1b1 expression. Therefore, most DBA+/tdTomato+ cells were Aldh1b1−, whereas tdTomato+ endocrine or acinar cells were always Aldh1b1− (SI Appendix, Fig. S3 E and F). Conversely, Aldh1b1+ cells retained their centroacinar morphology throughout this period. We then asked whether Aldh1b1-expressing cells could switch on expression of Aldh1a1. Immunofluorescence analysis of 215 TdTomato+ cells from 3 different 24-wk-old animals showed that 214/215 of these cells were Aldh1a1−, firmly establishing that the Aldh1b1+ and Aldh1a1+ centroacinar cells are distinct subpopulations (SI Appendix, Fig. S3M).
We then determined whether Aldh1b1-expressing cells self-renew in vivo. Despite the detection of differentiated progeny that were contributed by the Aldh1b1-expressing cells, the percentage of undifferentiated Aldh1b1+ cells, as assessed by immunofluorescence, centroacinar morphology, and localization outside of islets or ducts, remained comparable at 8 and 24 wk of age (1.8 ± 0.6% and 1.4 ± 0.9%, n = 3 and 4, respectively, an average of 170 Aldh1b1+ cells scored per pancreas) (SI Appendix, Fig. S3 I–K). Importantly, the percentage of Aldh1b1+ cells that were also tdTomato+ remained the same between 8 and 24 wk of age (81.4 ± 2.8% and 84.8 ± 5.2%, n = 3 and 4, respectively) (SI Appendix, Figs. S3 J, K, and O). These results indicated that Aldh1b1+ cells are not replenished from another source but self-renew in vivo. To confirm this, we compared the percentage of Aldh1b1+ cells that remain proliferative with that of the general pancreatic cell population at 8 and 24 wk of age by scoring at least 500,000 cells per pancreas (n = 3 for each time point and condition). Immunofluorescence for Ki67 expression, a marker strictly associated with proliferative cells (35), showed that while the percentage of proliferative Aldh1b1+ cells remained stable at 8 and 24 wk (1.9 ± 0.3% and 1.2 ± 0.3% respectively), the corresponding percentage of the general population was substantially lower (0.2 ± 0.02) at 8 wk and declined further (0.04 ± 0.01) at 24 wk (Fig. 3N).
Considered altogether, these data show that Aldh1b1-expressing cells contribute to all 3 pancreatic lineages under conditions of homeostasis and are maintained as a long-term self-renewing progenitor population.
Progenitor Cell Signature and Early Differentiation Routes.
Identification of a stem/progenitor cell population based solely on single-cell RNA sequencing (scRNA-seq) results is extremely difficult, particularly in tissues with rare such cells (36, 37). Our FACS analysis indicated that Aldh1b1-expressing adult pancreatic cells represent just 0.5% of the cells in the adult organ. We used the Aldh1b1CreERT2 lineage tracer in combination with scRNA-seq to establish the molecular signature of these rare cells and infer their early differentiation routes.
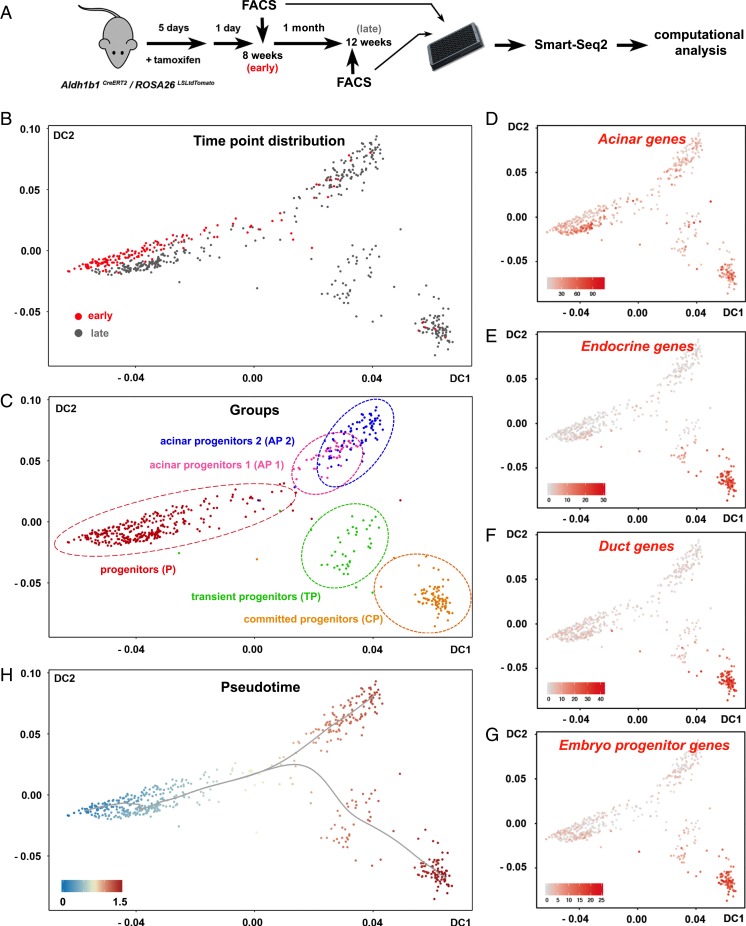
TdTomato+ cells were FACS-isolated 1 d after the completion of tamoxifen administration (early cells) and after 1 mo (late cells) and their transcriptomes were determined by an adapted Smart-Seq2 protocol for scRNA-seq (38) (Fig. 4A). This experimental design introduced a key criterion for the identification of the adult progenitor cell population because early cells would contribute primarily, if not exclusively, to the original progenitor population. After initial mapping and computation of unique reads, only cells that passed all quality control criteria were retained. This resulted in a total of 578 tdTomato+ cells, of which 164 and 414 originated from the 8-wk (early) and the 24-wk (late) time points, respectively (Fig. 4A) (39).
Fig. 4.
scRNA-seq of Aldh1b1-expressing cells and their early progeny. (A) Aldh1b1CreERT2/ROSA26LSLtdTomato double heterozygotes were treated with tamoxifen and labeled cells were FACS-isolated 1 d (early cells) or 1 mo later (late cells) and processed for Smart-Seq2 sequencing and computational analysis. (B and C) Early or late cells are depicted on the diffusion map as red or gray dots, respectively (B), and molecular marker analysis revealed the identity of the 5 identified clusters (C). (D–G) Mapping of the aggregate acinar (D), endocrine (E), duct (F), and progenitor (G) gene expression values. (H) DPT suggested 2 differentiation pathways (gray line) with a single branching point. Numbers on the axes of all diffusion maps (B–H) represent diffusion coefficients (DC) and the gray line (H) represents differentiation pathways. Values on the gene expression heat maps (D–G) correspond to the aggregate log10(sequencing counts per million) values.
To identify distinct cell groups we used K-means unsupervised clustering (SC3) (40) for early cells and all cells. The results were visualized through the generation of diffusion maps, a nonlinear dimensionality reduction method that strongly reduces noise and is able to represent data from differentiation routes with branching points (41). Early cells segregated in 2 groups, 1 large and 1 small (SI Appendix, Fig. S4A). Cells in the small group expressed both embryonic pancreas progenitor markers and markers of the endocrine, acinar, and ductal lineages, indicating that they are differentiating progenitors. In contrast, cells in the large group of progenitors retained primarily expression of only acinar markers (SI Appendix, Fig. S4 B and C and Table S1). To identify the progenitor population we proceeded with the analysis of all cells, isolated at both early and late time points. K-means unsupervised clustering identified 5 distinct groups (SI Appendix, Fig. S4D), the largest of which contained the large majority (90%) of early cells and a large number (41%) of late cells (compare Fig. 4 B and C with SI Appendix, Fig. S4E). Considering that early-time-point cells contribute primarily to the original progenitor population, we conclude that this group constitutes the progenitor cell group, the source of all other cells (Fig. 4 B and C and SI Appendix, Fig. S4E). Plotting the combined expression of either acinar, ductal, endocrine or embryonic progenitor marker genes (SI Appendix, Table S1) on the diffusion map showed that all cells retained some expression of acinar terminal markers. Embryonic pancreas progenitor, endocrine, and ductal markers were all activated in a single cluster, the committed progenitor cluster (Fig. 4 C–G). Apart from progenitor and committed progenitor populations, this analysis established the presence of acinar progenitors that do not activate other lineage or progenitor markers and the presence of a transient progenitor population. The latter was characterized by the expression of Dclk1, a recently identified marker of quiescent pancreatic progenitors (42) (Fig. 4C and SI Appendix, Fig. S4F). Consistent with an ongoing, long, differentiation process between the 2 time points at which cells were collected, the number of expressed terminal differentiation markers for each of the 3 lineages increased substantially in the late cell population (SI Appendix, Table S1).
To infer the early differentiation routes, we implemented pseudotemporal ordering of the cells using diffusion pseudotime (DPT) (43). DPT orders cells on the diffusion map by comparing their differentiation probabilities toward different cell fates and suggests the most probable differentiation routes including branching points (43). DPT analysis of all cells suggested 2 differentiation routes with a single branching point similar to the corresponding pattern in early cells (Fig. 4H and SI Appendix, Fig. S4G).
Differential gene expression analysis identified the molecular signatures of the adult pancreas progenitor cells and their early progeny (SI Appendix, Table S2). Differentially expressed genes include cell surface markers, signal receptors, and transcription factors that follow distinct expression patterns prefiguring the transitions between cell subpopulations (SI Appendix, Fig. S4 H and I). The information from these findings could be exploited for the formation of bona fide pancreatic organoids from adult progenitor cells in vitro, for directing adult pancreas regeneration in vivo, and for studies that would increase our understanding of the pathogenesis of pancreatic disorders. In this regard, we inquired whether this analysis could offer insights into the origin of pancreatic cancer, as Gene Ontology analysis of differentially expressed genes suggested a key role of small GTPases, including Kras, in the progenitor cells (SI Appendix, Table S3).
Aldh1b1 Is Required for the Development of Mouse PDAC.
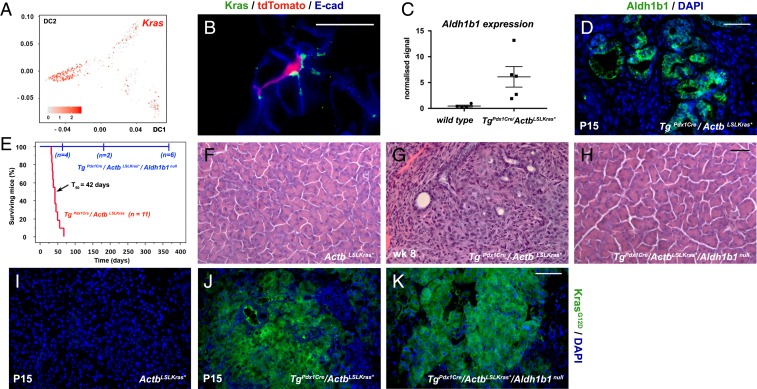
The scRNA-seq analysis indicated that Kras is preferentially expressed in the adult progenitor cells (Fig. 5A). This was confirmed by immunofluorescence with an antibody against wild-type Kras in sections of adult pancreata labeled and collected as in Fig. 2F. The results showed that the Kras+ cells are relatively rare (∼5%) and confined exclusively to the exocrine compartment exhibiting apparent centroacinar but not acinar morphology. Kras+ cells were particularly enriched in the Aldh1b1-expressing (tdTomato+) population as 31.7 ± 9.8% of the latter (n = 3) were Kras+ (Fig. 5B). This significant degree of Kras/Aldh1b1 coexpression was intriguing considering that ALDH1B1 protein expression was 12-fold higher than normal in human pancreatic cancers (30), while ALDH1B1 gene expression was 5-fold higher in human PDAC specimens (n = 96) (44). Additionally, there is a nearly 7-fold up-regulation of Aldh1b1 expression (n = 5), also reflected in immunofluorescence experiments (Fig. 5 C and D) in our fully validated TgPdx1Cre/ActbLSLKras* mouse model which faithfully simulates human PDAC (45, 46). This model shows stagewise, orderly, but rapid, progression to invasive PDAC (45) that was also confirmed during this work (n = 10). The mice first develop low-grade pancreatic intraepithelial neoplasias (PanIN) around postnatal day 10 (P10) and have a median survival time of 42 d after birth (45) (Fig. 5E). Pairwise comparisons of microarray gene expression profiles with either the TgPdx1Cre/KrasLSLKras* model (6) or with human PDAC showed a very high degree of statistically significant overlap (45). Despite nearly uniform expression of Kras* initiated during pancreas development of the TgPdx1Cre/ActbLSLKras* mice, Aldh1b1+ cells at P1 were still exceedingly rare, as in control mice (SI Appendix, Fig. S5 A and B), indicating that Aldh1b1 may not be downstream of Kras activation and that the number of Aldh1b1+ cells increases only postnatally, during cancer progression.
Fig. 5.
Aldh1b1 is strongly up-regulated in mouse PDAC and necessary for its initiation. (A) Kras is preferentially expressed in progenitor cells. Numbers on the axes represent diffusion coefficients and the values of the heat map correspond to the log10(normalized sequencing counts) values. (B) Immunofluorescence for Kras showed colocalization with tdTomato+ cells. (C and D) Aldh1b1 expression is dramatically up-regulated in the Pdx1Cre/ActbLSLKras* PDAC model at both the mRNA (microarray gene expression analysis) (C) and protein levels as detected by immunofluorescence (D). (E) Kaplan–Meier mouse survival curves showed that TgPdx1-Cre/ActbLSLKras* mice had a median survival rate (T50) of 42 d (red line). In contrast, Kras* was unable to exert its oncogenic action in TgPdx1Cre/ActbLSLKras*/Aldh1b1null mice (blue line) (log-rank P < 0.0001). From a cohort of 12 animals, 6 were killed at the time points indicated on the graph and histological analysis revealed that they were tumor- and PanIN-free. The remaining 6 animals were healthy and tumor-free until the age of 12 mo. (F–H) Normal pancreas morphology in ActbLSLKras* animals (F) and invasive PDAC in the Pdx1Cre/ActbLSLKras* PDAC model at P15 (G) which never appears in absence of functional Aldh1b1 (H). (I–K) Immunofluorescence showed that induction of Kras* (E and F) is not affected in the absence of Aldh1b1 (F). (Scale bars: 20 μm, B; 50 μm, D and F–H; 60 μm, I–K.) In C the mean (wide horizontal bar) and the SEM (narrow horizontal bar) are shown.
We then asked whether Aldh1b1 is required for Kras-driven tumor initiation in the TgPdx1Cre/ActbLSLKras* mouse model (45, 46). Strikingly, transferring the TgPdx1Cre/ActbLSLKras* transgenes into the Aldh1b1 null genetic background (33) completely abrogated cancer development and permitted normal lifespan. From a cohort of 12 animals, 6 were killed at the different time points indicated on the graph and histological analysis revealed that they were tumor- and PanIN-free. The remaining 6 animals were healthy and tumor-free until the age of 12 mo (Fig. 5 E–H). This occurred despite sustained strong Kras* expression in the pancreata of the TgPdx1Cre/ActbLSLKras*/Aldh1b1null rescued animals (Fig. 5 I–K), indicating a strong requirement for Aldh1b1 in the development of mouse pancreatic cancer.
We then addressed whether Kras* induction, specifically in the Aldh1b1-expressing cells, would result in development of PDAC. To this end, we activated the ActbLSLKras* transgene using our Aldh1b1CreERT2 Cre driver. We observed that upon induction of Kras* expression by tamoxifen injection in adult Aldh1b1CreERT2/ActbLSLKras*/ROSA26tdTomato triple heterozygotes, mice became cachectic and moribund, necessitating their being killed after less than 15 d. This early killing precluded the detection of PDAC that could have developed at a later time point. PanIN was not yet detectable histologically but this might not be surprising considering that in the TgPdx1Cre/ActbLSLKras* model cells have been exposed to Kras* activity for at least 10 d during development and an additional 10 d postnatally before the detection of low-grade PanIN (41). On the other hand, the autopsy revealed the presence of extensive intestinal tumors (to be analyzed in detail elsewhere), consistent with the strong Aldh1b1 expression in the intestinal crypts (SI Appendix, Fig. S5C). Importantly, it indicated that Aldh1b1-expressing cells in other tissues may be particularly susceptible to Kras*-driven oncogenic transformation. This information should be taken into consideration in future studies.
Our genetic analyses showed that the Aldh1b1-expressing adult pancreatic progenitors might be a prime target of Kras*-mediated pancreatic oncogenesis and identified Aldh1b1 function as a requirement for mouse pancreatic cancer initiation.
Discussion
Centroacinar cells have been prospectively isolated on the basis of their high Aldh activity using the live Aldh substrate Aldefluor, which detects several Aldh isoforms (23, 24). When cultured under specific conditions in suspension these cells generated pancreatospheres containing endocrine and acinar cells raising the possibility that these cells could carry a similar function in vivo. Gene expression analysis for the Aldh1a1, 1a2, 1a3, 1a7, and 8a1 genes suggested that Aldh1a1 was the main isoform expressed (26). However, this analysis covered only a small number of the 19 murine Aldh genes. It did not include Aldh1b1, which was at the time unknown (47). Due to the lack of definitive molecular markers, the in vivo potential of these cells has not been addressed since then. In this paper, we provide strong evidence that a subpopulation of Aldh+ cells, characterized by the expression of Aldh1b1, contribute to all 3 pancreatic lineages, self-renew in vivo and in vitro, and constitute the cells of origin of pancreatic organoids. Characterization of this population and its early descendants by scRNA-seq provided its molecular signature and suggested the preferential expression of Kras in Aldh1b1+ centroacinar cells. This was confirmed by immunofluorescence, while subsequent genetic experiments showed an absolute requirement of Aldh1b1 in the development of KrasG12D-driven pancreatic cancer. We think that, collectively, these findings reconcile a large number of seemingly disparate reports postulating that either ducts or acinar cells contain stem or progenitor cells that may give rise to PanIN and PDAC.
The Aldh1b1-expressing progenitor cells appear to be centroacinar cells, and this is consistent with many studies which suggested that progenitor cells are of ductal origin (13, 48, 49). Aldh+ centroacinar cells, as defined by location and morphology, are a heterogeneous population as indicated by the existence of centroacinar Aldh1a1+ cells that are clearly distinct from Aldh1b1+ cells. Previous studies suggested that Sox9+ or Hes1+ centroacinar cells could be a source of progenitor cells (20, 21, 26) but lineage tracing did not confirm this hypothesis (14, 15). This could be due to incomplete labeling and the heterogeneity of centroacinar cells described here could be an additional factor. Moreover, our scRNA-Seq analysis showed that Hes1 and Sox9 are highly, but variably, expressed in the committed progenitor population, but less so in the progenitor cells themselves (SI Appendix, Fig. S4 J and K).
Aldh1b1-expressing cells give rise to cells of all 3 lineages and self-renew in vivo for an extended period of time, while maintaining significantly higher proliferative capacity compared to the rest of the pancreas. On the other hand, and in line with the much lower regenerative capacity of the pancreas compared to that of the intestine, fewer than 2% of the Aldh1b1+ cells are proliferative as opposed to 90% of the Lgr5+ intestinal stem cells (50). Aldh1b1-expressing cells are distributed throughout the pancreas, sometimes in proximity to islets or large ducts. Their mode of migration into islets and ducts would be of interest, particularly given that they may constitute cells of origin of pancreatic cancer and therefore their migration mechanism could play a role in metastasis.
Lineage tracing analysis over a period of 4 mo showed a gradual increase in the number of tdTomato+ cells, while the number of differentiated cells increased significantly only at the later time point. Additionally, there was an increase in the number of islets containing labeled β cells. These findings are consistent with a model according to which Aldh1b1-expressing cells first expand and then differentiate. The question arises whether they are tripotent progenitors or a heterogeneous population consisting of acinar, endocrine, and duct progenitors. The scRNA-seq analysis favors the first hypothesis, considering that progenitors appear all in 1 cluster. Within this cluster there is some divergence based on whether cells are isolated early or late (Fig. 4B). At present, however, we cannot distinguish whether this reflects the genuine evolution of this population over time or is due to experimental variation. Collectively, these findings suggest the intriguing possibility that the population of Aldh1b1-expressing cells could include bona fide adult pancreatic stem cells, but additional molecular markers and clonal analysis are required to conclusively address this issue.
Interestingly, it was recently shown that among ductal cells only a subset, expressing the cell surface marker CD24, were able to form organoids (51). We have identified the same marker in the signature of committed progenitor cells (SI Appendix, Table S2). Because Aldh+ cells, in already formed human pancreatic organoids, have endocrine potential and have been identified as the self-renewing component of the same organoids (52), it will be important to identify the equivalent cells in the human pancreas.
The cell(s) of origin of pancreatic cancer remain(s) elusive. Since the demonstration that a Pdx1-Cre– or Ptf1a-Cre–driven expression of KrasG12D in all embryonic pancreas progenitor cells is sufficient to induce pancreatic cancer (3, 6), several Cre drivers have been used to identify adult cell populations that can undergo Kras*-driven oncogenic transformation. The use of acinar specific Cre drivers, such as Ptf1a-CreER, Mist1-Cre, and Ela-Cre, suggested that a small subset of acinar cells can give rise to pancreatic neoplasia and PDAC (5, 7, 53). However, not all acinar cells can undergo oncogenic transformation. Acinar cells are generally resistant to Kras*-driven transformation, possibly because the mature acinar genes Nr5a2, Ptf1a, and Mist1 are effectively acting as Kras* tumor suppressors (54–56). Thus, it is interesting that Aldh1b1-expressing progenitors express a number of acinar markers but not these tumor suppressor transcription factors (SI Appendix, Table S1). Other reports suggested that centroacinar and ductal cells may also give rise to pancreatic neoplasias, particularly after PTEN ablation (25, 27, 57, 58). Whereas Sox9 is necessary for the development of pancreatic cancer, Sox9-CreER–driven activation of KrasG12D expression was not sufficient to induce oncogenic transformation of ductal or centroacinar cells (7). This still leaves open the possibility that Aldh1b1+/Sox9− centroacinar cells may serve as TICs in the pancreas. A common limitation of these approaches is that, while they reveal the potential of a cell population for neoplastic transformation, they do not show that the cells under consideration actually function as TICs. We propose that Kras+ cells are more likely to undergo oncogenic transformation upon the acquisition of activating mutations in this gene. Our data suggested that Kras-expressing cells have centroacinar morphology and that Aldh1b1-expressing cells are enriched in Kras expression, making them strong candidates for TICs. Pancreatic cancer risk factors, such as chronic pancreatitis, may induce Kras expression in dedifferentiating acinar cells. Therefore, it is not unlikely that more than 1 cell type could be susceptible to neoplastic transformation but in such a scenario, Aldh1b1 expression would need to be induced de novo.
Survival rates of PDAC patients have not been improved for the last 40 y. This is due to late diagnosis, because the identified prevalent genetic mutations and genes essential for the early steps in disease progression are notoriously difficult to target (7, 59–61). Therefore, the identification of additional causal or necessary factors is an urgent endeavor. Thus, the association of high ALDH1B1 expression with increased PDAC invasiveness (30) and the up-regulation of several ALDH genes that has been detected in all human PDAC samples examined (44) (SI Appendix, Fig. S5D) are noteworthy. Additionally, high ALDH activity has been used as the most efficient means to identify rare tumor-initiating cells (62, 63) but an essential role of this enzymatic activity in pancreatic progenitor cells and cancer had not been demonstrated. Small GTPase expression, and Kras expression in particular, is a hallmark of Aldh1b1+ cells and our study has revealed that the mitochondrial Aldh1b1 is necessary for Kras*-induced PDAC in mice. We think, therefore, that the Aldh1b1+ cell population, which already expresses Kras and maintains significantly higher proliferative capacity compared to the rest of the pancreas, would be particularly vulnerable to oncogenic transformation through the acquisition of Kras-activating mutations. These findings render this population a strong candidate for the cells, or 1 type of the cells, of origin of pancreatic cancer.
The mechanism by which Aldh1b1 activity enables Kras*-mediated oncogenic transformation requires investigation beyond the scope of this study. Aldh1b1 may enhance cell metabolism to promote cell proliferation, but we also note that Kras* triggers a tumorigenic pathway that enhances reactive oxygen species (ROS) production through the alteration of mitochondrial metabolism (64). To sustain tumorigenesis, ROS levels should be regulated to remain below toxic levels. Thus, as an example of another potential mechanism, this could be attained through the antioxidant action of ALDHs (65) and, more specifically, Aldh1b1. In the context of these hypotheses, we view the Aldh1b1 function not as oncogenic per se but as a precondition for Kras*-induced PDAC.
The findings reported here suggest possibilities for pancreas regeneration and a potential therapeutic target in pancreatic cancer.
Materials and Methods
The Aldh1b1tm2(CreERT2)Agav (Aldh1b1CreERT2) line was generated by inserting the bicistronic Aldh1b1/P2A/CreERT2 transgene followed by the FRT-PGK-neo-pA-FRT selection cassette in the endogenous Aldh1b1 locus (SI Appendix, Fig. S4). Activation of the CreERT2 was obtained by intraperitoneal injection for 5 consecutive days of corn oil-dissolved tamoxifen (20 mg/mL; Sigma) as described in SI Appendix.
For FACS isolation of Aldh+ cells and progeny of Aldh1b1-expressing cells, adult mouse pancreata were dissected, minced into small pieces, and digested at 37 °C by 1 mg/mL collagenase D (11088858001; Roche). Aldh+ cells were isolated based on their Aldh activity using the Aldefluor reagent (Aldefluor kit 01700; Stem Cell Technologies), whereas Aldh1b1-expressing cells and their progeny were isolated from Aldh1b1CreERT2/ROSA26LSLtdTomato double-heterozygous mice by virtue of their tdTomato expression.
For the 3D culture of adult pancreas progenitor organoids, FACS-isolated Aldh+ or tdTomato+ cells were seeded in growth factor-reduced Matrigel (356231; BO Biosciences) to a final density of 500 or 400 cells per μL, in a 96-well plate. After gelation of the Matrigel, culture medium was added as described in SI Appendix and replenished every 3 d. Organoids usually formed on the third day after seeding cells in culture and were passaged for the first time after 10 to 14 d.
Single-cell library preparation, RNA sequencing, bioinformatics analysis as well as quantitative flow cytometry, time lapse fluorescent microscopy, and other nonspecialized methods are described in SI Appendix, along with a list of mouse strains, antibodies, genotyping primers, and other biological reagents used.
Animal maintenance and experimentation were conducted in accordance with the Federation of European Laboratory Animal Science Associations recommendations and the ethical and practical guidelines for the care and use of laboratory animals set by the competent veterinary authorities in the authors’ institutions.
Supplementary Material
Acknowledgments
We thank Heiko Lickert, Mina Gouti, Lori Sussel, and Gaetano Gargiulo for critically reading the manuscript and providing comments; Chris Wright, Michael Ray, and Tetsuo Sudo for reagents; the CRTD FACS core facility and the MPI-CBG Dresden transgenic core for expert technical assistance; and the personnel of the animal houses at the Center for Regenerative Therapies, Dresden and Biomedical Research Foundation of the Academy of Athens (BRFAA) for help with animal husbandry. Research in A.E.’s laboratory was supported by funds provided to BRFAA by a Krauss Maffei Wegmann GmbH and Co Offset Program. Research in A.G.’s laboratory was supported by the German Center for Diabetes Research (DZD e.V.), which is supported by the German Ministry for Education and Research (BMBF), German Research Foundation Grant SFB 655, Center for Regenerative Therapies Dresden Seed Grant 2014, German Research Foundation Project Grant GA 2004/3-1, and a Technische Universität Dresden Graduate Academy Bridging Grant to E.M.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The single-cell RNA Seq data reported in this paper have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE110283).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901075116/-/DCSupplemental.
References
- 1.Kopp J. L., Grompe M., Sander M., Stem cells versus plasticity in liver and pancreas regeneration. Nat. Cell Biol. 18, 238–245 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Murtaugh L. C., Keefe M. D., Regeneration and repair of the exocrine pancreas. Annu. Rev. Physiol. 77, 229–249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguirre A. J., et al. , Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 17, 3112–3126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De La O J. P., et al. , Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc. Natl. Acad. Sci. U.S.A. 105, 18907–18912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habbe N., et al. , Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc. Natl. Acad. Sci. U.S.A. 105, 18913–18918 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hingorani S. R., et al. , Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Kopp J. L., et al. , Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storz P., Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 14, 296–304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baeyens L., et al. , Transient cytokine treatment induces acinar cell reprogramming and regenerates functional beta cell mass in diabetic mice. Nat. Biotechnol. 32, 76–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Cavelti-Weder C., et al. , Hyperglycaemia attenuates in vivo reprogramming of pancreatic exocrine cells to beta cells in mice. Diabetologia 59, 522–532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan F. C., et al. , Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development 140, 751–764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A., In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455, 627–632 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inada A., et al. , Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc. Natl. Acad. Sci. U.S.A. 105, 19915–19919 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopinke D., et al. , Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development 138, 431–441 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopp J. L., et al. , Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development 138, 653–665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solar M., et al. , Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev. Cell 17, 849–860 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Sancho R., Gruber R., Gu G., Behrens A., Loss of Fbw7 reprograms adult pancreatic ductal cells into α, δ, and β cells. Cell Stem Cell 15, 139–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huch M., et al. , Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J., et al. , Expansion and conversion of human pancreatic ductal cells into insulin-secreting endocrine cells. eLife 2, e00940 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cleveland M. H., Sawyer J. M., Afelik S., Jensen J., Leach S. D., Exocrine ontogenies: On the development of pancreatic acinar, ductal and centroacinar cells. Semin. Cell Dev. Biol. 23, 711–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beer R. L., Parsons M. J., Rovira M., Centroacinar cells: At the center of pancreas regeneration. Dev. Biol. 413, 8–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaspre F., et al. , Centroacinar cells are progenitors that contribute to endocrine pancreas regeneration. Diabetes 64, 3499–3509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balber A. E., Concise review: Aldehyde dehydrogenase bright stem and progenitor cell populations from normal tissues: Characteristics, activities, and emerging uses in regenerative medicine. Stem Cells 29, 570–575 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Zhou L., et al. , Identification of cancer-type specific expression patterns for active aldehyde dehydrogenase (ALDH) isoforms in ALDEFLUOR assay. Cell Biol. Toxicol. 35, 161–177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto Y., et al. , Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 3, 565–576 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Rovira M., et al. , Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc. Natl. Acad. Sci. U.S.A. 107, 75–80 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanger B. Z., et al. , Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell 8, 185–195 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Jackson B., et al. , Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genomics 5, 283–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ioannou M., et al. , ALDH1B1 is a potential stem/progenitor marker for multiple pancreas progenitor pools. Dev. Biol. 374, 153–163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh S., et al. , Aldehyde dehydrogenase 1B1 as a modulator of pancreatic adenocarcinoma. Pancreas 45, 117–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broutier L., et al. , Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724–1743 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto D., et al. , Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anastasiou V., et al. , Aldehyde dehydrogenase activity is necessary for beta cell development and functionality in mice. Diabetologia 59, 139–150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giannios I., et al. , Protein methyltransferase inhibition decreases endocrine specification through the upregulation of Aldh1b1 expression. Stem Cells 37, 640–651 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchins J. R., et al. , Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baron M., et al. , A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360.e4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wollny D., et al. , Single-cell analysis uncovers clonal acinar cell heterogeneity in the adult pancreas. Dev. Cell 39, 289–301 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Picelli S., et al. , Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Mameishvili E., Lesche M., Reinhardt S., Dahl A., Gavalas A., Single-cell RNA-seq of adult mouse pancreatic progenitor cells. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110283. Deposited 7 February 2018.
- 40.Kiselev V. Y., et al. , SC3: Consensus clustering of single-cell RNA-seq data. Nat. Methods 14, 483–486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Angerer P., et al. , Destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Westphalen C. B., et al. , Dclk1 defines quiescent pancreatic progenitors that promote injury-induced regeneration and tumorigenesis. Cell Stem Cell 18, 441–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haghverdi L., Büttner M., Wolf F. A., Buettner F., Theis F. J., Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Bailey P., et al. ; Australian Pancreatic Cancer Genome Initiative , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Stellas D., et al. , Therapeutic effects of an anti-Myc drug on mouse pancreatic cancer. J. Natl. Cancer Inst. 106, dju320 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Chio I. I., Yordanov G., Tuveson D., MAX-ing out MYC: A novel small molecule inhibitor against MYC-dependent tumors. J. Natl. Cancer Inst. 106, dju365 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Stagos D., et al. , Aldehyde dehydrogenase 1B1: Molecular cloning and characterization of a novel mitochondrial acetaldehyde-metabolizing enzyme. Drug Metab. Dispos. 38, 1679–1687 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sancho P., et al. , MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 22, 590–605 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Xu X., et al. , Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell 132, 197–207 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Basak O., et al. , Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 33, 2057–2068 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rezanejad H., et al. , Heterogeneity of SOX9 and HNF1β in pancreatic ducts is dynamic. Stem Cell Rep. 10, 725–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loomans C. J. M., et al. , Expansion of adult human pancreatic tissue yields organoids harboring progenitor cells with endocrine differentiation potential. Stem Cell Rep. 10, 712–724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimura Y., et al. , ARID1A maintains differentiation of pancreatic ductal cells and inhibits development of pancreatic ductal adenocarcinoma in mice. Gastroenterology 155, 194–209.e2 (2018). [DOI] [PubMed] [Google Scholar]
- 54.von Figura G., Morris J. P. 4th, Wright C. V., Hebrok M., Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut 63, 656–664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krah N. M., et al. , The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. eLife 4, e07125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi G., et al. , Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology 136, 1368–1378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guerra C., et al. , Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Kopp J. L., et al. , Loss of pten and activation of Kras synergistically induce formation of intraductal papillary mucinous neoplasia from pancreatic ductal cells in mice. Gastroenterology 154, 1509–1523.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fox R. G., et al. , Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature 534, 407–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleeff J., et al. , Pancreatic cancer. Nat. Rev. Dis. Primers 2, 16022 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Saborowski M., et al. , A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev. 28, 85–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ishizawa K., et al. , Tumor-initiating cells are rare in many human tumors. Cell Stem Cell 7, 279–282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim M. P., et al. , ALDH activity selectively defines an enhanced tumor-initiating cell population relative to CD133 expression in human pancreatic adenocarcinoma. PLoS One 6, e20636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liou G. Y., et al. , Mutant KRas-induced mitochondrial oxidative stress in acinar cells upregulates EGFR signaling to drive formation of pancreatic precancerous lesions. Cell Rep. 14, 2325–2336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh S., et al. , Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 56, 89–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.