Significance
Nipah and Hendra viruses are pathogenic zoonotic henipaviruses, transmitted from wildlife to humans, and potential epidemic threats. Zoonoses from wildlife hosts to humans require overcoming barriers that limit cross-species transmission. Successful usage of virus entry receptors represents a bottleneck to zoonosis. Conserved ephrins are recognized entry receptors for henipaviruses like Nipah virus. Here, we characterize the ephrin entry receptor usage of Cedar virus (CedV), a related henipavirus with unknown zoonotic potential that is nonpathogenic in animals known to be susceptible to Nipah and Hendra disease. We discovered that CedV utilizes 4 different ephrins, 3 of which are unique receptors for CedV, and determined that a single natural amino acid difference between human and mouse ephrin-A1 can dictate functional receptor usage.
Keywords: Cedar virus, henipaviruses, ephrins, virus receptors, entry
Abstract
Cedar virus (CedV) is a bat-borne henipavirus related to Nipah virus (NiV) and Hendra virus (HeV), zoonotic agents of fatal human disease. CedV receptor-binding protein (G) shares only ∼30% sequence identity with those of NiV and HeV, although they can all use ephrin-B2 as an entry receptor. We demonstrate that CedV also enters cells through additional B- and A-class ephrins (ephrin-B1, ephrin-A2, and ephrin-A5) and report the crystal structure of the CedV G ectodomain alone and in complex with ephrin-B1 or ephrin-B2. The CedV G receptor-binding site is structurally distinct from other henipaviruses, underlying its capability to accommodate additional ephrin receptors. We also show that CedV can enter cells through mouse ephrin-A1 but not human ephrin-A1, which differ by 1 residue in the key contact region. This is evidence of species specific ephrin receptor usage by a henipavirus, and implicates additional ephrin receptors in potential zoonotic transmission.
Hendra virus (HeV) and Nipah virus (NiV) are the prototypical zoonotic henipaviruses, and they exhibit a broad mammalian host range with evidence of natural infection of humans, bats, horses, pigs, cattle, goats, and dogs as well as experimental infection of ferrets, hamsters, mice, cats, guinea pigs, and nonhuman primates (1, 2). HeV and NiV infection spillover events into domestic animals and humans cause severe and often fatal respiratory and neurological disease. These viruses are biosafety level-4–restricted pathogens (3), and both NiV and HeV remain threats to economically important livestock throughout South Asia and Australia (4). NiV and henipaviral diseases have been classified by the World Health Organization (WHO) as an epidemic threat needing urgent research and development action, and are included in the WHO R&D Blueprint list of priority pathogens with epidemic potential (5). The genus Henipavirus also includes Cedar virus (CedV) and 2 candidate sequences, those of Ghana virus (GhV) and Mòjiāng virus (MojV). CedV has several notable characteristics that separate it from HeV and NiV, including a nonpathogenic phenotype in mammalian disease models (6, 7).
HeV and NiV receptors, ephrin-B2 and -B3 (8, 9), are ubiquitous membrane proteins highly conserved across mammalian species (10). Ephrin receptor sequence conservation may facilitate cross-species transmission and mediate broad host tropism (11, 12). Henipaviruses express 2 membrane-anchored envelope glycoproteins: the fusion protein (F) and the receptor-binding protein (G) that is also responsible for triggering F-mediated membrane fusion (13–16). Henipavirus G proteins are tetramers (dimer of dimers), with each monomer composed of a 6-bladed β-propeller receptor-binding domain that attaches to the flexible, exposed ephrin-B2, -B3 G protein-binding (G-H) loop (8, 17–19). The ephrin-B2, -B3 G-H loop contains 4 key amino acids, which fit into the cavity of the henipavirus G-binding pocket in an induced-fit lock and key mechanism (19). Ephrin-B2 and -B3 are expressed in many cell and tissue types, including neurons (20–22), vascular endothelium (23), and respiratory epithelium (24), and their broad somatic distribution could account for the multisystemic vasculitis and central nervous system (CNS) symptoms (meningitis and encephalitis) observed during both HeV and NiV infection (25–27).
Receptor usage and receptor tissue distribution can greatly influence host range, cellular pathology, and disease outcome of emerging viruses (28, 29). Of the known henipaviruses, HeV and NiV interact with both ephrin-B2 and -B3, while CedV and GhV engage with ephrin-B2 but not ephrin-B3 (6, 30, 31). Although GhV is only a putative henipavirus, serological exposure to a henipavirus-like virus was documented in bat and human populations sampled in Cameroon, suggesting zoonotic spillover of an African henipavirus (32). The host range and pathology of emerging henipaviruses such as GhV/MojV are less defined than those of HeV/NiV; however, henipavirus receptor usage may be useful for the prediction of both (33, 34).
Here, we characterize CedV as a model of henipavirus receptor usage. We observed strong binding, cell–cell fusion, and cell entry with ephrin-B1. Additionally, we observed binding to ephrin-A2 and ephrin-A5 and utilization of these proteins for entry into host cells. We also determined the crystal structure of CedV G in complex with ephrin-B1 and -B2, which suggested why CedV G can interact with ephrin-B1 and some A-class ephrins, whereas HeV and NiV G cannot. Finally, we established that a single amino acid difference in an exposed loop of ephrin-A1 accounts for species specific cell entry of CedV.
Results
CedV G Binds to B- and A-Class Ephrins.
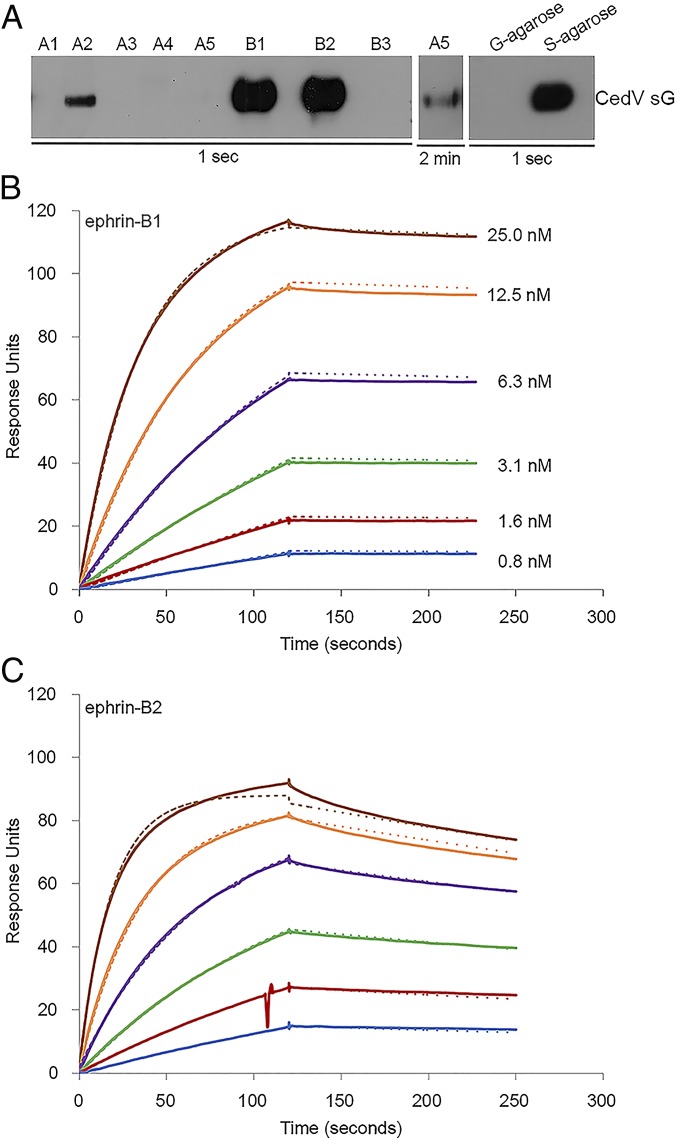
The 8 human ephrins are categorized into subclasses based on membrane anchors; ephrin-A1 to A5 are linked to the cell membrane by glycosylphosphatidylinositol (GPI), whereas ephrin-B1 to -B3 possess a transmembrane domain. Since we previously observed that replication-competent recombinant CedV (rCedV) can enter ephrin-B2–deficient HeLa-USU cells (30), we investigated whether CedV G could interact with other ephrins. We approached this question by coprecipitation of tetrameric soluble CedV G (sG) (35) with a panel of soluble ephrins. Fig. 1A documents that CedV sG coprecipitated ephrin-B2, but not ephrin-B3. Surprisingly, CedV sG coprecipitated ephrin-B1 as efficiently as ephrin-B2. We also detected weak interactions with the A-class ephrins, -A2 and -A5 (Fig. 1A).
Fig. 1.
CedV G protein interacts strongly with both ephrin-B1 and -B2 and weakly with ephrin-A2 and -A5. (A) Coprecipitation of purified S-tagged CedV sG with a panel of soluble Fc-tagged ephrin molecules: human ephrin-A1, mouse ephrin-A2, human ephrin-A3, human ephrin A4, human ephrin-A5, mouse ephrin-B1, mouse ephrin-B2, and human ephrin-B3. CedV sG controls were precipitated with S protein agarose beads (S) or G protein agarose beads (G) in the absence of soluble ephrins. An overexposure of protein blot was included for coprecipitation with soluble ephrin-A5 and CedV sG. SPR (BIAcore) sensorgrams record the interaction in response units, between sG proteins and soluble (B) mouse ephrin-B1 or (C) mouse ephrin-B2. Cycles of receptor association and dissociation performed at 6 different receptor concentrations are shown. The continuous lines represent the experimental data, and the receptor concentrations are color-coded as indicated. A 1:1 Langmuir interaction model was used to fit the data (dotted line).
Next, we used surface plasmon resonance (SPR) to quantify the interactions of ephrin-B1, -B2, -A2, and -A5. As expected, ephrin-B2 bound strongly to CedV sG (Fig. 1C and Table 1; with an equilibrium dissociation constant [Kd] = 0.56 nM). Remarkably, the affinity-binding constant of ephrin-B1 was similar to that of ephrin-B2 (Fig. 1B and Table 1; Kd = 0.24 nM). Ephrin-A2 and -A5 interacted with CedV sG at an ∼500-fold lower level than ephrin-B1 and -B2 (Table 1; Kd = 196 nM and Kd = 113 nM, respectively).
Table 1.
Affinity and kinetic rate constants for ephrin ligand binding to soluble G protein
| Ephrin ligand* | kon (M−1⋅s−1) × 103 | koff (s−1) × 10−4 | Kd, nM |
| B1 | 1,200.0 | 2.9 | 0.24 |
| B2 | 1,840.0 | 10.3 | 0.56 |
| A2 | 32.4 | 63.6 | 196.0 |
| A5 | 7.0 | 7.9 | 113.0 |
koff, dissociation reaction; kon, association reaction.
Ephrin-B1, -B2, and -A2 are of mouse origin; ephrin-A5 is of human origin.
CedV Glycoproteins Mediate Cell–Cell Fusion with GPI-Anchored and Transmembrane Ephrins.
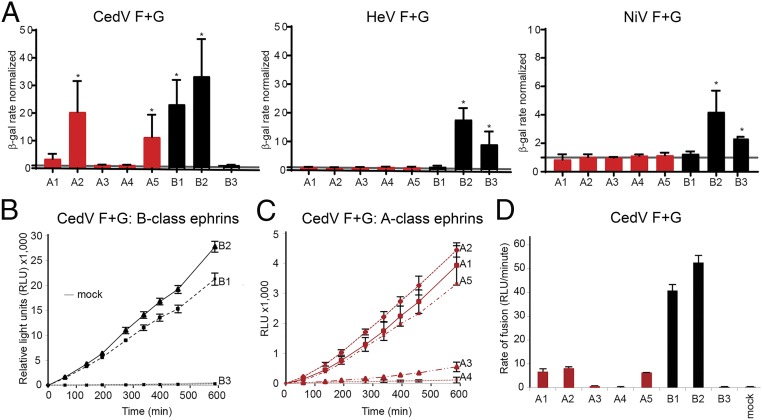
To assess whether all CedV sG-binding ephrins could support membrane fusion, we transfected CHO745 target cells (36, 37) with expression plasmids encoding each ephrin, and subsequently incubated with CHO745 effector cells transiently expressing CedV glycoproteins. Indeed, effector cells expressing CedV glycoproteins fused with target cells expressing ephrin-A2, -A5, -B1, and -B2 (Fig. 2 A, Left). We also noted minimal levels of fusion with ephrin-A1, but no fusion with ephrin-A3 and -A4. In line with previously published results, CedV glycoproteins were unable to support fusion with target cells expressing ephrin-B3 (6, 30). As controls, cells expressing HeV or NiV glycoproteins fused only with cells expressing ephrin-B2 and -B3 (Fig. 2 A, Center and Right).
Fig. 2.
CedV glycoproteins can use ephrin-B1, -B2, -A2, or -A5 to mediate cell–cell fusion. (A) β-Galactosidase (β-gal) reporter cell–cell fusion assay performed by coculturing CHO745 effector cells transiently expressing the indicated glycoproteins and target cells transiently expressing the indicated human ephrins or a control plasmid (mock). The fusion rate was normalized to mock-transfected CHO745 cells with the background fusion level indicated by a gray line. Graphs represent 3 independent experiments in technical duplicates (mean ± SD). *P < 0.05 by Student’s t test. (B and C) Split-luciferase reporter–based cell–cell fusion assay mediated by the CedV glycoproteins in CHO-K1 cells transiently expressing the indicated human ephrins or a control plasmid (mock). Data are a representation of 3 independent experiments (mean ± SD). (D) Rate of fusion was calculated based on the slope of the line in B and C. The graph represents 3 independent experiments in technical triplicates, and the error bars indicate SD.
To determine the kinetics of CedV glycoprotein-mediated cell–cell fusion with different ephrins, we used a split-luciferase reporter–based fusion assay (30). We cocultured CHO-K1 cells (SI Appendix, Fig. S1) expressing one of the ephrins with cells transiently expressing CedV glycoproteins. CHO-K1 cells expressing ephrin-B2 showed the highest fusion activity. Ephrin-B1 sustained similar levels of fusion activity as ephrin-B2, while ephrin-B3 did not support cell–cell fusion (Fig. 2B). Ephrin-A1, -A2, and -A5 also sustained cell–cell fusion, while ephrin-A3 and -A4 did not (Fig. 2C). The A-class ephrins were ∼5-fold less efficient at supporting fusion than the B-class ephrins, as calculated from the level of fusion over the linear portion of the time course (Fig. 2D). While human ephrin-A1 did not pull down sG (Fig. 1A), it did sustain low levels of fusion (Fig. 2 A and D). It should be noted that while we operated with the tagged ectodomain of ephrin-A1 for the binding assay, we used the authentic full-length protein for the cell–cell fusion assays. Moreover, the split-reporter–based fusion assay is very sensitive, even low-affinity interactions that allow fusion triggering and membrane merger may be detected, and the fusion levels remained consistent within technical replicates and across independent experiments (Fig. 2).
CedV Utilizes Ephrin-A2, -A5, and -B1 as Receptors.
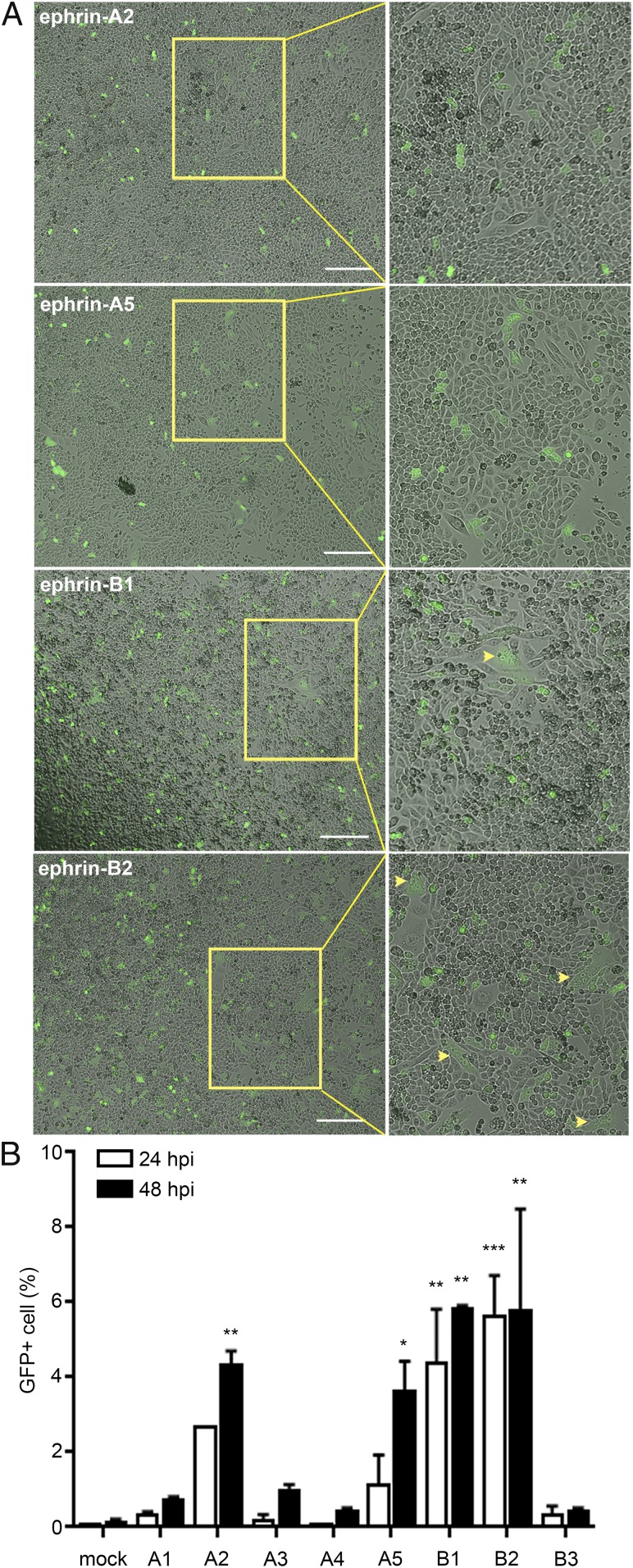
Next, we sought to determine whether the ephrins that mediated cell–cell fusion would also facilitate virus entry. We inoculated a recombinant CedV that expresses GFP (rCedV-GFP) (30) on CHO-K1 cells that had been transiently transfected with the relevant ephrins. We observed GFP expression, indicative of cell entry, in cells expressing ephrin-A2, -A5, -B1, or -B2 (Fig. 3A). More GFP-positive cells were monitored in CHO-K1 cells expressing ephrin-B1 and -B2 compared with cells expressing ephrin-A2 and -A5. In contrast to cell–cell fusion assays (Fig. 2 A and C), but in agreement with the coprecipitation data (Fig. 1A), we did not observe rCedV-GFP infection of ephrin-A1–expressing CHO-K1 cells. Interestingly, we only observed syncytia in CHO-K1 cells expressing ephrin-B1 or -B2 (Fig. 3A, yellow arrows), whereas only individual GFP-positive cells were observed with ephrin-A2 or -A5 expression (Fig. 3 A, Upper 2 panels). We quantified GFP-positive cells 24 and 48 h postinfection (hpi). CHO-K1 cells expressing ephrin-B1 and ephrin-B2 had significant levels of rCedV infection by 24 hpi (Fig. 3B), while it took longer for a similar number of ephrin-A2– or -A5–expressing cells to become infected (Fig. 3B; 48 hpi).
Fig. 3.
rCedV can utilize ephrin-A2, -A5, -B1, or -B2 to enter cells. (A) CHO-K1 cells transiently expressing the indicated human ephrins were infected with rCedV-GFP (MOI = 5.0) and examined for GFP expression and syncytia 24 hpi. Images were captured with a Zeiss Axio Observer A1 inverted microscope using a 5× objective. (Insets) Magnified areas within yellow boxes and arrowheads are used to indicate areas of syncytia. (Scale bars, 50 μm.) (B) Percentage of CHO-K1 cells expressing GFP after transient expression of the indicated ephrins and infection with rCedV-GFP (MOI = 2.0). GFP positivity was quantified by flow cytometry. All data are representative of 2 independent experiments (mean ± SD). *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA (1-way). Dunnett’s multiple comparisons are compared to mock hpi controls.
Structure of CedV G.
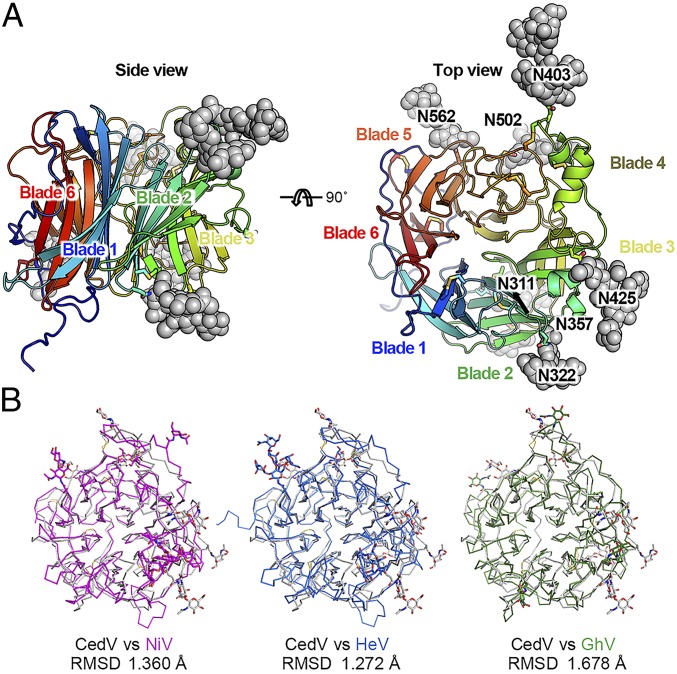
The unexpectedly broad receptor utilization by CedV G could be explained by a distinct structure of its receptor-binding site. To gain structural insights into the receptor recognition mechanism, we determined the structures of CedV G by itself and in complex with each of the 2 high-affinity receptors, ephrin-B1 and -B2. The CedV G ectodomain structure was determined at 3.3 Å resolution (SI Appendix, Table S1). Like the other henipavirus G proteins, the CedV G head domain adopts a 6-bladed β-propeller structure (Fig. 4A). To assess structural similarity, we determined the root-mean-square deviation (R.M.S.D) of all CedV G atoms when superimposed on the previously determined NiV G, HeV G, and GhV G structures (17, 18, 31). The R.M.S.D.s are 1.360 Å, 1.272 Å, and 1.678 Å, respectively (Fig. 4B). Interestingly, CedV G has more N-linked glycosylation modifications, and these modifications have different locations, than those of the other G proteins. We observed 8 N-linked glycan moieties at asparagine residues 311, 322, 357, 403, 425, 441, 502, and 562 (Fig. 4A and SI Appendix, Fig. S2), which is consistent with the prediction (NetNGlyc 1.0 Server). Moreover, we observed 8 disulfide bonds, including C239 to C263, C305 to C318, C310 to C376, C399 to C416, C404 to C520, C514 to C524, C586 to C595, and the long-range bond C212 to C622 (SI Appendix, Fig. S2). Compared with other henipavirus G proteins, an extra disulfide bond, C310 to C376, is located between blade2strand1 and blade3strand1′ of the β-propeller architecture.
Fig. 4.
Structure of CedV G and a comparison with other henipavirus G proteins. (A) Top and side views of CedV G structures in cartoon diagrams. Disulfide bonds and N-linked glycans are shown as sticks and spheres, respectively. (B) Superimposition of head domain structures from selected henipaviruses.
Structures of CedV G in Complex with Ephrin-B1 or -B2.
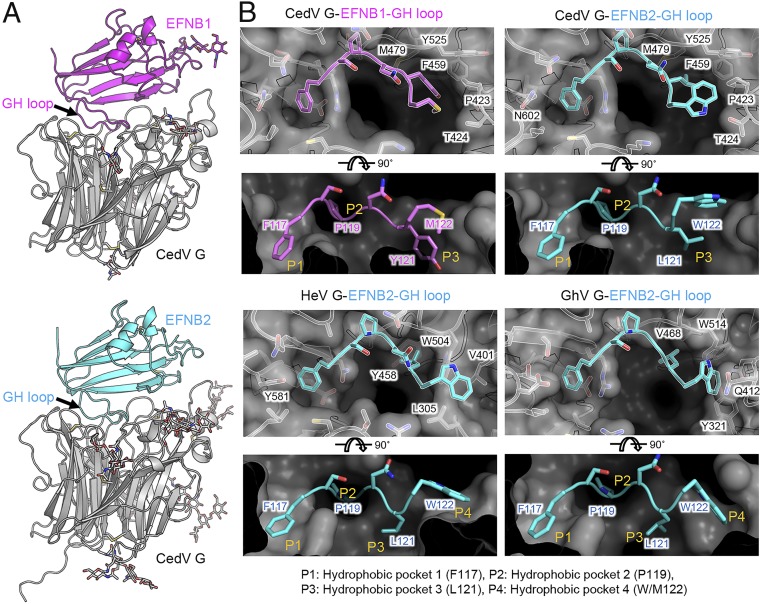
The CedV G–ephrin-B1 and CedV G–ephrin-B2 complex structures were determined at 3.50 Å and 2.85 Å resolution, respectively (Fig. 5A and SI Appendix, Table S1). The interface between CedV G protein and ephrin-B2 or ephrin-B1 buries surface areas of 1,412 Å2 and 1,425 Å2, respectively. A hydrophobic core and a hydrophilic rim, which were observed in other henipavirus–ephrin interfaces (18, 38), were also found in the CedV G–ephrin B1/B2 interfaces. The hydrophobic core was formed by inserting the tip of the ephrin G-H loop into the receptor-binding pocket of the G protein, whereas the hydrophilic rim involved multiple hydrogen bonds and electrostatic interactions between either residues K103 and K113 of ephrin-B2 or residues K59 and K113 of ephrin-B1, and residues E522, E576, and E554 of CedV G protein (SI Appendix, Figs. S3 and S4, respectively). All electrostatic interactions involved in ephrin-B2 receptor binding are conserved between the CedV G–ephrin-B2 complex and HeV G–ephrin-B2 complexes. By comparing the receptor-bound and unbound CedV G, we observed no significant conformational change, even in the receptor-binding pocket region (SI Appendix, Fig. S5A). Furthermore, CedV–G-ephrin complex structures resemble those of other reported henipavirus–G-ephrin complexes (19, 31) with a similar receptor G-H loop insertion angle (SI Appendix, Fig. S5B).
Fig. 5.
Structural basis of receptor recognition by CedV G. (A) Structures of CedV G in complexes with either ephrin-B1 or ephrin-B2. Structures are shown in a cartoon side view with CedV G in gray, ephrin-B1 in magenta, and ephrin-B2 in cyan. (B) Structural comparison of the receptor-binding pocket on G proteins of CedV, HeV, and GhV, and G-H loop insertion by ephrin-B1 or ephrin-B2. Ephrin residues 117 to 122 are shown in sticks. G protein receptor-binding sites are shown as surface, with critical contacting residues shown in sticks and labeled.
Distinct Receptor-Binding Pockets of CedV G Underlie Its Promiscuous Receptor Usage.
The residues at the tip of the G-H loop differ between ephrin family members (SI Appendix, Fig. S6) and are critical determinants of henipavirus receptor specificity (37). The central cavity of the receptor-binding site of henipavirus G is composed of 4 hydrophobic pockets, which are occupied by 4 critical residues at the tip of the ephrin G-H loop upon its insertion. Using HeV G–ephrin-B2 as an example, the 4 receptor-binding pockets (hereafter referred to as P1 to P4) are defined by the enclosed residues on the G-H loop, including F117, P119, L121, and W122, respectively (Fig. 5 B, Lower Left).
Here, we compared the receptor-binding pocket structures between CedV G and previously reported G proteins, including HeV G and GhV G. Distinct from all previously reported henipavirus G protein structures, the CedV G receptor-binding site only includes the first 3 pockets P1 to P3 (Fig. 5 B, Upper). The critical P4 pocket-forming residue W504HeV/W514GhV was substituted by Y525 in CedV G (Fig. 5B and SI Appendix, Fig. S2). This substitution allows Y525CedV side-chain stabilization by pi-stacking with residue F459CedV and swings out of the pocket region. In the vertical direction, another pocket P4 boundary-forming residue, L305HeV or Y321GhV, is replaced by D328CedV, which points away from the pocket due to the lack of hydrophobic interaction. These amino acid changes result in the loss of pocket P4 and the enlargement of pocket P3 in CedV G (Fig. 5B).
This structural alteration of the receptor-binding pockets allows accommodation of residues with larger side chains in the pocket P3, as evidenced by the fitting of residue Y121 in the CedV G–ephrin-B1 complex (Fig. 5 B, Upper Left). It is worth noting that Y121 sterically clashes with pocket P3 of other henipavirus G proteins, preventing ephrin-B1 from being utilized as their receptor (37). As a consequence of lacking pocket P4, the fourth G-H loop–inserting residue (e.g., W122ephrin-B2, M122ephrin-B1) inserts into or stays on top of pocket P3, and is stabilized by CedV G residues P423 and T424. An altered receptor-binding pocket structure therefore underlies CedV G’s extraordinary binding compatibility to various ephrin members with diverse G-H loop sequence composition.
The inability to utilize other ephrin members, such as ephrin-B3 and -A3, can also be attributed to the structural and sequence features of CedV G. Pi-stacking between Y581 in the NiV G and HeV G P1 pockets and the pocket-inserting aromatic residue (e.g., Y120 in ephrin-B3) is an important stabilizing contact for G-H loop insertion. The substitution of Y581 by N602 in CedV G (Fig. 5B) abolishes the pi-stacking, reducing the binding compatibility in the P1 pocket. For P1 pocket insertion, tyrosine residue is energetically less advantageous than phenylalanine residue due to addition of a hydroxyl group. Therefore, neither ephrin-B3 nor -A3 with tyrosine at this position of the G-H loop can be effectively utilized by CedV G. Similarly, ephrin-B3 cannot be used by GhV due to the substitution of Y518NiV by T591GhV.
Species Specific Use of Ephrin-A1 as a CedV Receptor.
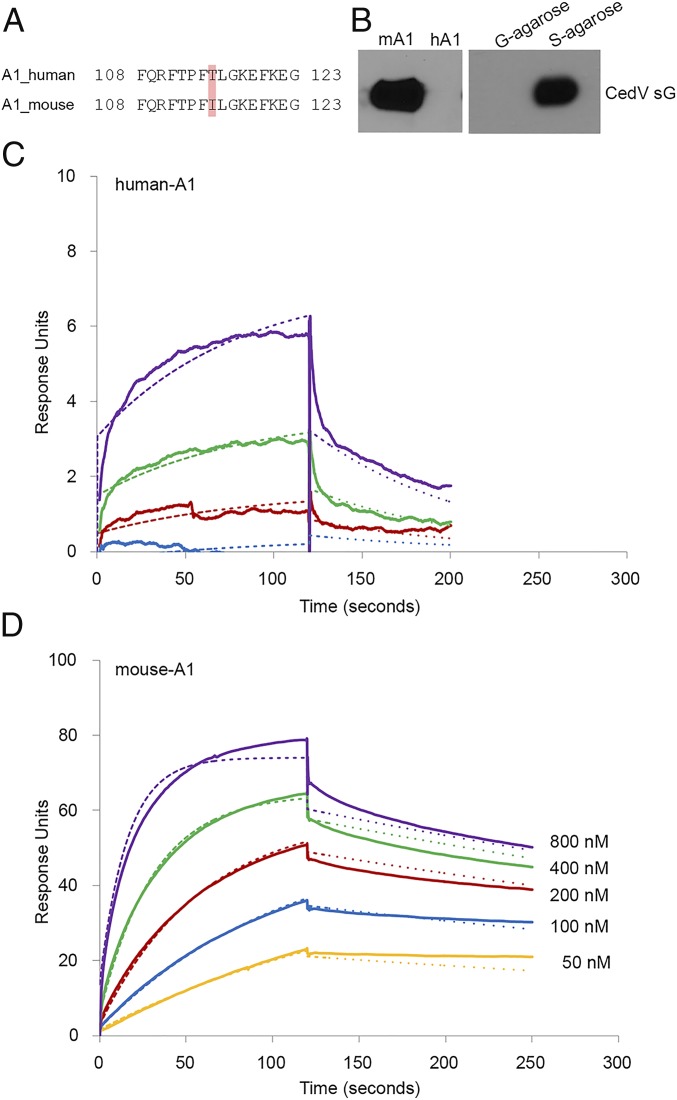
Fruit bats in the Pteropus genus are the natural reservoirs of CedV, HeV, and NiV (6, 39–41). Interestingly, the MojV sequence was isolated from a rodent (42), expanding the potential henipavirus reservoirs. The receptor for MojV is unknown, but ephrin-B2 or -B3 has been experimentally ruled out (43). Having observed that CedV uses several ephrins as receptors, we compared the 4 key contacting residues of the G-H loop between humans and rodents. Noting a single amino acid difference in the receptor-binding G-H loop between human (T115) and mouse (I115) ephrin-A1 (Fig. 6A), we assessed whether mouse ephrin-A1 functions as a henipavirus receptor. Indeed, CedV sG coprecipitates soluble mouse ephrin-A1, but not human ephrin-A1 (Fig. 6B). Further, we recorded minimal binding of CedV sG to human ephrin-A1 (Fig. 6C and Table 2; Kd = 4,210 nM), while binding to mouse ephrin-A1 was more than 100-fold stronger (Fig. 6D and Table 2; Kd = 24.5 nM).
Fig. 6.
Differential recognition of human and mouse ephrin-A1 by CedV G. (A) Sequence alignment of the G-H loop of mouse and human ephrin-A1. Highlighted residues are critical for G protein interaction. Residue 115, which differs between mouse and human ephrin-A1, is indicated. (B) Coprecipitation of purified S-tagged sG with human (hA1) or mouse (mA1) ephrin-A1. (C and D) SPR (BIAcore) sensorgrams recording the interaction (in response units) between sG proteins and the indicated ephrin-A1. Cycles of receptor association and dissociation performed at 5 different receptor concentrations are shown. The continuous lines represent the experimental data, and the receptor concentrations are color-coded as indicated. A 1:1 Langmuir interaction model was used to fit the data (dotted line).
Table 2.
Human versus mouse ephrin-A1 binding constants
| Ephrin ligand | kon (M−1⋅s−1) × 103 | koff (s−1) × 10−4 | Kd, nM |
| Human A1 | 2.5 | 98.1 | 4210.0 |
| Mouse A1 | 73.9 | 17.9 | 24.5 |
koff, dissociation reaction; kon, association reaction.
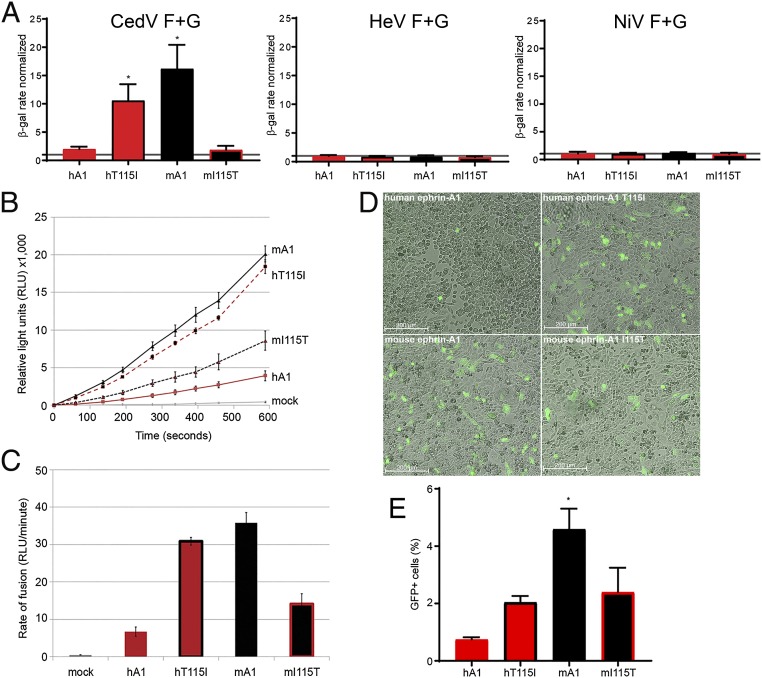
Next, we assessed whether the T115I substitution accounted for the differences in binding. Toward this, we constructed human ephrin-A1 (T115I) and mouse ephrin-A1 (I115T), and tested these mutants and the wild-type mouse and human ephrin-A1 in a fusion assay. Mouse ephrin-A1 supported robust cell–cell fusion, while human ephrin-A1 fusion was minimal (Fig. 7 A, Left; mouse A1 [mA1] and human A1 [hA1], respectively). The T115I mutation in human ephrin-A1 increased fusion efficiency, whereas the I115T mutation in mouse ephrin-A1 had the opposite effect (Fig. 7 A, Left; hT115I and mI115T, respectively). Neither HeV nor NiV could use any ephrin-A1 as a receptor (Fig. 7 A, Center and Right, respectively). We then used the split-luciferase reporter assay to quantify fusion function more precisely (Fig. 7B). Mouse ephrin-A1 supported a rate of fusion that was ∼7-fold faster than that supported by human ephrin-A1 (Fig. 7C). The T115I mutation in human ephrin-A1 increased cell–cell fusion to levels similar to those of mouse ephrin-A1 (Fig. 7 B and C, compare mA1 and hT115I). The I115T mutation in mouse ephrin-A1 reduced its fusion levels (Fig. 7 B and C, compare hA1 and mI115T).
Fig. 7.
Species specificity of ephrin-A1–dependent cell fusion and cell entry. (A) β-Galactosidase (β-gal) reporter cell–cell fusion assay mediated by the glycoproteins of CedV, NiV, and HeV interacting with either human or mouse ephrin-A1 or the indicated mutants. Mock-transfected CHO745 cells were used as a negative control (given a value of 1). The graphs represent 3 independent experiments in technical duplicates (mean ± SD). *P < 0.05. (B) Split-luciferase reporter–based fusion assay mediated by the CedV glycoproteins in CHO-K1 cells transiently expressing the indicated ephrin-A1 constructs. The graph is representative of 3 independent experiments (mean ± SD). (C) Rate of fusion calculated as the slope of the line in B. The graph represents 3 independent experiments in technical triplicates, and the error bars indicate SD. (D) Fluorescent microscopy images of CHO-K1 cells transfected with the indicated ephrin-A1 and infected with rCedV-GFP (MOI = 5.0) 24 hpi. (E) Measurement by flow cytometry of GFP-positive CHO-K1 cells, 24 and 48 hpi rCedV-GFP (MOI = 2.0). The graph represents 2 independent experiments (mean ± SD). *P < 0.05. hA1, human ephrin-A1; hT115I, human ephrin-A1 T115I; mA1, mouse ephrin-A1; mI115, mouse ephrin-A1 I115T.
Lastly, we investigated whether mouse ephrin-A1 can support CedV entry, and the effects of the Ile115 and Thr115 substitutions. Significantly more CedV-infected cells (as monitored by GFP expression) were observed after infection of cells transfected with mouse ephrin-A1 compared with human ephrin-A1 (Fig. 7 D, Lower Left and Upper Left, respectively). The T115I mutation in human ephrin-A1 resulted in increased cell entry (Fig. 7 D, Upper Right), while the I115T mutation in mouse ephrin-A1 decreased cell entry (Fig. 7 D, Lower Right). Quantification of GFP-positive cells by flow cytometry revealed that mouse ephrin-A1 supported the highest level of cell entry. The T115I mutation in human ephrin-A1 sustained increased rCedV-GFP entry, while the I115T mutation in mouse ephrin-A1 decreased rCedV-GFP entry, although a significant number of GFP positive cells were still monitored (Fig. 7E).
To understand the structural basis for the differential recognition between human and mouse ephrin-A1 by CedV G, we generated a homology model of the ephrin-A1–CedV G complex using our ephrin-B1–CedV G complex structure. The model suggested that isoleucine at position 115 is more energetically favorable for P3 pocket insertion due to its hydrophobicity and the filling of the empty binding cavity by its bulkier side chain (Fig. 8).
Fig. 8.
Structural basis for species specific usage of ephrin-A1. Structural modeling of residue T/I115 insertion into the CedV G receptor-binding pocket. CedV G is shown as green surface, while ephrin-A1 is shown in yellow. (Inset) I115 is shown as surface and sticks. Critical I115-contacting G protein residues are shown as sticks and labeled.
Discussion
Bat-borne henipaviruses NiV and HeV are broadly tropic at both the host and tissue levels, mediated through use of highly conserved virus receptors, ephrin-B2 and ephrin-B3, which are widely distributed throughout endothelial cells, vasculature, and the CNS (44). In this study, we show that CedV can use human ephrin-A2, -A5, and -B1 as receptors, in addition to the previously known ephrin-B2. This is unexpected since neither HeV nor NiV can engage ephrin-B1 or any of the A-class ephrins (37, 45).
Despite overall structural similarity in henipavirus G proteins, CedV G has distinct receptor-binding pocket architecture. The facts that HeV antisera is cross-reactive, but not cross-neutralizing, with CedV (6), and a human monoclonal antibody (m102.4) that competitively inhibits HeV/NiV G–ephrin-B2/B3 interactions (46–48) does not bind to CedV G (SI Appendix, Fig. S7) further highlight differences in the receptor-binding pocket. The enlarged CedV G-H loop-binding pocket P3 can accommodate amino acid side chains of various size, charge, and shape, including Leu121 in ephrin-B2, Tyr121 in ephrin-B1, Ile115 in mouse ephrin-A1, Ser137 in ephrin-A2, and Ser125 in ephrin-A5, which, moreover, allows sustained functional interaction between CedV G and these ephrins. The unique receptor-binding pocket structure in CedV G therefore underlies its broad ephrin receptor usage.
Physiologically, ephrin ligands interact with Eph receptors, members of the family of receptor protein-tyrosine kinases governing bidirectional intracellular signaling pathways that regulate developmental processes (49, 50). For the most part Eph–ephrin interactions are A- or B-class–dependent; however, Eph receptor–ephrin ligand promiscuity has been observed with EphB2 and EphA4 binding to both A- and B-class ephrins (51–53). Similar to these examples of Eph–ephrin interactions, CedV utilizes both ephrin-B1 and -B2 and GPI-linked ephrin-A2 and -A5 as functional receptors, although a delay was observed in infection with A-class ephrins. Thus, ephrin-A2 and -A5 appear to be less efficient CedV receptors compared with ephrin-B1 and -B2, consistent with affinity binding and cell–cell fusion rate analyses. GPI-linked proteins and lipid rafts have been identified as entry and egress sites for several viruses (reviewed in ref. 54); however, the in vivo relevance of A-class ephrin receptor use is unclear at this time as this evidence is being masked by the high-affinity ephrin-B1 and -B2 receptors.
Receptor tropism represents one of the barriers for virus spillover into susceptible hosts (55). Bat-borne viruses such as severe acute respiratory syndrome-coronavirus (CoV) and Middle Eastern respiratory syndrome-CoV provide well-studied examples of how mutations in virus receptors can influence tropism: Murine and human ACE-2 (56–59) and bat DDP4 orthologs (60), respectively, differ in key amino acids, which can influence virus cross-species adaptation. Functional receptor usage of mouse ephrin-A1 by CedV is evidence that henipaviruses utilize ephrin ligands in a species specific manner. A single substitution in the G-H loop (T115I) between human and mouse ephrin-A1 accounts for both binding to and functional use of mouse ephrin-A1 (Figs. 6 and 7) at levels similar to ephrin-B2. Comparable fusion activity between mouse ephrin-A1 and human ephrin-B2 further suggests that the decreased level of fusion activity with human ephrin-A2 and -A5 is a result of CedV G-binding affinities with ephrin-A2 and -A5, and not an effect of the GPI-linked anchor. Since the key henipavirus G-contacting residues in ephrin-B1 and -B2 are conserved in both human and mouse orthologs, disparities between mouse and human ephrin-A1 may not singularly promote a novel murine host switch.
Our discovery that ephrin-B2 and -B3 are not the only henipavirus receptors sheds light on the mode of interspecies transmission of these viruses. Ephrin ligand and Eph receptor interactions have major roles in developmental processes like cell migration and axon guidance, and, as a result, some ephrins have overlapping spatial and tissue expression. Notably, a distinct expression pattern of ephrin-B3 in the spinal cord may underlie CNS pathogenesis during HeV and NiV infection (12, 37, 45). Interestingly, ephrin-B1 is expressed in the hippocampus, cerebellum, and cortex (61), as well as in the spinal cord (62). However, HeV and NiV are unable to utilize ephrin-B1 as a receptor, while nonpathogenic CedV can efficiently utilize ephrin-B1. Structural comparisons of the GhV G receptor-binding pocket with HeV and CedV G, as presented here, and NiV G (31), additionally, suggest that GhV G would be incapable of accommodating ephrin-B1. Extended receptor usage could be postulated to enhance pathogenicity; however, this is not the case with CedV. Interestingly, the vaccine strain of measles virus has acquired broad tropism due to the use of the ubiquitous receptor CD46, which contributes to its attenuation (63, 64). Additionally, CedV does not express interferon antagonists V and W proteins, which are expressed by nearly all other paramyxoviruses. Thus, broad receptor usage and a lack of innate immune antagonism may both contribute to a lack of disease outcome from CedV infections. Therefore, both the identification of receptors that mediate cross-species transmission and the study of orthologous virulence factors (e.g., V protein, W protein) that suppress antiviral immune responses (65, 66) will greatly improve our ability to evaluate the public health risk of emerging henipaviruses.
Methods
Gene Synthesis and Protein Constructs.
CedV (GenBank accession no. NC_025351.1), HeV (GenBank accession no. NC_001906.3), and NiV (GenBank accession no. NC_002728.1) fusion (F) and attachment (G) glycoprotein open reading frames (ORFs) were codon-optimized and synthesized (Genscript). DNA plasmids of human ephrin-B1, -B2, and -B3; ephrin-A1, -A2, -A3, -A4, and -A5; and mouse ephrin-A1 full-length ORFs were purchased (Origene). A tetrameric soluble CedV G (sG) was constructed with an immunoglobulin (Ig) κ-leader sequence, S-tag, and tetrameric GCN4 motif at the N terminus of the coding region, replacing the cytoplasmic tail and transmembrane domain elements. CedV sG was produced from a stable-expressing cell line and purified by S-protein agarose affinity chromatography and size exclusion chromatography as previously described (8, 35). All ORFs were cloned into the expression vector pcDNA3.1+hygro+CMV (67); the expression plasmid encoding human ephrin-B3 has been previously described (68).
Cell Lines and Virus.
CHO-K1 (American Type Culture Collection [ATCC] CCL-61) cells and pgsA-745 (ATCC CRL-2242) cells, a derivative deficient in xylosyltransferase (UDP-D-xylose/serine-1,3-D-xylosyltransferase), referred to as CHO745 in this study, were maintained at 37 °C with 5% CO2 in Gibco F-12 Nutrient K (Kaighn’s Modification of Ham’s F-12) Medium (Thermo Fisher Scientific) supplemented with 10% HyClone Cosmic Calf Serum (Thermo Fisher Scientific) and 1× penicillin/streptomycin. The reverse genetics technique for the production of replication-competent, recombinant CedV was described previously (30). The rCedV antigenome plasmid was designed based on the sequence available in the GenBank (accession no. NC_025351.1).
Coprecipitation Assay.
S-tagged CedV sG purified protein was incubated in a 1:2 (microgram) ratio with a panel of soluble Fc-conjugated A-class and B-class ephrins, including human ephrin-A1 Fc (transiently expressed in 293 cells and purified) and mouse ephrin-A1, mouse ephrin-A2, human ephrin-A3, human ephrin-A4, human ephrin-A5, mouse ephrin-B1, mouse ephrin-B2, and human ephrin-B3 (R&D Systems) in the presence of 1× complete protease inhibitor (Roche Diagnostics) in radioimmunoprecipitation assay (RIPA) buffer at 4 °C overnight. This commercially available panel of soluble ephrins contains those derived from both mouse and human sequences. The key residues in the ephrin G-H loop that insert into the receptor-binding pocket of henipavirus G proteins are conserved between human and mouse orthologs of ephrin-A2, -B2, and -B3. The CedV sG–ephrin ligand complexes were precipitated using 50 μL of a 20% protein G agarose slurry, washed 3 times with RIPA buffer, boiled in 2× NuPage LDS sample buffer (Life Technologies) containing 5% 2-mercaptoethanol (Sigma–Aldrich), and run on a 4 to 12% NuPage Bis-Tris gel. Coprecipitated CedV sG proteins were detected by Western blot using horseradish peroxidase-conjugated rabbit anti–S-tag polyclonal antibody (Bethyl Laboratories, Inc.).
Receptor Interaction Analysis by SPR.
The interaction of CedV sG with different ephrins was monitored by SPR using a BIAcore T200 instrument and CM5 sensor chips (GE Healthcare) using adopted methods (69). Detailed experimental procedures can be found in SI Appendix, Supplementary Experimental Procedures.
Cell–Cell Fusion Assays.
The β-galactosidase reporter gene cell–cell fusion assay is well described (70–73) and was carried out as previously reported for henipavirus glycoproteins (8, 74). The split-luciferase–based kinetic fusion assay was adapted from a dual-split reporter assay previously described (30, 75). Detailed experimental procedures can be found in SI Appendix, Supplementary Experimental Procedures.
Virus Infections.
CHO-K1 cells seeded at 2.5 × 105 cells per well (12-well plates) were transfected with 2 μg of ephrin expression plasmids. Cells were infected with rCedV-GFP (multiplicity of infection [MOI] = 5.0) 48 h posttransfection. The formation of rCedV-GFP–induced syncytia was examined 24 hpi when images of viral GFP-positive syncytia were observed. To quantify virus entry, flow cytometry was utilized to measure the percentage of cells that were GFP-positive. CHO-K1 cells seeded at 1.25 × 105 cells per well (12-well plates) were transfected with the respective ephrin expression plasmids (2 μg). Cells were infected with rCedV-GFP (MOI = 2.0) 24 h posttransfection. Cell culture supernatant and cells were collected 24 and 48 hpi and centrifuged. The cell pellets were resuspended and fixed with 4% paraformaldehyde for 20 min at 37 °C. The fixed cells were washed, and GFP-positive cells were measured with an LSR II flow cytometer (BD Biosciences).
Protein Expression and Purification.
All proteins used for structure determination, including ephrin-B1 (UniProt Knowledgebase [UniProtKB] P52795, residues 29 to 170) eprhin-B2 (UniProtKB P52799, residues 27 to 170), and the globular domain of CedV G (GenBank accession no. YP_009094086, residues 193 to 622), were prepared as previously described (18). Essentially, proteins were expressed in Hi5 insect cells (Invitrogen) using the Baculovirus Expression System (Pharmagen). Recombinant protein purification was facilitated by fusion of all expression constructs to a thrombin cleavable Fc region of human IgG. Gel filtration in an SD-200 column (GE Healthcare) was used as the final purification step after Fc-tag removal by thrombin protease.
Crystallization and Structure Determination.
For complex formation, purified ephrins and CedV G were mixed in a 2:1 molar ratio in 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) at pH 7.2 and 150 mM NaCl buffer. Both ephrin-B1 and -B2 form stable complexes with CedV G in the gel-filtration column, and the purified complexes were collected as single peaks after size exclusion chromatography. Crystallization trials were conducted with a “Mosquito” robot (TTL Technology) using the sitting drop vapor diffusion method by mixing 100 nL of protein solution with 100 nL of well solution. Crystals of CedV G only and CedV G in complex with ephrin-B1 or ephrin-B2 were grown in wells containing 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6.0) with 15% polyethylene glycol (PEG) 3350, 100 mM Tris 8.5 with 20% PEG 6000, and 100 mM Tris 8.0 with 18% PEG 5000 monomethyl ether (MME), respectively. Well solution supplemented by 25% glycerol was used as a cryoprotectant for flash-freezing the crystals in liquid nitrogen. Diffraction datasets were collected remotely at beamline ID-24 of the Advanced Photon Source (Argonne National Laboratory). The datasets were processed with HKL2000 (76). Phases were calculated by molecular replacement in Phaser (77), using NiV G or its complex with ephrin-B3 (3D11 and 3D12) as a search model. Model building and refinement were performed by using Coot (78) and Phenix refine (79). Crystallographic statistics are presented in SI Appendix, Table S1. The structures have been deposited in the Protein Data Bank under ID codes 6P72, 6P7S, and 6P7Y.
Structural Modeling of Ephrin-A1 in Complex with CedV G.
Structural modeling was performed by homology modeling in the SWISS-MODEL server with the structure of the CedV G–ephrin-B1 complex.
Mutagenesis.
Point mutations in the human and mouse ephrin-A1 expression plasmids (T115I and I115T, respectively) were introduced by QuikChange site-directed mutagenesis (Agilent Technologies) and verified by sequencing in the vicinity of the mutation. At least 2 independent clones were tested for each mutation.
Illustrations.
All molecular representations were generated with PyMOL (Delano Scientific LLC). Figures were prepared using GraphPad Prism, Microsoft Excel, Adobe Illustrator, and Adobe Photoshop.
Sequence Alignments.
Human and mouse ephrin ligand sequences were analyzed with Clone Manager Suite9 software (Scientific & Educational Software). The henipavirus G proteins were aligned using Clustal Omega (80).
Supplementary Material
Acknowledgments
Funding for this study was provided from NIH Grants AI054715, AI077995, and AI137813 (to C.C.B.). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results. The views expressed in the manuscript are solely those of the authors, and they do not represent official views or opinions of the Department of Defense or the Uniformed Services University of the Health Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6P72, 6P7S, and 6P7Y).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911773116/-/DCSupplemental.
References
- 1.Geisbert T. W., Feldmann H., Broder C. C., Animal challenge models of henipavirus infection and pathogenesis. Curr. Top. Microbiol. Immunol. 359, 153–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Wit E., Munster V. J., Animal models of disease shed light on Nipah virus pathogenesis and transmission. J. Pathol. 235, 196–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broder C. C., Weir D. L., Reid P. A., Hendra virus and Nipah virus animal vaccines. Vaccine 34, 3525–3534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.United States Department of Agriculture , Henipavirus Gap Analysis Workshop Report (US Department of Agriculture, Agricultural Research Service, Washington, DC, 2018) http://go.usa.gov/xnHgR. Accessed 1 May 2019.
- 5.Sweileh W. M., Global research trends of World Health Organization’s top eight emerging pathogens. Global Health 13, 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh G. A., et al. , Cedar virus: A novel Henipavirus isolated from Australian bats. PLoS Pathog. 8, e1002836 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schountz T., et al. , Differential innate immune responses elicited by Nipah virus and Cedar virus correlate with disparate in vivo pathogenesis in hamsters. Viruses 11, E291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonaparte M. I., et al. , Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U.S.A. 102, 10652–10657 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negrete O. A., et al. , EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436, 401–405 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Lisabeth E. M., Falivelli G., Pasquale E. B., Eph receptor signaling and ephrins. Cold Spring Harb. Perspect. Biol. 5, a009159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton B. T., Broder C. C., Middleton D., Wang L. F., Hendra and Nipah viruses: Different and dangerous. Nat. Rev. Microbiol. 4, 23–35 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pernet O., Wang Y. E., Lee B., Henipavirus receptor usage and tropism. Curr. Top. Microbiol. Immunol. 359, 59–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee B., Ataman Z. A., Modes of paramyxovirus fusion: A Henipavirus perspective. Trends Microbiol. 19, 389–399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilar H. C., Iorio R. M., Henipavirus membrane fusion and viral entry. Curr. Top. Microbiol. Immunol. 359, 79–94 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Steffen D. L., Xu K., Nikolov D. B., Broder C. C., Henipavirus mediated membrane fusion, virus entry and targeted therapeutics. Viruses 4, 280–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bossart K. N., Fusco D. L., Broder C. C., Paramyxovirus entry. Adv. Exp. Med. Biol. 790, 95–127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden T. A., et al. , Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 15, 567–572 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Xu K., et al. , Host cell recognition by the henipaviruses: Crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U.S.A. 105, 9953–9958 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu K., Broder C. C., Nikolov D. B., Ephrin-B2 and ephrin-B3 as functional henipavirus receptors. Semin. Cell Dev. Biol. 23, 116–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kullander K., et al. , Role of EphA4 and EphrinB3 in local neuronal circuits that control walking. Science 299, 1889–1892 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Benson M. D., et al. , Ephrin-B3 is a myelin-based inhibitor of neurite outgrowth. Proc. Natl. Acad. Sci. U.S.A. 102, 10694–10699 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dines M., Lamprecht R., The role of Ephs and ephrins in memory formation. Int. J. Neuropsychopharmacol. 19, pyv106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gale N. W., et al. , Ephrin-B2 selectively marks arterial vessels and neovascularization sites in the adult, with expression in both endothelial and smooth-muscle cells. Dev. Biol. 230, 151–160 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Bennett K. M., et al. , Ephrin-B2 reverse signaling increases α5β1 integrin-mediated fibronectin deposition and reduces distal lung compliance. Am. J. Respir. Cell Mol. Biol. 49, 680–687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maisner A., Neufeld J., Weingartl H., Organ- and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb. Haemost. 102, 1014–1023 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Wong K. T., Ong K. C., Pathology of acute henipavirus infection in humans and animals. Pathol. Res. Int. 2011, 567248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong K. T., Tan C. T., Clinical and pathological manifestations of human henipavirus infection. Curr. Top. Microbiol. Immunol. 359, 95–104 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Shinya K., et al. , Avian flu: Influenza virus receptors in the human airway. Nature 440, 435–436 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Shelton H., et al. , Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J. Virol. 85, 1875–1880 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laing E. D., et al. , Rescue and characterization of recombinant cedar virus, a non-pathogenic Henipavirus species. Virol. J. 15, 56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee B., et al. , Molecular recognition of human ephrinB2 cell surface receptor by an emergent African henipavirus. Proc. Natl. Acad. Sci. U.S.A. 112, E2156–E2165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernet O., et al. , Evidence for henipavirus spillover into human populations in Africa. Nat. Commun. 5, 5342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeltina A., Bowden T. A., Lee B., Emerging paramyxoviruses: Receptor tropism and zoonotic potential. PLoS Pathog. 12, e1005390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thibault P. A., Watkinson R. E., Moreira-Soto A., Drexler J. F., Lee B., Zoonotic potential of emerging paramyxoviruses: Knowns and unknowns. Adv. Virus Res. 98, 1–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan L. S., Laing E. D., Broder C. C., Expression system for recombinant henipavirus glycoproteins. Methods Mol. Biol., in press. [Google Scholar]
- 36.Huynh-Do U., et al. , Ephrin-B1 transduces signals to activate integrin-mediated migration, attachment and angiogenesis. J. Cell Sci. 115, 3073–3081 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Negrete O. A., et al. , Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2, e7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu K., et al. , New insights into the Hendra virus attachment and entry process from structures of the virus G glycoprotein and its complex with ephrin-B2. PLoS One 7, e48742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halpin K., et al. ; Henipavirus Ecology Research Group , Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 85, 946–951 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halpin K., Young P. L., Field H. E., Mackenzie J. S., Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 81, 1927–1932 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Yob J. M., et al. , Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7, 439–441 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Z., et al. , Novel Henipa-like virus, Mojiang paramyxovirus, in rats, China, 2012. Emerg. Infect. Dis. 20, 1064–1066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rissanen I., et al. , Idiosyncratic Mòjiāng virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses. Nat. Commun. 8, 16060 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Himanen J. P., Nikolov D. B., Eph signaling: A structural view. Trends Neurosci. 26, 46–51 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Negrete O. A., Chu D., Aguilar H. C., Lee B., Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J. Virol. 81, 10804–10814 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu K., et al. , Crystal structure of the Hendra virus attachment G glycoprotein bound to a potent cross-reactive neutralizing human monoclonal antibody. PLoS Pathog. 9, e1003684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bossart K. N., et al. , Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 79, 6690–6702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Z., et al. , Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J. Infect. Dis. 197, 846–853 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Himanen J. P., Saha N., Nikolov D. B., Cell-cell signaling via Eph receptors and ephrins. Curr. Opin. Cell Biol. 19, 534–542 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasquale E. B., Eph-ephrin bidirectional signaling in physiology and disease. Cell 133, 38–52 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Gale N. W., et al. , Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron 17, 9–19 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Smith A., Robinson V., Patel K., Wilkinson D. G., The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr. Biol. 7, 561–570 (1997). [DOI] [PubMed] [Google Scholar]
- 53.Frisén J., Holmberg J., Barbacid M., Ephrins and their Eph receptors: Multitalented directors of embryonic development. EMBO J. 18, 5159–5165 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metzner C., Salmons B., Günzburg W. H., Dangerfield J. A., Rafts, anchors and viruses–A role for glycosylphosphatidylinositol anchored proteins in the modification of enveloped viruses and viral vectors. Virology 382, 125–131 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Parrish C. R., et al. , Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72, 457–470 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W., et al. , Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 78, 11429–11433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li W., et al. , Animal origins of the severe acute respiratory syndrome coronavirus: Insight from ACE2-S-protein interactions. J. Virol. 80, 4211–4219 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li W., et al. , Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24, 1634–1643 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Doremalen N., et al. , Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 88, 9220–9232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Letko M., et al. , Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 24, 1730–1737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Migani P., Bartlett C., Dunlop S., Beazley L., Rodger J., Regional and cellular distribution of ephrin-B1 in adult mouse brain. Brain Res. 1247, 50–61 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Jevince A. R., Kadison S. R., Pittman A. J., Chien C.-B., Kaprielian Z., Distribution of EphB receptors and ephrin-B1 in the developing vertebrate spinal cord. J. Comp. Neurol. 497, 734–750 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Condack C., Grivel J. C., Devaux P., Margolis L., Cattaneo R., Measles virus vaccine attenuation: Suboptimal infection of lymphatic tissue and tropism alteration. J. Infect. Dis. 196, 541–549 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Mateo M., Navaratnarajah C. K., Cattaneo R., Structural basis of efficient contagion: Measles variations on a theme by parainfluenza viruses. Curr. Opin. Virol. 5, 16–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Satterfield B. A., et al. , The immunomodulating V and W proteins of Nipah virus determine disease course. Nat. Commun. 6, 7483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Satterfield B. A., et al. , Nipah virus C and W proteins contribute to respiratory disease in ferrets. J. Virol. 90, 6326–6343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan Y. P., Yan L., Feng Y. R., Broder C. C., Preparation of recombinant viral glycoproteins for novel and therapeutic antibody discovery. Methods Mol. Biol. 525, 31–58, xiii (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bossart K. N., et al. , Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology 372, 357–371 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Navaratnarajah C. K., et al. , Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150). J. Biol. Chem. 283, 11763–11771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Broder C. C., Berger E. A., Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc. Natl. Acad. Sci. U.S.A. 92, 9004–9008 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng Y., Broder C. C., Kennedy P. E., Berger E. A., HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 872–877 (1996). [DOI] [PubMed] [Google Scholar]
- 72.Nussbaum O., Broder C. C., Berger E. A., Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68, 5411–5422 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nussbaum O., et al. , Functional and structural interactions between measles virus hemagglutinin and CD46. J. Virol. 69, 3341–3349 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bossart K. N., Broder C. C., Viral glycoprotein-mediated cell fusion assays using vaccinia virus vectors. Methods Mol. Biol. 269, 309–332 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Navaratnarajah C. K., Rosemarie Q., Cattaneo R., A structurally unresolved head segment of defined length favors proper measles virus hemagglutinin tetramerization and efficient membrane fusion triggering. J. Virol. 90, 68–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Otwinowski Z., Minor W., “Processing of X-ray Diffraction Data Collected in Oscillation Mode” in Methods in Enzymology, Carter C. W. Jr, Sweet R. M., Eds. (Academic Press, New York, 1997), pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 77.McCoy A. J., Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D. Biol. Crystallogr. 63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.