Significance
A major challenge in biology is characterizing the structural flexibility of intrinsically disordered proteins (IDPs). Ensemble-averaged experimental data do not provide the underlying protein structures. Here, we performed independently small-angle neutron and X-ray scattering experiments and unbiased molecular dynamics simulations to probe the solution structure of an IDP. We report that enhancing the sampling of the simulations can generate an ensemble of IDP structures in quantitative agreement with scattering and NMR, without the need for biasing the simulation or reweighting the results. The demonstration of established simulation technology that produces accurate physical models of flexible biosystems may pave the way to relating conformational flexibility to biological function.
Keywords: intrinsically disordered protein, MD simulation, small-angle scattering, conformational ensemble, transient helices
Abstract
Intrinsically disordered proteins (IDPs) are abundant in eukaryotic proteomes, play a major role in cell signaling, and are associated with human diseases. To understand IDP function it is critical to determine their configurational ensemble, i.e., the collection of 3-dimensional structures they adopt, and this remains an immense challenge in structural biology. Attempts to determine this ensemble computationally have been hitherto hampered by the necessity of reweighting molecular dynamics (MD) results or biasing simulation in order to match ensemble-averaged experimental observables, operations that reduce the precision of the generated model because different structural ensembles may yield the same experimental observable. Here, by employing enhanced sampling MD we reproduce the experimental small-angle neutron and X-ray scattering profiles and the NMR chemical shifts of the disordered N terminal (SH4UD) of c-Src kinase without reweighting or constraining the simulations. The unbiased simulation results reveal a weakly funneled and rugged free energy landscape of SH4UD, which gives rise to a heterogeneous ensemble of structures that cannot be described by simple polymer theory. SH4UD adopts transient helices, which are found away from known phosphorylation sites and could play a key role in the stabilization of structural regions necessary for phosphorylation. Our findings indicate that adequately sampled molecular simulations can be performed to provide accurate physical models of flexible biosystems, thus rationalizing their biological function.
Intrinsically disordered proteins (IDPs) are structurally flexible and lack stable secondary structures in physiological conditions. They play a critical role in many cellular functions, such as signal transduction, transcriptional regulation, cell growth, binding, gene expression, and homeostasis (1–5). IDPs are also associated with multiple diseases, such as cancers (6, 7), amyloidosis, diabetes, cardiovascular problems, and neurodegenerative disorders (8, 9).
To understand the disorder–function relationship IDPs present and to elucidate the mechanism by which their pathological mutations lead to human diseases (2), we need to determine their configurational ensemble: the collection of 3-dimensional (3D) structures they adopt (10). Several experimental techniques have been applied to study the structures of IDPs, including NMR spectroscopy (6, 11–18), single-molecule Förster resonance energy transfer (sm-FRET) (17, 19, 20), cryoelectron microscopy (21, 22), and small-angle X-ray and neutron scattering (SAXS and SANS) (6, 15, 17, 23–28). However, the information obtainable from these experiments is limited and they cannot provide a high-resolution description of IDP ensembles (29–34). Further, most techniques employed to study IDPs suffer from a conundrum: The experimental observables represent an average over the conformational ensemble, but the ensemble itself cannot be unequivocally inferred from the experiments (10, 29, 35). For SAXS and SANS, unambiguous structural interpretation is made more difficult by the inherent orientational averaging.
Molecular dynamics (MD) simulation can, in principle, provide the structural ensemble of biomolecules. However, MD faces 2 distinct, yet related, challenges when applied to IDPs: the accuracy of the physics-based molecular mechanics force fields that model the interactions in the system and the need for adequate sampling of the large configurational space of an IDP and its associated solvent. Recent progress has considerably improved molecular mechanics force fields for IDPs (36–43). Nevertheless, the accuracy of simulations, as assessed by comparison to experiments, was found to depend on the specific IDP, the employed force field, and configurational sampling in a weakly funneled energy landscape (36–38, 40, 42).
The current state-of-the-art approach to structurally characterizing IDPs at atomic detail requires either restraining the MD or reweighting (44–50) the MD-generated ensemble to match the experimental data (35, 51). This creates overfitting issues and makes it difficult to judge the accuracy of the models because of a degeneracy problem: Different ensembles may yield the same experimental observable (29, 52, 53). Despite recent successes of Bayesian methods when refining MD-derived structures against SAXS data (54, 55), reproducing experiments without the need to modifying simulation results presents one of the most important challenges in structural biology of IDPs. Addressing this challenge would lead to increased confidence that the obtained models accurately describe the physical behavior of the systems.
Here, we combine MD simulation with SAXS, SANS, and NMR experiments to elucidate the conformational ensemble of the 95-residue intrinsically disordered N terminus of the proto-oncogene nonreceptor human tyrosine kinase c-Src, which contains the Src homology 4 (SH4) and unique domains. c-Src plays a major role in cellular transduction pathways such as cell growth, differentiation, transcription, proliferation, adhesion, and survival (12, 56) and is associated with a variety of human cancers (57). SH4 regulates c-Src kinase localization in response to cellular stimulation (56) by binding/anchoring to the cell membrane, whereas the unique domain (UD) mediates the interaction of Src with lipids, specific receptor and protein targets (12, 13, 58).
In the present work a heterogeneous ensemble of structures of SH4UD was generated by enhancing the sampling of unbiased atomistic simulations in an explicit solvent by using Hamiltonian replica exchange MD (HREMD) (59–61). Critically, the SAXS, SANS, and NMR observables of SH4UD thus generated are found to be in excellent agreement with the experimental data without the need for biasing or reweighting. The use of HREMD is critical in achieving this agreement. The simulations reveal a weakly funneled and rugged free energy landscape and predict 3 transient helical regions that do not coincide with the known phosphorylation sites but are proximal to a lipid-binding region. We hypothesize that such transient structures act as a molecular recognition feature for phosphorylation-mediated regulator of enzyme activity and for membrane binding.
Results
Agreement between Experiments and Simulations.
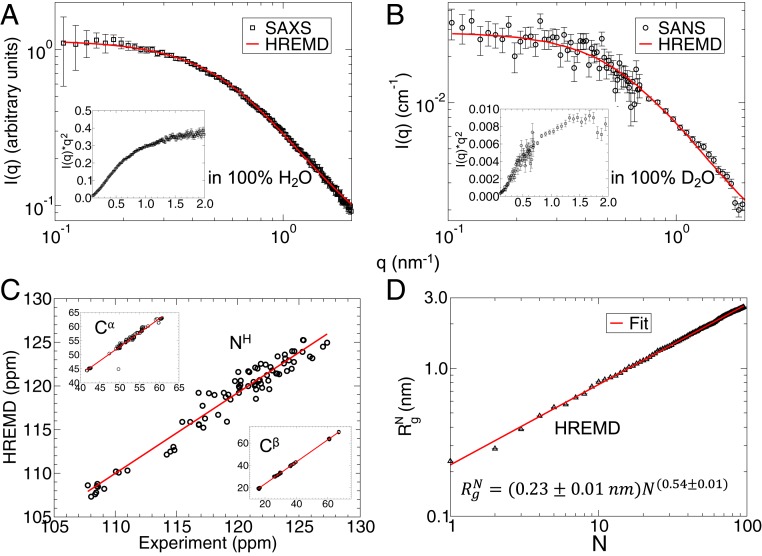
We conducted SAXS (Fig. 1A) and SANS (Fig. 1B) experiments of SH4UD. The Guinier [ln(I) vs. q2] and Zimm [1/I(q) vs. q2] plots follow a linear behavior at low q, as expected for monodisperse samples (SI Appendix, Fig. S1). The upturn at high q in the Kratky plot (Fig. 1, Insets) confirms the disordered nature of SH4UD. The distribution of atom-pair distances, P(r), is asymmetric with an extended shoulder at distances r ≥ 6 nm (SI Appendix, Fig. S2). This suggests the average shape of SH4UD is elongated, where the interpretation is consistent with previous studies (62, 63).
Fig. 1.
(A and B) Experimental (A) SAXS in 100% H2O and (B) SANS in 100% D2O profiles are shown in black squares and circles, respectively. Experimental Kratky plots, I(q) × q2 vs. q shown in the Insets confirm the unfolded structure of SH4UD. The ensemble-averaged profiles calculated from ∼510-ns HREMD trajectory are shown as a red line. (C) Comparison between experimental and calculated (HREMD) NMR chemical shifts, expressed in parts per million (ppm), of the backbone NH, Cα, and Cβ atoms. The NMR experimental data are taken from Biological Magnetic Resonance Data Bank entry 15563 (12) and the theoretical chemical shifts were calculated using SHIFTX2 from ∼51,000 structures (122). (D) Radius of gyration of a protein segment consisting of N residues calculated from the HREMD simulation. The red line is a fit of Eq. 1 to the data.
SANS and SAXS provide different information because the 2 techniques are sensitive to different atomic properties: neutrons interact with atomic nuclei, whereas X-rays interact with electron clouds. For example, there is significantly different contrast between a protein and its hydration shell in SAXS and SANS (64–66). This is illustrated in our data when obtaining the radius of gyration (Rg) by taking the second moment of the P(r). We obtain = 2.52 ± 0.10 nm for SANS and = 2.71 ± 0.04 nm for SAXS of the same sample. The difference in Rg between SAXS and SANS reported here is consistent with scattering measurements from globular proteins, in which Rg determined from SANS in 100% D2O is smaller than that obtained from SAXS data of the same sample in 100% H2O (65, 67).
The ensemble-averaged SAXS and SAXS profiles calculated directly from the trajectory of the lowest-rank replica of the HREMD simulation are found to be in excellent agreement with experiment: for SAXS and for SANS (see Eq. 2) (Fig. 1 and SI Appendix, Figs. S3, S5, and S6). This agreement is obtained by an unbiased/unrestrained simulation, free from reweighting and using explicit water molecules when calculating SAXS/SANS curves, thus avoiding the need for optimized free parameters to account for the contribution of the hydration shell.
We also calculated the ensemble average NMR chemical shifts of the backbone atoms (NH, Cα, and Cβ) of SH4UD from the HREMD simulation and compared them to previously published experimental values (12). The excellent agreement between NMR and HREMD is reflected by the regression coefficients R2 > 0.93 (Fig. 1C and SI Appendix, Fig. S7).
The unbiased HREMD simulations are thus consistent with 3 independent experimental probes of global and local protein structure: SAXS, SANS, and NMR. The underlying conformational ensemble contains all of the structures from the ∼510-ns-long trajectory and has a broad distribution of Rg, as reflected in the density plot of the theoretical I(q) (SI Appendix, Fig. S3). The use of HREMD is critical in obtaining good agreement with scattering and NMR experiments (SI Appendix, Figs. S3 and S7). The convergence of the calculations is shown in detail in SI Appendix, Figs. S5–S9.
Chain Statistics.
The degree of compaction of a polypeptide chain in aqueous solution can be quantified by the Flory exponent, ν, which is determined by the relative strengths of the protein–solvent and intraprotein interactions (17, 24, 25, 68). Polymer theory predicts only 3 values of ν for polymers: When intraprotein interactions are favorable, ν = 0.333 and the protein adopts a collapsed conformation, for balanced interactions, ν = 0.5 and the protein is a Gaussian random coil, whereas for favorable protein–solvent interactions ν = 0.588 and the protein adopts a self-avoiding random coil conformation (24).
Here, the Flory exponent was calculated from the atomic coordinates using the relation (24, 69, 70),
| [1] |
where is the radius of gyration of a protein segment consisting of N residues and R0 is a constant. Fitting Eq. 1 to the simulation data (Fig. 1D) gives ν = 0.54 ± 0.01, suggesting the overall SH4UD–water interaction is favorable and SH4UD chain statistics lie between Gaussian and self-avoiding random coil behavior. Several studies have indicated similar chain statistics of other IDPs (17, 24, 25, 68).
Weakly Funneled Free Energy Landscape.
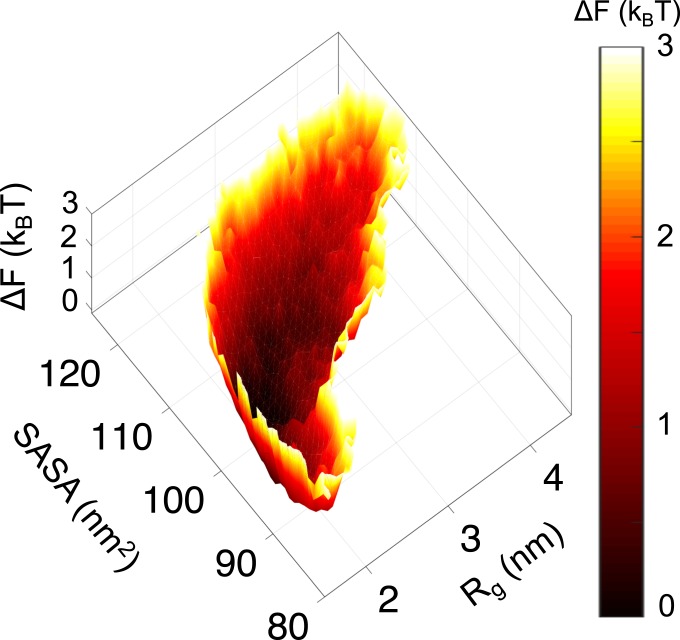
Rg and the solvent-accessible surface area (SASA) quantify important global protein characteristics: the overall size and solvent exposure, respectively. As such, they are used here as collective variables for describing the conformations of SH4UD by projecting the free energy onto them. SH4UD depicts a weakly funneled and rugged free energy landscape (Fig. 2). We note that barrier heights derived from 2D histogram analysis of HREMD simulations may carry large errors. The multiple shallow minima indicate the large number of conformational substates demonstrating a conformational heterogeneity. This structural heterogeneity is one of the distinctive characteristics of IDPs (25, 71–73).
Fig. 2.
Free energy landscape of SH4UD as a function of its Rg and SASA from HREMD simulations showing a large number of the conformational substates sampled in the simulation.
Secondary Structure Propensity.
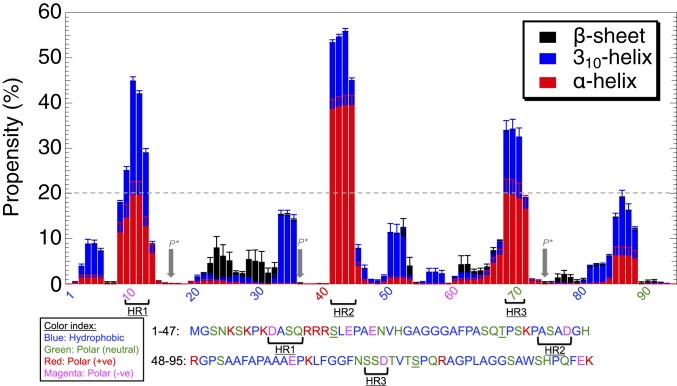
The propensity of residues to form secondary structure, calculated from the HREMD, indicates that SH4UD mainly adopts coil structures with almost negligible beta-sheet content, in agreement with NMR experiments (12). However, 3 short sequence segments that display occasional helical structures were found, defined here as having helical propensity higher than 20% (Fig. 3): helical region 1 (HR1), residues D10-A11-S12-Q13; HR2, A42-S43-A44-D45; and HR3, S69-S70-D71.
Fig. 3.
Propensity of each residue to form secondary structures calculated from the HREMD using DSSP (119). Arrows at underlined polar (neutral) residues S17, T37, and S75 are the known phosphorylation sites (P*) located between the predicted transient helical regions (HR1, HR2, and HR3).
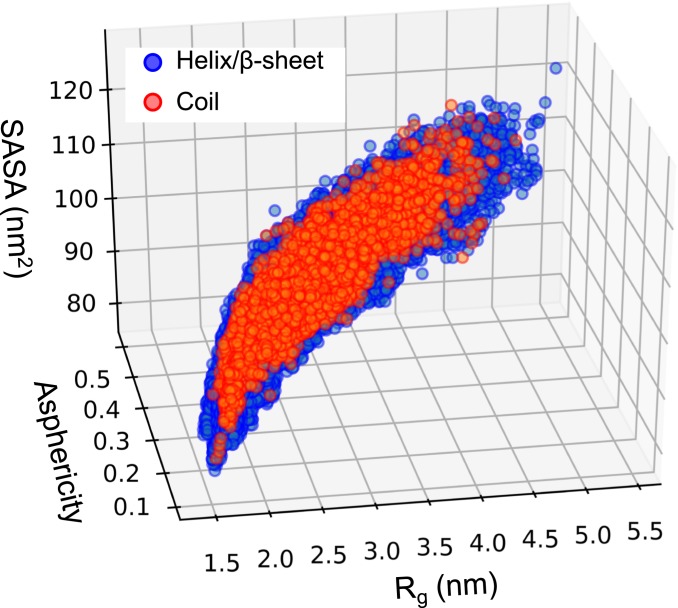
The conformations sampled by HREMD are of varying degree of compactness and asphericity (70) (Fig. 4). The broad distribution of Rg, SASA, and asphericity indicate the continuous and heterogeneous nature of the structures comprising the ensemble. Coil conformations with no secondary structure represent only ∼6% of the HREMD trajectory and are evenly distributed in the (Rg, asphericity, SASA) space (SI Appendix, Fig. S10). The chain statistics (Eq. 1) of the coil structures are similar to those of the entire ensemble and yield a ν = 0.54 ± 0.01 Flory exponent (SI Appendix, Fig. S10).
Fig. 4.
SH4UD structures projected on Rg, SASA, and asphericity. Frames are separated into those that have only coil (red) and those that have at least 1 helix or beta-sheet (blue) in their sequence.
Hydration Shell Structure.
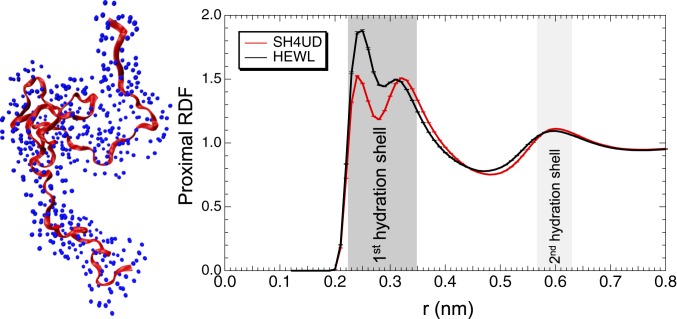
The hydration shell structure was quantified by calculating the proximal radial distribution function (pRDF) (74, 75) of water oxygen atoms around the nonhydrogen atoms of protein (Fig. 5). The 2 peaks of the pRDF at a distance r ∼ 0.30 nm from the protein surface correspond to the first hydration shell, which is found here to have a higher density than the bulk. The peak at r ∼ 0.25 nm arises from correlation distances involving protein oxygen and nitrogen atoms with water oxygen atom, whereas the peak at r ∼ 0.30 nm corresponds to carbon atoms (SI Appendix, Fig. S11).
Fig. 5.
(Left) A snapshot of SH4UD (red) and water oxygen atoms (blue) within a distance of 0.3 nm from protein’s surface. (Right) The pRDF as a function of the distance r between an oxygen atom of water and the nonhydrogen atoms of HEWL (black dashed curve) and SH4UD (solid red curve) proteins.
We compare the pRDFs of hydration water around SH4UD and hen egg white lysozyme, HEWL, a globular folded protein. While the pRDFs are similar for distances >0.35 nm, the first hydration shell is less dense for SH4UD than for HEWL, as shown by the lower peak heights, especially for the water oxygen–protein nitrogen interactions (SI Appendix, Fig. S11). This result indicates that the IDP hydration shell is more similar to bulk than for folded proteins. A smaller fraction of hydrophobic residues on the surface of HEWL compared to SH4UD were found that might explain the smaller hydration shell density of SH4UD (SI Appendix, Tables S1 and S2).
Discussion
The biological significance of IDPs and their abundance in many genomes have been increasingly realized (3). IDPs represent ∼30% of the human genome, play important cellular functions, and are associated with many diseases. Understanding their biological function in detail requires resolving the heterogeneous 3D ensembles of their structures, which remains a challenge because of the ensemble-averaged nature of most experimental data.
To determine IDP ensembles MD simulation is frequently combined with SAXS experiments (6, 24, 76, 77). However, it has hitherto been necessary to bias the populations of conformations obtained by MD to produce agreement with experiment (35, 46). In the present work we conducted unbiased HREMD simulations and obtained SAXS, SANS, and NMR observables in quantitative agreement with experiment. We calculated theoretical SAXS and SANS profiles considering explicitly water molecules in the simulation, avoiding the use of fitting parameters to account implicitly for the hydration shell (49). Critically, we overcame the need to reweight the simulation trajectories by enhancing the configurational sampling using HREMD. The sampling provided by HREMD, as compared to 10 μs of standard MD simulation, was crucial for obtaining good agreement with experiment (SI Appendix, Fig. S3).
The generation of a configurational ensemble in agreement with experiment suggests that details of the generated ensemble can be examined with some confidence. SH4UD is found to occupy numerous conformational substates in a weakly funneled and rugged free energy landscape (Fig. 2), which differentiates it from the strongly funneled free energy landscape of folded globular proteins (18, 71–73).
Low mean hydrophobicity and high mean net charge are considered as characteristics of IDPs (78–82). However, SH4UD has an abundance of hydrophobic residues that make its sequence more similar to folded globular proteins. Indeed, SH4UD lies near the border line of the charge-hydrophobicity plot commonly used to identify IDP propensity (78, 80, 81) (SI Appendix, Fig. S12). Based on the fraction of positively and negatively charged residues (SI Appendix, Fig. S13), SH4UD would be predicted to adopt collapsed structures (68, 83). However, perhaps somewhat surprisingly, SH4UD is found to adopt extended conformations. This is understood to occur from the distribution (“mixing”) of charged and polar residues across the entire sequence of SH4UD (68) (Fig. 3). The extended coil conformation of SH4UD is consistent with the finding that many hydrophobic IDPs are expanded in water (24).
The Flory exponent we obtained for SH4UD (ν = 0.54; Fig. 1D) lies in between the 2 values predicted for coils from polymer theory: ν = 0.5 for a Gaussian ideal coil and ν = 0.588 for a self-avoiding random coil. The value of ν = 0.54 has been reported for other IDPs studied with SAXS and sm-FRET (19, 24, 68). Thus, SH4UD cannot be accurately described by polymer theory. The Flory exponent of ν = 0.54 is found both for SH4UD structures that contain transient secondary structure elements and for pure coils (SI Appendix, Fig. S10).
SH4UD contains 3 known phosphorylation sites (residues S17, T37, and S75) that regulate Src activity (84, 85) and that do not coincide with the 3 transient helical regions we identified (HR1, HR2, and HR3). The present findings suggest that the higher solvent accessibility of the phosphorylation sites afforded by the absence of secondary structure may facilitate their modification by kinases. Further, we hypothesize that the transient helices provide an inherent stability near phosphorylation sites in SH4UD, consistent with the finding of short and transient helical structures near the phosphorylation sites of the (unphosphorylated) disordered region of the sodium protein exchanger 1 (86).
A recent NMR study found SH4UD has lipid-binding regions that include residues S51, A53, and A55 (13), which have ∼10% helical propensity in the HREMD (Fig. 3). We speculate that the transient helices might facilitate binding of c-Src to the cell membrane. Similar molecular recognition and stabilization due to the presence of transient secondary structures have been reported for other IDPs (16, 77, 86–94).
The hydration shells of IDPs have been studied less than those of folded proteins (65, 95–97). We found the hydration water around SH4UD to be less dense compared to folded HEWL. This result contrasts with previous simulation studies of 2 IDPs, alpha-synuclein and beta-amyloid (98), which, however, employed the Amber ff99SB and TIP3P force fields that may not accurately account for the interactions of IDPs and water (37, 38, 99–101).
Conclusions
We have obtained an accurate, atomic-detail physical model of the heterogeneous ensemble of structures of the intrinsically disordered SH4UD terminus of c-Src kinase. The model is based on structures generated by unbiased enhanced sampling MD simulations and is validated by calculation of SAXS, SANS, and NMR observables and comparison to experiment without constraining or reweighting the simulations. This physical model cannot be obtained alone by experiment, which provides either sparse or low-resolution information. We show that SH4UD explores heterogeneous conformational substates, from extended to partially collapsed in a weakly funneled and rugged free energy landscape. The conformations of SH4UD do not obey simple polymer theory. Transient helices are located away from known phosphorylation sites. We suggest that the transient helices are preconfigured structures, which localize c-Src by binding to the cell membrane and may facilitate phosphorylation-mediated regulation of c-Src. Our results suggest accurate physical models of flexible biomolecular systems, such as IDPs and proteins consisting of multiple domains connected by linkers, are within reach, and may pave the way to relating conformational flexibility to biological function.
Materials and Methods
Sample Preparation.
A gene encoding a 95-amino acid SH4UD construct with the sequence MGSNKSKPKDASQRRRSLEPAENVHGAGGGAFPASQTPSKPASADGHRGPSAAFAPAAAEPKLFGGFNSSDTVTSPQRAGPLAGGSAWSHPQFEK that included a C-terminal Strep tag (SAWSHPQFEK) was cloned into the pET-14b vector with NcoI and BamHI sites. Briefly, the SH4UD was expressed overnight in Escherichia coli Rosetta BL21 (DE3) at 20 °C and the cells were lysed by sonication in 100 mM Tris and 150 mM NaCl, pH 8.0. The lysate was passed over a strepavidin Sepharose column, washed with 20 column volumes of 100 mM Tris and 150 mm NaCl, pH 8.0, and eluted with 10 mM desthiobiotin. The eluted protein was then concentrated and purified to homogeneity using size-exclusion chromatography (Superdex 75; GE Healthcare).
SAXS.
The peak fraction of SH4UD obtained after size-exclusion chromatography, as described above, was analyzed using SAXS. The SAXS experiments were conducted on a Rigaku BioSAXS 2000 instrument equipped with a Pilatus 100 K detector (Rigaku Americas). The sample-to-detector distance and beam center were calibrated using a silver behenate standard. The samples were automatically loaded into a Julabo temperature-controlled flow cell from a Julabo temperature-controlled 96-well plate. SAXS data of SH4UD were collected at a concentration of 1.5 mg/mL at room temperature. The data were reduced using instrument reduction software to obtain the scattering intensity, I(q), as a function of wave vector, q [= 4π sin(θ)/λ, where 2θ is the scattering angle and λ is the wavelength], for sample and buffer. Subsequently, the buffer background was subtracted, and the resulting sample scattering profile was employed for further data analysis. The pair distance distribution function [P(r)] was calculated from the indirect Fourier transform of I(q) using GNOM/ATSAS (102).
SANS.
SANS measurements were performed at the EQ-SANS instrument in the facility Spallation Neutron Source located at the Oak Ridge National Laboratory (103). The spallation source was operating at 60 Hz. A sample aperture of 10 mm was used for beam collimation. Two different configurations were used to obtain a large q-range, to cover a combined q-range of 0.05 nm−1 < q < 7 nm−1 · q = (4π/λ) sin(θ) is the magnitude of the scattering vector, where 2θ is the scattering angle. For these 2 configurations, the sample-to-detector distances were 2.5 m and 4 m with a minimum neutron wavelengths of 2.5 Å and 10 Å, respectively. Data reduction with I(q) vs. q as output was performed according to standard procedures implemented in the Mantid software (104), and the reduced intensities were placed on an absolute scale using a calibrated porous silica standard with a scattering intensity of 450 cm−1 at very small q. All SANS data were measured at low temperatures (∼10 °C) using 1-mm-thick banjo Hellma cells. For a spatially homogenous temperature over the sample area, these Hellma cells were loaded into titanium sample holders using the banjo cell adaptors.
Atomistic Model.
MD simulations were performed on the same 95-amino acid residue SH4UD protein sequence (discussed above) that was analyzed using SAXS and SANS. The initial disordered 3D structure was obtained using I-TASSER (105) and a starting structure was chosen after equilibrating with MD simulation. The protein was solvated in a cubic box of 12.6-nm edge, and 4 chlorine ions were added to neutralize the system, which contained a total of ∼250,000 atoms. The Amber ff03ws (39, 106) protein force field and the TIP4P/2005s water model (107) were employed in all of the simulations.
MD Simulations.
We performed unbiased MD simulations using GROMACS 5.1.2 (108–113). All bonds involving hydrogen atoms were constrained using LINCS algorithm (114). The Verlet leapfrog algorithm was used to numerically integrate the equation of motions with a time step of 2 fs. A cutoff of 1.2 nm was used for short-range electrostatic and Lenard-Jones interactions. Long-range electrostatic interactions were calculated by particle-mesh Ewald (115) summation with a fourth-order interpolation and a grid spacing of 0.16 nm. The solute and solvent were coupled separately to a temperature bath of 300 K using a modified Berendsen thermostat with a relaxation time of 0.1 ps. The pressure coupling was fixed at 1 bar using the Parrinello–Rahman algorithm (116) with a relaxation time of 2 ps and isothermal compressibility of 4.5 × 10−5 bar−1. The energy of each system was minimized using the 1,000 steepest decent steps followed by 1-ns equilibration at NVT ensemble. The production runs were carried out in the NPT ensemble.
HREMD Simulations.
To enhance the conformational sampling, replica exchange with solute tempering 2 (REST2) (59, 60), a HREMD method, was employed. In HREMD, interaction potentials of part of the system can be scaled so as to promote large-scale conformational change. REST2 was chosen because it is computationally cheaper than temperature REMD and because it has been shown to efficiently explore conformational space in proteins (59–61, 117). This method scales intraprotein interactions by a factor λ, and the protein–water interaction by √λ, while water–water interactions are unaltered. Here, λ = Teff,0/Teff,i, where Ti is the effective temperature of ith replica and lowest temperature replica has order 0. Using λmax = 1 and λmin = 0.75, 20 replicas were simulated (SI Appendix), each for ∼510 ns, with protein snapshots saved every 10 ps. Exchange of configurations between neighboring replicas was attempted every 400 MD steps, with an average exchange probability of 0.6. The trajectory from the lowest rank replica (i.e., T = 300 K) was used for analysis. REST2 was implemented in GROMACS 5.1.4 (108–113) patched with PLUMED 2.3.4 (118). More details on the HREMD simulation are provided in SI Appendix. The analysis of MD trajectories was performed using GROMACS. The secondary structure prediction was calculated with DSSP (119). The asphericity was determined by calculating the gyration tensor of the protein (70). Convergence of the HREMD is discussed in detail in SI Appendix.
Calculation of Scattering Profiles.
The theoretical SAXS profiles were calculated from the HREMD trajectory using SWAXS (120), which considers the protein hydration shell explicitly. The theoretical SANS profiles were calculated using SASSENA (121), a program that takes explicitly into account the solvent molecules in the simulation. The water and protein exchangeable hydrogen atoms were assigned the deuterium scattering length density, while the protein nonexchangeable hydrogen atoms were assigned that of hydrogen. As such, the hydration shell density and thickness are determined directly from the simulation, avoiding overfitting issues encountered in implicit hydration shell methods (36).
The agreement between experiment and simulation was evaluated with the following χ2 score:
| [2] |
where <Iexpt(q)> and <Isim(q)> are the ensemble averaged experimental and theoretical SAXS or SANS data, respectively, N is the number of experimental q points, c is a scaling factor, bgd is a constant background, and σexpt is the experimental error.
The 2 fitting parameters in Eq. 2 are almost inevitably used when comparing experiment and theory. The scaling factor c is required because experimental data are often expressed in arbitrary or absolute units (depending on the data normalization method at the instrument), making it necessary to scale the theoretical SAXS/SANS data to the same units as experiments. c has no effect on the shape of SAXS/SANS profiles. A background (bgd) term is required to incorporate the uncertainty due to mismatch in buffer subtraction at higher q-values (24).
Supplementary Material
Acknowledgments
This work is supported by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle LLC and by project ERKP300 funded by the Office of Biological & Environmental Research in the Department of Energy (DOE) Office of Science (BER). Use of the Center for Structural Molecular Biology resources is supported by BER. This research used the resources of 3 DOE user facilities: The National Energy Research Scientific Computing Center (contract no. DE-AC02-05CH11231), the Oak Ridge Leadership Computing Facility (contract no. DE-AC05-00OR22725), and the Spallation Neutron Source. This manuscript has been authored by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the US DOE. The US government retains and the publisher, by accepting the article for publication, acknowledges that the US government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for US government purposes. DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (https://www.energy.gov/downloads/doe-public-access-plan).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907251116/-/DCSupplemental.
References
- 1.Mollica L., et al. , Binding mechanisms of intrinsically disordered proteins: Theory, simulation, and experiment. Front. Mol. Biosci. 3, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latysheva N. S., Flock T., Weatheritt R. J., Chavali S., Babu M. M., How do disordered regions achieve comparable functions to structured domains? Protein Sci. 24, 909–922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habchi J., Tompa P., Longhi S., Uversky V. N., Introducing protein intrinsic disorder. Chem. Rev. 114, 6561–6588 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Keul N. D., et al. , The entropic force generated by intrinsically disordered segments tunes protein function. Nature 563, 584–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells M., et al. , Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 105, 5762–5767 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uversky V. N., Roman A., Oldfield C. J., Dunker A. K., Protein intrinsic disorder and human papillomaviruses: Increased amount of disorder in E6 and E7 oncoproteins from high risk HPVs. J. Proteome Res. 5, 1829–1842 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Knowles T. P. J., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Uversky V. N., Oldfield C. J., Dunker A. K., Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 37, 215–246 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Sormanni P., et al. , Simultaneous quantification of protein order and disorder. Nat. Chem. Biol. 13, 339–342 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Robustelli P., Stafford K. A., Palmer A. G. 3rd, Interpreting protein structural dynamics from NMR chemical shifts. J. Am. Chem. Soc. 134, 6365–6374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez Y., Gairí M., Pons M., Bernadó P., Structural characterization of the natively unfolded N-terminal domain of human c-Src kinase: Insights into the role of phosphorylation of the unique domain. J. Mol. Biol. 391, 136–148 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Pérez Y., et al. , Lipid binding by the Unique and SH3 domains of c-Src suggests a new regulatory mechanism. Sci. Rep. 3, 1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C., Kim D.-H., Lee S.-H., Su J., Han K.-H., Structural investigation on the intrinsically disordered N-terminal region of HPV16 E7 protein. BMB Rep. 49, 431–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arbesú M., et al. , The unique domain forms a fuzzy intramolecular complex in src family kinases. Structure 25, 630–640.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Arai M., Sugase K., Dyson H. J., Wright P. E., Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc. Natl. Acad. Sci. U.S.A. 112, 9614–9619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuertes G., et al. , Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proc. Natl. Acad. Sci. U.S.A. 114, E6342–E6351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen M. R., Zweckstetter M., Huang J. R., Blackledge M., Exploring free-energy landscapes of intrinsically disordered proteins at atomic resolution using NMR spectroscopy. Chem. Rev. 114, 6632–6660 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Hofmann H., et al. , Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 16155–16160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuler B., Soranno A., Hofmann H., Nettels D., Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Annu. Rev. Biophys. 45, 207–231 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Busch D. J., et al. , Intrinsically disordered proteins drive membrane curvature. Nat. Commun. 6, 7875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine Z. A., Larini L., LaPointe N. E., Feinstein S. C., Shea J. E., Regulation and aggregation of intrinsically disordered peptides. Proc. Natl. Acad. Sci. U.S.A. 112, 2758–2763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Receveur-Bréchot V., Durand D., How random are intrinsically disordered proteins? A small angle scattering perspective. Curr. Protein Pept. Sci. 13, 55–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riback J. A., et al. , Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water. Science 358, 238–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruskamo S., et al. , Juxtanodin is an intrinsically disordered F-actin-binding protein. Sci. Rep. 2, 899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansen D., Jeffries C. M. J., Hammouda B., Trewhella J., Goldenberg D. P., Effects of macromolecular crowding on an intrinsically disordered protein characterized by small-angle neutron scattering with contrast matching. Biophys. J. 100, 1120–1128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldenberg D. P., Argyle B., Minimal effects of macromolecular crowding on an intrinsically disordered protein: A small-angle neutron scattering study. Biophys. J. 106, 905–914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banks A., Qin S., Weiss K. L., Stanley C. B., Zhou H. X., Intrinsically disordered protein exhibits both compaction and expansion under macromolecular crowding. Biophys. J. 114, 1067–1079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trewhella J., Small-angle scattering and 3D structure interpretation. Curr. Opin. Struct. Biol. 40, 1–7 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Drulyte I., et al. , Approaches to altering particle distributions in cryo-electron microscopy sample preparation. Acta Crystallogr. D Struct. Biol. 74, 560–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammel M., Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS). Eur. Biophys. J. 41, 789–799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riback J. A., et al. , Response to comment on “Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water”. Science 361, eaar7949 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Best R. B., et al. , Comment on “Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water”. Science 361, eaar7101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien E. P., Morrison G., Brooks B. R., Thirumalai D., How accurate are polymer models in the analysis of Förster resonance energy transfer experiments on proteins? J. Chem. Phys. 130, 124903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Best R. B., Computational and theoretical advances in studies of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 42, 147–154 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Henriques J., Arleth L., Lindorff-Larsen K., Skepö M., On the calculation of SAXS profiles of folded and intrinsically disordered proteins from computer simulations. J. Mol. Biol. 430, 2521–2539 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Henriques J., Cragnell C., Skepö M., Molecular dynamics simulations of intrinsically disordered proteins: Force field evaluation and comparison with experiment. J. Chem. Theory Comput. 11, 3420–3431 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Henriques J., Skepö M., Molecular dynamics simulations of intrinsically disordered proteins: On the accuracy of the TIP4P-D water model and the representativeness of protein disorder models. J. Chem. Theory Comput. 12, 3407–3415 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Best R. B., Zheng W., Mittal J., Balanced protein-water interactions improve properties of disordered proteins and non-specific protein association. J. Chem. Theory Comput. 10, 5113–5124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robustelli P., Piana S., Shaw D. E., Developing a molecular dynamics force field for both folded and disordered protein states. Proc. Natl. Acad. Sci. U.S.A. 115, E4758–E4766 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J., et al. , CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J., MacKerell A. D. Jr, Force field development and simulations of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 48, 40–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson M. J., Tirado-Rives J., Jorgensen W. L., Improved peptide and protein torsional energetics with the OPLSAA force field. J. Chem. Theory Comput. 11, 3499–3509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hummer G., Köfinger J., Bayesian ensemble refinement by replica simulations and reweighting. J. Chem. Phys. 143, 243150 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Bernadó P., Svergun D. I., Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol. Biosyst. 8, 151–167 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I., Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Roux B., Weare J., On the statistical equivalence of restrained-ensemble simulations with the maximum entropy method. J. Chem. Phys. 138, 084107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camilloni C., Vendruscolo M., Statistical mechanics of the denatured state of a protein using replica-averaged metadynamics. J. Am. Chem. Soc. 136, 8982–8991 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Cheng P., Peng J., Zhang Z., SAXS-oriented ensemble refinement of flexible biomolecules. Biophys. J. 112, 1295–1301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang S., Blachowicz L., Makowski L., Roux B., Multidomain assembled states of Hck tyrosine kinase in solution. Proc. Natl. Acad. Sci. U.S.A. 107, 15757–15762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bottaro S., Lindorff-Larsen K., Biophysical experiments and biomolecular simulations: A perfect match? Science 361, 355–360 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Boomsma W., Ferkinghoff-Borg J., Lindorff-Larsen K., Combining experiments and simulations using the maximum entropy principle. PLoS Comput. Biol. 10, e1003406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rieping W., Habeck M., Nilges M., Inferential structure determination. Science 309, 303–306 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Shevchuk R., Hub J. S., Bayesian refinement of protein structures and ensembles against SAXS data using molecular dynamics. PLoS Comput. Biol. 13, e1005800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hub J. S., Interpreting solution X-ray scattering data using molecular simulations. Curr. Opin. Struct. Biol. 49, 18–26 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Tatosyan A. G., Mizenina O. A., Kinases of the Src family: Structure and functions. Biochemistry (Mosc.) 65, 49–58 (2000). [PubMed] [Google Scholar]
- 57.Wheeler D. L., Iida M., Dunn E. F., The role of Src in solid tumors. Oncologist 14, 667–678 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kathiriya J. J., et al. , Presence and utility of intrinsically disordered regions in kinases. Mol. Biosyst. 10, 2876–2888 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Bussi G., Hamiltonian replica exchange in GROMACS: A flexible implementation. Mol. Phys. 112, 379–384 (2013). [Google Scholar]
- 60.Wang L., Friesner R. A., Berne B. J., Replica exchange with solute scaling: A more efficient version of replica exchange with solute tempering (REST2). J. Phys. Chem. B 115, 9431–9438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugita Y., Okamoto Y., Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314, 141–151 (1999). [Google Scholar]
- 62.Nors Perdersen M., et al. , Direct correlation between ligand-induced α-synuclein oligomers and amyloid-like fibril growth. Sci. Rep. 5, 10422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durand D., et al. , Small-angle X-ray scattering reveals an extended organization for the autoinhibitory resting state of the p47(phox) modular protein. Biochemistry 45, 7185–7193 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Svergun D. I., et al. , Protein hydration in solution: Experimental observation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. U.S.A. 95, 2267–2272 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merzel F., Smith J. C., Is the first hydration shell of lysozyme of higher density than bulk water? Proc. Natl. Acad. Sci. U.S.A. 99, 5378–5383 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang F., et al. , Hydration and interactions in protein solutions containing concentrated electrolytes studied by small-angle scattering. Phys. Chem. Chem. Phys. 14, 2483–2493 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Stanley C., Krueger S., Parsegian V. A., Rau D. C., Protein structure and hydration probed by SANS and osmotic stress. Biophys. J. 94, 2777–2789 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Das R. K., Pappu R. V., Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 110, 13392–13397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kohn J. E., et al. , Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12491–12496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petridis L., Schulz R., Smith J. C., Simulation analysis of the temperature dependence of lignin structure and dynamics. J. Am. Chem. Soc. 133, 20277–20287 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Chebaro Y., Ballard A. J., Chakraborty D., Wales D. J., Intrinsically disordered energy landscapes. Sci. Rep. 5, 10386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zerze G. H., Miller C. M., Granata D., Mittal J., Free energy surface of an intrinsically disordered protein: Comparison between temperature replica exchange molecular dynamics and bias-exchange metadynamics. J. Chem. Theory Comput. 11, 2776–2782 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Granata D., et al. , The inverted free energy landscape of an intrinsically disordered peptide by simulations and experiments. Sci. Rep. 5, 15449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin B., Pettitt B. M., Note: On the universality of proximal radial distribution functions of proteins. J. Chem. Phys. 134, 106101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen B. L., Pettitt B. M., Effects of acids, bases, and heteroatoms on proximal radial distribution functions for proteins. J. Chem. Theory Comput. 11, 1399–1409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oroguchi T., Ikeguchi M., Sato M., Towards the structural characterization of intrinsically disordered proteins by SAXS and MD simulation. J. Phys. Conf. Ser. 272, 012005 (2011). [Google Scholar]
- 77.Battisti A., Ciasca G., Tenenbaum A., Transient tertiary structures in tau, an intrinsically disordered protein. Mol. Simul. 39, 1084–1092 (2013). [Google Scholar]
- 78.Uversky V. N., Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 43, 1090–1103 (2011). [DOI] [PubMed] [Google Scholar]
- 79.van der Lee R., et al. , Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Uversky V. N., Gillespie J. R., Fink A. L., Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41, 415–427 (2000). [DOI] [PubMed] [Google Scholar]
- 81.Uversky V. N., Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 11, 739–756 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Müller-Späth S., et al. , From the cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 14609–14614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holehouse A. S., Das R. K., Ahad J. N., Richardson M. O., Pappu R. V., CIDER: Resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 112, 16–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Obara Y., Labudda K., Dillon T. J., Stork P. J., PKA phosphorylation of Src mediates Rap1 activation in NGF and cAMP signaling in PC12 cells. J. Cell Sci. 117, 6085–6094 (2004). [DOI] [PubMed] [Google Scholar]
- 85.Shenoy S., Chackalaparampil I., Bagrodia S., Lin P. H., Shalloway D., Role of p34cdc2-mediated phosphorylations in two-step activation of pp60c-src during mitosis. Proc. Natl. Acad. Sci. U.S.A. 89, 7237–7241 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hendus-Altenburger R., et al. , A phosphorylation-motif for tuneable helix stabilisation in intrinsically disordered proteins–Lessons from the sodium proton exchanger 1 (NHE1). Cell. Signal. 37, 40–51 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Battisti A., Ciasca G., Grottesi A., Bianconi A., Tenenbaum A., Temporary secondary structures in tau, an intrinsically disordered protein. Mol. Simul. 38, 525–533 (2012). [Google Scholar]
- 88.Rosenlöw J., Isaksson L., Mayzel M., Lengqvist J., Orekhov V. Y., Tyrosine phosphorylation within the intrinsically disordered cytosolic domains of the B-cell receptor: An NMR-based structural analysis. PLoS One 9, e96199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song J., et al. , Intrinsically disordered gamma-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc. Natl. Acad. Sci. U.S.A. 105, 1505–1510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ytreberg F. M., Borcherds W., Wu H., Daughdrill G. W., Using chemical shifts to generate structural ensembles for intrinsically disordered proteins with converged distributions of secondary structure. Intrinsically Disord. Proteins 3, e984565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sólyom Z., et al. , The disordered region of the HCV protein NS5A: Conformational dynamics, SH3 binding, and phosphorylation. Biophys. J. 109, 1483–1496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim D. H., Han K. H., Transient secondary structures as general target-binding motifs in intrinsically disordered proteins. Int. J. Mol. Sci. 19, E3614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kennedy J. A., Daughdrill G. W., Schmidt K. H., A transient α-helical molecular recognition element in the disordered N-terminus of the Sgs1 helicase is critical for chromosome stability and binding of Top3/Rmi1. Nucleic Acids Res. 41, 10215–10227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ithuralde R. E., Turjanski A. G., Phosphorylation regulates the bound structure of an intrinsically disordered protein: The p53-TAZ2 case. PLoS One 11, e0144284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venu K., Svensson L. A., Halle B., Orientational order and dynamics of hydration water in a single crystal of bovine pancreatic trypsin inhibitor. Biophys. J. 77, 1074–1085 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barbosa R. d. C., Barbosa M. C., Hydration shell of the TS-Kappa protein: Higher density than bulk water. Physica A 439, 48–58 (2015). [Google Scholar]
- 97.Russo D., Ollivier J., Teixeira J., Water hydrogen bond analysis on hydrophilic and hydrophobic biomolecule sites. Phys. Chem. Chem. Phys. 10, 4968–4974 (2008). [DOI] [PubMed] [Google Scholar]
- 98.Rani P., Biswas P., Local structure and dynamics of hydration water in intrinsically disordered proteins. J. Phys. Chem. B 119, 10858–10867 (2015). [DOI] [PubMed] [Google Scholar]
- 99.Chong S.-H., Chatterjee P., Ham S., Computer simulations of intrinsically disordered proteins. Annu. Rev. Phys. Chem. 68, 117–134 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Rauscher S., et al. , Structural ensembles of intrinsically disordered proteins depend strongly on force field: A comparison to experiment. J. Chem. Theory Comput. 11, 5513–5524 (2015). [DOI] [PubMed] [Google Scholar]
- 101.Piana S., Donchev A. G., Robustelli P., Shaw D. E., Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B 119, 5113–5123 (2015). [DOI] [PubMed] [Google Scholar]
- 102.Franke D., et al. , ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 50, 1212–1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao J. K., Gao C. Y., Liu D., The extended Q-range small-angle neutron scattering diffractometer at the SNS. J. Appl. Cryst. 43, 1068–1077 (2010). [Google Scholar]
- 104.Arnold O., et al. , Mantid—Data analysis and visualization package for neutron scattering and SR experiments. Nucl. Instrum. Methods Phys. Res. Sect. A 764, 156–166 (2014). [Google Scholar]
- 105.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Best R. B., Mittal J., Protein simulations with an optimized water model: Cooperative helix formation and temperature-induced unfolded state collapse. J. Phys. Chem. B 114, 14916–14923 (2010). [DOI] [PubMed] [Google Scholar]
- 107.Abascal J. L., Vega C., A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys. 123, 234505 (2005). [DOI] [PubMed] [Google Scholar]
- 108.Berendsen H. J. C., van der Spoel D., van Drunen R., GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995). [Google Scholar]
- 109.Lindahl E., Hess B., van der Spoel D., GROMACS 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 7, 306–317 (2001). [Google Scholar]
- 110.Van Der Spoel D., et al. , GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 111.Suardíaz R., Pérez C., Crespo-Otero R., García de la Vega J. M., Fabián J. S., Influence of density functionals and basis sets on one-bond carbon-carbon NMR spin-spin coupling constants. J. Chem. Theory Comput. 4, 448–456 (2008). [DOI] [PubMed] [Google Scholar]
- 112.Pronk S., et al. , GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abraham M. J., et al. , GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 114.Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
- 115.Darden T., York D., Pedersen L., Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 116.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 117.Peng E., Todorova N., Yarovsky I., Effects of forcefield and sampling method in all-atom simulations of inherently disordered proteins: Application to conformational preferences of human amylin. PLoS One 12, e0186219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bonomi M., et al. , PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 180, 1961–1972 (2009). [Google Scholar]
- 119.Kabsch W., Sander C., Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983). [DOI] [PubMed] [Google Scholar]
- 120.Chen P. C., Hub J. S., Validating solution ensembles from molecular dynamics simulation by wide-angle X-ray scattering data. Biophys. J. 107, 435–447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lindner B., Smith J. C., Sassena - X-ray and neutron scattering calculated from molecular dynamics trajectories using massively parallel computers. Comput. Phys. Commun. 183, 1491–1501 (2012). [Google Scholar]
- 122.Han B., Liu Y., Ginzinger S. W., Wishart D. S., SHIFTX2: Significantly improved protein chemical shift prediction. J. Biomol. NMR 50, 43–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.