Significance
The highly toxic cadmium ion can cause destructive hazards to living systems by nonspecific and tight binding on functional macromolecules. However, most of the developed cadmium detoxification systems are not sufficient to recognize or detoxify cadmium ions, specifically due to the similar coordination behavior of heavy metal ions in thiolate-rich sites. Here we report that the ultraspecific cadmium regulator CadR has evolved 2 distinct types of functional recognition sites rather than a mono-type thiolate-rich site to achieve outstanding selectivity. The thiolate-rich site I and the adjacent histidine-rich recognition site II are highly associated with transcription activation. This cooperative binding mechanism could improve our understanding of the relationship between the structural dynamics and biological function of metalloregulators.
Keywords: cadmium transcriptional regulator, cooperative binding, MerR family, protein crystallography, NMR spectroscopy
Abstract
Detoxification of the highly toxic cadmium element is essential for the survival of living organisms. Pseudomonas putida CadR, a MerR family transcriptional regulator, has been reported to exhibit an ultraspecific response to the cadmium ion. Our crystallographic and spectroscopic studies reveal that the extra cadmium selectivity of CadR is mediated by the unexpected cooperation of thiolate-rich site I and histidine-rich site II. Cadmium binding in site I mediates the reorientation of protein domains and facilitates the assembly of site II. Subsequently, site II bridge-links 2 DNA binding domains through ligands His140/His145 in the C-terminal histidine-rich tail. With dynamic transit between 2 conformational states, this bridge could stabilize the regulator into an optimal conformation that is critical for enhancing the transcriptional activity of the cadmium detoxification system. Our results provide dynamic insight into how nature utilizes the unique cooperative binding mechanism in multisite proteins to recognize cadmium ions specifically.
Cadmium pollution in the food chain has become a monumental public health concern since the emergence of the first serious cadmium-related disease (Itai-Itai disease) in Japan (1–4). The toxicity of cadmium is due mainly to its interference with the metabolism of zinc and other essential metal ions in the organism (5–7). Examination of a rare biological function of cadmium showed that the cadmium ion acts as a catalysis center of carbonic anhydrase in marine diatoms at low zinc levels (8, 9). Organisms have evolved multiple detoxification systems to eliminate the toxicity of cadmium. Metallothionein (MT), widely present in organisms, binds multiple cadmium ions in clusters, which can be divided into an α-domain Cd4S11 cluster and a β-domain Cd3S9 cluster in mammalian ΜΤ subtypes (10–12). In bacteria, 2 ArsR/SmtB family homologs, CadC and CmtR, with functional CdS4 or CdS3O allosteric sites have been reported to regulate the cadmium efflux system (13–16). These detoxification systems can detoxify other heavy metal ions besides cadmium, such as lead or mercury. In addition, MT is involved in the metabolism of zinc and copper (17, 18). Therefore, this indicates that the thiolate-rich sites in the reported cadmium-binding metalloproteins cannot readily distinguish cadmium ion from other heavy metals.
Pseudomonas putida CadR belongs to the MerR family of transcriptional regulators, which regulates its own transcription and the transcription of a cadmium efflux P-type ATPase CadA (19). It is the most specific natural cadmium-binding protein reported to date, as the cadR promoter is induced only by cadmium, and the induction of the cadA promoter by lead and zinc is much weaker than that by cadmium (19). The molecular basis underlying the specific cadmium selectivity of CadR remains elusive, however. The previously reported MerR family homologs possess remarkable metal sensitivity and selectivity; for instance, CueR can respond to free Cu(I) at zeptomolar concentrations (20). These metal-binding properties are typically achieved by the unique coordination geometry in a binding site, such as the linear CuS2 in CueR (20), the planer triangle HgS3 in MerR (21, 22), and the triangular pyramid PbS3 in PbrR691 (23, 24). Along with its outstanding metal-binding ability, the MerR family protein has a unique allosteric transcriptional regulation mechanism. These dual-function, σ70-dependent regulators control transcription very tightly by changing the overall topology of the promoter with an elongated 19- to 20-bp spacer between the −10 element and the −35 element, and they act as a repressor in the effector-free form or as an activator in the effector-binding form (25–27).
In the present study, we characterized the CadR protein by crystallography and spectroscopy. These methods provide dynamic insight into the specific cadmium selectivity of the CadR protein. The crystal structures of CadR in 3 different states—apo-state, metal-free DNA-bound state, and cadmium-substituted DNA-bound state—show that CadR has 2 distinct types of functional sites. Both the thiolate-rich allosteric site I, which is commonly used by heavy-metal ion metalloregulators, and the additional recognition site II are critical to enhance the transcriptional activity of the cadmium detoxification system. The unique cooperation of both metal-binding sites is the key factor for elucidating the extremely specific cadmium selectivity of CadR.
Results
Overall Structure.
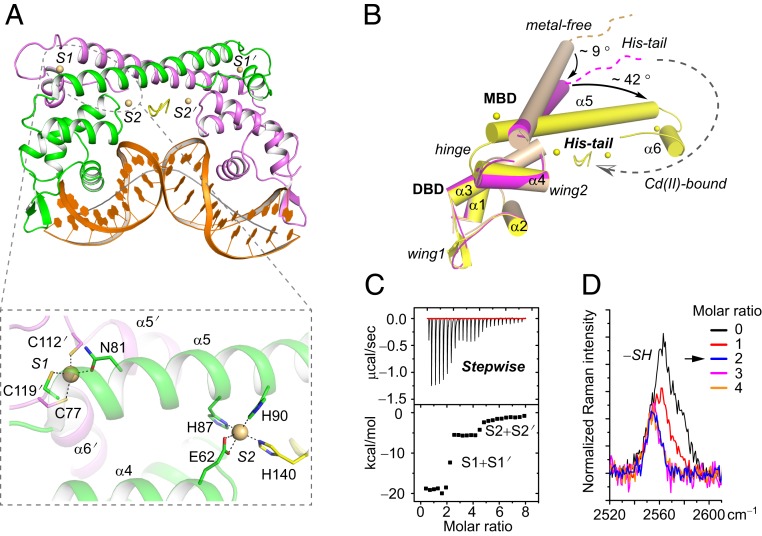
To reveal the structural basis underlying the cadmium selectivity of CadR, 3 different states of CadR structure were resolved: CadR in the apo form (apo-CadR), metal-free CadR and promoter binary complex (CadR/DNA), and cadmium-bound CadR/DNA ternary complex (Cd/CadR/DNA) (Fig. 1A and SI Appendix, Fig. S1). The structures show that CadR contains 2 major domains as other MerR homologs: the N-terminal DNA-binding domain (DBD; residues 1 to 71) and the C-terminal metal-binding domain (MBD; residues 76 to 147), which are connected by the flexible hinge (residues 72 to 75). In addition, CadR has a unique extended C-terminal histidine-rich region (His-tail; residues 138 to 147) containing His138, His140, His145, and His147 (SI Appendix, Fig. S2A). Although the MerR family proteins share a similar topological arrangement, the C-terminal MBD is flexible, and the orientation between the MBD and DBD differs (SI Appendix, Fig. S2B).
Fig. 1.
Two types of cadmium-binding sites and the allosteric effects triggered by cadmium binding. (A) Cartoon presentations of the Cd/CadR/DNA complex. DNA is colored in orange. The helical axis of DNA is represented as a solid gray line. Cadmium ions are shown as spheres. Close-up view of the first coordination shell of metal-binding sites is shown within the dashed box. (B) Superimposition of crystal structures based on the DBD of a CadR molecule of the metal-free CadR/DNA complex. For clarity, the partner CadR molecule and DNA are omitted. The 3 structures—apo-CadR, metal-free CadR/DNA complex, and Cd/CadR/DNA complex—are in wheat, magenta, and yellow, respectively. The allosteric movements triggered by cadmium binding are represented by dashed and solid arrows. (C) ITC curves showing that CadR has a clear, stepwise binding model toward the cadmium ion. (D) Raman peak of sulfhydryl group (∼2,563 cm−1) in CadR with a different molar ratio of Cd(II) ions.
The DBD of CadR is composed of 4 helices (α1 to α4) with loops connecting them and is involved in the specific recognition of promoter DNA, which was mapped by a footprint and electrophoretic mobility shift assay (EMSA) (SI Appendix, Fig. S3). There are 3 motifs involved in protein–DNA interactions (SI Appendix, Fig. S4): the helix-turn-helix (HTH) motif (α1-turn-α2), wing 1 motif (loop region between α2 and α3), and wing 2 motif (loop region between α3 and α4). The rmsd values of Cα atoms of DBD in the 3 different states are <0.525 Å, implying that the DBD backbone is rigid (Fig. 1B). The DBD is involved in interdomain motion with the MBD on DNA and cadmium binding. In contrast, the MBD of CadR can adopt flexible conformations depending on the states of the structures. Residues 79 to 126 form a long-extended helix (α5) in apo-CadR, which bends approximately 9° and becomes more flexible in the CadR-DNA complex. When bound to metal ions, residues 111 to 119 transfer from helix to coil, and residues 120 to 126 form an independent α6 helix. Meanwhile, the remaining residues 76 to 110 of α5 helix bends another 42°. Importantly, the His-tail crosses over the molecular center and contributes to metal binding in the Cd/CadR/DNA complex, while it is disordered in other structures (Fig. 1B).
CadR Contains 2 Types of Functional Cadmium-Binding Sites.
Interestingly, different from other MerR family proteins, CadR has 2 distinct types of metal-binding sites, and each CadR dimer binds to 4 cadmium ions (Fig. 1A and SI Appendix, Fig. S5 A–C). The metal-binding site I (S1 and S1′) is located at each end of the dimerization helices (α5 and α5′). The site is composed of 3 cysteines (Cys77, Cys112′, and Cys119′) and 1 asparagine residue (Asn81). Cys77 comes from the same chain as Asn81, while Cys112′ and Cys119′ come from the partner chain. The cysteine-rich environment of site I is conserved in the MerR family proteins. The metal-binding site II (S2 and S2′) is located in the middle of the molecule and formed by 2 histidines (His87 and His90 in the α5 helix), 1 glutamic acid (Glu62 in the α4 helix), and a variable last ligand from the His-tail region. S2 and S2′ are asymmetric such that His140 coordinates to S2 and His145 coordinates to S2′. These metal-binding ligands are highly conserved among CadR homologs (SI Appendix, Fig. S5D). Although the sequences of the extended C terminus are variable, a conserved HX4H motif is seen.
To investigate the thermodynamics of these 2 cadmium-binding sites, we performed a series of titration experiments. CadRWT binds cadmium ions with a stepwise-binding model, as the isothermal titration calorimetry (ITC) result shows 2 steps clearly separated at N ∼2 and 4 (Fig. 1C). Moreover, the Raman spectrum shows that the peak of the sulfhydryl group (SH stretch; ∼2,563 cm−1) reaches a minimum at a Cd(II)/CadRdimer ratio of ∼2 in the titration system (Fig. 1D). Three cysteine residues (Cys77, Cys112, and Cys119) are blocked with the metal binding in site I, while the other 2 (Cys12 and Cys53) remain free. Based on these observations, we propose that the cadmium ion first binds to the cysteine-rich site I (S1 + S1′) stoichiometrically and then binds to the histidine-rich site II (S2 + S2′). The binding affinity of site I is significantly stronger than that of site II, and the apparent binding affinities are fitted to K(Cd, site I) = 3.5 ± 0.1 × 1013 M−1 and K(Cd, site II) = 1.0 ± 0.4 × 107 M−1, respectively (SI Appendix, Fig. S6).
Site I Is a Conserved Allosteric Site.
As discussed above, the thiolate-rich binding site I is conserved among the MerR family metalloproteins. The crystal structures show that the conformation of MBD itself and the interaction network between MBD and DBD changed dramatically on the binding of cadmium ions (SI Appendix, Fig. S7 A and B). Residues of site I disperse from one another in the metal-free structures. Cys77 is disordered, and the distance between Cys112′ and Cys119′ is >10 Å in the crystal structure. Asn81 forms a hydrogen bond with Asn113′. On cadmium ion binding to site I, Cys119′ moves toward the coordination center, and the newly formed α6′ helix bends toward and interacts with the DBD. Meanwhile, Cys77 becomes ordered and coordinates with the cadmium ion. Within the interface of the α3, α4, α5, α5′, and α6′ helices bundle, a vast network of hydrophobic interactions is formed to stabilize the protein conformation (SI Appendix, Fig. S7C).
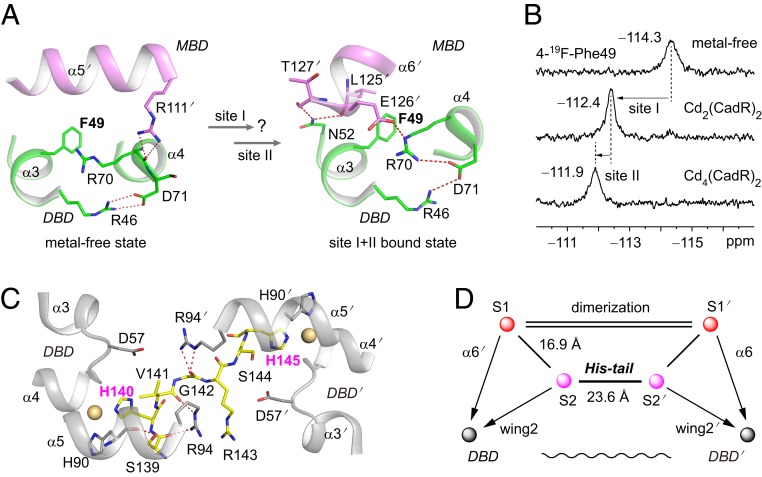
To gain more insight into structural function of the metal-binding sites, we used 19F NMR spectroscopy (28). Fortunately, a sole phenylalanine residue (Phe49) is located at the interface between the DBD and the MBD that exhibits a significant conformational change on cadmium binding (Fig. 2A). In metal-free structures, the DBD directly interacts with the α5′ helix of the partner molecule, in which Arg70 of the α4 helix makes a hydrogen bond with Arg111′ of the α5′ helix. In the cadmium-bound structures, Arg70 makes hydrogen bonds with Glu126′ in the α6′ helix. In addition, Asn52 in the α3 helix forms 2 hydrogen bonds with Leu125′ and Thr127′ in the α6′ helix. Thus, 19F-labeled Phe49 is an ideal probe for detecting the allosteric motion of CadR. Three clearly separated peaks—δ = −114.3, −112.4, and −111.9 ppm—are observed with the titration of cadmium ions (Fig. 2B). Compared with the ITC result, which shows a stepwise-binding model (Fig. 1C), these peaks can be assigned to the metal-free state, site I bound state in Cd2(CadR)2 and the sites I and II bound state in Cd4(CadR)2, respectively. Cadmium binding in site I triggers a 1.9-ppm downfield shift. Therefore, NMR spectroscopy also suggests that site I is an allosteric site.
Fig. 2.
Site I and site II are both allosteric sites. (A) The phenylalanine biomarker, Phe49, located at the interface between the DBD and MBD for probing the allosteric motion triggered by cadmium binding in site I and site II. Hydrogen bonds are represented by red dashed lines. (B) NMR spectra of the 19F-labeled protein titrated by cadmium ion. (C) Close up view of the interaction network around the His-tail region. The cadmium ions in sites S2 and S2′ are shown as spheres. The His-tail region (residues 139 to 145, SHVGRSH) is shown as yellow sticks. (D) Schematic model representing the topological arrangement of metal-binding sites and the DBD. The metal-binding sites are connected by the dimerization helices and the His-tail. The dimerization helices (α5 and α5′) are represented by double lines. The allosteric regulation pathways are indicated by arrows. The long-range communication of 2 DBDs through the His-tail is indicated as a wavy line.
Site II Bridging 2 DBDs by the Extended C-Terminal His-Tail.
The foregoing 19F NMR data also show that further cadmium binding in site II triggers a 0.5-ppm downfield shift of Phe49 (Fig. 2B). This suggests that site II acts as an additional allosteric site to further alter the conformation of the DBD. The orientations of the α4 and α5 helices are stabilized by the strong coordination bond among cadmium, Glu62 of the α4 helix, and His87/His90 of the α5 helix. In the second coordination shell, residues Glu62 and His87 also form hydrogen bonds with Thr59 and Leu56 in wing2, respectively (SI Appendix, Fig. S7 A and B). Part of the His-tail region (residues 139 to 145) is ordered in such a way that His140 coordinates with cadmium ions at site S2 and Ser139 contributes hydrogen bonds with His90 and Arg94.
In particular, the cadmium ion of site S2′ coordinates with His145 instead of His140′ from the partner molecule (Fig. 2C). The Cd/CadR/DNA complex is asymmetrical, in which residues 141 to 145 cross over the molecular center once His140 participates in coordination. Various interactions form during this process. Hydrogen bonds occur between Val141 and Arg94 and between Gly142 and Arg94′, polar contact occurs between Asp57′ and Ser144, and hydrophobic interactions include those among Val141, Val91, and Leu98′. The residue His145 will reach the vicinity of site S2′ and coordinate to the cadmium ion. Consequently, the 2 DBDs in the Cd/CadR/DNA complex are bridge-linked through the His-tail (Fig. 2D). The length of this bridge is 23.6 Å as measured by the distance between the cadmium ions at sites S2 and S2′.
NMR Spectroscopy Verified the Extended His-Tail Region Contributes to Cadmium Binding in Solution.
To further study properties of the His-tail region and metal-binding sites, we performed a series of NMR experiments. 19F-NMR spectroscopy was applied to directly observe the solution state of the His-tail region. A phenylalanine His-tail variant, G142F, was designed. The signals of 4-19F-Phe49 and 4-19F-Phe142 were detected (Fig. 3A). Fortunately, the peaks of 4-19F-Phe142 can be readily assigned by site-directed mutation. In the CadRF49L/G142F variant, the signal of 4-19F-Phe49 disappears (SI Appendix, Fig. S8A). The peak height of 19F-Phe142 at δ = −114.8 ppm in the metal-free state decreases greatly and shifts to δ = −114.7 ppm. The peak width increases along with titration of the cadmium ion, which indicates that the His-tail region has a restrained position on cadmium binding, with the crystallography data showing that the His-tail transforms from a disordered state to a loop state packing in the center of the CadR homodimer (Fig. 1B). A new peak contributed by 19F-Phe142 (δ = −113.1 ppm) is observed at a Cd/CadR ratio of 4, which can be assigned to another state of the His-tail region. To better illustrate the conformation states of the His-tail region, the peak at -114.7 ppm is assigned to a His-tail “off” state in which His140/145 does not participate in coordination (Fig. 3A), while the peak at −113.1 ppm is assigned to a His-tail “on” state in which His140/145 coordinates to the cadmium ion.
Fig. 3.
NMR spectroscopy verifying that the His-tail region contributes to cadmium binding in solution. (A) NMR spectra of the 19F-labeled His-tail variant (CadRG142F) titrated by Cd(II) at 298 K. The peaks contributed by 4-19F-Phe49 and 4-19F-Phe142 are assigned as black and red arrows, respectively. The His-tail region is represented by red dashed lines (“off” state) or arrows (“on” state). (B) NMR spectra of full 113Cd-bound CadRWT (Left) and CadRWT-DNA (Right) performed on a 500-MHz spectrometer at 308.5 K in phosphate buffer (relaxation delay, 10 s; number of scans, 8,000). The 113Cd-loaded protein sample is divided into 2 equal parts. One part has the added 25-bp promoter DNA (113Cd-CadRWT-DNA), while the other does not (113Cd-CadRWT). The chemical shift of cadmium-binding sites and the ratio of relative integrated intensity of peaks are indicated by the numbers in black.
Meanwhile, 113Cd NMR spectroscopy is applied to directly observe the solution state of the cadmium-binding sites. The NMR spectrum of full 113Cd-substituted CadRWT shows a strong resonance peak at δ = 526 ppm (Fig. 3B). Site I, which contains 3 cysteine ligands, contributes to this peak because the thiolate ligands are more deshielding relative to the oxygen and nitrogen ligands (29). Unexpectedly, the signal of site II is weak and broad in the 113Cd-CadRWT sample. Two peaks, 1 at δ = 241 ppm and the other at δ = 252 ppm, contributed by sites S2 and S2′, appeared in the spectrum (Fig. 3 B, Left). The coordination spheres of sites S2 and S2′ are very similar but not equivalent (His140 in S2 and His145 in S2′, based on crystallography), which may lead to subtle differences between the 2 peaks on the spectrum.
As CadR is a homodimer, there are 2 equivalent C-terminal regions. The His-tail and His-tail′ have equal probability to coordinate to sites S2 and S2′ (SI Appendix, Fig. S8B). Therefore, it is reasonable to propose that the flexible His-tail region can switch between the “on” and “off” states dynamically. Sites S2 and S2′ present a chemical exchange process among multiple states, with which the “off” state S2* is barely detected in the 113Cd NMR spectrum. To confirm our speculation, we performed 2 experiments. First, we acquired another NMR spectrum of the 113Cd-CadRWT-DNA sample. An equal ratio of promoter DNA was added into the 113Cd-CadRWT sample to stabilize the conformation of the CadR molecule as inspired by the crystallography data. As expected, peak intensities became stronger in the presence of promoter DNA (Fig. 3 B, Right). The ratio of integrated intensity of site I to site II was ∼1.0:0.96, while that in the 113Cd(II)-CadR sample without promoter DNA was ∼1.0:0.60. These results indicate that the His-tail “on” state is overwhelmingly preferred with the addition of promoter DNA. Those “off” states transform to the His-tail “on” state with an optimal promoter-binding conformation. Thus, the peak intensities of sites S2 and S2′ increase.
In addition, the line shape of the spectrum can change along with temperature if chemical exchange exists (30). The 113Cd NMR spectra are measured at different temperatures (SI Appendix, Fig. S8C). The faster the exchange rate (i.e., at higher temperature), the smaller the chemical shift difference between sites S2 and S2′. The observed difference is in agreement with the exchange speculation, increasing from 1.2 kHz to 2.1 kHz when temperature decreases from 308.5 K to 278.5 K. Moreover, the 19F NMR spectra show that the peak of Phe142 shifts at lower temperature, which also indicates the exchange of the His-tail (SI Appendix, Fig. S8A). Taken together, these data verify that the flexible His-tail region contributes to cadmium binding on site II in solution.
Site I and Site II Are Both Functionally Important and Bind the Cadmium Ion Cooperatively.
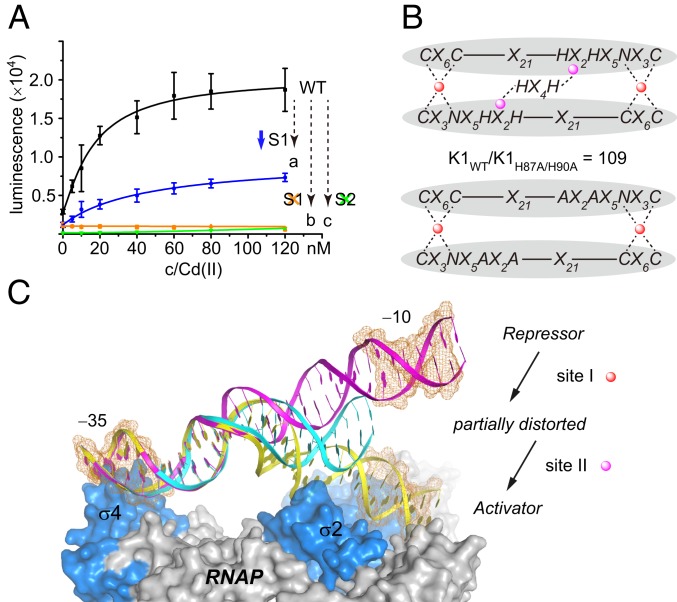
To confirm the functional importance of site I and site II in cells, a luxAB reporter system was constructed to measure cadmium sensitivity (Fig. 4A). We designed 3 variants with mutated metal-binding sites: CadRN81A, CadRC77S/C112S/C119S, and CadRH87A/H90A. CadRN81A had a 5-fold weaker site I, with an apparent binding affinity fitted to K(Cd, site I) of 6.6 ± 0.7 × 1012 M−1 (SI Appendix, Fig. S9). The cadmium sensitivity of the CadRN81A variant was decreased by >50%. Moreover, the CadRH87A/H90A variant with mutated site II (SI Appendix, Fig. S10) and the CadRC77S/C112S/C119S variant with mutated site I (SI Appendix, Fig. S11) could hardly respond to the cadmium ion in the nanomolar range, which infers the essentiality of the conserved site I and an additional site II in CadR.
Fig. 4.
Cooperation of 2 sites triggers the specific selectivity of cadmium ions. (A) Cadmium sensitivity study using the luxAB reporter system in vivo. CadRWT (black) and its variants, CadRN81A (a, blue), CadRC77S/C112S/C119S (b, orange), and CadRH87A/H90A (c, green) are tested. Error bars represent SD. (B) The scaffold of the MBD in CadRWT and CadRH87A/H90A. Glu62 in the DBD is omitted for clarity. Site I and site II are shown as red and magenta circles, respectively. The coordination bonds are shown as dashed lines. (C) A putative model of the partially distorted state promoter (cyan) between the repressor state (magenta) and activator state (yellow). The σ factor and core enzyme of RNAP are colored in marine and gray, respectively. The −10 and −35 elements are indicated by the orange mesh.
Further 113Cd NMR spectroscopy analysis suggested that site I of the CadRH87A/H90A variant retains a CdS3O coordination sphere that is highly similar to the wild-type protein without the presence of site II. However, UV-vis titration results show that the binding affinity of site I was decreased by 2 orders of magnitude (Fig. 4B and SI Appendix, Fig. S10). Meanwhile, the ITC results show that cadmium ion can bind to the CadRC77S/C112S/C119S variant with a similar affinity (106−7) to site II of CadRWT (107) but with a different binding model (SI Appendix, Fig. S11). We performed 113Cd NMR spectroscopy to characterize the cadmium-binding center in this variant. To our surprise, no specific peaks were detected in the spectra. This implies that site I is also essential for the successful assembly of site II, as the structural alignment shows that His87, His90, and the His-tail region are reoriented (SI Appendix, Fig. S12). Taken together, these data suggest that site I and site II are in fact dependent on each other to attain biological function (SI Appendix, Fig. S13). Thus, we conclude that CadR binds cadmium ion cooperatively.
Cooperation Between Site I and Site II Contributes to Specific Cadmium Selectivity vs. Lead and Zinc Ions.
The foregoing results and analysis inspired us to examine whether the cooperation of site I and site II is a key factor for elucidating specific cadmium selectivity. Cadmium binding to the allosteric site I (S1 + S1′) triggers orientation of the DBD, changing the promoter from a repressor state to a partially distorted state. With further Cd(II) binding to the additional allosteric site II (S2 + S2′), CadR is stabilized to an optimal DNA-binding conformation and transforms to a stable activator state (Fig. 4C). The activated promoter, with a shortened distance (SI Appendix, Fig. S14) and reduced phase angle between the −10 and −35 elements (SI Appendix, Fig. S15) as in other MerR-type activator state promoters, is suitable for interacting with RNA polymerase (RNAP), similar to the situation for CueR (26, 31, 32).
Using the aforementioned cooperative binding mechanism, CadR can distinguish cadmium ion from other metal ions, such as zinc and lead ions. Compared with the cadmium ion, there is only 1 set of lead-binding sites, as suggested by the ITC titration curve (SI Appendix, Fig. S16A). Meanwhile, the UV-vis spectrum of Pb(II)-bound CadR shows a specific peak appearing at λ = 340 nm (SI Appendix, Fig. S16B), which was contributed by the PbS3 center (23, 24). These results show that the lead ion can only bind to the thiolate-rich site I, and that the apparent binding affinity is fitted to K(Pb) = 4.3 ± 0.3 × 1012 M−1. Although site I is only 8-fold weaker than that of Cd(II), the Pb(II)-CadR complex definitely decreases the total binding affinity, since the lead ion could not bind to site II. The promoter DNA would only shift to a partially distorted state. To verify the speculation of the partially distorted promoter in the presence of lead ion, we synthetized a DNA probe (33, 34), in which the fluorescence intensity of the probe is positively coupled with the distortion level of promoter DNA (SI Appendix, Fig. S17). As expected, the fluorescence intensity of the Pb(II)-bound probe is much weaker than that of the Cd(II)-bound probe. It indicates that the promoter DNA is indeed less distorted by Pb(II)-CadR with only site I occupied.
In the case of the zinc ion, the ITC experiment and Raman spectroscopy suggest that its binding model differs from that for the cadmium ion (SI Appendix, Fig. S18). Further UV-vis titration data show that the zinc-binding mechanism of CadRWT is more complicated than the cadmium-binding mechanism due to the cooperation and similar binding affinity of sites I and II (SI Appendix, Fig. S19). A plausible 4-step binding model is proposed based on the Raman results, and the binding affinities are fitted. This model shows that the affinity of the thiolate-rich site I (∼1010) is 3 orders of magnitude weaker than that of cadmium ion (∼1013).
To better understand the molecular basis of the weak zinc sensitivity of CadR in vivo (19), we solved the crystal structure of the Zn(II)-bound CadR/DNA complex (SI Appendix, Fig. S20). Although the zinc ion binds to the same sites as the cadmium ion, the Asn81 residue clearly does not coordinate to the zinc ion, because it is smaller than the cadmium ion. The structural alignment indicates that zinc substitution in site I can induce a 10-degree overall shift of the MBD and a 4-Å displacement of the DBD′ compared with the Cd/CadR/DNA structure. Consequently, the promoter is less distorted, in agreement with the fluorescent DNA titration data (SI Appendix, Fig. S17C). Taking into account a weaker site I and a less distorted promoter, the transcription activity of the zinc ion is much weaker than that of the cadmium ion (19).
Discussion
Bacteria have developed a specific and effective cadmium-detoxification system that facilitates their survival in cadmium-polluted environments through their rapid evolutionary rate (35). The CadR-regulated system is one of the most successful examples of this system. Compared with previously characterized MerR family homologs or other natural cadmium-binding proteins (SI Appendix, Fig. S21), CadR adopts 2 types of functional metal-binding sites rather than 1 type to achieve this remarkable capability. The selective cadmium recognition is mediated not only by the specific chemical environment of each site, but also by their structural function and cooperation. In summary, the strong cadmium-binding site I is the allosteric site to reorient protein domains and DNA promoter; the additional allosteric site II further alters and bridge-links the 2 DBDs and subsequently stabilizes the regulator-promoter system in activator form.
The first coordination shell in site I is most likely a distorted tetrahedron S3O with an oxygen ligand contributed by the Oδ atom of asparagine. Despite the Cd/CadR/DNA structure’s low resolution (2.7 Å), it shows a reasonable Cd-O bond length at 2.3 to 2.4 Å (SI Appendix, Fig. S22). The shell is different from the cadmium-substituted CmtR (16, 36) and TRI peptide (37–41), where an external water molecule contributes the oxygen ligand. The 113Cd chemical shift of site I is 96 ppm upfield of that of 113Cd-substituted Staphylococcus aureus pI258 CadC (13) and 46 ppm downfield of 113Cd-substituted Mycobacterium tuberculosis CmtR (16). The cadmium binding affinity of site I is similar to that in CmtR and CadC (∼1012−13 M−1), which shows that the natural cadmium-binding protein exhibits subpicomolar sensitivity in the thiolate-rich sites. The residue Asn81 is important to specific cadmium binding, as site-directed mutagenesis from Asn81 to Ala81 decreases the binding affinity of site I by ∼5-fold and the transcriptional activity by 50%. The pocket size of site I is also highly optimized for cadmium ions vs. zinc ions. The residue Asn81 does not coordinate to zinc ions because of the smaller ion radius. The change in the coordination shell alters the orientation between the MBD and the DBD and subsequently influences the conformation of the DNA promoter. Our results suggest that the reduced affinity in site I and the less distorted promoter can be the factors that weaken the zinc sensitivity of CadR in vivo (19).
The metal-binding site II contains 3 histidine residues and 1 glutamic acid residue arranged in a tetrahedral geometry. It can bind to cadmium ions and zinc ions but not to lead ions. Mutating site II will cause abrogation of the transcriptional activity in vivo. The most striking feature of site II is its dynamic and asymmetric environment. The flexible C-terminal His-tail region contributes to cadmium binding in site II. Our present NMR studies reveal that the His-tail switches between the “on” and “off” states in solution. The switching motion is highly associated with the sequence-specific promoter binding, as 113Cd NMR and crystallography both show that the “on” state is overwhelmingly preferred in the presence of promoter DNA.
These 2 types of functional sites represent a positive cooperative binding mechanism in which binding in site I facilitates the assembly of site II and the presence of site II enhances the binding affinity of site I by 2 orders of magnitude without altering the cadmium coordination sphere. In terms of allosteric ensembles (42), most of the reported MerR family proteins adopt a backbone dynamics allosteric regulation pattern induced by 1 type of effector-binding site (SI Appendix, Fig. S21A). The end of the dimerization helix bends toward DBD on metal ion binding, subsequently altering the conformation of DBD and promoter. Here we show that the disordered C-terminal tail in CadR can fold into a loop structure with an allosteric regulation function. Thus, we classify the allosteric regulation pattern of CadR as a new type with a combination of transcription factor backbone dynamics and a local unfolding mechanism (42). We believe that this cooperative regulation system with dynamic C-termini can expand our understanding of the allosteric regulation pattern of metalloregulators.
Materials and Methods
The materials and methods used in this study are described in detail in SI Appendix. Oligonucleotides are listed in SI Appendix, Table S1; data collection and refinement statistics of CadR and its complex are reported in SI Appendix, Table S2; and plasmids are listed in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank the staff of beamlines BL17U and BL19U at the Shanghai Synchrotron Radiation Facility for their assistance with crystallographic data collection, and the staff of the Nuclear Magnetic Resonance System at the National Facility for Protein Science in Shanghai, Zhangjiang Laboratory. We also thank Changlin Tian and Yangzhong Liu for their helpful suggestions on NMR spectroscopy. This work was supported by the Ministry of Science and Technology of the People’s Republic of China Key Project 2015CB856303 and 2012CB933802 (to H.C.) and the National Natural Science Foundation of China 21071077, 91013009, and 21778032 (to H.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.V.O. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.wwpdb.org/ (PDB ID codes 6JGF, 6JGV, 6JGW, 6JGX, and 6JNI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908610116/-/DCSupplemental.
References
- 1.Nordberg G. F., Health hazards of environmental cadmium pollution. Ambio 3, 55–66 (1974). [Google Scholar]
- 2.Järup L., Åkesson A., Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. 238, 201–208 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Nawrot T. S., et al. , Cadmium exposure in the population: From health risks to strategies of prevention. Biometals 23, 769–782 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Nordberg G. F., Nogawa K., Nordberg M., “Cadmium” in Handbook on the Toxicology of Metals, Nordberg G. F., Fowler B A., Nordberg M., Eds. (Academic Press, New York, ed. 4, 2015), pp. 667–716. [Google Scholar]
- 5.Goyer R. A., Toxic and essential metal interactions. Annu. Rev. Nutr. 17, 37–50 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Bertin G., Averbeck D., Cadmium: Cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review). Biochimie 88, 1549–1559 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Sigel A., Sigel H., Sigel R. K., Cadmium: From Toxicity to Essentiality (Springer, New York, 2013). [Google Scholar]
- 8.Xu Y., Feng L., Jeffrey P. D., Shi Y., Morel F. M., Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature 452, 56–61 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Lane T. W., et al. , Biochemistry: A cadmium enzyme from a marine diatom. Nature 435, 42 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Klaassen C. D., Liu J., Choudhuri S., Metallothionein: An intracellular protein to protect against cadmium toxicity. Annu. Rev. Pharmacol. Toxicol. 39, 267–294 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Otvos J. D., Armitage I. M., Structure of the metal clusters in rabbit liver metallothionein. Proc. Natl. Acad. Sci. U.S.A. 77, 7094–7098 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zangger K., Oz G., Otvos J. D., Armitage I. M., Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy. Protein Sci. 8, 2630–2638 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busenlehner L. S., et al. , Spectroscopic properties of the metalloregulatory Cd(II) and Pb(II) sites of S. aureus pI258 CadC. Biochemistry 40, 4426–4436 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Sun Y., Wong M. D., Rosen B. P., Role of cysteinyl residues in sensing Pb(II), Cd(II), and Zn(II) by the plasmid pI258 CadC repressor. J. Biol. Chem. 276, 14955–14960 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Cavet J. S., Graham A. I., Meng W., Robinson N. J., A cadmium-lead-sensing ArsR-SmtB repressor with novel sensory sites. Complementary metal discrimination by NmtR AND CmtR in a common cytosol. J. Biol. Chem. 278, 44560–44566 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Hemmingsen L., Giedroc D. P., Structural and functional characterization of Mycobacterium tuberculosis CmtR, a PbII/CdII-sensing SmtB/ArsR metalloregulatory repressor. Biochemistry 44, 8976–8988 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Coyle P., Philcox J. C., Carey L. C., Rofe A. M., Metallothionein: The multipurpose protein. Cell. Mol. Life Sci. 59, 627–647 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sigel A., Sigel H., Sigel R. K., Metallothioneins and Related Chelators (Royal Society of Chemistry, London, 2009), vol. 5. [Google Scholar]
- 19.Lee S.-W., Glickmann E., Cooksey D. A., Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl. Environ. Microbiol. 67, 1437–1444 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Changela A., et al. , Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Chang C.-C., Lin L.-Y., Zou X.-W., Huang C.-C., Chan N.-L., Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR. Nucleic Acids Res. 43, 7612–7623 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D., et al. , Structural analysis of the Hg(II)-regulatory protein Tn501 MerR from Pseudomonas aeruginosa. Sci. Rep. 6, 33391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen P. R., et al. , Spectroscopic insights into lead(II) coordination by the selective lead(II)-binding protein PbrR691. J. Am. Chem. Soc. 129, 12350–12351 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Huang S., et al. , Structural basis for the selective Pb(II) recognition of metalloregulatory protein PbrR691. Inorg. Chem. 55, 12516–12519 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Ansari A. Z., Chael M. L., O’Halloran T. V., Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature 355, 87–89 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Philips S. J., et al. , Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science 349, 877–881 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heldwein E. E. Z., Brennan R. G., Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature 409, 378–382 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Li H., Frieden C., NMR studies of 4-19F-phenylalanine-labeled intestinal fatty acid binding protein: Evidence for conformational heterogeneity in the native state. Biochemistry 44, 2369–2377 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Coleman J. E., Cadmium-113 nuclear magnetic resonance applied to metalloproteins. Methods Enzymol. 227, 16–43 (1993). [DOI] [PubMed] [Google Scholar]
- 30.Rule G. S., Hitchens T. K., Fundamentals of Protein NMR Spectroscopy (Springer Science & Business Media, Berlin/Heidelberg, 2006), Vol. 5. [Google Scholar]
- 31.Newberry K. J., Brennan R. G., The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J. Biol. Chem. 279, 20356–20362 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Watanabe S., Kita A., Kobayashi K., Miki K., Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 105, 4121–4126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen P., He C., A general strategy to convert the MerR family proteins into highly sensitive and selective fluorescent biosensors for metal ions. J. Am. Chem. Soc. 126, 728–729 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Chen P., et al. , An exceptionally selective lead(II)-regulatory protein from Ralstonia metallidurans: Development of a fluorescent lead(II) probe. Angew. Chem. Int. Ed. Engl. 44, 2715–2719 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Denef V. J., Banfield J. F., In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science 336, 462–466 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Banci L., et al. , NMR structural analysis of cadmium sensing by winged helix repressor CmtR. J. Biol. Chem. 282, 30181–30188 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Matzapetakis M., et al. , Comparison of the binding of cadmium(II), mercury(II), and arsenic(III) to the de novo designed peptides TRI L12C and TRI L16C. J. Am. Chem. Soc. 124, 8042–8054 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Matzapetakis M., Pecoraro V. L., Site-selective metal binding by designed α-helical peptides. J. Am. Chem. Soc. 127, 18229–18233 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Lee K. H., Cabello C., Hemmingsen L., Marsh E. N. G., Pecoraro V. L., Using nonnatural amino acids to control metal-coordination number in three-stranded coiled coils. Angew. Chem. Int. Ed. Engl. 45, 2864–2868 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Peacock A. F., Hemmingsen L., Pecoraro V. L., Using diastereopeptides to control metal ion coordination in proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 16566–16571 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stachura M., et al. , Direct observation of nanosecond water exchange dynamics at a protein metal site. J. Am. Chem. Soc. 139, 79–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motlagh H. N., Wrabl J. O., Li J., Hilser V. J., The ensemble nature of allostery. Nature 508, 331–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.