Significance
Aerenchyma formation has a crucial role in conferring abiotic stress tolerance to plants, including agronomically important crops. In rice, root aerenchyma constitutively forms under aerobic conditions and is further induced under oxygen deficiency. Although ethylene is involved in inducible aerenchyma formation, the factors involved in constitutive aerenchyma formation remain unclear. Here we show that AUX/IAA-ARF-dependent auxin signaling is required for constitutive aerenchyma formation in rice roots. We identify a LBD transcription factor whose expression is regulated by an AUX/IAA-ARF complex as a key regulator of both constitutive aerenchyma and lateral root formation. However, their formation is differently controlled during root development. Our findings reveal a previously unreported role of auxin in plant development and plant responses to abiotic stresses.
Keywords: aerenchyma, auxin response factor, AUX/IAA protein, lateral organ boundary domain, lateral roots
Abstract
Lateral roots (LRs) are derived from a parental root and contribute to water and nutrient uptake from the soil. Auxin/indole-3-acetic acid protein (AUX/IAA; IAA) and auxin response factor (ARF)-mediated signaling are essential for LR formation. Lysigenous aerenchyma, a gas space created by cortical cell death, aids internal oxygen transport within plants. Rice (Oryza sativa) forms lysigenous aerenchyma constitutively under aerobic conditions and increases its formation under oxygen-deficient conditions; however, the molecular mechanisms regulating constitutive aerenchyma (CA) formation remain unclear. LR number is reduced by the dominant-negative effect of a mutated AUX/IAA protein in the iaa13 mutant. We found that CA formation is also reduced in iaa13. We have identified ARF19 as an interactor of IAA13 and identified a lateral organ boundary domain (LBD)-containing protein (LBD1-8) as a target of ARF19. IAA13, ARF19, and LBD1-8 were highly expressed in the cortex and LR primordia, suggesting that these genes function in the initiation of CA and LR formation. Restoration of LBD1-8 expression recovered aerenchyma formation and partly recovered LR formation in the iaa13 background, in which LBD1-8 expression was reduced. An auxin transport inhibitor suppressed CA and LR formation, and a natural auxin stimulated CA formation in the presence of the auxin transport inhibitor. Our findings suggest that CA and LR formation are both regulated through AUX/IAA- and ARF-dependent auxin signaling. The initiation of CA formation lagged that of LR formation, which indicates that the formation of CA and LR are regulated differently by auxin signaling during root development in rice.
Lysigenous aerenchyma is an internal gas space created by programmed cell death (PCD) and subsequent lysis of root cortical cells (1). Many plant species, including agronomically important crops, form lysigenous aerenchyma in the roots (2). Aerenchyma aids the internal transport from shoot to root tips and simultaneously reduces the respiratory costs of roots in waterlogged soils (3). Aerenchyma also reduces respiratory costs and stimulates root growth under drought or nutrient-deficient conditions (4).
Under aerobic conditions, roots of rice (Oryza sativa) form aerenchyma constitutively (5, 6), and they further enhance its formation in response to oxygen-deficient conditions (7–9). The former and latter are defined as constitutive aerenchyma (CA) and inducible aerenchyma (IA), respectively (7). Some barley (Hordeum vulgare) cultivars form aerenchyma (root cortical scenesence) under aerobic conditions (10), but generally upland crops, such as wheat (Triticum aestivum) and maize (Zea mays ssp. mays), form little CA under well-controlled aerobic conditions (11), and thus sudden oxygen-deficiency severely damages their roots (7). Therefore, CA in roots is considered to help plants cope with oxygen-deficient conditions by supplying their roots with oxygen immediately after waterlogging (7–9). However, the molecular mechanisms regulating CA formation remain unclear (9).
PCD during aerenchyma formation under waterlogged conditions (i.e., IA formation) is stimulated by the gaseous phytohormone ethylene (7–9, 12, 13). We recently found that ethylene-induced reactive oxygen species (ROS) generation is required for IA formation in rice roots (14). Aerenchyma forms to some extent when roots are treated with an ethylene perception inhibitor (15) and when a gene involved in ROS generation is knocked out, even under aerobic conditions (14), showing that other factors must be involved in aerenchyma formation.
Auxin signaling is mediated by a family of B3 domain-containing transcription factors called auxin response factors (ARFs), which bind to a type of cis-element, auxin response element (AuxRE), on the promoters of auxin-responsive genes (16). ARFs are classified into activators or repressors, and the activator-type ARFs contain a glutamine (Q)-rich middle region (17). ARF-dependent transcriptional regulation is repressed by auxin/indole-3-acetic acid proteins (AUX/IAAs; IAAs), which have 4 conserved amino acid sequence motifs: AUX/IAA domains I, II, III, and IV (18). Although IAA proteins do not have a nuclear localization signal, domain I mediates the transcriptional repression through its interaction with ARF (19). Domain II is required for the proteolysis of IAA proteins (20). IAA and ARF share domains III and IV, which allow heterodimerization between these proteins (21).
The development of lateral roots (LRs) and adventitious (crown) roots is regulated through auxin (22). In Arabidopsis, IAA14, which interacts with ARF7 and ARF19, is involved in LR initiation (23). Plants have a family of transcription factors called lateral organ boundary domain (LBD)-containing proteins (24). In Arabidopsis, overexpression of LBD16 and LBD29 can partly restore LR formation in the arf7arf19 double knockout mutant (25), indicating that these LBDs act downstream of ARF7 and ARF19 during LR formation. In rice, the dominant-negative iaa13 mutant, which has a single amino acid substitution in the auxin-dependent degradation domain (i.e., AUX/IAA domain II) of IAA13, has fewer LRs than the WT with the same genetic background (26). Because IAA13 is the rice IAA homolog closest to Arabidopsis IAA14 (27), genetic control of LR development might be common to rice and Arabidopsis.
Here we examined the involvement of AUX/IAA‐ and ARF-dependent signaling in CA formation in rice roots. We analyzed CA formation in adventitious roots of the iaa13 mutant and found that it was reduced in iaa13. We identified ARF19 as an interactor with IAA13 and identified another transcription factor (LBD1-8) as a target of ARF19. Restoration of LBD1-8 expression in iaa13, in which LBD1-8 expression was suppressed, recovered the levels of CA and LR formation. Moreover, we show that the formation of CA and LR are regulated independently, even though both are regulated by auxin signaling.
Results
AUX/IAA-Mediated Auxin Signaling Is Involved in CA and LR Formation.
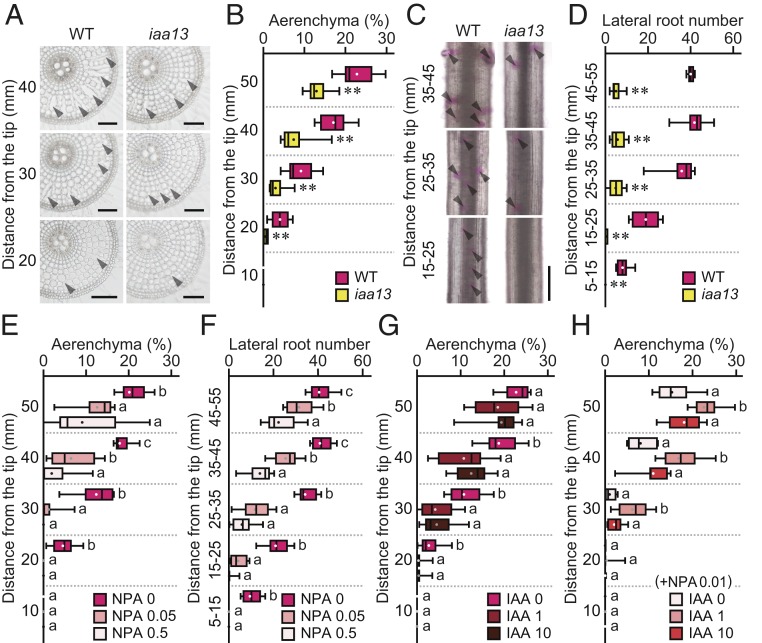
To evaluate the involvement of auxin signaling in CA formation, 20-d-old aerobically grown iaa13 mutant and its background WT rice (cv. Taichung 65; T65) were further grown under aerated conditions for 48 h, after which transverse sections along the adventitious roots were prepared (Fig. 1A). In the WT, CA formation was initiated at 20 mm from the root tips and gradually increased toward the basal part of the roots (Fig. 1B). The percentages of aerenchyma in iaa13 at all positions were lower than those in the WT (Fig. 1B).
Fig. 1.
Analyses of aerenchyma and lateral root formation in roots of the WT and iaa13 mutant. (A–D) Twenty-day-old aerobically grown WT and iaa13 seedlings were further grown under aerated conditions for 48 h. (A) Cross-sections at 20, 30, and 40 mm from the tips of adventitious roots. Aerenchyma is indicated by arrowheads. (Scale bars, 100 μm.) (B) Percentages of aerenchyma in root cross-sectional area at 10, 20, 30, 40, and 50 mm. (C) Pictures of lateral roots at 15 to 25, 25 to 35, and 35 to 45 mm. Lateral root primordia visualized by Feulgen staining are indicated by arrowheads. (Scale bar, 500 μm.) (D) Numbers of lateral roots at 5 to 15, 15 to 25, 25 to 35, 35 to 45, and 45 to 55 mm. In B and D, significant differences between the genotypes at **P < 0.01 (2-sample t test). (E–H) Twenty-day-old aerobically grown WT seedlings were further grown under aerated conditions with 0, 0.05, or 0.5 µM NPA (E and F), and with 0, 1, or 10 µM IAA without (G) or with 0.01 µM NPA (H) for 48 h. Percentages of aerenchyma (E, G, and H) and numbers of lateral roots (F) are shown. Boxplots show the median (horizontal lines), 25th to 75th percentiles (edges of the boxes), minimum to maximum (edges of the whiskers), and mean values (dots in the boxes) (n = 9). In E–H, different lowercase letters denote significant differences among the conditions (P < 0.05, one-way ANOVA followed by Tukey’s test for multiple comparisons).
To confirm that the decreased CA in iaa13 roots is caused by the mutation in the IAA13 gene (SI Appendix, Fig. S1A) (26), the mutated IAA13 was expressed in the WT background. The dominant-negative (ProIAA13::iaa13) transgenic plants showed a dwarf phenotype (SI Appendix, Fig. S1B), and their roots had fewer CA compared with roots of the vector control (ProIAA13::GUS) (SI Appendix, Fig. S1C). Subsequently, lateral root tips of the WT and iaa13 were visualized by Feulgen staining (Fig. 1C), and the LRs were counted (Fig. 1D). LRs were first detected at 5 to 15 mm in the WT, compared with 25 to 35 mm in iaa13 (Fig. 1D). At 35 to 45 mm, the number of LRs was ∼7-fold lower in iaa13 compared with the WT (Fig. 1D).
To confirm the involvement of auxin in the formation of CA and LR in rice roots, 20-d-old WT seedlings were treated with the polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA; 0, 0.05, or 0.5 µM) (Fig. 1 E and F). As a result, NPA dose-dependently decreased the formation of CA (Fig. 1E) and LR (Fig. 1F). Subsequently, the WT was treated with a natural auxin IAA (0, 1, or 10 µM) under aerated conditions for 48 h (Fig. 1G). Because both 1 and 10 µM IAA suppressed CA formation (Fig. 1G), the WT was treated with 0, 1, or 10 µM IAA together with 0.01 µM NPA for 48 h (Fig. 1H). As a result, 1 µM IAA restored CA formation in the presence of NPA, whereas 10 µM IAA suppressed its formation (Fig. 1H).
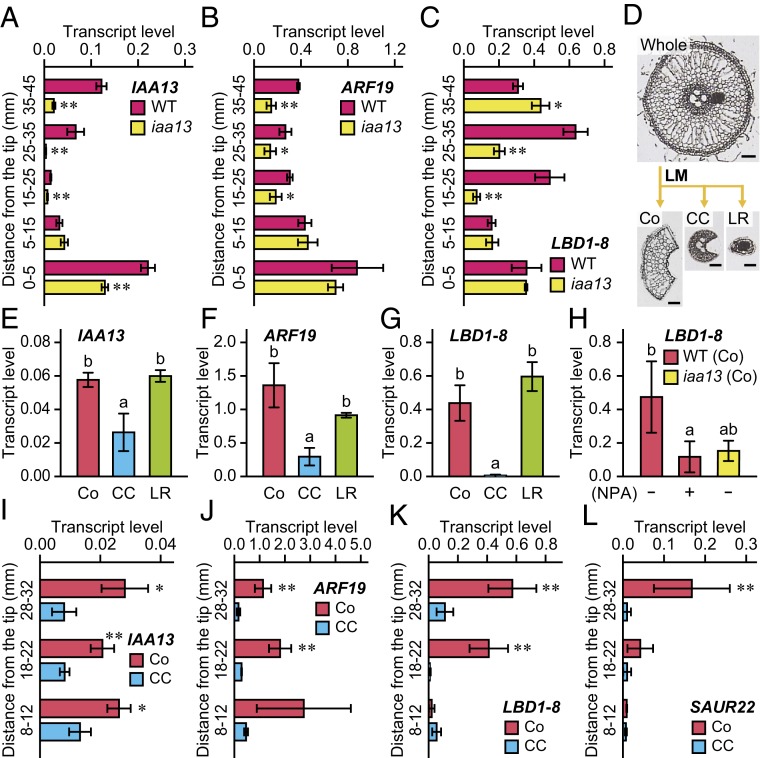
To understand where IAA13 functions in rice roots, the transcript level of IAA13 was analyzed by quantitative reverse-transcription PCR (qRT-PCR). In the WT, the transcript level was highest at 0 to 5 mm from the root tips, gradually decreased to a minimum at 15 to 25 mm, and then gradually increased toward the basal part of the roots (Fig. 2A). In iaa13, the transcript level of the mutated IAA13 was highest at 0 to 5 mm and then gradually decreased toward the basal part of the roots (Fig. 2A). Therefore, iaa13 might function mainly in the apical part of iaa13 roots, and this might cause the gradual increase of CA formation toward the basal part of the roots (Fig. 1B). If this were the case, then CA formation should be completely suppressed by long-term NPA treatment. To test this hypothesis, 20-d-old WT and iaa13 seedlings were treated with 0.5 µM NPA for 72 h. During this NPA treatment, elongation of adventitious roots of the WT and iaa13 were comparable (∼60 mm; SI Appendix, Fig. S2A), and the treatment blocked CA formation almost completely (SI Appendix, Fig. S2 B and C).
Fig. 2.
Expression analysis of auxin-related genes in roots of the WT and iaa13 mutant. (A–G) Twenty-day-old aerobically grown WT and iaa13 seedlings were further grown under aerated conditions for 48 h. Shown are relative transcript levels of IAA13 (A), ARF19 (B), and LBD1-8 (C) at 0 to 5, 5 to 15, 15 to 25, 25 to 35, and 35 to 45 mm from the tips of adventitious roots of the WT and iaa13. (D) Isolation of the cortex (Co), central cylinder (CC), and lateral root (LR) by laser microdissection (LM). (Scale bars, 50 μm.) Relative transcript levels of IAA13 (E), ARF19 (F), and LBD1-8 (G) in the cortex, central cylinder, and LR at 18 to 22 mm of the WT roots. (H) Relative transcript levels of LBD1-8 in the cortex at 18 to 22 mm of the WT roots under aerated conditions with or without 0.5 µM NPA and of iaa13 without NPA for 48 h. (I–L) Twenty-day-old aerobically grown WT seedlings were further grown under aerated conditions for 48 h. Shown are relative transcript levels of IAA13 (I), ARF19 (J), LBD1-8 (K), and SAUR22 (L) in the cortex and central cylinder at 8 to 12, 18 to 22, and 28 to 32 mm. The gene encoding transcription initiation factor IIE served as a control. Values are mean ± SD (n = 3). Significant differences between the genotypes (A–C) or tissues (I–L) at **P < 0.01 and *P < 0.05 (2-sample t test). Different lowercase letters denote significant differences among the tissues (E–G), or genotypes and conditions (H) (P < 0.05, one-way ANOVA followed by Tukey’s test for multiple comparisons).
Transcription of LBD1-8 Is Regulated through AUX/IAA-Mediated Signaling.
To identify the interactor of IAA13, transcript levels of 25 ARF genes in adventitious roots of rice were analyzed using microarray data obtained in our previous study (28). Among these 25 genes, ARF19 encoding an activator-type ARF had the highest signal intensity (SI Appendix, Fig. S3). Interestingly, the transcript accumulation pattern of ARF19 was similar to that of IAA13 (Fig. 2 A and B).
To identify genes regulated by AUX/IAA-mediated signaling, transcript levels at 15 to 25 mm were compared between the WT and iaa13 and also between the WT without NPA (WT –NPA) and with 0.5 µM NPA (WT +NPA) by microarray analyses. Among the genes in the array, 432 genes had signal intensities at least 2-fold higher (P < 0.05) in the WT and WT –NPA than in the iaa13 and WT +NPA (SI Appendix, Fig. S4). In these genes, we focused on 2 LBD genes (LBD1-8 and LBD5-3), because some LBD genes are regulated by ARFs (25, 29). At 15 to 25 mm, the transcript levels of LBD1-8 and LBD5-3 were strongly increased in the WT roots, whereas they were significantly lower in iaa13 (Fig. 2C and SI Appendix, Fig. S5A). The absolute transcript level of LBD1-8 was ∼5-fold higher than that of LBD5-3 (SI Appendix, Fig. S5B).
The tissue-specific transcript levels of IAA13, ARF19, and 2 LBD genes were investigated using RNA obtained from cortex (Co), central cylinder (CC), and LR collected from roots of the WT at 18 to 22 mm by laser microdissection (Fig. 2D). The transcript levels of IAA13, ARF19, and LBD1-8 were higher in the cortex and LR than in the central cylinder (Fig. 2 E–G), whereas the transcript level of LBD5-3 was much higher in the LR than in the cortex and central cylinder (SI Appendix, Fig. S5C). In the WT, 0.5 µM NPA reduced the transcript level of LBD1-8 in the cortex (Fig. 2H). The transcript level of LBD1-8 in the cortex was lower in iaa13 than in the WT in the absence of NPA (Fig. 2H). To understand the longitudinal pattern of LBD1-8 expression in the cortex, we compared the transcript level of LBD1-8 with the levels of IAA13 and ARF19 in the cortex and central cylinder at 8 to 12, 18 to 22, and 28 to 32 mm (Fig. 2 I–K). The latter 2 levels were higher in the cortex than in the central cylinder (Fig. 2 I and J).
Interestingly, the transcript level of LBD1-8 in the cortex at 8 to 12 mm, where little aerenchyma was detected (at 10 mm) (Fig. 1B), was comparable to that in the central cylinder but gradually increased toward the basal part of the roots (Fig. 2K). The transcript level of LBD5-3 in the cortex also gradually increased toward the basal part (SI Appendix, Fig. S5D). In addition, the transcript accumulation pattern of the auxin-responsive SMALL AUXIN UP RNA (SAUR22) gene (SI Appendix, Fig. S4) was similar to that of LBD1-8 (Fig. 2 K and L). Taken together, these results are consistent with the possibility that LBD1-8 and LBD5-3 are transcriptionally regulated through IAA13- and ARF19-dependent signaling in the cortex.
ARF19 Interacts with IAA13 and Binds to AuxRE on the LBD1-8 Promoter.
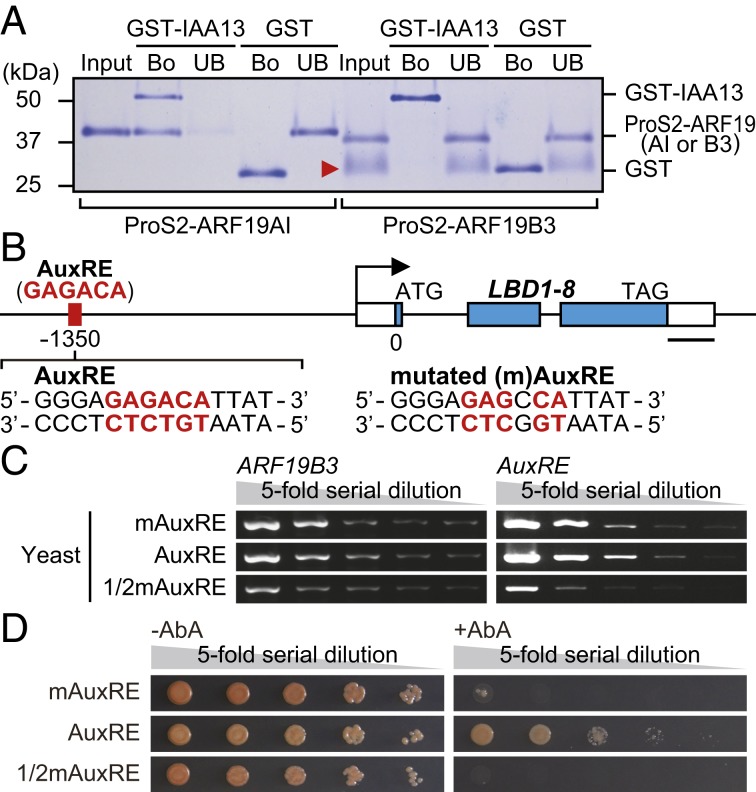
The transcript accumulation patterns of ARF19 and IAA13 were well associated with each other (Fig. 2 E and F), suggesting that auxin signaling in rice roots is regulated through the interaction between ARF19 and IAA13. To test this hypothesis, GST-tagged IAA13, AUX/IAA domains III and IV of ARF19 (ARF19AI), and the B3 DNA-binding domain of ARF19 (ARF19B3) were expressed in Escherichia coli, and in vitro interactions between IAA13 and ARF19AI and between IAA13 and ARF19B3 were tested by pull-down assays. As a result, in vitro binding of GST-IAA13 to ARF19AI, but not to ARF19B3, was detected (Fig. 3A).
Fig. 3.
Analysis of ARF19- and IAA13-mediated transcriptional regulation of LBD1-8. (A) Pull-down assays to analyze the interaction between ARF19 and IAA13. Immobilized GST-IAA13 and GST were incubated with ProS2-ARF19 (AI, AUX/IAA domain; B3, B3 DNA-binding domain). Bound (Bo) and unbound (UB) proteins were analyzed by SDS/PAGE and Coomassie blue staining. Molecular mass: GST-IAA13, 54.5 kDa; GST, 29.5 kDa; ProS2-ARF19AI, 38.6 kDa; ProS2-ARF19B3, 37.5 kDa. The red arrowhead indicates the degraded protein. (B) AuxRE cis-element on the LBD1-8 promoter. Sequences of the AuxRE and mutated mAuxRE used for the yeast one-hybrid assays are shown. (Scale bar, 200 bp.) (C) PCR analyses of ARF19B3 on the prey vector and AuxRE on the bait vector. Yeast cultures (mAuxRE and AuxRE, Ab600 = 0.5; 1/2mAuxRE, Ab600 = 0.25) were serially diluted (1:5) and used for the PCR analyses. (D) Yeast one-hybrid assays to analyze the binding activity of ARF19B3 to the AuxRE or mAuxRE. The serially diluted yeast cultures were spotted onto SD/−Ura plates with (Right) or without (Left) 100 mg L−1 aureobasidin A (AbA), and the colonies were photographed at 3 d after the spotting.
The LBD1-8 promoter has a single auxin response element (AuxRE) in the reverse orientation (GAGACA) (Fig. 3B). Thus, we tested the binding of ARF19 to this element by yeast one-hybrid assays. The DNA fragment from −1,357 to −1,344 bp of the LBD1-8 promoter, which contains the AuxRE (TGTCTC) with 4-bp flanking sequences (Fig. 3B), was inserted upstream of the Aureobasidin A (AbA) resistance gene (AUR1-C), and another was similarly inserted with the mutated mAuxRE (TGGCTC). Yeast strains containing the activation domain of yeast GAL4 fused with ARF19B3 and the AuxRE- or mAuxRE-inserted AUR1-C cassette were generated. The yeast cultures were serially diluted, and the amount of yeast in each dilution was checked by PCR analyses for the ARF19B3 and AuxRE regions (Fig. 3C). Subsequently, binding of ARF19B3 to the AuxRE or mAuxRE was tested on uracil-minus synthetic dextrose (SD/−Ura) medium containing 0 or 100 mg L−1 AbA (Fig. 3D). Because the yeast strain containing the mAuxRE grew faster than the yeast strain containing the AuxRE, 2-fold diluted mAuxRE (1/2mAuxRE) was also tested (Fig. 3 C and D). In the presence of AbA, the yeast strain containing ARF19B3 with the AuxRE grew faster than the strain with the mAuxRE (Fig. 3D).
LBD1-8 Stimulates CA and LR Formation in Rice Roots.
To evaluate the role of LBD1-8, expression of LBD1-8 was restored in the iaa13 background. We used the IAA13 promoter (ProIAA13::LBD1-8) by which LBD1-8 expression can be restored when the dominant-negative iaa13 suppressed LBD1-8 expression. Regenerated shoots of the ProIAA13::LBD1-8 (pIL1 to 5) and the vector control (ProIAA13::GUS) were grown to obtain the T1 progenies. Unexpectedly, pIL3 to 5 and the vector control grew very slowly and were sterile (SI Appendix, Fig. S6).
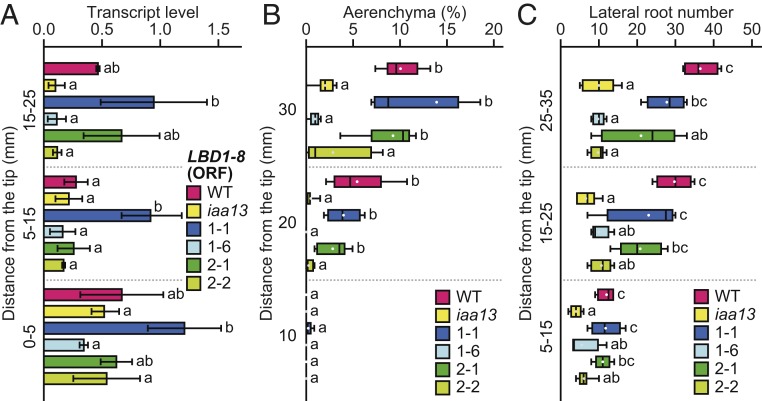
To check the segregation of the ProIAA13::LBD1-8 T-DNA region in the genomic DNA of the pIL1 and 2 (T1 generation), the relative copy numbers were evaluated by qRT-PCR (SI Appendix, Fig. S7A). The copy numbers of pIL1–1, 1–2, and 2–1 were ∼2-fold higher than those in pIL1–3 to 1–5, and pIL1–6 and 2–2 had no T-DNA copies (SI Appendix, Fig. S7A). To evaluate the levels of LBD1-8 expression in the pIL lines, qRT-PCR primers were designed to amplify the ORF reflecting the transcriptions of the endogenous LBD1-8 gene and LBD1-8 transgene. The transcript levels of LBD1-8 (ORF) at 15 to 25 mm in pIL1–1, 1–2, and 2–1 were ∼2-fold higher than the level in the WT, but the levels in pIL1–3 to 1–5 were comparable to the level in the WT, and the levels of pIL1–6 and 2–2 were comparable to the level in iaa13 (SI Appendix, Fig. S7B). These results suggest that pIL1–1 and 2–1 are homozygous, whereas pIL1–6 and 2–2 are the segregated WT. Indeed, transcriptions of the LBD1-8 transgene were not detected in pIL1–6 and 2–2 (SI Appendix, Fig. S8A).
The transcript levels of LBD1-8 (ORF) were significantly higher in pIL1–1 than in iaa13 at all positions of the roots, whereas the levels in pIL2–1 at 0 to 5 mm and 5 to 15 mm were comparable to those in iaa13 and gradually increased toward the basal part of the roots (Fig. 4A). The percentages of aerenchyma at 20 and 30 mm in pIL1–1 and 2–1 were comparable to those in the WT (Fig. 4B). In contrast, CA formation in pIL1–6 and 2–2 was comparable with that in iaa13 (Fig. 4B). The numbers of LRs at 5 to 15 mm in pIL1–1 and 2–1 were comparable to the number in the WT, whereas the numbers of LRs at 15 to 25 mm and 25 to 35 mm in pIL1–1 and 2–1 were intermediate between the numbers in the WT and iaa13 (Fig. 4C). The transcript levels of LBD5-3 at 15 to 25 mm in pIL1–1 and 2–1 were comparable with the level in iaa13 (SI Appendix, Fig. S8B). Because the transcript level of LBD5-3 was ∼4-fold higher in the LR than in the cortex (SI Appendix, Fig. S5C), we cannot rule out the possibility that the smaller average LR numbers in pIL1–1 and 2–1 than in the WT (Fig. 4C) are caused by the lower LBD5-3 expression in the pIL lines (SI Appendix, Fig. S8B).
Fig. 4.
Phenotyping of LBD1-8 transgenic plants. (A–C) Twenty-day-old aerobically grown WT, iaa13, and ProIAA13::LBD1-8 transgenic lines were further grown under aerated conditions for 48 h. (A) Relative transcript levels of the LBD1-8 (ORF) at 0 to 5, 5 to 15, and 15 to 25 mm from the tips of adventitious roots. The gene encoding transcription initiation factor IIE served as a control. Values are mean ± SD (n = 3). (B and C) Percentages of aerenchyma at 10, 20, and 30 mm (B), and numbers of lateral roots at 5 to 15, 15 to 25, and 25 to 35 mm (C). Boxplots show the median (horizontal lines), 25th to 75th percentiles (edges of the boxes), minimum to maximum values (edges of the whiskers), and mean values (dots in the boxes) (n = 4 to 6). Different lowercase letters denote significant differences among the genotypes (P < 0.05, one-way ANOVA followed by Tukey’s test for multiple comparisons).
Discussion
In this study, we found that aerenchyma formation under aerobic conditions (i.e., CA formation) was reduced in adventitious roots of the rice iaa13 mutant (Fig. 1B), which has a single amino acid substitution in the degradation domain of IAA13 (Fig. 5A and SI Appendix, Fig. S1A). We also found that the auxin transport inhibitor NPA almost completely suppressed CA formation (Fig. 1E and SI Appendix, Fig. S2C), and that the natural auxin IAA partly restored CA formation in the presence of NPA (Fig. 1H). Moreover, a high concentration of IAA suppressed CA formation in the presence of NPA (Fig. 1H), suggesting that there is an optimal auxin level for the initiation of CA formation in rice roots. Although ethylene and ROS are involved in inducible aerenchyma (IA) formation under oxygen-deficient conditions (8, 9), aerenchyma forms to some extent in rice roots even when ethylene- or ROS-mediated IA formation is blocked (14, 15). The present results demonstrate that another phytohormone, auxin, is a key regulator of CA formation in rice roots.
Fig. 5.
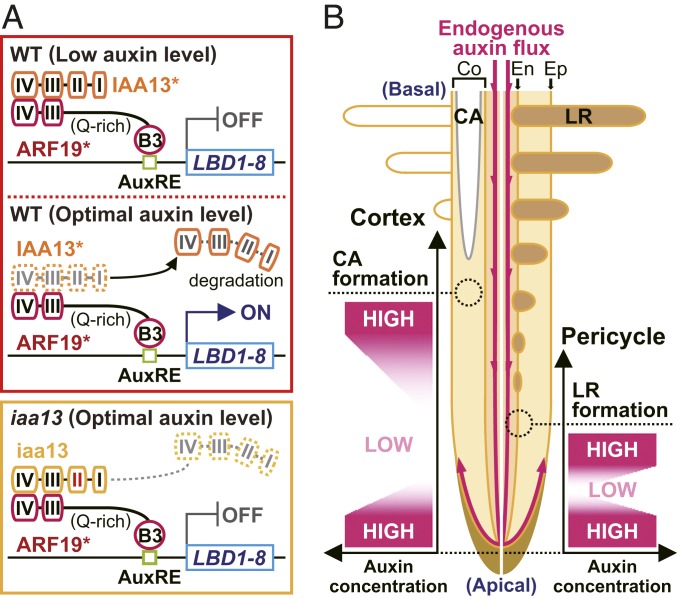
Model of constitutive aerenchyma and lateral root formation. (A) Transcriptional regulation of LBD1-8 in roots of the WT or iaa13 mutant. At low auxin levels, IAA13 or 1 of its homologs (indicated by IAA13* in the figure) binds to ARF19 or 1 of its homologs (indicated by ARF19*) through AUX/IAA domains III and IV, repressing the activation of LBD1-8 transcription. At optimal auxin levels, IAA13 is degraded by proteolysis, and ARF19 activates LBD1-8 transcription. In iaa13, the mutated IAA13 (iaa13) prevents its own degradation. (B) Possible auxin concentration gradients in the cortical cells and pericycle cells where CA and LR are formed, respectively. Co, cortex; En, endodermis; Ep, epidermis.
We identified ARF19 as an interactor of IAA13 and identified LBD1-8 as a target of ARF19 (Figs. 2 and 3). ARF19 is classified into the same clade as ARF7 and ARF19 in Arabidopsis (30), both of which interact with IAA14 (23), and which is the closest homolog to rice IAA13 (27). Indeed, rice IAA13 interacts with ARF19 through AUX/IAA domains III and IV (Figs. 3A and 5A). We also found that the B3 DNA-binding domain of ARF19 binds to the AuxRE on the LBD1-8 promoter (Figs. 3D and 5A). This is also consistent with the finding that ARF7 binds to the AuxREs on the LBD16 and LBD29 promoters in Arabidopsis (25). Several of our findings suggest the involvement of IAA13- and ARF19-mediated transcriptional regulation of LBD1-8 in CA formation. These include (i) IAA13, ARF19, and LBD1-8 were highly expressed in the cortex (Fig. 2 E–G); (ii) LBD1-8 expression was lower in the cortex of iaa13 and the NPA-treated WT than in the cortex of the NPA-untreated WT (Fig. 2H); and (iii) restoration of LBD1-8 expression in the iaa13 background recovered CA formation (Fig. 4 A and B).
The number of LRs was decreased in iaa13 (Fig. 1D) (26). Similarly, LR formation is defective in a rice mutant containing a dominant-negative IAA11 gene (31), suggesting that IAA13 and other IAAs coordinately regulate auxin signaling in rice roots. In Arabidopsis, LR formation is almost completely suppressed in the arf7arf19 double knockout mutant (25). Therefore, the function of rice ARF19 in LR formation is conserved in its closest homologs, (i.e., ARF7 and ARF19) in Arabidopsis. In agreement with the idea that rice ARF19 has a role in LR formation, ARF19 deficiency decreases LR formation to some extent but not completely, because of the redundant function of homologous ARFs (32). Multiple mutant analyses are required to fully understand the functions of ARFs in LR formation as well as in CA formation. LBD1-8 is not highly homologous to Arabidopsis LBD16, LBD18, and LBD29 (33), all of which have roles in LR formation (25, 34). The finding that restoration of LBD1-8 expression partly recovered LR formation in iaa13 (Fig. 4 A and C) indicates that LBD1-8 also functions in LR formation in rice roots. We cannot rule out the possibility that LBD5-3, which is highly expressed in LRs (SI Appendix, Fig. S5C), also has a role in LR formation.
The dominant-negative effect of iaa13 at the apical part (Fig. 2A) suppressed LR formation along iaa13 roots (Fig. 1D), suggesting that LR formation is initiated predominantly at the apical part of rice roots. In Arabidopsis, LR formation is initiated at the end of the elongation zone of primary roots, where the auxin level is lower than that in the most apical part of the roots (35). When LR cap cells reach the start of the elongation zone, they undergo cyclic PCD, which provides a local, cyclic auxin source that in turn stimulates cyclic LR formation (36). Taken together, these observations suggest that a reduction and subsequent increase in the auxin level in pericycle cells are required for the initiation of LR formation (Fig. 5B). Indeed, in a rice mutant defective in the auxin efflux carrier protein pin-formed 2 (PIN2), LR initiation is retarded by the elevated auxin level in the apical part of the roots (37).
In the cortical cells, both NPA and IAA suppressed CA formation (Fig. 1 E and G), suggesting that a local concentration gradient of auxin is also essential for CA formation in rice roots. The expression level of LBD1-8 was reduced at 5 to 15 mm (Fig. 2C), while the expression levels of LBD1-8 and auxin-responsive gene SAUR22 in the cortex gradually increased toward the basal part of the roots (Fig. 2 K and L). In Arabidopsis roots, when the auxin level in the transition zone reaches a minimum, it triggers root cells to switch from cell division to cell differentiation (38). Similarly, a reduction and subsequent increase of the auxin level in the cortical cells might be required for CA formation. On the other hand, increased CA formation and increased LBD1-8 expression in the cortex were both detected at ∼20 mm from the root tips (Figs. 1B and 2K), indicating that the local concentration gradients of auxin in the cortical cells and pericycle cells are regulated differently at the beginning of CA and LR formation in rice roots (Fig. 5B).
Based on the foregoing results, we propose a model for auxin-dependent CA and LR development in rice roots (Fig. 5). IAA13- and ARF19-dependent signaling stimulates the transcription of LBD1-8, which then allows the auxin responses, including the formation of CA and LR (Fig. 5A). At present, we cannot rule out the possibility that other IAAs and ARFs are involved in regulating these processes. A reduction and subsequent increase of auxin levels are required for CA formation in cortical cells and LR formation in pericycle cells (Fig. 5B). LR formation, which is stimulated by cyclic PCD of lateral root cap cells in Arabidopsis (36), is initiated at the more apical part of rice roots compared with CA formation. A possible auxin source in cortical cells is auxin that diffuses from the tips of LR primordia (39), which emerge before CA formation. Further studies are needed to identify the auxin source in cortical cells that initiates CA formation in rice roots.
Under excess-water, low-water, or nutrient-deficient conditions, aerenchyma formation reduces the respiratory costs and nutrient requirements of roots (3, 4), but it can also reduce the radial transport of water and nutrients (4). In view of this trade-off, it might be disadvantageous for plants to form CA at the most apical part of the roots, where water and nutrients are efficiently taken up (40). On the other hand, having more LRs at the apical part of the primary or adventitious (crown) root would improve the uptake of water and nutrients from the soil. In conclusion, by allowing AUX/IAA and ARF-dependent signaling to control the positions of CA and LR development independently, rice can better adapt to the soil environment.
Materials and Methods
Detailed information on plant materials and growth conditions, chemical treatments, Feulgen staining, qRT-PCR and microarray analyses, laser microdissection, protein expression and purification, pull-down assays, yeast one-hybrid assays, generation of transgenic rice plants, and statistical analyses are provided in SI Appendix, Materials and Methods. A complete set of microarray data has been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE130131.
Supplementary Material
Acknowledgments
We thank Y. Nagamura for assistance with microarray analyses; Y. Kitomi for providing the ProIAA13::iaa13 vector; H. Ichikawa for providing the pSMAHdN636L-GateA binary vector; M. Fujimoto for advice on yeast one-hybrid experiment; and T. D. Colmer, O. Pedersen, A. I. Marik, H. Takahashi, S. Nishiuchi, and K. Watanabe for stimulating discussions. This work was partly supported by the Japan Society for the Promotion of Science (KAKENHI Grant 18H02175) (to M.N.) and the Japan Science and Technology Agency PRESTO Grants JPMJPR17Q8 (to T.Y.) and JPMJPR15Q3 (to Y.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: A complete set of microarray data has been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE130131).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907181116/-/DCSupplemental.
References
- 1.Jackson M. B., Armstrong W., Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol. 1, 274–287 (1999). [Google Scholar]
- 2.Justin S. H. F. W., Armstrong W., The anatomical characteristics of roots and plant response to soil flooding. New Phytol. 106, 465–495 (1987). [Google Scholar]
- 3.Colmer T. D., Long-distance transport of gases in plants: A perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ. 26, 17–36 (2003). [Google Scholar]
- 4.Lynch J. P., Root phenes that reduce the metabolic costs of soil exploration: Opportunities for 21st century agriculture. Plant Cell Environ. 38, 1775–1784 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Jackson M. B., Fenning T. M., Jenkins W., Aerenchyma (gas-space) formation in adventitious roots of rice (Oryza sativa L.) is not controlled by ethylene or small partial pressures of oxygen. J. Exp. Bot. 36, 1566–1572 (1985). [Google Scholar]
- 6.Justin S. H. F. W., Armstrong W., Evidence for the involvement of ethene in aerenchyma formation in adventitious roots of rice (Oryza sativa L.). New Phytol. 118, 49–62 (1991). [Google Scholar]
- 7.Colmer T. D., Voesenek L. A. C. J., Flooding tolerance: Suites of plant traits in variable environments. Funct. Plant Biol. 36, 665–681 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Voesenek L. A. C. J., Bailey-Serres J., Flood adaptive traits and processes: An overview. New Phytol. 206, 57–73 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Yamauchi T., Colmer T. D., Pedersen O., Nakazono M., Regulation of root traits for internal aeration and tolerance to soil waterlogging-flooding stress. Plant Physiol. 176, 1118–1130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider H. M., et al. , Root cortical senescence decreases root respiration, nutrient content and radial water and nutrient transport in barley. Plant Cell Environ. 40, 1392–1408 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Yamauchi T., Abe F., Tsutsumi N., Nakazono M., Root cortex provides a venue for gas-space formation and is essential for plant adaptation to waterlogging. Front. Plant Sci. 10, 259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mustroph A., Steffens B., Sasidharan R., Signalling interactions in flooding tolerance. Annu. Plant Rev. 1, 1–42 (2018). [Google Scholar]
- 13.Sasidharan R., et al. , Signal dynamics and interactions during flooding stress. Plant Physiol. 176, 1106–1117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamauchi T., et al. , An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 29, 775–790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamauchi T., et al. , Ethylene-dependent aerenchyma formation in adventitious roots is regulated differently in rice and maize. Plant Cell Environ. 39, 2145–2157 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Ulmasov T., Hagen G., Guilfoyle T. J., ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Ulmasov T., Hagen G., Guilfoyle T. J., Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. U.S.A. 96, 5844–5849 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouse D., Mackay P., Stirnberg P., Estelle M., Leyser O., Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Tiwari S. B., Hagen G., Guilfoyle T. J., Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray W. M., Kepinski S., Rouse D., Leyser O., Estelle M., Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Kim J., Harter K., Theologis A., Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 11786–11791 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coudert Y., Périn C., Courtois B., Khong N. G., Gantet P., Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15, 219–226 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Fukaki H., Nakao Y., Okushima Y., Theologis A., Tasaka M., Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44, 382–395 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Majer C., Hochholdinger F., Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M., ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitomi Y., Inahashi H., Takehisa H., Sato Y., Inukai Y., OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 190, 116–122 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Jain M., et al. , Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6, 47–59 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kulichikhin K., Yamauchi T., Watanabe K., Nakazono M., Biochemical and molecular characterization of rice (Oryza sativa L.) roots forming a barrier to radial oxygen loss. Plant Cell Environ. 37, 2406–2420 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Inukai Y., et al. , Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17, 1387–1396 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D., et al. , Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Zhu Z. X., et al. , A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol. Plant 5, 154–161 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Zhang S., et al. , The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Coudert Y., Dievart A., Droc G., Gantet P., ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol. Biol. Evol. 30, 569–572 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Lee H. W., Kim N. Y., Lee D. J., Kim J., LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151, 1377–1389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubrovsky J. G., et al. , Auxin minimum defines a developmental window for lateral root initiation. New Phytol. 191, 970–983 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Xuan W., et al. , Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351, 384–387 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Inahashi H., et al. , OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant. 164, 216–225 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Di Mambro R., et al. , Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 114, E7641–E7649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Q., et al. , A MAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral root emergence. Nat. Plants 5, 414–423 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Geldner N., The endodermis. Annu. Rev. Plant Biol. 64, 531–558 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.