Summary
In the rice blast fungus Magnaporthe oryzae, the cell wall integrity (CWI) signalling pathway governs cell wall changes in response to external cues and normal CWI signalling is critical for appressorium function and pathogenicity. We previously characterized the mitogen-activated protein kinase (MAPK) kinase MoMkk1 as an integral component of the CWI pathway. Using the affinity purification approach, we have identified MoMkk1-interacting MoPpe1 as a homologue of Saccharomyces cerevisiae serine/threonine protein phosphatase Sit4/Ppe1. We found that MoPpe1 is required for vegetative growth, conidiation and full virulence. In addition, we found that MoPpe1 interacts with MoSap1, a protein with functions similar to MoPpe1. Intriguingly, we found that MoPpe1–MoSap1 interaction is related to CWI and target of rapamycin (TOR) pathways. We presented evidence suggesting that MoPpe1 and MoSap1 function as an adaptor complex linking CWI and TOR signalling and that the activation of the TOR pathway leads to suppression of CWI signalling, resulting in defects in appressorium function and pathogenicity. Taken together, our studies not only reveal important functions of MoMkk1–MoPpe1–MoSap1 interactions in growth and pathogenicity of the blast fungus, but also highlight the complexity of regulatory networks involving conserved yet novel regulatory mechanisms of CWI and TOR signalling.
Introduction
Magnaporthe oryzae causes devastating rice blast worldwide and is also a widely adopted model organism for studying the plant–pathogen interaction (Wilson and Talbot, 2009; Zhang et al., 2016). In Magnaporthe oryzae signalling pathways, including the cAMP-PKA pathway and MoPmk1 and MoMps1 MAP kinase signalling pathways, sense and transduce extracellular signals to regulate growth, differentiation and pathogenicity (Adachi and Hamer, 1998; Marroquin-Guzman and Wilson, 2015; Li et al., 2017c). The cell wall integrity (CWI) MAP kinase pathway, consisting of MoMck1, MoMkk1 and MoMps1, is also important for appressorium function and virulence (Jeon et al., 2008; Yin et al., 2016). Moreover, the highly conserved target of rapamycin (TOR) pathway controls vegetative growth and differentiation of fungus by regulating metabolic processes in response to various nitrogen sources (Beck and Hall, 1999; Yu et al., 2014). Elucidation of crosstalk among these important pathways, therefore, is important to study virulence mechanisms of the blast fungus.
We have previously shown that the high-affinity phosphodiesterase MoPdeH is involved in modulating intracellular cAMP levels and MoPdeH directly interacts with MoMck1 (Yin et al., 2016). Intriguingly, overexpression of MoMCK1 in a ΔMopdeH mutant partially suppressed the defects in hyphal autolysis and pathogenicity. This finding suggested that MoPdeH links cAMP-PKA and CWI signalling (Liu et al., 2016; Yin et al., 2016). Recently, a study demonstrated that rapamycin inhibited TOR signalling enhanced appressorium formation in a ΔMocpka mutant (Marroquin-Guzman and Wilson, 2015), suggesting additional linkage mechanisms present in this fungus.
In this study, we have provided evidence to support a mechanism that the MoPpe1–MoSap1 complex functions as adaptor to mediate CWI and TOR signalling. In light of crosstalk identified between the TOR and CWI pathways of Saccharomyces. cerevisiae based on the evidence that Rho1 GTPase binds directly to the TOR complex and rapamycin induces a rapid activation of Rho1 (Yan et al., 2012a,b), our studies presented a different mechanism that the protein phosphatase MoPpe1 binds with MoMkk1 to positively regulate CWI. We also showed that the ΔMoppe1 and ΔMosap1 mutants were defective in the utilization of nonpreferred nutrients but with the increased utilization of preferred nutrients, which indicated a negative regulation of TOR signalling. Finally, we provided evidence to support the hypothesis that enhanced TOR signalling suppresses CWI resulting in impaired appressorium function and pathogenicity.
Results
The identification of MoPPE1 in M. oryzae
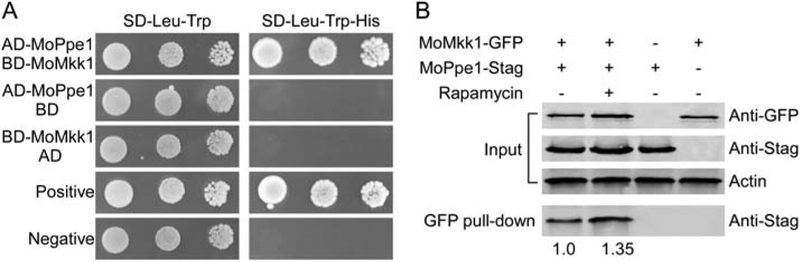
Previously, we found the MAP kinase kinase MoMkk1 as a component of the MAP kinase cascade that regulates CWI signalling and that MoMkk1 is required for pathogenicity in M. oryzae (Yin et al., 2016). To investigate the underlying functional mechanisms, we utilized the affinity purification approach to identify M. oryzae proteins that interact with MoMkk1. We introduced the MoMkk1–3xFLAG construct into the wild-type strain and verified the presence of an expected 60 kDa band by Western blot analysis. Following affinity purification, proteins bound to anti-FLAG M2 beads were eluted and analysed by mass spectrometry (MS; Supporting Information Table S1). From them, we identified the serine/threonine protein phosphatase MoPpe1 that shares amino acid sequence homologue to S. cerevisiae Sit4/Ppe1. The introduction of the MoPPE1 gene completely suppressed the growth defect of the Δsit4 yeast mutant at 30 °C and 37 °C (Supporting Information Fig. S1). In addition, the interaction between MoMkk1 and MoPpe1 was verified by yeast two-hybrid (Y2H) and co-immunoprecipitation (Co-IP) analyses. Furthermore, the presence of rapamycin also enhanced this interaction (Fig. 1).
Fig. 1.
MoMkk1 interacts with MoPpe1. A. Yeast two hybrid assay for the interaction between MoMkk1 and MoPpe1. The AD-MoPpe1 and BDMoMkk1 vectors were co-introduced into the yeast strain AH109, and the transformants were plated with serial dilutions of yeast cells on SDLeu-Trp for 3 days and on selective SD-Leu-Trp-His added with 2 mM 3-AT (3-amino-1,2,4-triazole) for 10 days. B. co-immunoprecipitation (Co-IP) assays. Following growth in liquid complete media (CM) for 48 h, mycelia were treated with or without 30 ng/ml rapamycin for 4 h and then harvested for protein extraction. Total proteins (input) of each strain were subjected to SDS-PAGE analysis and immunoblots were incubated with anti-Stag and anti-GFP antibodies. In addition, each protein sample was pulled down using anti-GFP agarose and further detected with the monoclonal anti-GFP antibody. Input samples were also incubated with the actin antibody as a reference. MoPpe1-Stag interacted with MoMkk1-GFP and this interaction was increased with the presence of rapamycin.
MoPpe1 is involved in vegetative growth and conidiation
To test functions of the interaction between MoMkk1 and MoPpe1, we first characterized MoPpe1 by generating the ΔMoppe1 mutants (Supporting Information Fig. S2). Since the two independent mutants exhibit similar phenotypes, only one was selected for further analysis. We found that the ΔMoppe1 mutant exhibited significantly growth reduction on complete medium (CM), minimal medium (MM), straw decoction and corn agar (SDC) and oatmeal medium (OM) after incubation at 28 °C for 7 days when compared with the wild-type strain Guy11 and the complemented strain (ΔMoppe1/MoPPE1; Supporting Information Fig. S3A and B).
Since conidia play a significant role in the infection cycle of M. oryzae, we examined conidiation and found that it was markedly reduced by 80% in the ΔMoppe1 mutant in comparison with the wild-type and complemented strains (Supporting Information Fig. S3C and D). We further examined the expression of genes essential for conidiation and found that the expression of MoCOM1, MoHOX2, MoCOS1 and MoSTUA were all reduced in the ΔMoppe1 mutant when compared with the wild-type strain (Kim et al., 2009; Nishimura et al., 2009; Zhou et al., 2009; Yang et al., 2010; Tang et al., 2015) (Supporting Information Fig. S4). Intriguingly, there was no obvious difference found between the mutant and the control strains in conidial morphology (Supporting Information Fig. S5).
MoPpe1 is required for host infection
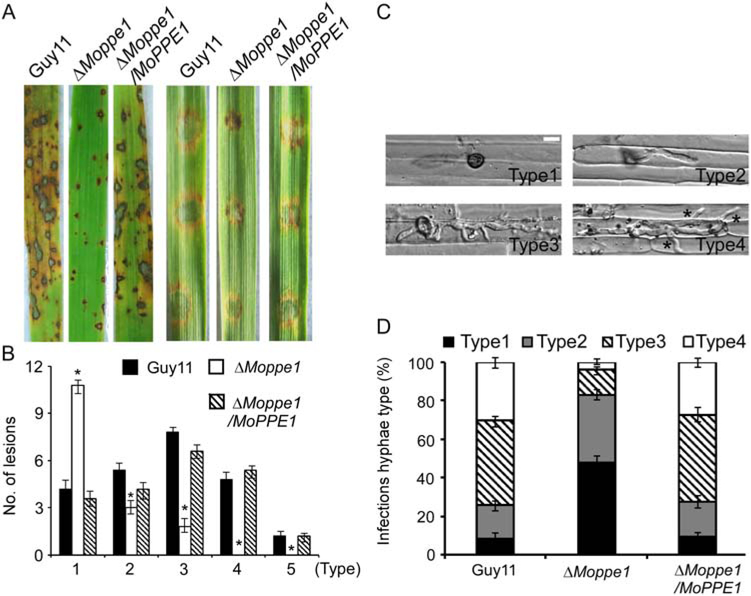
To examine the role of MoPpe1 in pathogenicity, conidial suspensions (5 × 104 conidia/ml) were sprayed onto susceptible 14 day-old rice seedlings (Oryza sativa cv. CO-39). The ΔMoppe1 mutant only produced small and restricted disease lesions 7 days postinoculation (dpi) compared with the wild-type and complemented strains (Fig. 2A). Lesion type scoring assay (Zhong et al., 2016) showed that lesions produced by the ΔMoppe1 mutant were mostly at Types 1 and 2, with few Type 3 and none Type 4 or Type 5, which was in sharp contrast to the lesions produced by Guy11 and the complemented strains (Fig. 2A and B). Similar results were also observed on detached barley leaves with the restricted expansion of the ΔMoppe1 mutant (Fig. 2A).
Fig. 2.
MoPpe1 is important for full virulence. A. Two week-old rice seedlings (Oryza sativa cv. CO-39) were sprayed with 4 ml conidial suspension (5 × 104 conidia/ml) and examined at 7 dpi. Separated barley leaves were drop-inoculated with same conidial suspensions and examined at 5 dpi. Experiments were repeated for three times with similar results. B. Lesions were measured at 7 dpi, numbers within an area of 1.5 cm2 were counted, quantification of lesion types (0, no lesion; 1, pinhead-sized brown specks; 2, 1.5 mm brown spots; 3, 2–3 mm grey spots with brown margins; 4, many elliptical grey spots longer than 3 mm; 5, coalesced lesions infecting 50% or more of the leaf area. Error bars represent the standard deviations. Asterisks represent significant differences (Duncan’s new multiple range test, p < 0.01). C. Conidial suspensions of three different strains were injected into separate rice sheaths and observed at 36 h postinoculation (hpi) (Type 1, no penetration; Type2, only with a single invasive hypha (IH) without branches; Type 3, with 1–3 branches but restricted in one cell; Type 4, more than three branches and extended to neighbouring cell. Asterisks indicate IH extended to surrounding cells. Bar = 5 μm. D. Percentages of different types of infectious hyphae in rice cells. Sample size (n) =50. Error bars represent the standard deviations.
We also carried out rice sheath penetration assay to detect the penetration ability of appressoria. The ΔMoppe1 mutant produced normally appressoria (Supporting Information Table S3), however, the appressoria were not effective in the penetration of the epidermal cells (Fig. 2C) and the invasive hyphae displayed extremely poor growth at 36 h (Fig. 2D). Invasive hyphae type assay (Wang et al., 2017a) showed that those produced by the ΔMoppe1 mutant were mainly Type 1 (48%), in contrast to Type 3 and Type 4 (70%) produced by the control strains. The above data suggested that the virulence defect of the ΔMoppe1 result from restricted appressorium penetration and defective infectious hyphal growth.
Previously, studies in M. oryzae showed that the nuclear division is critical in appressorium function (Marroquin-Guzman et al., 2017). To further understand whether the deletion of MoPPE1 caused a nuclear division defect that may account for restricted invasion, a histone H1 protein fused to a red fluorescent protein (H1-RFP) construct was introduced into the wild-type strain and ΔMoppe1 mutant (Li et al., 2017a,b; Marroquin-Guzman et al., 2017). However, the nuclear number were of no significant difference between the ΔMoppe1 mutant and the wild-type strains at 0, 4, 12, 24 h and infection phase when observed, suggesting that MoPpe1 may be dispensable for nuclear division which was in coordination with cell growth (Supporting Information Fig. S6).
MoSap1 associates with MoPpe1 and exhibits similar functions
In S. cerevisiae, Sit4 interacts with four high-molecular-mass proteins known as Sit4-associated proteins (Saps) which are associated and functionally positive with the Sit4 phosphatase (Sutton et al., 1991; Luke et al., 1996; Kapitzky et al., 2010; Barreto et al., 2011). To examine if any such proteins also present in M. oryzae, we used a MoPpe1–3xFLAG construct and identified only one such protein we named MoSap1. MoSap1 was able to complement the defects of all four yeast Sap mutants (Supporting Information Fig. S1).
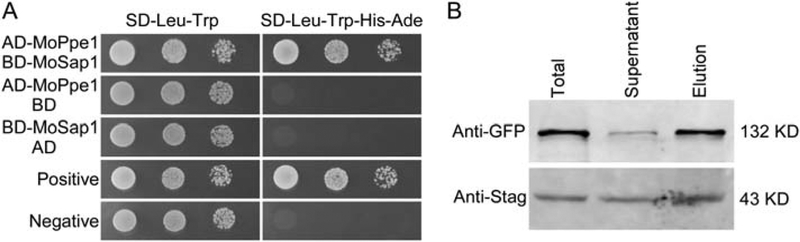
The interaction between MoPpe1 and MoSap1 was verified by Y2H and Co-IP assays (Fig. 3A and B). To investigate the functions of MoSap1 and MoPpe1– MoSap1 interaction, the ΔMosap1 single and ΔMoppe1ΔMosap1 double mutants were obtained (Supporting Information Fig. S2). Both mutants exhibited phenotypes similar to that of the ΔMoppe1 mutant, including significant reduction in hyphal growth, conidiation and the expression of MoCOM1, MoHOX2, MoCOS1 and MoSTUA (Supporting Information Figs S4, S7A and B), and no obvious difference in conidial morphology (Supporting Information Fig. S5). These findings indicated that MoPpe1 and MoSap1 may function as a complex important in hyphal growth and conidiation.
Fig. 3.
MoPpe1 interacts with MoSap1. A. Yeast two hybrid assays for the interaction between MoPpe1 and MoSap1. The AD-MoPpe1 and BDMoSap1 vectors were co-transformed into yeast strain AH109, the transformants were plated with serial dilutions of yeast cells on SD-Leu-Trp for 3 days and on selective SD-Leu-Trp-His-Ade for 10 days and photographed. B. Co-IP assays. Total proteins extracted from Guy11, MoPpe1-Stag and MoSap1-GFP strains. Proteins were eluted from anti-GFP agarose and detected with the anti-Stag and anti-GFP antibodies respectively. Proteins in the supernatant and total proteins served as controls.
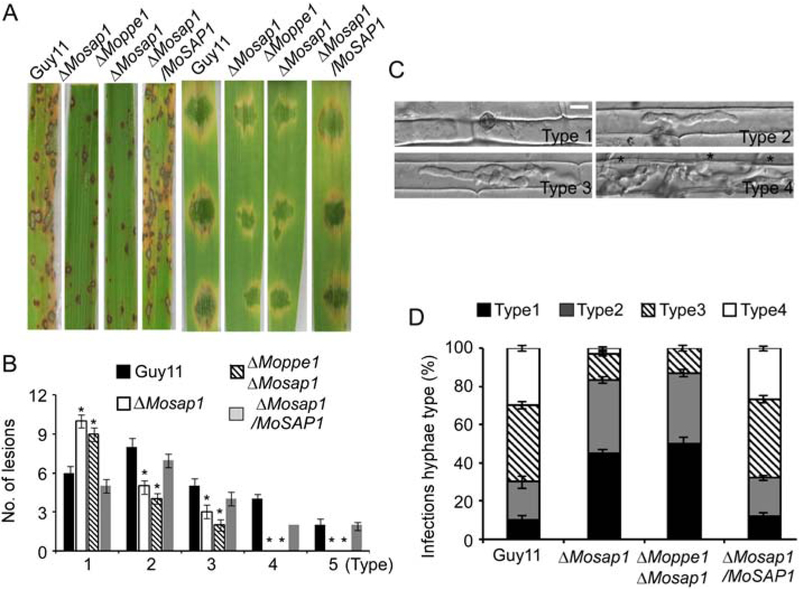
We continued with virulence assessment and found that both the ΔMosap1 single and ΔMoppe1 ΔMosap1 double mutants caused fewer and more restricted lesions than the wild-type strain (Fig. 4A and B). We found that about 45% and 50% of the appressoria formed by the ΔMosap1 single mutant and the ΔMoppe1 ΔMosap1 double mutant, respectively, failed to penetrate rice sheath cells and only about 15% of penetration sites extended to Types 3 and 4 lesions (Fig. 4C and D). These results implied that MoSap1 and MoPpe1– MoSap1 association play important roles in virulence by impacting host penetration and invasive hyphal growth.
Fig. 4.
ΔMosap1 and ΔMoppe1 ΔMosap1 double mutants exhibit reduced pathogenicity. A. Two-week-old rice seedlings were sprayed with a conidial suspension (5 × 104 conidia/ml) and examined at 7 dpi. Separated barley leaves were drop-inoculated with conidial suspensions and examined at 5 dpi. Experiments were repeated three times all showed similar results. B. Lesions were measured at 7 dpi, numbers within an area of 1.5 cm2 were counted, quantification of lesion types as described before and error bars represent the standard deviations. Asterisks denote statistical significances (p < 0.01). C. Conidial suspensions of three different strains were injected in separate rice sheaths at 36 hpi. Sample size, 50 (n = 50), asterisks indicate IH extended to surrounding cells. Bar = 5 μm. D. Percentages of different types of infectious hyphae in rice cells, error bars represent the standard deviations.
MoPpe1 and MoSap1 are important for scavenging host-derived reactive oxygen species
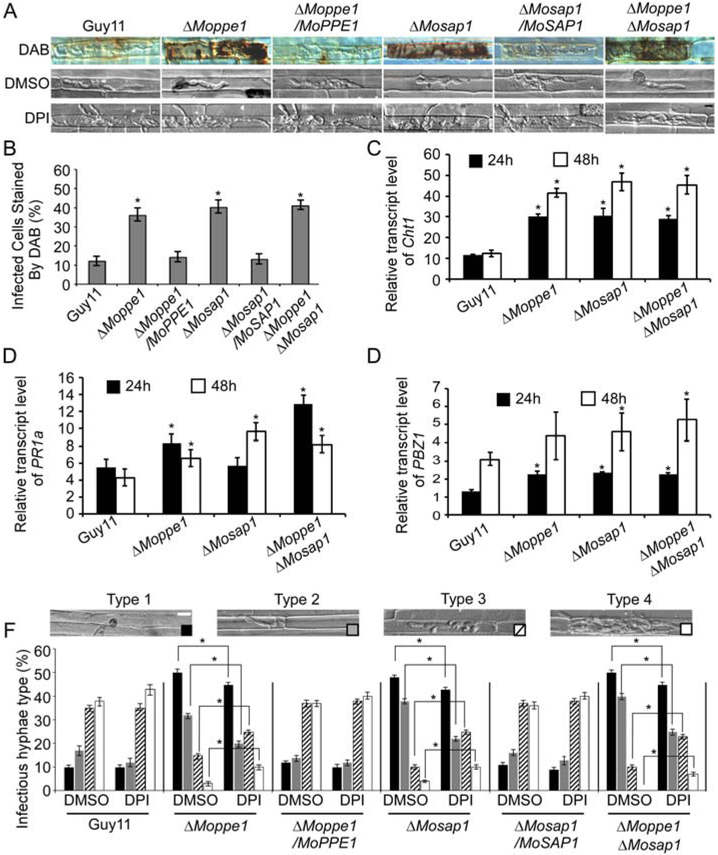
Reactive oxygen species (ROS) are antimicrobial compounds that also serve as stimulators and by-products of plant defence reactions (Guo et al., 2011; Huang et al., 2011). Since ΔMoppe1, ΔMosap1 and ΔMoppe1 ΔMosap1 mutants all caused restricted lesions and severely suppressed infectious hyphal growth, we tested the hypothesis that these defects may due to the high ROS levels generated by the host. We, therefore, examined ROS with DAB (3, 3′-diaminobenzidine) staining (Guo et al., 2010, 2011) and found scant stains in rice cells infected with Guy11. However, intense stains of reddish-brown precipitate surrounding the infection sites were found in cells infected with the ΔMoppe1, ΔMosap1 and ΔMoppe1 ΔMosap1 double mutant, suggesting that these mutants failed to scavenge ROS during infection phase (Fig. 5A and B). As diphenyleneiodonium (DPI) inhibits the activity of plant NADPH oxidases that are responsible for ROS generation (Bolwell et al., 1998; Grant et al., 2000; Zhang et al., 2009), we suppressed the NADPH oxidase activity with 0.5 μM DPI and found that the invasive hyphae of all three mutants produced improved extensions than DMSO controls (Fig. 5A and F).
Fig. 5.
MoPpe1 and MoSap1 are required for scavenging host-derived reactive oxygen species (ROS). A. Conidial suspensions of Guy11, three mutant strains and complement strains were injected in separate rice sheaths, treated with or without 0.5 μM diphenyleneiodonium (DPI). At 24 hpi, 3, 3′- diaminobenzidine (DAB) was used to stain the sheaths for 8 h and the results were observed at 36 h. B. Percentages of cells with infectious hyphae were dyed by DAB. Bar = 5 μm. Sample size, (n = 50). C–E. Transcriptions of CHT1, PR1a and PBZ1 in the infected host were assayed using quantitative real-time polymerase chain reaction (qRT-PCR). The histogram results were of the three independent biological experiments with three replicates (p < 0.05). F. Percentages of different types of infectious hyphae in rice cells treated with DPI or DMSO. The rice sheath treated with DMSO was used as a negative control. Bar = 5 μm. Error bars represent the standard deviations. Asterisks represent significant differences (p < 0.01).
Moreover, when used real-time quantitative PCR (qRT-PCR) to assess transcription levels of several pathogenesis-related (PR) genes, including CHT1, PR1a and PBZ1, we found that rice infected with the ΔMoppe1, ΔMosap1 and ΔMoppe1 ΔMosap1 mutants showed high expression levels than the control at 24 post-inoculation (hpi) and 48 hpi (Fig. 5C–E). Based on these observations, we proposed that MoPpe1 and MoSap1 are important for the suppression of host innate immunity during infection.
MoPpe1 and MoSap1 are required for CWI
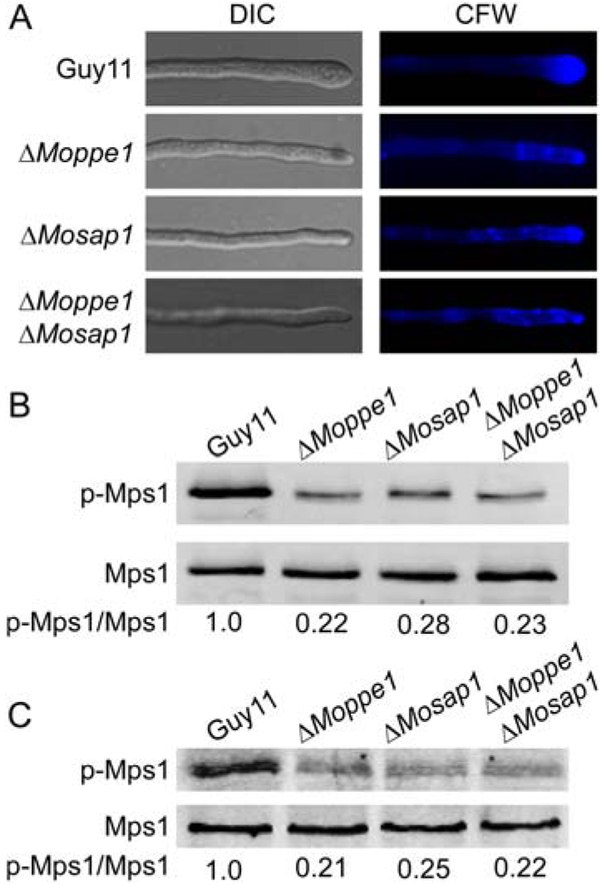
Based on the important cellular functions of MoPpe1 and MoSap1, we hypothesized that the MoMkk1 and MoPpe1 interaction could play a role in the regulation of the CWI pathway. To test this hypothesis, we first compared the stress response to various cell wall stressors. The ΔMoppe1 and ΔMosap1 mutants exhibited hypersensitivity to Calcofluor white (CFW) and Congo red (CR), in comparison with the wild-type strain (Supporting Information Figs S8 and S9). Compared with Guy11, the chitin distributions (CFW stain) of the mutants were apparently uneven, which were not restricted to growing apices (Fig. 6A). Meanwhile, with the available antibodies to MoMps1p (Liu et al., 2016; Yin et al., 2016), we found that the MoMps1 phosphorylation level was severely impaired in the ΔMoppe1, ΔMosap1 and ΔMoppe1 ΔMosap1 mutants at mycelium and appressoria phases (Fig. 6B and C). These results supported that MoPpe1 and MoSap1 are involved in regulating cell wall integrity pathway.
Fig. 6.
MoPpe1 and MoSap1 play important roles in cell wall integrity. A. Deletion of MoPPE1 and MoSAP1 altered the distribution of chitin in the cell wall. The wild-type strain Guy11 and mutant hyphae were stained with 10 μg/ml Calcofluor white (CFW) for 3 min without light before being photographed. The experiment was repeated several times with triple replications that yielded similar results. B. Phosphorylation of Mps1 in the Guy11, ΔMoppe1 mutant, ΔMosap1 mutant and ΔMoppe1 ΔMosap1 double mutant strains. Total protein was isolated from mycelia of the strains listed above. MoMps1 and phosphorylated MoMps1 protein was detected using the yeast Mpk1 (yN-19) antibody and phospho-p44/42 MAP kinase antibody respectively. Three independent experiments were replicated that showed similar results. C. Mps1 phosphorylation level analysis with proteins extracted from appressoria incubated on hydrophobic surfaces for 24 h (Li et al., 2017b). The phosphorylation levels of Mps1 (44 kDa) were detected using the phosphor-MAPK antibody (upper panel). The endogenous Mps1 was detected using the MAPK antibody (lower panel).
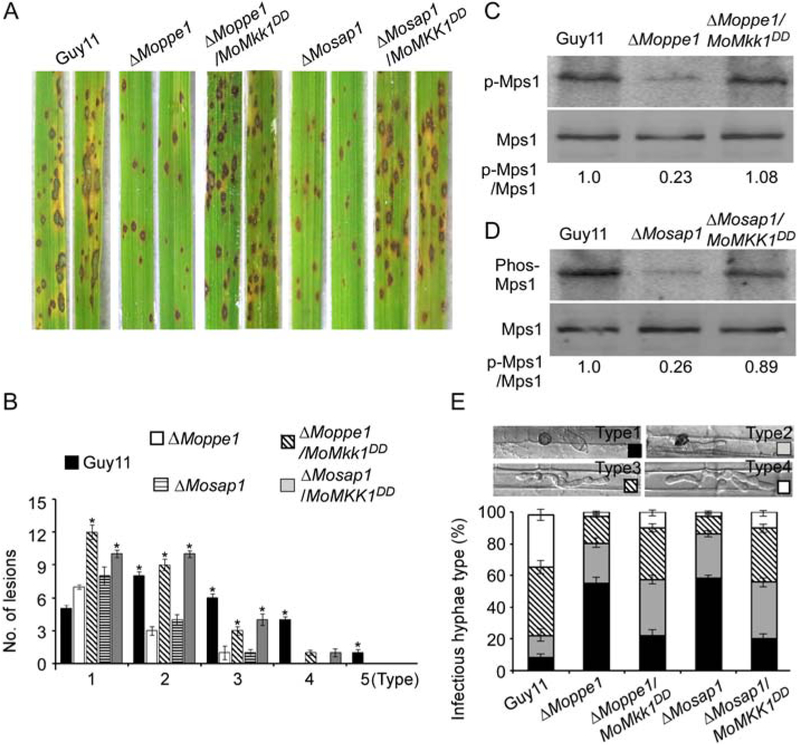
In M. oryzae, the CWI MAP kinase pathway plays significant roles in pathogenicity through the regulation of appressoria function (Li et al., 2012). We, therefore, proposed that the poor penetration of the mutants may result from the severely suppressed CWI pathway, as evidenced by the sharp decrease in MoMps1 phosphorylation levels. To test this proposition, we introduced the constitutively activated MoMKK1T369D, T375D allele into the ΔMoppe1 and ΔMosap1 mutants (Yin et al., 2016) and verified that MoMps1 phosphorylation was restored in the ΔMoppe1/MoMKK1DD and ΔMosap1/MoMKK1DD strains. Virulence testing using rice spray inoculation revealed a partially suppression. The ‘lesion type’ scoring assay showed that the activated strain now produced more Types 1–3 lesions than the ΔMoppe1 and ΔMosap1 mutants did. The ‘relative fungal growth’ assay by qRT-PCR revealed that the biomass of activated strains was more abundant than the mutant strains (Fig. 7A and B; Supporting Information Fig. S10). In the penetration assay, the ΔMoppe1/MoMKK1DD and ΔMosap1/MoMKK1DD strains also displayed increased penetration and better expansion (Fig. 7E). Taken together, these results suggested that MoPpe1 and MoSap1 contribute to appressorium penetration and full virulence by regulating MoMps1 phosphorylation levels in M. oryzae.
Fig. 7.
Pathogenicity assessment of ΔMoppe1 and ΔMosap1 mutants with constitutively activated MoMps1 through expressing MKK1T369D, T375D. A. Rice spraying assay as described in Fig. 2, experiments were performed three times independently and exhibit similar results. B. Quantification of lesion types. The error bars represent the standard deviations and asterisks represent significant difference (p < 0.01). C. Phosphorylation test of MoMps1 in the Guy11, ΔMoppe1 mutant and ΔMoppe1/MoMkk1DD strains. D. Phosphorylation levels of MoMps1 in Guy11, ΔMosap1 and ΔMosap1/MoMkk1DD strains. E. Observation of infection in rice sheaths. Excised rice leaf sheaths harvested from 4 week-old rice seedlings were inoculated with a conidial suspension (1 × 105 spores/ml each), and the invasive hyphae growth was examined at 36 hpi. Percentages of different types of infectious hyphae in rice cells (n = 50), error bars represent the standard deviations.
MoPpe1 phosphatase activity contributes to CWI regulation
As indicated above, MoPpe1 functions as a phosphatase that may dephosphorylate target proteins. So, what lead to the decreased phosphorylation of MoMps1 in disruption of MoPpe1? We speculated MoPpe1 may positive regulate the CWI pathway through its protein phosphatase activity. We screened the putative negative regulator from the interacting proteins data of MoPpe1 and MoMkk1 and found a tyrosine-protein phosphatase, MoPmp1 (Wang et al., 2017b). Yeast two-hybrid assays indicated that MoPmp1 functions as an adaptor between MoPpe1 and MoMkk1 (Supporting Information Fig. S11A and B). In our further studies, we found the phosphorylation of MoPmp1 was increased in the ΔMoppe1 mutant, and MoPmp1 is required for the dephosphorylation of MoMkk1 (Supporting Information Fig. S11C and D). Moreover, MoPmp1 functions as a CWI negative regulator with increased MoMps1 phosphorylation levels detected in ΔMopmp1 mutant and enhanced resistant to cell wall stressors of the ΔMopmp1 mutant (Supporting Information Fig. S11).
Evidence of MoPpe1 and MoSap1 function in TOR signalling
In S. cerevisiae, the TOR signalling pathway is activated by the presence of abundant preferred nutrients and Tap42 phosphorylation, which leads to the binding and inhibition of Sit4 function (Beck and Hall, 1999; Yan et al., 2006; Loewith and Hall, 2011). In contrast, upon the treatment with rapamycin or under nitrogen starvation conditions, Sit4 disassociates from Tap42, then dephosphorylate Gln3, a GATA transcriptional activator downstream target of TOR and required for nonpreferred nutrient utilization (Rohde et al., 2008). The dephosphorylated Gln3, then, enters into the nucleus to activate the transcription of nitrogen-regulated genes (Froeliger and Carpenter, 1996).
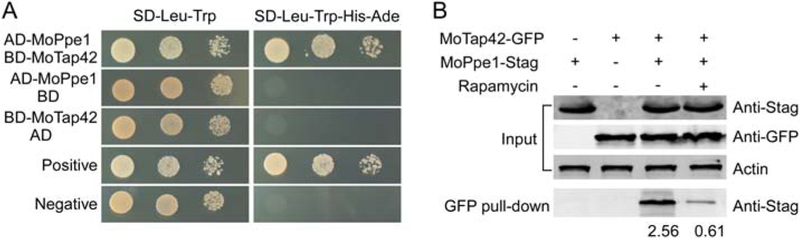
To determine whether a similar process occurs in M. oryzae, we first assessed the interaction between MoPpe1 and MoTap42 by Y2H and Co-IP assays with or without rapamycin. The results showed that the two proteins also have strong interactions and rapamycin induces a sharp decrease of the interaction (Fig. 8A and B). In addition, the MoPpe1 phosphatase activity was reduced in the presence of MoTap42 (Supporting Information Fig. S12).
Fig. 8.
MoPpe1 is involved in the TOR pathway through interacting with MoTap42. A. Yeast two hybrid assays for the interaction between MoPpe1 and MoTap42. The AD-MoPpe1 and BD-MoTap42 vectors were co-transformed into yeast strain AH109, and the transformants were plated with serial dilutions of yeast cells on SD-Leu-Trp for 3 days and on selective SD-Leu-Trp-His-Ade for 5 days. B. Rapamycin weakened the interaction between MoPpe1 and MoTap42. Mycelia from transformants co-expressing the MoPPE1-Stag and MoTAP42-GFP were treated with or without 30 ng/ml rapamycin for 4 h and then harvested for protein extraction. Then, the proteins eluted from anti-GFP agarose were detected with anti-Stag and anti-GFP antibodies respectively. Proteins in the supernatant and total proteins served as controls.
MoPpe1 and MoSap1 are required for nutrient utilization and affects the subcellular localization of MoNut1
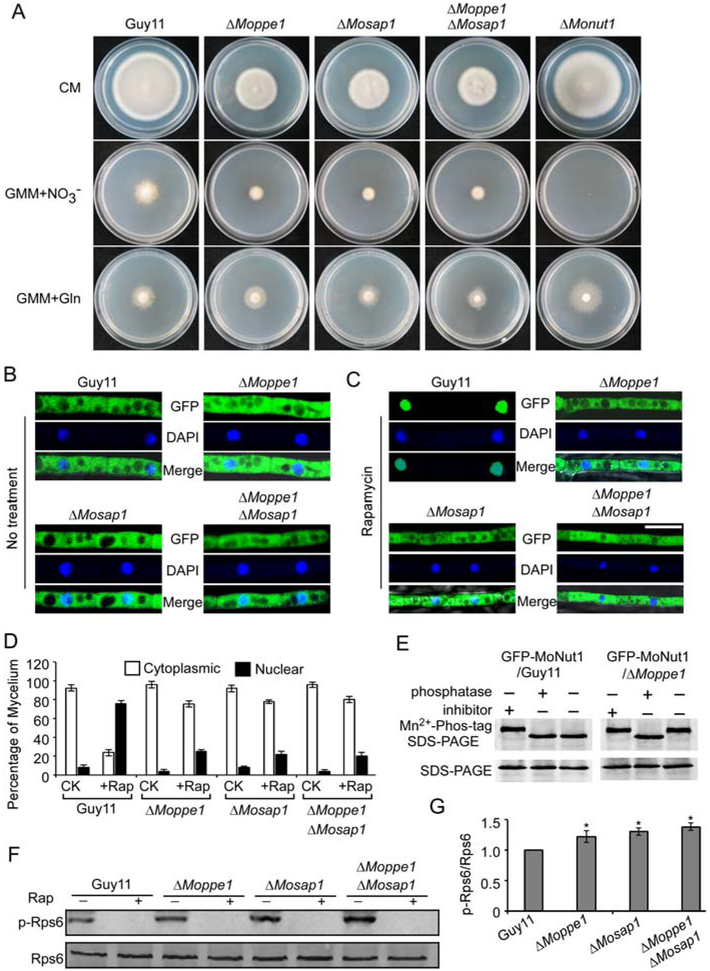
Since MoPpe1 interacts with MoTap42 to function in the TOR pathway, we hypothesized that MoPpe1 and MoSap1 could be involved in nutrient utilization. We then tested the growth rate of the ΔMoppe1 and ΔMosap1 mutants in various nitrogen sources (Supporting Information Table S4). The mutants exhibited significant reduction in growth when supplemented with nonpreferred nutrient sources, such as nitrate as the sole nitrogen source. However, when provided with preferred nutrient source, including l-glutamine, an agonist of TOR, the ΔMoppe1 and ΔMosap1 mutants showed the increased utilization rate (Fig. 9A and Supporting Information Table S4). These results indicated that MoPpe1 and MoSap1 are required for the utilization of nonpreferred nutrient sources but not the utilization of preferred nutrition.
Fig. 9.
MoPpe1 and MoSap1 function in the TOR pathway. A. The ΔMoppe1, ΔMosap1, ΔMoppe1 ΔMosap1 and ΔMonut1 mutants showed defects in utilizing nitrate as sole nitrogen sources increased utilization of glutamine when compared with wild-type. B, C. Subcellular localization of GFP-MoNut1 in the Guy11, ΔMoppe1, ΔMosap1 and ΔMoppe1 ΔMosap1 mutant strains with or without rapamycin (30 ng/ml) 40 min, nuclei were stained with DAPI, Bar = 5 μm. D. Statistics graph about the fusion protein GFP-MoNut1 in the cytoplasm and nucleus of each strain. E. GFP::MoNut1 proteins treated with a phosphatase inhibitor or phosphatase were separated by Mn2+-Phos-tag SDS-PAGE and normal SDS-PAGE, respectively, probed with the GFP antibody. F. The TOR activity was tested using the phospho-S6 specific antibody (ribosomal protein S6), phosphorylated ribosomal protein S6 (upper panel) and the endogenous S6 (lower panel). Rapamycin treatment was added as a control 30 ng/ml 4 h. G. The steady-state phosphor-S6/total S6 ratio in different strains. Three independent experiments were repeated. Error bars represent SD and asterisk represents significant difference (p < 0.05). The results were repeated three times.
In S. cerevisiae, Sit4/Ppe1 controls the nonpreferred nutrient utilization through Gln3 (Yan et al., 2006). We, therefore, speculated that the nonpreferred nutrient utilization defects may also due to the function of MoNnt1, a Gln3 homologue of M. oryzae (Froeliger and Carpenter, 1996). When transformed into a Δgln3 mutant of yeast (YER040W), MoNnt1 could partially suppress the growth defect of the mutant at 37 °C (Supporting Information Fig. S1). Since the nutrient affects the subcellular localization of Gln3, we monitored the localization of the GFPMoNut1 fusion protein in the ΔMoppe1, ΔMosap1, ΔMoppe1 ΔMosap1 and wild-type strains under different nutrient conditions. When the nutrient was abundant, GFP-MoNut1 was mostly localized to the cytoplasm (Fig. 9B). When rapamycin was added to simulate nitrogen stress, the most GFP-MoNut1 protein was translocated to the nucleus in the wild-type strain, but not the mutants (Fig. 9C). Finally, we confirmed that MoPpe1 is required for the direct dephosphorylation of MoNut1 upon nitrogen stress (Fig. 9C–E).
Our evidence so far suggested that MoPpe1 and MoSap1 act as the negative regulatory components of the TOR pathway in M. oryzae. With this finding, we further examined the TOR activity by detecting the phosphorylation level of the ribosomal protein S6 (Rps6), a TOR substrate demonstrated as a valuable tool to study the TOR activity (Gonzalez et al., 2015). Western blot analysis showed the phosphorylation level of Rps6 was increased in the mutants when compared with Guy11 (Fig. 9F and G), further demonstrating that MoPpe1 and MoSap1 are negative regulators of the TOR pathway in M. oryzae.
Inhibition of TOR signalling alleviates the suppression of the CWI pathway and the defect in host penetration
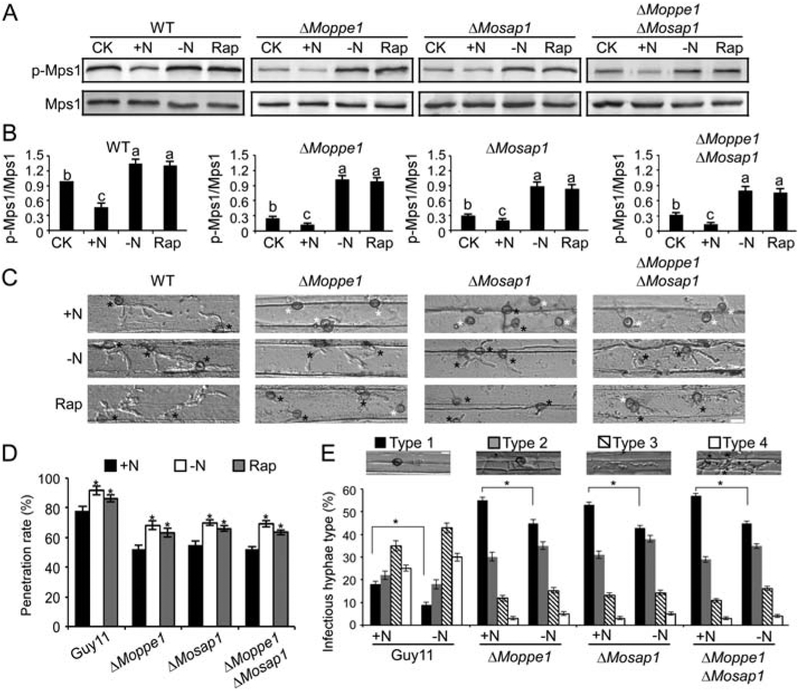
We have so far presented evidence suggesting that MoPpe1 and MoSap1 regulate the CWI pathway in a positive manner but act as a negative component of the TOR pathway. We, then, wondered whether there exist certain regulatory mechanisms between these two pathways. We examined the MoMps1 phosphorylation level with different nutrient treatments and found that they were restored in MoMps1 phosphorylation levels under nitrogen starvation conditions but not when supplied with abundant nutrients (Fig. 10A and B). We then explored whether there were also any differences in plant penetration under different nutrient conditions. A more efficient penetration symptom was observed on the back of barley leaves with nitrogen limitation condition than abundant nitrogen conditions (Fig. 10C and D). Similar results were obtained in the rice penetration assay in which appressoria in the nitrogen depletion condition were of better ability in penetrating than those with a good nitrogen source (Fig. 10E). In addition, experiments also showed that rapamycin affects the susceptibility to cell wall stressors in the wild-type and mutant strains. When added with different concentrations of rapamycin under the same CFW stress. The inhibition line chart of the test rose first then fell following the addition of 5 and 10 ng/ml rapamycin (Supporting Information Fig. S13).
Fig. 10.
Nitrogen limitation condition contributes to improve the MoMps1 phosphorylation level and enhances appressorium penetration. A. MoMps1 phosphorylation level test with different nutrient treatments for 4 h. MoMps1 phosphorylation levels were detected by Western blot analysis (+N: treated with 50 mM (NH4)2SO4;-N: with MM-N treat; Rap: 10 ng/ml rapamycin). B. The ratio of phosphorylated MoMps1/endogenous MoMps1 in different strains with different treatments. Three independent experiments were repeated. Values on the bars followed by the same letter are not significantly different (p < 0.05). C. Disease symptoms on the back of detached barley that were drop-inoculated with conidial suspensions and examined at 24 hpi. Bar = 10 μm. Black asterisks donate appressorium which has penetrated with invasive hyphae, asterisks in white represent appressoria with no penetration. D. Penetration rates of the appressoria on the back of detached barley with different treatment assays observed at 24 hpi, for each sample, (+N) treatment, another the spores treated with (–N) treatment appressorium penetration sites (n = 100) error bars represent the standard deviations. Asterisks indicate significant differences at (p < 0.05). E. Statistics of different types of infectious hyphae in rice cells one group with (+N) treatment, another the spores treated with (–N) treatment. n = 50. Error bars represent the standard deviations. Classification criteria were same as Fig. 2C. Asterisks indicate significant differences at (p < 0.01). Statistics were calculated from three repeated.
Discussion
For host infections, the rice blast fungus M. oryzae has evolved several well-conserved genetic mechanisms to regulate different developmental and infectious processes (Li et al., 2012; Zhang et al., 2016). In this study, we demonstrated that MoPpe1 interacts with MoMkk1 and, in conjunction with MoSap1, to positively regulate the CWI signalling pathway. We further revealed this regulation by MoPpe1 and MoSap1 is linked to TOR signalling and an activated TOR pathway inhibits CWI signalling and affects appressoria function and pathogenicity. In addition, we presented the link between nutrients and virulence, as well as the underlying regulatory mechanisms. Our novel findings should enhance the understanding of functions between nutrients and virulence and provide new insights into infection-related processes for the control of rice blast.
In S. cerevisiae, Sit4/Ppe1 encodes the catalytic subunit of a type 2A-related protein phosphatase and Sit4/Ppe1-associated SAP proteins function as regulatory subunit components. SAP proteins are not functional in the absence of Sit4/Ppe1 and, likewise, Sit4/Ppe1 is not functional in the absence of the SAPs (Luke et al., 1996). In this study, we revealed that MoSap1 could complement all four yeast Sap mutants and by generating MoPpe1 and MoSap1 deletion mutants, we found that they are important for growth, conidiation and virulence. Moreover, the ΔMoppe1ΔMosap1 double mutant exhibited similar defects as single mutants, together with their interaction relations, we propose that MoPpe1 and MoSap1 function as a complex in M. oryzae.
The reduced virulence of the ΔMoppe1 and ΔMosap1 mutants were owing to multiple aspects. First, the vegetative growth rate of mutants was significantly reduced than the wild-type strain on different media. Consistent with these results, the decreased growth of the ΔMoppe1 and ΔMosap1 mutant strains usually led to attenuated virulence in pathogenic fungi, including Candidia albicans and Fusarium graminearum (Lee et al., 2004; Yun et al., 2014). Second, the ΔMoppe1 and ΔMosap1 mutants showed defects in ROS scavenging, which is regarded as one of the first responses to fungal invasion (Mellersh et al., 2002; Guo et al., 2011). We indicated here that the impaired hyphal extension of the ΔMoppe1 and ΔMosap1 mutants in host cells is due to a defect in ROS scavenging, and the impairment was partially restored when DPI was added to suppress the ROS production. Third, we revealed that the repressed CWI pathway causes attenuated virulence in the ΔMoppe1 and ΔMosap1 mutants, as the CWI pathway is well conserved during the invasion of host cells (Li et al., 2012; Yin et al., 2016). Following the constitutive activation of MoMps1 with MoMKK1DD in mutants, the reactivated strains exhibited improved penetration in rice sheaths and increased numbers of diseased lesions on rice leaves were also found, indicating that CWI repression was a primary cause of the penetration defects in the mutants.
Interestingly, MoPpe1 functions as a protein phosphatase but we found a decreased phosphorylation level of MoMps1 in the ΔMoppe1 mutant, suggesting that MoPpe1 may indirectly regulate the CWI pathway through certain negative regulators of CWI pathway. Here, our results found the phosphorylation level of MoPmp1 was increased in the ΔMoppe1 mutant and that MoPmp1 regulates the dephosphorylation process of MoMkk1 and MoMps1 (Wang et al., 2017b). Thereby, there were possibilities that MoPpe1 positive regulates CWI via modulating the phosphorylation level of MoPmp1. In addition, MoPpe1 binding of MoMkk1 may affect its dephosphorylation mediated by MoPmp1.
Previous research has indicated that the crosstalk between the CWI signalling network and TORC1-dependent processes is mediated by the CWI pathway central component GTPase Rho1 (Yan et al., 2012b). In this study, we provided the evidence that the MoPpe1–MoSap1 complex mediates crosstalk between CWI and TOR signalling. When rapamycin or nitrogen starvation was employed to inhibit the TOR activity, MoMps1 phosphorylation was restored, along with improved appressoria penetration efficiency. This finding is in accordance with previous evidence that nitrogen limitation is a key signal for the early infection process of M. oryzae (Lopez-Berges et al., 2010), and here, we provide another new clues to support this finding.
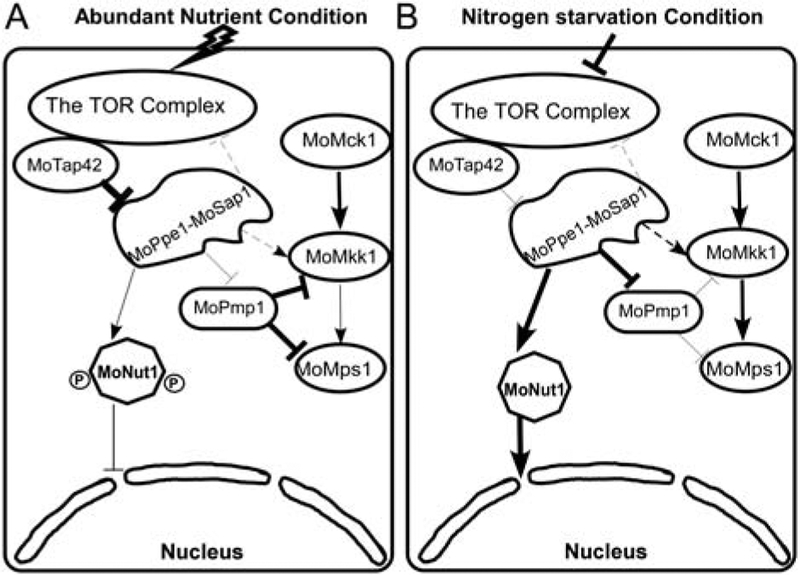
We proposed a working model for the MoPpe1– MoSap1 complex that functions as an adaptor for the negative regulation of the CWI pathway by TOR signalling (Fig. 11). This model shows that TOR signalling is activated under abundant nutrient conditions. MoTap42 strongly inhibits MoPpe1 function and results in the repression of the CWI pathway that is subjected to dephosphorylation regulation by MoPmp1. MoNut1 is hyperphosphorylated in the cytoplasm. However, when faced with nitrogen limitation condition or in the presence of rapamycin, the TOR complex and the MoTap42 are both severely inhibited. MoPpe1 is activated that may dephosphorylate MoPmp1 to repress its regulation on CWI components and MoNut1 is also dephosphorylated prior to entering into the nucleus.
Fig. 11.
A proposed working model of TOR and CWI pathways medicated by the MoPpe1–MoSap1 complex. Thick lines indicate strengthened function, and thin lines indicate attenuated regulation. A. With abundant nutrients, the TOR signalling pathway is activated, leading to MoTap42 binding with MoPpe1 to inhibit its function while the phosphorylated MoNut1 remains cytoplasmic bound. The CWI pathway repression is caused by the dephosphorylation function MoPmp1 on CWI components. B. When treated with TOR specific inhibitor rapamycin or in nutrient starvation condition, TOR signalling was repressed. The activated MoPpe1–MoSap1 complex plays a dual role as a regulator to dephosphorylate MoNut1 and an activator of the CWI pathway through the strengthened repression on MoPmp1 and enhanced interaction with MoMkk1.
In summary, we identified MoPpe1 that interacts with MoMkk1 and its associated protein MoSap1. We demonstrated that they regulate vegetative growth, conidia formation and pathogenicity in M. oryzae. The reduced virulence of these mutants was due to defects in penetration and suppression of ROS scavenging. The penetration defects were due to the severe repression of the CWI pathway. We further revealed that this suppression was regulated by the TOR signalling pathway. We proposed that the MoPpe1–MoSap1 complex acts as one of the signal adaptors between the CWI pathway and the TOR pathway. These findings provided a novel viewpoint for understanding pathways that regulate pathogenicity and promote the discovery of new ways to manage the rice blast.
Methods
Strains and culture conditions
Guy11 was used as wild-type of M. oryzae for transformation in this study. Mycelia harvested from liquid CM was used for DNA and RNA extractions. For vegetative growth, small squares of mycelia were cut from the edge of 7 day-old strains and placed onto the media (CM, MM, OM and SDC) and incubated in the dark at 28 °C for another 7 days. For conidia collection, mycelial blocks were inoculated on SDC (100 g of straw, 40 g of corn powder, 15 g of a gar in 1 l of distilled water) agar media (Zhang et al., 2011) maintain at 28 °C for 7 days in the dark followed 3 days continuous illumination under fluorescent light.
Gene deletion and complementation
The ΔMoppe1 mutant was generated by the one-step gene replacement strategy. Two fragments with 1.0 kb of sequences flanking the targeted gene were PCR amplified with primer pairs (Table S2, see Supporting Information). The resulting PCR products were ligated to the hygromycin resistance cassette released from pCX62 (Chen et al., 2017). The 3.4 kb fragment, which included the flanking sequences and the HPH cassette transformed into Guy11 protoplasts. Putative mutants were screened by PCR and confirmed by Southern blotting analysis (Supporting Information Fig. S1). For the complement strains which contains the entire MoPpe1 gene coding region and its native promoter region, was amplified by PCR with primers (Supporting Information Table S2) and inserted into pYF11 (bleomycin resistance). The MoSAP1 and MoNUT1 gene deletion mutants and the complement strains were obtained by the same strategy.
Assays for pathogenicity and penetration
Pathogenicity test was performed as described (Qi et al., 2016). Conidia was collected from 10 day-old SDC agar cultures and resuspended by 0.2% (w/v) gelatin solution to a concentration of 5 × 104 spores/ml. For spraying assay, 2 week-old seedlings of rice (O. sativa cv. CO39) were sprayed with 4 ml of conidial suspension of each treatment and kept in a growth chamber at 25 °C with 90% humidity in the dark for the first 24 h, followed by a 12 h-light and 12 h-dark cycle. Lesion formation was daily checked and photographed after 7 days inoculation. The 7 day-old barley leaves were drop-inoculated with three droplets (20 μl) of conidial suspension and photographs were taken 5 days after infection. For ‘relative fungal growth’ assay, total DNA was extracted from 1.5 g disease leaves and test by qRT-PCR (HiScript II Reverse Transcriptase, Vazyme Biotech Co., Nanjing, China) with 28S/Rubq1 primers (Supporting Information Table S2) (Zhong et al., 2016). For rice sheaths penetration and invasive hyphae expansion, conidial suspension (1 × 105 spores/ml) was inoculated into the sheaths. After incubation for about 36 h at 28 °C, the sheath cuticle cells were observed under Zeiss Axio Observer A1 inverted microscope.
Western blot analysis for protein phosphorylation
The mutants and wild-type Guy11 strains were cultured in liquid CM for 2 days then left untreated or treated with 30 ng/ml rapamycin 4 h before the mycelia of were harvested and added with 1 ml lysis buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 5 mM EDTA, 1% Triton X-100, 2 mM PMSF) and 10ul of protease inhibitor cocktail (Sangon, Shanghai, China). After homogenization with a vortex shaker, the lysate was centrifuged at 12 000 r.p.m. for 10 min at 4 °C twice. Then, 200 μl of supernatant was mixed with an equal volume of 50 μl loading buffer and boiled for 5 min. The proteins separated on SDS-PAGE gels were transferred onto a polyvinylidene fluoride membrane with a Bio-Rad blotting apparatus. The intensity of the signal corresponding to phosphorylated Mps1 was detected by binding of an antiphospho-p44/42 MAP kinase antibody (Cell Signalling Technology, Boston, MA, USA), with the Mpk1 antibody (N-terminal anti-Mpk1) from Santa Cruz Biotechnology (Santa Cruz, CA, USA) used as a control. The TOR antibodies are as follows: phospho-Ser235/Ser236-S6 (#2211, Cell Signalling Technology), RPS6 (#ab40820, Abcam).
Affinity purification and mass spectrometry analysis
First construct the MoMKK1-3xFLAG and MoPPE1-3xFLAG construct and introduced it into the wild-type stain by transformation. Total proteins extracted from transformants MoMKK1-Flag/Guy11 and MoPPE1-Flag/Guy11 respectively. Approximately 30 μl of anti-FLAG agarose (Abmart, Shanghai, China) was added into 1 ml diluted total protein (Total protein/Lysis Buffer = 1: 4) samples to capture MoMkk1 and MoPpe1 interacting proteins, following the manufacturer’s instructions. After incubation at 4 °C overnight, the agarose was washed three times with 500 μl of TBS (20 mM Tris–HCl, 500 mM NaCl, pH 7.5). Proteins binding to the beads were immediately eluted with 45 μl elution buffer (0.2 M glycine, pH 2.5). The eluent was immediately neutralized with 5 μl of neutralization buffer (1.5 M Tris, pH 9.0). The eluted protein samples were send to (Beijing Protein Innovation Co., Ltd) for mass spectrometry analysis.
Co-immunoprecipitation (Co-IP) assay
MoPPE1 was amplified and cloned into pXY203 by the yeast gap repair approach to generate the Stag fusion constructs. A similar approach was employed to generate the GFP fusion construct pYF11 (bleomycin resistance) for the MoTap42 and MoSap1. The resulting fusion constructs were transformed into the Guy11 strain. Transformants expressing the fusion constructs were confirmed by Western blot analysis. For Co-IP assays, total proteins were isolated and incubated with the anti-GFP agarose. Proteins eluted from agarose were analysed by Western blot detection with the anti-Stag and anti-GFP antibodies (Abmart, Shanghai, China).
Yeast two-hybrid assays
The bait constructs were generated by cloning MoMKK1 and MoSAP1 full-length cDNAs into pGBKT7 respectively. The cDNAs of MoPPE1 was cloned into pGADT7 as the prey construct (see primers in Supporting Information Table S2). The resulting prey and bait constructs were co-transformed into the yeast strain AH109 as per the description of BD library construction & screening kit (Clontech, USA). The transformants from SD-Trp-Leu plates were isolated and assayed for growth on SD-Trp-Leu-His and SD-Trp-Leu-His-Ade medium. Yeast stains for positive and negative controls were provided by the BD library construction and screening kit.
DAPI staining
To observe the cellular localization of MoNut1 in various strains with rapamycin treatment, MoNUT1 fused with a GFP tag in the N-terminus and transformed into all these strains respectively. The strains were cultured in liquid complete medium (CM) for 24 h and the mycelium was observed under a confocal fluorescence microscope (Zeiss LSM710, 63x oil). In addition, the nucleus was stained with 10 μl/ml 4′6-diamidino-2-phenylindole (DAPI, Sigma).
Phosphatase activity assay
Phosphatase assays were performed according to the manufacturer’s instructions as follows: 20 μg of recombinant Ppe1 protein samples was incubated with 100 μl of p-nitrophenylphosphate (pNPP) substrate solution (NEB). The colorimetric pNPP substrate turns yellow upon the release of a phosphate group by the phosphatase. The pNPP reaction was stopped after 15 min by the addition of 50 μl of 3 M NaOH solution. Activity was measured by spectrophotometer as absorbance at 405 nm. All assays were performed in biological duplicate and technical quadruplicate.
Phosphorylation analysis with phos-tag gel
The GFP-MoNut1 fusion construct was introduced into ΔMoppe1 and ΔMonut1 mutants respectively. The proteins extracted from mycelium were resolved on 8% SDS-polyacrylamide gels prepared with 50 μM acrylamide-dependent Phos-tagligand and 100 μM MnCl2 as described (Li et al., 2017a,b). Gel electrophoresis was run at 60 V for 6 ± 8 h. Prior to transfer, gels were equilibrated in transfer buffer containing 5 mM EDTA for 30 min two times and then in transfer buffer without EDTA for 15 min. Protein transfer from the Mn2+-phos-tag acrylamide gel to the PVDF membrane was performed overnight at 80 V, then the membrane was analysed by western blotting using the anti-GFP antibody.
Accession number
Gene sequences can be found in the GenBank database under the following accession numbers: XP_003719961.1 (MoPPE1), XP_003716542.1 (MoSAP1), XP_003720998.1 (MoNUT1), XP_003717079.1 (MoMKK1).
Supplementary Material
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Fig. S1. Complementation of the yeast mutants with Magnaporthe oryzae gene counterparts. (A) MoPPE1 could completely suppress the growth defect of yeast SIT4 deletion mutant at 37 °C. The yeast SIT4 mutant was complemented with MoPPE1 cDNA to generate the strain BY4741Δsit4 + pYES2-MoPPE1. The WT strain BY4741 and SIT4 mutant transformed with empty pYES2 vector, respectively, were used as controls. Serial dilutions of cell suspension of each strain were spotted under different stresses as indicated in the Fig. (B) MoNut1 partially repressed the S. cerevisiae GLN3 mutant. (C) MoSAP1 could completely suppress the yeast mutant SAP190 in 1 mM tunicamycin and could partially repress the yeast mutant SAP185 which exhibit an increased resistance in 300 μg/ml hygromycin B, SAP155 which was sensitive to 200 μg/ml hygromycin B and SAP4 with the stress of 1 mM H2O2 that mutant is more resistant.
Fig. S2. Targeted genes knockout strategy and confirmation by Southern blot analysis. (A) Strategy of knocking out target genes in M. oryzae genome. Thin lines below the arrows indicate the probe sequence of each gene. (B) Southern blot analysis was used to confirm the MoPPE1 deletion and the copy of the HPH gene. The genomic DNA of Guy11 and ΔMoppe1 mutant was digested with EcoR I and hybridized with probes. (C-F) Strategies of knocking out MoSAP1 and MoNUT1 and verified by southern blot. The genomic DNA of Guy11 and ΔMosap1 mutant was digested with Xba I and ΔMonut1 mutant was digested with Kpn I. (G) The ΔMoppe1 ΔMosap1 double mutant strain was generated with a 3.4 kb fragment, which included the flanking sequences of MoPPE1 and the bleomycin sequence transformed into ΔMosap1 mutant protoplasts then using the PCR to identify the putative double mutant. (H) The PCR results of verification the double mutant, number #1 and #12 were the double mutants. (I) The southern blot used to detect the copy of the bleomycin gene in different strains.
Fig. S3. MoPpe1 is involved in the vegetative growth and conidiation. (A) Guy11, ΔMoppe1 mutant and complemented strain were inoculated on CM, MM, OM and SDC media cultured at 28 °C for 7 days, then photographed. (B) Statistical analyses of the colony diameter from wild-type Guy11, ΔMoppe1 mutant and the complemented strain on different medium. Error bars represent the standard deviations; Asterisks denote statistical significances (p < 0.01). (C) Conidia were observed under a light microscope after illumination for 24 h then photographed. (D) The conidia were harvested from the Guy11, ΔMoppe1 mutant and complemented strain incubated on SDC medium for 7 days. The number of conidia were calculated and analysed. Error bars represent the standard deviations. Asterisks represent significant difference (p < 0.01).
Fig. S4. Expression levels of conidiation-related genes. The expression results of MoCOM1, MoHOX2, MoCON2, MoCOS1 and MoSTUA genes that were shown previously to be important in the process of conidial development. Histogram shown the results of three biology repeats, error bars denote standard errors of three biology experiments. Asterisk denote values that are not significantly different at (p < 0.05).
Fig. S5. There is no great difference in conidial morphology between wild-type and mutants. Conidia were harvested from different mutants the observed by light microscopy. Bars = 10 μm.
Fig. S6. MoPpe1 is dispensable function in nuclear division in M. oryzae. (A) Nucleus was viewed, photograph and calculated during appressorium formation at 0, 4, 12, 24 h time point and infection phase with the transformation of H1-RFP into ΔMoppe1 mutant and Guy11 respectively. The merged image shows H1: RFP and DIC. Bars = 10 μm.
Fig. S7. MoPpe1 and MoSap1 are important for the vegetative growth and conidia formation of M. oryzae. (A) Guy11, ΔMosap1 single mutant, ΔMoppe1 ΔMosap1 double mutant and ΔMosap1 complemented strain were inoculated on CM, MM, OM and SDC media cultured at 28 °C for 7 days, then photographed. (B) Statistical analyses of the colony diameter of four different strains on different medium. Error bars represent the standard deviations, asterisks denote statistical significances (p < 0.01). (C) The conidia were photographed under a light microscope after illumination for 24 h. (D) Conidia production of Guy11, ΔMosap1 single mutant, complemented strain and ΔMoppe1 ΔMosap1 double mutant were collected after 7 days on SDC medium, then calculated and analysed. Error bars represent the standard deviations. Asterisks denote statistical significances (p < 0.01).
Fig. S8. MoPpe1 is involved in the cell wall stress response of M. oryzae. (A) Guy11, ΔMoppe1 mutant and the complemented strain were incubated on complete medium (CM) plates containing different concentrations of Congo Red (CR), Calcofluor white (CFW) and sodium dodecyl sulfate (SDS) at 28 °C for 7 days. (B) The inhibition rate was determined by plotting the percentage of colonies in the presence of various concentrations of CR, CFW and SDS against regular CM. The asterisks denote statistical significances (p < 0.01)
Fig. S9. MoSap1 is important for cell wall stress responses of M. oryzae. (A) The wild-type strain, ΔMosap1 mutant and the complemented strain were incubated on complete medium (CM) plates with different concentrations of Congo Red (CR), Calcofluor white (CFW) and sodium dodecyl sulfate (SDS) at 28 °C for 7 days. (B) The inhibition rate was determined by plotting the percentage of colonies in the presence of various concentrations of CR, CFW and SDS against regular CM, asterisks denote statistical significances (p < 0.01)
Fig. S10. The relative fungal growth assay. Diseased rice leaves were collected after 7 days inoculation. Total DNA was extracted from per 1.5 g disease leaves and test by qRT-PCR (HiScript II Reverse Transcriptase, Vazyme Biotech Co., Nanjing, China) with 28S/Rubq1 primers. The results were of three biology repeats. Single asterisks denote statistical significances (p < 0.05), double asterisks represent statistical significances (p < 0.01)
Fig. S11. MoPpe1 regulates the CWI pathway via MoPmp1. (A) Yeast two hybrid assay for the interaction between MoPpe1 and MoPmp1. The AD-MoPmp1 and BD-MoPpe1 vectors were co-introduced into yeast strain AH109, and the transformants were plated with serial dilutions of yeast cells on SD-Leu-Trp for 3 days and on selective SD-Leu-Trp-His added with 2 mM 3-AT (3-amino-1,2,4-triazole) for 10 days. (B) Interaction between MoMkk1 and MoPmp1. The AD-MoPmp1 and BD-MoMkk1 vectors were co-introduced into yeast strain AH109, and the transformants were plated with serial dilutions of yeast cells on SD-Leu-Trp for 3 days and on selective SD-Leu-Trp-His added with 2.5 mM 3-AT (3-amino-1,2,4-triazole) for 12 days. (C) MoPmp1 was hyperphosphorylated in ΔMoppe1 mutant. (D) MoPmp1 dephosphorylate the MoMkk1 in M. oryzae. (E) The MoMps1 phosphorylayion increased in ΔMopmp1 mutant. (F) The ΔMopmp1 mutant exhibited increased resistance to cell wall stress.
Fig. S12. Protein phosphatase MoPpe1 possesses the phosphatase activity that is reduced upon MoTap42 addition. Recombinant His-tagged MoPpe1 and MoTap42 was expressed in bacteria and purified. Phosphatase activity was determined using the indicated proteins and p-nitrophenyl phosphate (pNPP) as a substrate.
Fig. S13. Rapamycin treatment affects the fungal susceptibility to calcofluor white. (A) The wild-type strain, ΔMoppe1, ΔMosap1 and ΔMoppe1 ΔMosap1 mutant were incubated with 150 μg/ml Calcofluor white (CFW) stress then added with different concentrations of rapamycin 5, 10, 20 ng/ml, respectively, the strains on complete media (CM) as a control. (B) The broken line graph of each strain’s inhibition rate with different treatment. Detailed inhibition rate along with positive and negative SD was shown in each graph.
Table S1. MoMkk1 and MoPpe1 putative interacting proteins identified by affinity capture assays
Table S2. Total of primers were used in this study
Table S3. Comparison of mycological characters
Table S4. Utilization rate of the strains with different nitrogen source
Acknowledgements
This research was supported by Natural Science Foundation of China (Grant number 31470248, XZ), the Fundamental Research Funds for the Central Universities, No. KYT Z201604, and Innovation Team Program for Jiangsu Universities (2017). The Wang lab research was supported by US NIH grants AI121451 and AI121460. We thank Dr. Yongheng Liang of Nanjing Agricultural University for providing yeast mutants and Dr. Youliang Peng of China Agricultural University for providing ΔMopmp1 mutant of M. oryzae.
References
- Adachi K, and Hamer JE (1998) Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell 10: 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto L, Canadell D, Petrezselyova S, Navarrete C, Maresova L, Perez-Valle J, et al. (2011) A genomewide screen for tolerance to cationic drugs reveals genes important for potassium homeostasis in Saccharomyces cerevisiae. Eukaryot Cell 10: 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T, and Hall MN (1999) The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402: 689–692. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Davies DR, Gerrish C, Auh CK, and Murphy TM (1998) Comparative biochemistry of the oxidative burst produced by rose and french bean cells reveals two distinct mechanisms. Plant Physiol 116: 1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Le X, Sun Y, Li M, Zhang H, Tan X, et al. (2017) MoYcp4 is required for growth, conidiogenesis and pathogenicity in Magnaporthe oryzae. Mol Plant Pathol 18: 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froeliger EH, and Carpenter BE (1996) NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol Gen Genet 251: 647–656. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Shimobayashi M, Eisenberg T, Merle DA, Pendl T, Hall MN, and Moustafa T (2015) TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae. PLoS One 10: e0120250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, and Mansfield J (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23: 441–450. [DOI] [PubMed] [Google Scholar]
- Guo M, Guo W, Chen Y, Dong S, Zhang X, Zhang H, et al. (2010) The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe Interact 23: 1053–1068. [DOI] [PubMed] [Google Scholar]
- Guo M, Chen Y, Du Y, Dong YH, Guo W, Zhai S, et al. (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the Rice blast fungus Magnaporthe oryzae. PLoS Pathog 7: e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Czymmek KJ, Caplan JL, Sweigard JA, and Donofrio NM (2011) HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog 7: e1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Goh J, Yoo S, Chi MH, Choi J, Rho HS, et al. (2008) A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol Plant Microbe Interact 21: 525–534. [DOI] [PubMed] [Google Scholar]
- Kapitzky L, Beltrao P, Berens TJ, Gassner N, Zhou C, Wuster A, et al. (2010) Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol 6: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park SY, Kim KS, Rho HS, Chi MH, Choi J, et al. (2009) Homeobox transcription factors are required for Conidiation and Appressorium development in the Rice blast fungus Magnaporthe oryzae. PLoS Genet 5: e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Nantel A, Jiang L, Whiteway M, and Shen SH (2004) The serine/threonine protein phosphatase SIT4 modulates yeast-to-hypha morphogenesis and virulence in Candida albicans. Mol Microbiol 51: 691– 709. [DOI] [PubMed] [Google Scholar]
- Li G, Zhou X, and Xu JR (2012) Genetic control of infection-related development in Magnaporthe oryzae. Curr Opin Microbiol 15: 678–684. [DOI] [PubMed] [Google Scholar]
- Li L, Chen X, Zhang S, Yang J, Chen D, Liu M, et al. (2017a) MoCAP proteins regulated by MoArk1-mediated phosphorylation coordinate endocytosis and actin dynamics to govern development and virulence of Magnaporthe oryzae. PLoS Genet 13: e1006814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao C, Li L, Liu M, Yin Z, Zhang H, et al. (2017b) MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae. PLoS Pathog 13: e1006449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang X, Hu S, Liu HQ, and Xu JR (2017c) PKA activity is essential for relieving the suppression of hyphal growth and appressorium formation by MoSfl1 in Magnaporthe oryzae. PLoS Genet 13: e1006954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Qian B, Gao C, Huang S, Cai Y, Zhang H, et al. (2016) The putative protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate the development and pathogenicity in Magnaporthe oryzae. Mol Plant Microbe Interact 29: 496–507. [DOI] [PubMed] [Google Scholar]
- Loewith R, and Hall MN (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Berges MS, Rispail N, Prados-Rosales RC, and Di Pietro A (2010) A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 22: 2459–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke MM, DellaSeta F, DiComo CJ, Sugimoto H, Kobayashi R, and Arndt KT (1996) The SAPs, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol 16: 2744–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroquin-Guzman M, and Wilson RA (2015) GATA-dependent Glutaminolysis drives Appressorium formation in Magnaporthe oryzae by suppressing TOR inhibition of cAMP/PKA signaling. PLoS Pathog 11: e1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marroquin-Guzman M, Sun G, and Wilson RA (2017) Glucose-ABL1-TOR signaling modulates cell cycle tuning to control terminal Appressorial cell differentiation. PLoS Genet 13: e1006557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh DG, Foulds IV, Higgins VJ, and Heath MC (2002) H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J 29: 257–268. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Fukada J, Moriwaki A, Fujikawa T, Ohashi M, Hibi T, and Hayashi N (2009) Mstu1, an APSES transcription factor, is required for Appressorium-mediated infection in Magnaporthe grisea. Biosci Biotechnol Biochem 73: 1779–1786. [DOI] [PubMed] [Google Scholar]
- Qi Z, Liu M, Dong Y, Zhu Q, Li L, Li B, et al. (2016) The syntaxin protein (MoSyn8) mediates intracellular trafficking to regulate conidiogenesis and pathogenicity of rice blast fungus. New Phytol 209: 1655–1667. [DOI] [PubMed] [Google Scholar]
- Rohde JR, Bastidas R, Puria R, and Cardenas ME (2008) Nutritional control via tor signaling in Saccharomyces cerevisiae. Curr Opin Microbiol 11: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Immanuel D, and Arndt KT (1991) The Sit4 protein phosphatase functions in late G1 for progression into S-phase. Mol Cell Biol 11: 2133–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Ru Y, Hong L, Zhu Q, Zuo R, Guo X, et al. (2015) System-wide characterization of bZIP transcription factor proteins involved in infection-related morphogenesis of Magnaporthe oryzae. Environ Microbiol 17: 1377–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yin Z, Tang W, Cai X, Gao C, Zhang H, et al. (2017a) The thioredoxin MoTrx2 protein mediates reactive oxygen species (ROS) balance and controls pathogenicity as a target of the transcription factor MoAP1 in Magnaporthe oryzae. Mol Plant Pathol 18: 1199–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RJ, Peng JB, Li QX, and Peng YL (2017b) Phosphorylation-mediated regulatory networks in mycelia of Pyricularia oryzae revealed by Phosphoproteomic analyses. Mol Cell Proteomics 16: 1669–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RA, and Talbot NJ (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7: 185–195. [DOI] [PubMed] [Google Scholar]
- Yan GH, Shen XY, and Jiang Y (2006) Rapamycin activates Tap42-associated phosphatases by abrogating their association with tor complex 1. EMBO J 25: 3546–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Lai Y, and Jiang Y (2012a) TOR under stress: targeting TORC1 by Rho1 GTPase. Cell Cycle 11: 3384–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan GH, Lai YM, and Jiang Y (2012b) The TOR Complex 1 is a direct target of Rho1 GTPase. Mol Cell 45: 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao XY, Sun J, Kang ZS, Ding SL, Xu JR, and Peng YL (2010) A novel protein Com1 is required for Normal Conidium morphology and full virulence in Magnaporthe oryzae. Mol Plant Microbe Interact 23: 112–123. [DOI] [PubMed] [Google Scholar]
- Yin ZY, Tang W, Wang JZ, Liu XY, Yang LN, Gao CY, et al. (2016) Phosphodiesterase MoPdeH targets MoMck1 of the conserved mitogen-activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae. Mol Plant Pathol 17: 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Gu Q, Yun Y, Yin Y, Xu JR, Shim WB, and Ma Z (2014) The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol 203: 219–232. [DOI] [PubMed] [Google Scholar]
- Yun Y, Liu Z, Zhang J, Shim WB, Chen Y, and Ma Z (2014) The MAPKK FgMkk1 of Fusarium graminearum regulates vegetative differentiation, multiple stress response, and virulence via the cell wall integrity and high-osmolarity glycerol signaling pathways. Environ Microbiol 16: 2023–2037. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhao Q, Liu K, Zhang Z, Wang Y, and Zheng X (2009) MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol Lett 293: 160–169. [DOI] [PubMed] [Google Scholar]
- Zhang H, Tang W, Liu K, Huang Q, Zhang X, Yan X, et al. (2011) Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog 7: e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Zheng XB, and Zhang ZG (2016) The Magnaporthe grisea species complex and plant pathogenesis. Mol Plant Pathol 17: 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong K, Li X, Le X, Kong X, Zhang H, Zheng X, et al. (2016) MoDnm1 dynamin mediating Peroxisomal and mitochondrial fission in complex with MoFis1 and MoMdv1 is important for development of functional Appressorium in Magnaporthe oryzae. PLoS Pathog 12: e1005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZZ, Li GH, Lin CH, and He CZ (2009) Conidiophore stalk-less1 encodes a putative zinc-finger protein involved in the early stage of Conidiation and mycelial infection in Magnaporthe oryzae. Mol Plant Microbe Interact 22: 402–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Fig. S1. Complementation of the yeast mutants with Magnaporthe oryzae gene counterparts. (A) MoPPE1 could completely suppress the growth defect of yeast SIT4 deletion mutant at 37 °C. The yeast SIT4 mutant was complemented with MoPPE1 cDNA to generate the strain BY4741Δsit4 + pYES2-MoPPE1. The WT strain BY4741 and SIT4 mutant transformed with empty pYES2 vector, respectively, were used as controls. Serial dilutions of cell suspension of each strain were spotted under different stresses as indicated in the Fig. (B) MoNut1 partially repressed the S. cerevisiae GLN3 mutant. (C) MoSAP1 could completely suppress the yeast mutant SAP190 in 1 mM tunicamycin and could partially repress the yeast mutant SAP185 which exhibit an increased resistance in 300 μg/ml hygromycin B, SAP155 which was sensitive to 200 μg/ml hygromycin B and SAP4 with the stress of 1 mM H2O2 that mutant is more resistant.
Fig. S2. Targeted genes knockout strategy and confirmation by Southern blot analysis. (A) Strategy of knocking out target genes in M. oryzae genome. Thin lines below the arrows indicate the probe sequence of each gene. (B) Southern blot analysis was used to confirm the MoPPE1 deletion and the copy of the HPH gene. The genomic DNA of Guy11 and ΔMoppe1 mutant was digested with EcoR I and hybridized with probes. (C-F) Strategies of knocking out MoSAP1 and MoNUT1 and verified by southern blot. The genomic DNA of Guy11 and ΔMosap1 mutant was digested with Xba I and ΔMonut1 mutant was digested with Kpn I. (G) The ΔMoppe1 ΔMosap1 double mutant strain was generated with a 3.4 kb fragment, which included the flanking sequences of MoPPE1 and the bleomycin sequence transformed into ΔMosap1 mutant protoplasts then using the PCR to identify the putative double mutant. (H) The PCR results of verification the double mutant, number #1 and #12 were the double mutants. (I) The southern blot used to detect the copy of the bleomycin gene in different strains.
Fig. S3. MoPpe1 is involved in the vegetative growth and conidiation. (A) Guy11, ΔMoppe1 mutant and complemented strain were inoculated on CM, MM, OM and SDC media cultured at 28 °C for 7 days, then photographed. (B) Statistical analyses of the colony diameter from wild-type Guy11, ΔMoppe1 mutant and the complemented strain on different medium. Error bars represent the standard deviations; Asterisks denote statistical significances (p < 0.01). (C) Conidia were observed under a light microscope after illumination for 24 h then photographed. (D) The conidia were harvested from the Guy11, ΔMoppe1 mutant and complemented strain incubated on SDC medium for 7 days. The number of conidia were calculated and analysed. Error bars represent the standard deviations. Asterisks represent significant difference (p < 0.01).
Fig. S4. Expression levels of conidiation-related genes. The expression results of MoCOM1, MoHOX2, MoCON2, MoCOS1 and MoSTUA genes that were shown previously to be important in the process of conidial development. Histogram shown the results of three biology repeats, error bars denote standard errors of three biology experiments. Asterisk denote values that are not significantly different at (p < 0.05).
Fig. S5. There is no great difference in conidial morphology between wild-type and mutants. Conidia were harvested from different mutants the observed by light microscopy. Bars = 10 μm.
Fig. S6. MoPpe1 is dispensable function in nuclear division in M. oryzae. (A) Nucleus was viewed, photograph and calculated during appressorium formation at 0, 4, 12, 24 h time point and infection phase with the transformation of H1-RFP into ΔMoppe1 mutant and Guy11 respectively. The merged image shows H1: RFP and DIC. Bars = 10 μm.
Fig. S7. MoPpe1 and MoSap1 are important for the vegetative growth and conidia formation of M. oryzae. (A) Guy11, ΔMosap1 single mutant, ΔMoppe1 ΔMosap1 double mutant and ΔMosap1 complemented strain were inoculated on CM, MM, OM and SDC media cultured at 28 °C for 7 days, then photographed. (B) Statistical analyses of the colony diameter of four different strains on different medium. Error bars represent the standard deviations, asterisks denote statistical significances (p < 0.01). (C) The conidia were photographed under a light microscope after illumination for 24 h. (D) Conidia production of Guy11, ΔMosap1 single mutant, complemented strain and ΔMoppe1 ΔMosap1 double mutant were collected after 7 days on SDC medium, then calculated and analysed. Error bars represent the standard deviations. Asterisks denote statistical significances (p < 0.01).
Fig. S8. MoPpe1 is involved in the cell wall stress response of M. oryzae. (A) Guy11, ΔMoppe1 mutant and the complemented strain were incubated on complete medium (CM) plates containing different concentrations of Congo Red (CR), Calcofluor white (CFW) and sodium dodecyl sulfate (SDS) at 28 °C for 7 days. (B) The inhibition rate was determined by plotting the percentage of colonies in the presence of various concentrations of CR, CFW and SDS against regular CM. The asterisks denote statistical significances (p < 0.01)
Fig. S9. MoSap1 is important for cell wall stress responses of M. oryzae. (A) The wild-type strain, ΔMosap1 mutant and the complemented strain were incubated on complete medium (CM) plates with different concentrations of Congo Red (CR), Calcofluor white (CFW) and sodium dodecyl sulfate (SDS) at 28 °C for 7 days. (B) The inhibition rate was determined by plotting the percentage of colonies in the presence of various concentrations of CR, CFW and SDS against regular CM, asterisks denote statistical significances (p < 0.01)
Fig. S10. The relative fungal growth assay. Diseased rice leaves were collected after 7 days inoculation. Total DNA was extracted from per 1.5 g disease leaves and test by qRT-PCR (HiScript II Reverse Transcriptase, Vazyme Biotech Co., Nanjing, China) with 28S/Rubq1 primers. The results were of three biology repeats. Single asterisks denote statistical significances (p < 0.05), double asterisks represent statistical significances (p < 0.01)
Fig. S11. MoPpe1 regulates the CWI pathway via MoPmp1. (A) Yeast two hybrid assay for the interaction between MoPpe1 and MoPmp1. The AD-MoPmp1 and BD-MoPpe1 vectors were co-introduced into yeast strain AH109, and the transformants were plated with serial dilutions of yeast cells on SD-Leu-Trp for 3 days and on selective SD-Leu-Trp-His added with 2 mM 3-AT (3-amino-1,2,4-triazole) for 10 days. (B) Interaction between MoMkk1 and MoPmp1. The AD-MoPmp1 and BD-MoMkk1 vectors were co-introduced into yeast strain AH109, and the transformants were plated with serial dilutions of yeast cells on SD-Leu-Trp for 3 days and on selective SD-Leu-Trp-His added with 2.5 mM 3-AT (3-amino-1,2,4-triazole) for 12 days. (C) MoPmp1 was hyperphosphorylated in ΔMoppe1 mutant. (D) MoPmp1 dephosphorylate the MoMkk1 in M. oryzae. (E) The MoMps1 phosphorylayion increased in ΔMopmp1 mutant. (F) The ΔMopmp1 mutant exhibited increased resistance to cell wall stress.
Fig. S12. Protein phosphatase MoPpe1 possesses the phosphatase activity that is reduced upon MoTap42 addition. Recombinant His-tagged MoPpe1 and MoTap42 was expressed in bacteria and purified. Phosphatase activity was determined using the indicated proteins and p-nitrophenyl phosphate (pNPP) as a substrate.
Fig. S13. Rapamycin treatment affects the fungal susceptibility to calcofluor white. (A) The wild-type strain, ΔMoppe1, ΔMosap1 and ΔMoppe1 ΔMosap1 mutant were incubated with 150 μg/ml Calcofluor white (CFW) stress then added with different concentrations of rapamycin 5, 10, 20 ng/ml, respectively, the strains on complete media (CM) as a control. (B) The broken line graph of each strain’s inhibition rate with different treatment. Detailed inhibition rate along with positive and negative SD was shown in each graph.
Table S1. MoMkk1 and MoPpe1 putative interacting proteins identified by affinity capture assays
Table S2. Total of primers were used in this study
Table S3. Comparison of mycological characters
Table S4. Utilization rate of the strains with different nitrogen source