Abstract
The hepatic endoplasmic reticulum (ER)–anchored monotopic proteins, cytochromes P450 (P450s), are enzymes that metabolize endobiotics (physiologically active steroids and fatty acids), as well as xenobiotics including therapeutic/chemotherapeutic drugs, nutrients, carcinogens, and toxins. Alterations of hepatic P450 content through synthesis, inactivation, or proteolytic turnover influence their metabolic function. P450 proteolytic turnover occurs via ER-associated degradation (ERAD) involving ubiquitin (Ub)-dependent proteasomal degradation (UPD) as a major pathway. UPD critically involves P450 protein ubiquitination by E2/E3 Ub-ligase complexes. We have previously identified the ER-polytopic gp78/AMFR (autocrine motility factor receptor) as a relevant E3 in CYP3A4, CYP3A23, and CYP2E1 UPD. We now document that liver-conditional genetic ablation of gp78/AMFR in male mice disrupts P450 ERAD, resulting in statistically significant stabilization of Cyp2a5 and Cyp2c, in addition to that of Cyp3a and Cyp2e1. More importantly, we establish that such stabilization is of the functionally active P450 proteins, leading to corresponding statistically significant enhancement of their drug-metabolizing capacities. Our findings, with clinically relevant therapeutic drugs (nicotine, coumarin, chlorzoxazone, and acetaminophen) and the prodrug (tamoxifen) as P450 substrates, reveal that P450 ERAD disruption could influence therapeutic drug response and/or toxicity, warranting serious consideration as a potential source of clinically relevant drug-drug interactions (DDIs). Because gp78/AMFR is not only an E3 Ub-ligase, but also a cell-surface prometastatic oncogene that is upregulated in various malignant cancers, our finding that hepatic gp78/AMFR knockout can enhance P450-dependent bioactivation of relevant cancer chemotherapeutic prodrugs is of therapeutic relevance and noteworthy in prospective drug design and development.
SIGNIFICANCE STATEMENT
The cell-surface and ER transmembrane protein gp78/AMFR, a receptor for the prometastatic autocrine motility factor (AMF), as well as an E3 ubiquitin-ligase involved in the ER-associated degradation (ERAD) of not only the tumor metastatic suppressor KAI1 but also of hepatic cytochromes P450, is upregulated in various human cancers, enhancing their invasiveness, metastatic potential, and poor prognosis. Liver-specific gp78/AMFR genetic ablation results in functional protein stabilization of several hepatic P450s and consequently enhanced drug and prodrug metabolism, a feature that could be therapeutically exploited in the bioactivation of chemotherapeutic prodrugs through design and development of novel short-term gp78/AMFR chemical inhibitors.
Introduction
Luminal and membrane-bound endoplasmic reticulum (ER) proteins incur proteolytic turnover via ubiquitin (Ub)-dependent proteasomal degradation (UPD) in a physiologic process termed “ER-associated degradation” (ERAD) (Olzmann et al., 2013; Christianson and Ye, 2014; Preston and Brodsky, 2017; Needham et al., 2019). This vital process is critical for the protein quality control required to mitigate the unfolded protein response triggered by ER stress and/or other cellular stresses, as well as for the normal physiologic ER-protein turnover (Olzmann et al., 2013; Christianson and Ye, 2014; Preston and Brodsky, 2017). Accordingly, we have found that the hepatic monotopic ER–anchored cytochromes P450 (P450s; CYPs) incur ERAD not just upon their mechanism-based inactivation that structurally damages the proteins but also in the course of their physiologic turnover (Correia et al., 1992; Wang et al., 1999; Faouzi et al., 2007). Human hepatic P450s are responsible for the metabolism and elimination of approx. 74% of therapeutically relevant drugs and participate in many drug-drug interactions (DDIs) and drug-related toxicities (Guengerich, 2015; Correia, 2018). The ERAD of human and rat hepatic P450s CYP3A4, CYP3A23, and CYP2E1 involves an initial Ser/Thr-phosphorylation by protein kinases A and C (Eliasson et al., 1992, 1994; Wang et al., 2001, 2009, 2011, 2012, 2015), their ubiquitination via Ub-activation enzymes E1 and E2/E3-Ub conjugation complexes (Morishima et al., 2005; Pabarcus et al., 2009; Wang et al., 2009, 2011, 2012; Kim et al., 2010), their extraction from the ER by p97/Ufd1/Npl4-AAA ATPase complex, and subsequent delivery to the 26S proteasome (Liao et al., 2006; Faouzi et al., 2007; Acharya et al., 2011).
In vitro functional reconstitution studies (Morishima et al., 2005; Pabarcus et al., 2009; Kim et al., 2010) of E1/E2/E3-mediated CYP3A4 and CYP2E1 ubiquitination have led to the identification of UbcH5a/Hsp70/CHIP and UBC7/gp78/AMFR complexes as two relevant E2/E3 systems in CYP3A4 and CYP2E1 ubiquitination: CHIP (carboxy-terminus of Hsc70-interacting protein), a cytoplasmic Hsc70-cochaperone, functions with its cognate UbcH5a E2 and Hsc70/Hsp70 in substrate ubiquitination (Ballinger at al., 1999; Connell et al., 2001; Jiang et al., 2001; Murata et al., 2001). gp78/AMFR (autocrine motility factor receptor) is a polytopic, transmembrane cell surface protein (Nabi et al., 1990), as well as an ER-integral protein with a cytoplasmic domain containing a RING-finger and other subdomains critical to its recruitment of its cognate E2 (UBC7 or Ube2G2) and catalytic E3-ligase–mediated ubiquitination role (Fang et al., 2001; Zhong et al., 2004; Chen et al., 2006, 2012; Joshi et al., 2017).
Our lentiviral short-hairpin RNA interference (shRNAi) analyses targeted individually against CHIP and gp78 verified their roles in CYP3A and CYP2E1 ubiquitination and ERAD in cultured rat hepatocytes (Kim et al., 2010). Thus, upon CHIP knockdown, CYP3A was stabilized largely as the parent (55-kDa) species along with a minor CYP3A fraction consisting of its high molecular mass ubiquitinated species. Similar stabilization of CYP3A parent species was also found upon gp78 knockdown, albeit with an appreciable fraction as its ubiquitinated species. Nevertheless, upon either E3 knockdown of CHIP or gp78, the functionally active CYP3A fraction, albeit proportional to the relative amount of the stabilized parent protein, was statistically significantly greater than that in the corresponding control (nontargeting) shRNA-treated hepatocytes (Kim et al., 2010). Furthermore, in vitro reconstitution studies of each CYP3A4 E2/E3-ubiquitination system, individually and in combination, revealed that although each system by itself was capable of CYP3A4 ubiquitination, this was both greatly accelerated and potentiated when both systems were present simultaneously (Wang et al., 2015). Thus, the sequential introduction of each E2/E3 in the reconstituted CYP3A4-ubiquitination system revealed that UbcH5a/CHIP most probably serves as the E3, whereas UBC7/gp78 serves as the E4 involved in the elongation of CYP3A4 polyUb chains (Wang et al., 2015).
Consistent with shRNAi findings, Cyp2e1 was also stabilized in a functionally active form upon mouse CHIP knockout (KO) (Kim et al., 2016a). This functional Cyp2e1 stabilization was associated with increased hepatic lipid peroxidation, microvesicular steatosis, and eventually, upon aging, with macrovesicular steatosis and liver injury characteristic of the nonalcoholic steatohepatitis (NASH) syndrome (Kim et al., 2016a).
To determine whether genetic ablation of gp78, the other E3 Ub-ligase identified as relevant to hepatic CYP3A4 and CYP2E1 ERAD, had similar consequences, we generated a mouse liver-conditional gp78-KO (gp78−/−) model. This gp78−/− model enabled us not only to examine the role of gp78-mediated ubiquitination in the ERAD and consequent physiologic function of Cyps 3a and Cyp2e1 but also to identify additional hepatic P450s as potential gp78-ubiquitination targets and to determine their therapeutic/toxicological consequences. Our findings are described herein.
Materials and Methods
Generation of a Liver-Conditional gp78-KO Mouse.
Cryopreserved mouse embryos carrying a conditional amfrtma1(Komp)Wtsi gene-trap knockout-first allele inserted into amfr intron 2 with loxP sites flanking exon 3 of amfr were generated as described (Skarnes et al., 2011; Bradley et al., 2012), resuscitated, cultured, and transplanted into oviducts of pseudopregnant females of C57BL/6J-Tyrc-Brd; C57BL/6N background, at the UC Davis Knockout Mouse Project Facility. Upon weaning, the heterozygous conditional gene-trap (cgt) genotype of the pups was confirmed by polymerase chain reaction (PCR) analyses. Mice aged 6 weeks were shipped to UCSF LARC Transgenic Barrier Facility, where the loxP-flanking sequence was deleted through mating of 8-week-old heterozygous females with transgenic male mice (C57BL/6J-background) carrying Flpe (flippase) recombinase driven by the β-actin promoter (Flpe-deleter; β-actin-Flp), and of corresponding heterozygous males with female Flpe-mice. After several intercrosses, homozygous gp78-floxed (flp/flp) mice were obtained. To generate liver-specific gp78 KOs, flp/flp mice were crossbred with Alb-Cre-recombinase (Cre)/Alb-Cre (B6.Cg-Tg(Alb-cre)21Mgn/J) mice (Postic and Magnuson, 2000). The resulting flp/+, Alb-Cre/+ mice were then crossed again with flp/flp mice to generate flp/flp, Alb-Cre/+ mice as liver-specific gp78 KOs. The primers (Tables 1 and 2) used for the tail-clip genotyping were located as indicated by the arrows (Fig. 1A). Mice were fed a standard chow diet and maintained under 12-hour light/dark cycle. All animal experimental protocols were approved by the UCSF/Institutional Animal Care and Use Committee.
TABLE 1.
Summary of the primer sequences used for genotyping
| Primer Name | Nucleotide Sequence (5′-3′) |
|---|---|
| 1: CSD‐ LoxP‐common forward | GAGATGGCGCAACGCAATTAAT |
| 2: Gene‐specific reverse (SR1) | CAGAGATTCTACCTCCTGAGTGCTGG |
| 3: WT‐F | GGATCTGAAATTCACTGAGCAGTGC |
| 4: WT‐TTR | TGTACATGTTCCTTCATGTGAGGAAG |
| 5: CSD-LoxF | GAGATGGCGCAACGCAATTAATG |
| 6: CSD-Amfr-R | ACCTCCTGAGTGCTGGGATTAAAGG |
| flpF | CACCTAAGGTCCTGGTTCGTCA |
| flpR | CCCAGATGCTTTCACCCTCACT |
| creF | CATTTGGGCCAGCTAAACAT |
| creR | TAAGCAATCCCCAGAAATGC |
cre, Cre-recombinase; F, forward; R, reverse.
TABLE 2.
Summary of the PCR product (amplicon) size for each primer pair and the corresponding genotype represented
| Primer Pairs | Band Size (bp) | Genotype |
|---|---|---|
| 1+2 | 337 | Cgt (conditional gene-trap cassette) |
| 3+4 | 200 | Wild type (WT) |
| flpF+flpR | 435 | With flpe recombinase (flpe) |
| 5+6 | 327 | Floxed (fl) |
| creF+creR | 233 | With Cre recombinase |
cre, Cre-recombinase; F, forward; R, reverse.
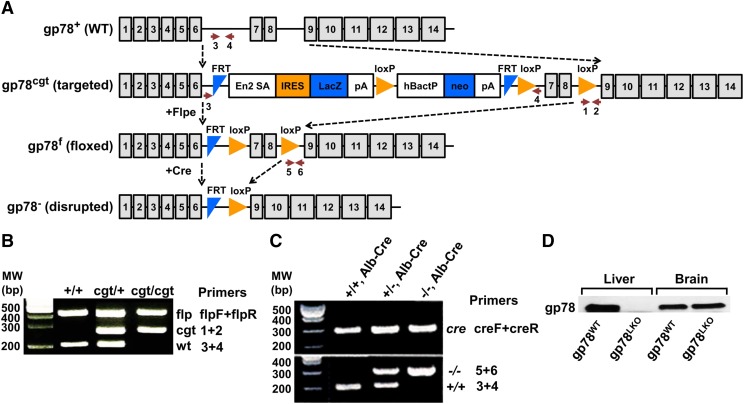
Fig. 1.
Strategy for generating the liver-conditional gp78/AMFR-KO mouse model and corresponding validation. (A) Conditional gene trap (cgt) strategy to develop gp78-floxed (gp78f) mice and, subsequently, liver-conditional gp78-KO mice (gp78−). Arrows indicate the sites of the genotyping primers employed. (B and C) PCR genotype analyses of genomic DNA isolated via tail-clipping of mice at various stages of the strategy. See Materials and Methods for experimental details. The primer pairs for each genotyping product and the corresponding amplicon size are indicated. flpF/flpR primers used to verify the amplicon obtained upon Flpe recombinase in WT (+/+); cgt/+, heterozygous with the cgt-cassette insert; and cgt/cgt, homozygous with the cgt-cassette insert. Amplicons verifying WT gp78 (+/+), Alb/Cre, heterozygous gp78-KO (+/−), and Alb-Cre and homozygous gp78-KO (−/−), each Alb-Cre carrying an Alb-Cre allele upon mating with Alb-Cre/Alb-Cre mice. (D) IB analyses of liver and brain lysates from WT and KO mice with a rabbit anti-mouse gp78-antibody, verifying the liver-specific gp78 KO.
Primary Mouse Hepatocyte Cultures.
Male mice, 8 to 9 weeks of age, were used throughout because in our preliminary studies, specific hepatic Cyp3a isoforms exhibited sex-dependent differences in content upon liver-conditional gp78-ablation (S.M. Kim and M.A. Correia, unpublished findings). Hepatocyte isolation from male gp78+/+:Alb-Cre [wild-type (WT); gp78+/+] and gp78−/−:Alb-Cre (KO; gp78−/−) mice was carried out by in situ liver perfusion with collagenase and purification by Percoll-gradient centrifugation by the UCSF Liver Center Cell Biology Core as described previously (Han et al., 2005). In a few studies, freshly isolated primary hepatocytes were used for monitoring the basal, hepatic constitutive P450 isoform content (see below). Alternatively, freshly isolated hepatocytes were cultured (3.0 × 106 cells/plate) on 60-mm Permanox plates (Thermo Fisher Scientific, Waltham, MA) coated with Type I collagen, and incubated in William’s E Medium containing 2 mM l-glutamine, penicillin-streptomycin, insulin-transferrin-selenium, 0.1% bovine serum albumin fraction V, and 0.1 μM dexamethasone (DEX). Cells were allowed to attach to the plates for 6 hours, before Matrigel (Matrigel Matrix; Corning Inc., New York) was overlaid and cell cultures allowed to stabilize for 2 days, during which the initial basal P450 content was almost completely lost. To restore functional P450 cellular content, after a 2-day stabilization, cultured primary hepatocytes were treated for 3 days with the following P450-inducers: β-naphthoflavone (β-NF; 25 μM; Cyp1a2), phenobarbital (PB; 1 mM; Cyps 2a, 2b and 2c), DEX (10 μM; Cyps 3a), and isoniazid (INH; 1 mM, Cyp2e1).
Determination of Relative P450 Protein Content in Freshly Isolated or Cultured gp78-KO and WT Mouse Hepatocytes.
In certain studies, freshly isolated hepatocytes were lysed in cell lysis buffer (Cell Signaling Technology, Inc., Danvers, MA) and directly used for immunoblotting (IB) analyses of constitutive P450s. In others, upon inducer treatment, cultured cells were harvested in the same cell lysis buffer. In either case, cell lysates were cleared by centrifugation (14,000g, 10 minutes, 4°C), and P450 proteins in the supernatant were subjected to Western IB analyses. Commercial primary antibodies were used for detecting Cyp1a2 (mouse monoclonal, cat. no. 14719; Cell Signaling Technology, Inc.), Cyp2a (rabbit polyclonal, cat. no. ab3570; Abcam, Cambridge, UK), and Cyp2d (rabbit polyclonal, cat. no. GTX56286; Genetex, Inc. Irvine, CA). Purified antibodies from rabbit (Cyp2b, Cyp2c, and Cyp2e1) and from goat (Cyps 3a) sera were also used as previously described (Kim et al., 2016a,b). Immunoblots were densitometrically quantified through NIH Image J software analyses.
Relative P450 mRNA Expression in Cultured WT and gp78-KO Mouse Hepatocytes.
βNF- and INH-pretreated cultured WT and gp78-KO hepatocytes were used for Cyp1a2 and Cyp2e1 mRNA level determination, respectively; PB-pretreated hepatocytes were used for monitoring Cyp2a5, 2b10, 2c29, 2c39, and 2d10 mRNA levels, whereas DEX-pretreated hepatocytes were used for monitoring Cyp3a11 and Cyp3a13 mRNA levels. On day 4, total RNA was extracted from the cultured hepatocytes with RNeasy mini-kit (Qiagen, Hilden, Germany), and cDNA was synthesized using SuperScript IV VILO Master Mix (Thermo Fisher Scientific). Hepatic P450 mRNA expression was determined by quantitative real-time (qRT)-PCR analyses, using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), and monitored by Agilent Mx3005P System (Agilent Technologies, Santa Clara, CA). Relative gene expression level of each P450 was determined by normalizing its threshold cycle (Ct) to that of GAPDH Ct. Primers used are listed (Table 3).
TABLE 3.
Primers used for qRT-PCR analyses of P450 mRNA expression
| Gene | Sequence 5′ → 3′ |
|---|---|
| Cyp1a2 | ATGATGAGAAGCAGTGGAAAGA CAGGATGGCTAAGAAGAGGAAG |
| Cyp2a5 | CACCCAGATATTGAGGCCAA CCCAGCATAGGAAACACTTCA |
| Cyp2b10 | AGGAGGAAGCCCAATGTTTAG CTCAAACATCTGGCTGGAGAA |
| Cyp2c29 | GTCACAGCTAAAGTCCAGGAAG TCATGCAGCACTGATGACAG |
| Cyp2c39 | GCAGGTCTGCAATAATTTCCC GTTGGATGTGATTTGCCTGTT |
| Cyp2d10 | CTGGCAGAGATAGAGAAGGTAAAG CCCTATGACCGCATCGATTT |
| Cyp2e1 | TCCTCATAGAGATGGAGAAGGAAA GGCCCAATAACCCTGTCAAT |
| Cyp3a11 | GTGAACGTGGATTCCCTCAA CTCGGTGCTTCTGCTTAGAAT |
| Cyp3a13 | TACCCTGCTACAGATGGAGTAT CTGATTCTTCTTGCTGAACCTTTC |
| gapdh | ACCACAGTCCATGCCATCAC CACCACCCTGTTGCTGTAGCC |
Cyp2a Ubiquitination and Degradation Analyses.
Cultured PB-pretreated hepatocytes were subjected to cycloheximide-chase analyses by treatment on day 4 at 0 hour with cycloheximide 50 μg/ml with or without the proteasome inhibitor bortezomib (10 μM), or the autophagy-lysosomal degradation (ALD) inhibitors, 3-methyladenine (3-MA; 5 mM/NH4Cl; 50 mM) for 0, 8, and 24 hours. Cells were harvested and the lysates were used for Cyp2a IB analyses.
Cyp2a5 ubiquitination analyses were conducted following P450 immunoprecipitation from the lysates (200 μg protein) of hepatocytes treated with or without bortezomib (10 μM) for 8 hours. Immunoprecipitates were incubated with Cyp2a antibody at 4°C overnight. The antibody-antigen complexes were captured by incubation with Dynabeads protein G (Thermo Fisher Scientific) at room temperature for 2 hours and then eluted by heating at 95°C for 10 minutes in 2× volume of SDS-PAGE loading buffer. The ubiquitinated P450s were detected by Western IB analyses with a mouse monoclonal Ub-antibody (cat. no. ab7254; Abcam plc), as described previously (Wang et al., 1999, 2015; Kim et al., 2010).
Fluorescence-Based Screening of P450 Functional Markers in Cultured Mouse Hepatocytes.
P450 functional activities in cultured primary hepatocytes were determined by the method of Donato et al. (2004), as previously described (Kim et al., 2010). On day 4 after P450-inducer pretreatment, hepatocytes were incubated with P450 substrates, 3-cyano-7-ethoxycoumarin (CEC, 30 μM; Cyp1a2; cat. no. 451014.; Corning Inc.), coumarin (CM, 50 μM; Cyp2a5; cat. no., C4261; Sigma-Aldrich Corp., St. Louis, MO), 7-ethoxy-4-(trifluoromethyl)-coumarin (EFC, 30 μM; Cyp2b; cat. no. T2803; Sigma-Aldrich Corp.), dibenzylfluorescein (DBF, 10 μM; Cyp2c; cat. no. 451740; Corning Inc.), 3-[2-(N,N-diethyl-N-methylammonium)ethyl]-7-methoxy-4-methylcoumarin (AMMC, 100 μM; Cyp2d; cat. no. 451700; Corning Inc.), 7-methoxy-4-(trifluoromethyl)-coumarin (MFC, 10 μM; Cyp2e1; cat. no. 451740; Corning Inc.), or 7-benzyloxy-4-trifluoromethylcoumarin (BFC, 100 μM; Cyp3a; cat. no. B5057; Sigma-Aldrich Corp.) at time 0 hour. The media containing CEC metabolite 3-cyano-7-hydroxycoumarin (CHC; cat. no. 451015; Corning Inc.), CM metabolite 7-hydroxycoumarin (7-HC; cat. no. H24003; Sigma-Aldrich Corp.), BFC, EFC, and MFC metabolite 7-hydroxy-4-trifluoromethylcoumarin (HFC; cat. no. 451731; Corning Inc.), or DBF metabolite fluorescein (cat. no. 46955; Sigma-Aldrich Corp.), along with their glucuronide and sulfate conjugates formed during the assay were collected at defined time points. The AMMC metabolite AMHC was commercially unavailable, and thus 3-[2-(diethylamino)ethyl]-7-hydroxy-4-methylcoumarin (AHMC; cat. no. 188611; Sigma-Aldrich Corp.) was used instead as previously described for the fluorescence quantification (Donato et al., 2004). The conjugates were hydrolyzed by incubation of the media with glucuronidase/arylsulfatase (150 Fishman units/ml and 1200 Roy units/ml, respectively) for 2 hours at 37°C. The medium concentrations of CEC, 7-HC, HFC, fluorescein, and AHMC were fluorometrically monitored at the wavelength [excitation (nanometer)/emission (nanometer)] of 408/455, 355/460, 410/510, 485/538, and 390/460, respectively, using the commercially purchased metabolites as authentic standards.
Nicotine Metabolism in Cultured Mouse Hepatocytes.
WT and gp78-KO hepatocytes were PB-pretreated as described above. On day 4, (–)-nicotine (500 μM) was added at time 0 hour and the cells and media were collected at 2, 4, 8, and 24 hours, and incubated with glucuronidase/arylsulfatase for 2 hours at 37°C. The reaction was stopped by the addition of ice-cold 30% perchloric acid, and the precipitated proteins were sedimented by centrifugation (15,000g, 10 minutes, 4°C). Nicotine metabolites (cotinine and trans-3′-hydroxycotinine, nornicotine, norcotinine, nicotine-N-oxide, and cotinine-N-oxide) were extracted from the supernatant and quantified by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses as described for cotinine and trans-3′-hydroxycotinine (Jacob et al., 2011), with minor modifications for the additional analytes (Benowitz et al., 2010, Wang et al., 2018a). The metabolites were found largely in the media, with relatively much smaller levels detected in the cell lysates.
Tamoxifen Metabolism in Cultured Mouse Hepatocytes.
Cyp3a was induced by DEX-pretreatment of primary hepatocytes as described above. On day 4, cultured hepatocytes were treated with tamoxifen (40 μM) for 2, 4, and 8 hours. The concentrations of tamoxifen and its metabolites, 4-hydroxytamoxifen, N-desmethyltamoxifen, and endoxifen, in the media were determined using a high-performance liquid chromatography–mass spectrometric (HPLC-MS) assay (Dahmane et al., 2010; Binkhorst et al., 2011). Toremifen was employed as an internal standard, and spiked into the media at 1 μM. The medium was incubated with glucuronidase/arylsulfatase for 2 hours at 37°C and the reactions terminated by the addition of 2× volume of ice-cold acetonitrile containing 0.1% formic acid. Precipitated proteins were removed by sedimentation at 10,000g for 10 minutes at 4°C, and an aliquot (300 μl) of the supernatant was mixed with 50 mM glycine buffer (pH 11.5; 700 μl). An aliquot (50 μl) of this mixture was injected into the HPLC system (Waters Separation Module 2695; Waters Corporation, Milford, MA), equipped with a C18 reversed-phase column (100 × 2.1 mm, 3 μm, Hypersil Gold aQ; Thermo Fisher Scientific), and maintained at a temperature of 30°C. An aqueous solution containing 0.1% formic acid was used as the mobile phase A, and acetonitrile containing 0.1% formic acid was used as the mobile phase B. Mobile phases A (30%) and B (70%) were applied isocratically at a flow rate of 0.2 ml/min. The peaks were detected at 254 nm by Waters dual absorbance detector 2487 (Waters Corporation). The retention times of endoxifen, 4-hydroxytamoxifen, toremifen, N-desmethyltamoxifen, and tamoxifen were 9.26, 11.16, 22.17, 23.77, and 26.92 minutes, respectively. The HPLC system was connected to a Waters Micromass ZQ single quadrupole mass analyzer (Waters Micromass Corporation, Manchester, U.K.), equipped with an electrospray ionization probe operated in the positive ionization mode. The ion-spray and cone voltage were 3 kV and 45 V, respectively. The protonated molecular ion [M+H]+ masses of N-desmethyltamoxifen, tamoxifen, endoxifen, 4-hydroxytamoxifen, and toremifen were found to be 358.2, 372.2, 374.2, 388.2, and 406.2 amu, respectively.
Chlorzoxazone Metabolism in Cultured Mouse Hepatocytes.
Cultured primary WT and KO mouse hepatocytes were INH-pretreated to induce Cyp2e1, as described above. On day 4, at time 0 hour, chlorzoxazone (400 μM) was added to the hepatocytes for 3 hours. The 6-hydroxychlorzoxazone (6-OH CZX) formed by Cyp2e1 was monitored by HPLC as described (Girre et al., 1994) with a slight modification. The collected medium was first incubated with glucuronidase/arylsulfatase for 2 hours at 37°C, and the reaction stopped with 40% perchloric acid. After sedimentation of the precipitated protein, the supernatant was mixed with 1.5× volume of ethyl acetate by vigorous vortex mixing for 20 minutes. The upper organic phase was collected and evaporated at 40°C. The resulting residue was reconstituted with 100 μl of mobile phase, acetonitrile, and 0.5% acetic acid (30:70, v/v, pH 3), and a 50-μl aliquot was injected into a Varian Prostar HPLC (Varian Inc., Palo Alto, CA) system, equipped with a Grace Prosphere 100 C18, 5 μm, 4.6- × 150-mm column (Thermo Fisher Scientific) for metabolite separation at a flow rate of 1.1 ml/min. of 6-OH-CZX (retention time of 7.6 minutes) was detected at 287 nm with a Varian Prostar 335 UV detector (Varian Inc.).
Acetaminophen-Induced Hepatotoxicity.
Cultured mouse hepatocytes pretreated with INH (1 mM) for 3 days were used for the acetaminophen (APAP) toxicity assay. On day 4, hepatocytes were incubated with APAP (1 or 5 mM) and the medium was collected at 2, 4, 8, and 24 hours. The medium alanine aminotransferase (ALT) activity was quantified using the ALT assay kit (MAK052; Sigma-Aldrich Corp.).
In Vivo APAP-Induced Hepatotoxicity in Liver-Specific gp78-KO Mice.
Male 8 week-old C57BL/6 mice (WT, n = 5; gp78 KO, n = 5) were fasted overnight (12 hours). APAP (200 mg/kg, i.p.) dissolved in 0.9% NaCl solution (10 ml/kg) was injected into the mice at time 0 hour. Blood was collected from the facial vein at 0, 6, and 12 hours after the injection, and the mice were killed at 12 hours to collect the liver tissue. Serum was isolated from the whole blood and ALT levels were monitored as described above. Hepatic malondialdehyde (MDA) levels were measured using liver homogenates by the TBARS assay kit (cat. no. 10009055; Cayman Chemical, Ann Arbor, MI). Cyp2e1 and Cyp3a protein levels in liver homogenate (5 μg) were determined by Western IB analyses using corresponding antibodies as indicated above. Fresh liver tissues were stored in 10% neutral buffered formalin, and the tissue sections were stained with hematoxylin and eosin. Microscopic histopathologic examination was performed by the UCSF Liver Center Pathology, Histology and Light Microscopy Cores.
Statistical Analyses.
In a strict statistical sense, this study is exploratory by nature and accordingly, all statistical analyses, employing the Student’s t test, should be considered descriptive and not hypothesis-testing. Our sample size was planned to be at least three male mice, but final sample sizes depended on how many male mice were in a given litter.
Results
Verification of the Liver-Conditional gp78-KO Mouse Genotype
The cgt-targeting strategy employed is shown (Fig. 1A). This strategy is different from that reported previously (Liu et al., 2012) in that the cgt cassette targeted only seven to eight exons (vs. five to eight), and it also employed resuscitated cryopreserved embryos rather than genetically engineered embryonic stem cells as the starting material. Verification of the genomic DNA by tail-clip genotyping via PCR analyses of wild-type (WT; gp78+/+) flp/flp-mice, cgt/+, gp78+/−, and gp78−/− mice, respectively, is shown (Fig. 1, B and C). Parallel IB analyses of liver and brain tissue lysates from WT and gp78−/−-mice provided unequivocal evidence of liver-conditional gp78 KO (Fig. 1D).
Identification of Hepatic P450s as gp78 Targets
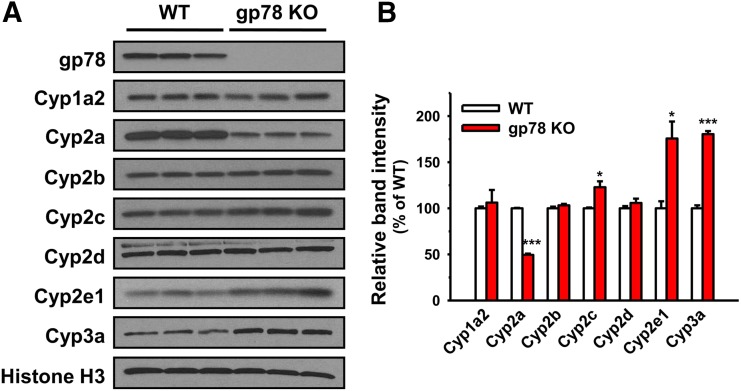
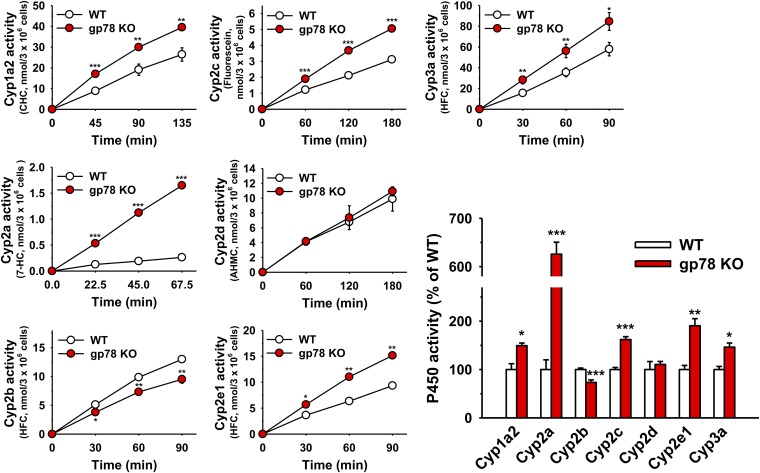
IB analyses of lysates from untreated, freshly isolated hepatocytes revealed the statistically significant stabilization of constitutive Cyp2e1 and Cyps 3a upon liver-conditional gp78 KO relative to corresponding age/strain-matched WT mice (Fig. 2). A slight, albeit statistically significant relative stabilization of constitutive Cyps 2c was also likewise detected (Fig. 2). By contrast, the expression of constitutive Cyps 2a, for reasons currently unclear, was statistically significantly decreased upon gp78 KO (Fig. 2). Furthermore, consistent with our gp78-shRNAi findings in cultured rat hepatocytes, IB analyses of lysates from cultured mouse hepatocytes verified that DEX-inducible Cyps 3a and INH-inducible Cyp2e1 were also statistically significantly stabilized upon gp78 KO (gp78−/−) relative to those in the WT (Fig. 3). Corresponding diagnostic functional probes revealed that this stabilization was largely of the functionally active forms (Fig. 4). Similar conclusions were also drawn for PB-inducible Cyps 2c (Figs. 3 and 4). The Cyp1a2 increase in gp78−/− hepatocytes relative to the WT hepatocytes, on the other hand, may be partially attributable to transcriptional induction (Fig. 5). By contrast, the relative hepatic content and corresponding functional activity of Cyps 2d were unaffected, whereas those of Cyps 2b were even decreased upon gp78 KO (Figs. 3 and 4). Surprisingly, in contrast to the statistically significant reduction of constitutive Cyps 2a (Fig. 2), a marked stabilization of PB-inducible Cyps 2a was observed in gp78−/− over gp78+/+ hepatocytes (Fig. 3), with a correspondingly marked relative increase of coumarin-7-hydroxylase in gp78−/− over gp78+/+ hepatocytes (Fig. 4). Thus, in addition to Cyps 3a and Cyp2e1, these studies identified Cyps 2a and Cyps 2c as gp78 targets, with their diagnostic functional probes attesting to their corresponding statistically significant functional increases. Furthermore, on the basis of: 1) the observed PB inducibility of the Cyp2a enzyme; 2) its close functional correlation with the elevated coumarin-7-hydroxylase activity, an excellent Cyp2a5 but not Cyp2a4-diagnostic probe (Chang and Waxman, 1996), coupled with 3) its ability for cotinine and trans-3′-hydroxycotinine formation (see below; Zhou et al., 2010), lead us to propose Cyp2a5 as the Cyp2a isoform most probably stabilized.
Fig. 2.
IB analyses of constitutive hepatic P450s from WT and gp78-KO mice. (A) IB analyses of a representative mouse sample. (B) Densitometric quantification of the approx. 50–55 kDa P450 band from hepatocytes freshly isolated and uncultured from three individual WT and gp78-KO mice. See Materials and Methods for details. Values (mean ± S.D.) of three individual experiments, each employing two culture plates (technical replicates) of hepatocytes isolated from each of the three mice. Statistical significance determined by the Student’s t test, *,***P < 0.05, and 0.001 vs. WT, respectively.
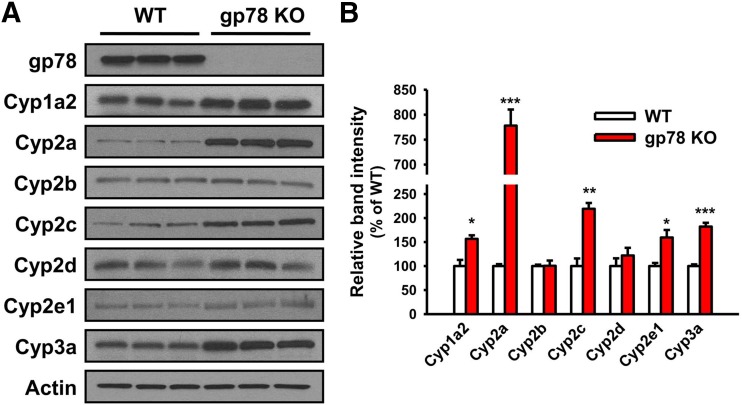
Fig. 3.
IB analyses of hepatic P450s from cultured WT and gp78-KO mouse hepatocytes. (A) IB analyses of a representative mouse sample. (B) Densitometric quantification of the approx. 50- to 55-kDa P450 band from hepatocytes isolated from three individual WT and gp78-KO mice, cultured and pretreated with P450-inducers. See Materials and Methods for details. Values (mean ± S.D.) of three individual experiments, each employing two culture plates (technical replicates) of hepatocytes isolated from each of the three mice. Statistical significance determined by the Student’s t test, *,**,***P < 0.05, 0.01, and 0.001 vs. WT, respectively.
Fig. 4.
P450 functional screening using diagnostic fluorescent probes. Hepatocytes from three WT and gp78-KO mice were cultured separately, pretreated with P450-inducers, and then incubated with individual diagnostic functional probes at time 0 hour. See Materials and Methods for details. Values (mean ± S.D.) of three individual experiments each employing two culture plates (technical replicates) of hepatocytes isolated from each of the three mice. Statistical significance determined by the Student’s t test, *,**,***P < 0.05, 0.01, and 0.001 vs. WT, respectively.
Fig. 5.
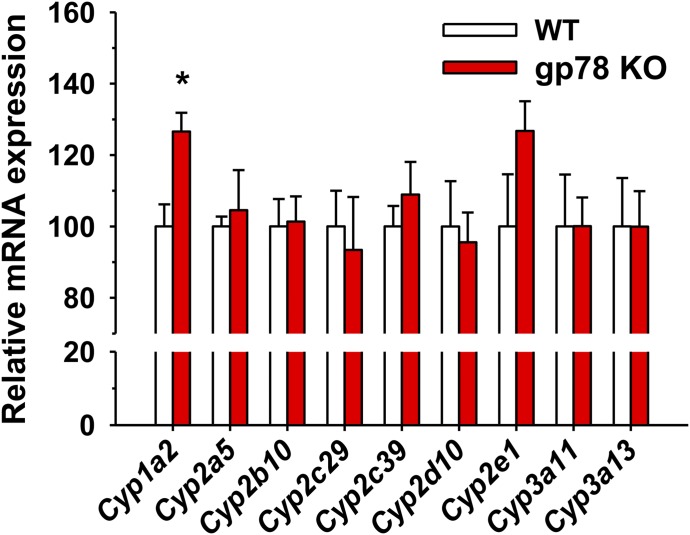
Relative P450 mRNA expression in cultured WT and gp78-KO mouse hepatocytes. Total RNA was extracted on day 4 from cultured hepatocytes, untreated or pretreated with P450 inducers for 3 days. Hepatic P450 mRNA expression was determined by qRT-PCR analyses as detailed in Materials and Methods. Values (mean ± S.D.) are derived from three individual cultures pretreated with P450 inducers as detailed. Statistical significance determined by the Student’s t test, *P < 0.05 vs. WT. GAPDH mRNA Ct levels (mean ± S.D.) between P450-inducer pretreated WT and gp78-KO cultured hepatocytes were as follows: βNF: 100 ± 1.92 (WT)/101.49 ± 1.75 (gp78 KO); PB: 100 ± 1.67 (WT)/100.33 ± 1.61 (gp78 KO); INH: 100 ± 1.99 (WT)/99.72 ± 1.66 (gp78 KO); DEX: 100 ± 2.19 (WT)/102.85 ± 3.94 (gp78 KO). No statistically significant differences were noted by the Student’s t test in GAPDH Ct values between WT and gp78-KO cultured hepatocytes pretreated with each of the four P450 inducers. Ct, threshold cycle.
Functional P450 Enhancement Results from Corresponding P450 Protein Stabilization upon gp78 KO.
We have previously reported that the CYP3A stabilization upon lentiviral shRNAi-elicited gp78 knockdown in cultured rat hepatocytes is attributable to its protein stabilization rather than increased CYP3A mRNA expression (Kim et al., 2010). Relative hepatic P450 mRNA expression analyses by qRT-PCR analyses of total RNA isolated from gp78−/− and gp78+/+ hepatocytes after each P450-inducer pretreatment coupled with the use of P450 isoform-specific probes revealed that the observed relative P450 content and functional increases (except in the case of Cyp1a2) were not caused by their transcriptional mRNA induction (Fig. 5).
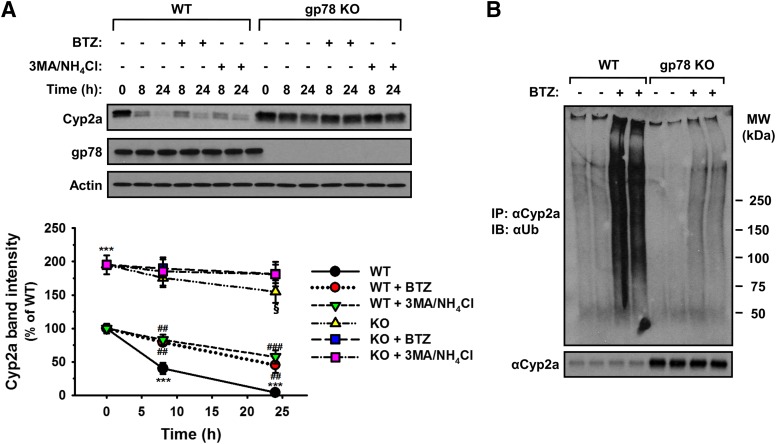
Cycloheximide-chase analyses of gp78−/− and gp78+/+ hepatocytes coupled with concurrent treatment with either the proteasome inhibitor bortezomib or the ALD inhibitors (3-MA/NH4Cl) revealed that Cyp2a5 degradation was considerably slower in gp78−/− hepatocytes relative to gp78+/+ hepatocytes but involved both UPD and ALD (Fig. 6A). Cyp2a5 immunoprecipitation from PB-pretreated gp78−/− and gp78+/+ hepatocytes and protein ubiquitination analyses directly verified that Cyp2a5 was primarily a gp78 target, as its ubiquitinated level was markedly reduced upon gp78 KO, and bortezomib-mediated inhibition of its proteasomal degradation failed to enhance it (Fig. 6B). Thus, gp78-mediated Cyp2a5 ubiquitination targets the protein for both UPD and ALD.
Fig. 6.
Hepatic Cyp2a5 UPD and ALD: a major role of gp78 in Cyp2a5 ubiquitination. (A) Cycloheximide-chase analyses of hepatocytes from three individual WT and gp78-KO mice were cultured, PB-pretreated for 3 days, and on day 4 treated with or without the proteasomal inhibitor bortezomib (BTZ) or the ALD-inhibitors 3-MA/NH4Cl for 0, 8, and 24 hours before cell harvesting. Cell lysates were immunoblotted for Cyp2a or gp78 with actin as a loading control. Densitometric quantification (mean ± S.D.) of individual cultures from three mice. Statistical significance determined by the Student’s t test, ***P < 0.001 vs. WT at 0 hour; ##, ###P < 0.01, 0.001 vs. WT+ CHX; §P < 0.05 vs. KO at 0 hour. (B) Representative Ub-immunoblotting analyses of Cyp2a-immunoprecipitates from aliquots of PB-pretreated cultured cell lysates obtained from two of the three different mice treated with or without BTZ as in (A).
Pharmacological Relevance of Hepatic P450 Stabilization upon gp78 KO
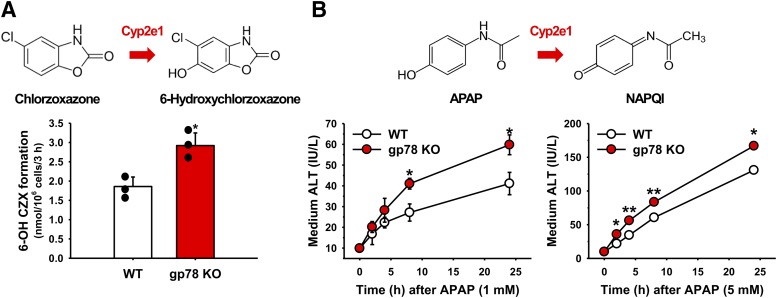
With the exception of coumarin, the above functional analyses employed readily assayable fluorescent diagnostic probes rather than therapeutic and/or pharmacologically active drugs as substrates. To determine the potential therapeutic relevance of CYP2e1 stabilization upon gp78 KO, we examined in gp78−/− and gp78+/+ hepatocytes the Cyp2e1-dependent 6-hydroxylation of chlorzoxazone, a clinically prescribed skeletal muscle relaxant and an established Cyp2e1 functional probe, (Fig. 7A). A statistically significant relative increase was detected in 6-OH-chlorzoxazone formation in gp78−/− over gp78+/+ hepatocytes (Fig. 7A), comparable to that observed with its other diagnostic Cyp2e1 probe (MFC) (Fig. 4), thus underscoring the potential therapeutic significance of these findings.
Fig. 7.
Relative chlorzoxazone metabolism and APAP-elicited liver injury in cultured WT and gp78-KO mouse hepatocytes. (A) INH-pretreated hepatocytes (three individual cultures) from WT and gp78-KO mice were incubated on day 4 with chlorzoxazone for 3 hours and 6-hydroxychlorzoxazone assayed as detailed in Materials and Methods. Values (mean ± S.D.) of three individual cultures. Statistical significance determined by the Student’s t test, *P < 0.05 vs. WT. (B) WT and gp78-KO mouse hepatocyte cultures were INH-pretreated, and on day 4 incubated with APAP (1 or 5 mM) for 0–24 hours. Aliquots of media were taken at the time points indicated and assayed for ALT activity as detailed in Materials and Methods. Values (mean ± S.D.) of three individual cultures. Statistical significance determined by the Student’s t test, *P < 0.05 vs. WT, **P < 0.01 vs. WT.
Cyp2e1 largely and Cyps 3a to a lesser extent are also involved in the metabolism of the clinically prescribed nonsteroidal analgesic acetaminophen (APAP) to its reactive nucleophilic species N-acetylparaquinoneimine (NAPQI) (Lee et al., 1996; Wolf et al., 2007). This pathway becomes increasingly hepatotoxigenic when liver concentrations of ingested APAP exceed the detoxification capability of its hepatic glucuronidation and sulfation pathways (Gillette et al., 1981; Nelson, 1990). More of the parent drug is then shunted into the reactive NAPQI pathway with consequent liver damage that can be clinically fatal (Nelson, 1990). To determine the potentially hepatotoxicological relevance of Cyp2e1 functional stabilization upon gp78 KO, we examined INH-pretreated gp78−/− and gp78+/+ hepatocytes treated with APAP at two different concentrations: 5 mM, the concentration routinely employed to trigger APAP-induced liver damage (Burcham and Harman, 1991; Bajt et al., 2004), and 1 mM, one-fifth the toxic concentration, over 24 hours of culture. Although, as expected, the 5-mM concentration led to statistically significantly increased injury to gp78+/+ hepatocytes, as monitored by the extracellular spillage into the cell medium of the liver cytoplasm-specific ALT enzyme, this was statistically significantly greater in gp78−/− hepatocytes (Fig. 7B). At the 1-mM concentration, however, although APAP produced a mild extracellular ALT rise in gp78+/+ hepatocytes, this was considerably accelerated and pronouncedly greater in gp78−/− hepatocytes (Fig. 7B). These findings thus attest to the relevance of P450 stabilization upon gp78 KO to drugs whose hepatotoxigenic potential largely depends on their Cyp2e1-mediated bioactivation.
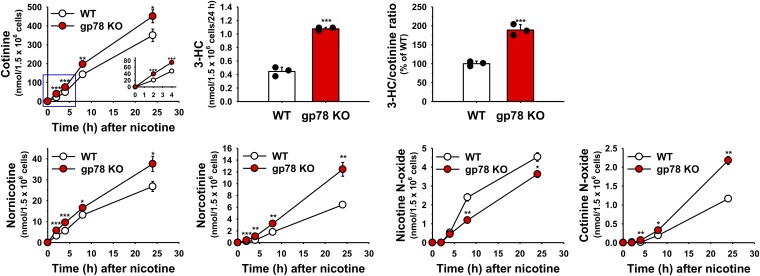
Furthermore, to determine the pharmacological relevance of the relatively marked functional Cyp2a5-stabilization, we also determined its effects on the metabolism of its prototype drug substrate nicotine, a pharmacologically active drug (Fig. 8). As expected, nicotine was metabolized to cotinine (via the nicotine-iminium ion and subsequent oxidation by aldehyde oxidase), nornicotine, norcotinine, trans-3′-hydroxycotinine, nicotine-N-oxide, and cotinine-N-oxide (Cashman et al., 1992; Nakajima et al., 1996; Hukkanen et al., 2005; Siu et al., 2006; Zhou et al., 2010). Whereas the nicotine-N-oxide is reportedly generated by flavin-monooxygenase (FMO3; Cashman et al., 1992), the other four metabolites are generated by various P450s, of which Cyp2a5 is the major catalyst (Nakajima et al., 1996; Siu et al., 2006; Zhou et al., 2010). On the other hand, trans-3′-hydroxycotinine is selectively generated by CYP2A6, a Cyp2a5 ortholog (Nakajima et al., 1996; Siu et al., 2006; Zhou et al., 2010). Our results revealed that, indeed, from 2 hours onwards, consistent with the marked increase in Cyp2a5, there is a statistically significant, accelerated increase in cotinine formation, with a 2-fold increase in trans-3′-hydroxycotinine formation detected at 24 hours in gp78−/− over gp78+/+ hepatocytes (Fig. 8)—thus, not quite as high as the marked functional fold-increases observed with the diagnostic Cyp2a5 functional marker (coumarin) (Fig. 4) but statistically significant nonetheless. One issue with monitoring secondary drug metabolism in isolated cultured hepatocytes in general is that the primary metabolite (cotinine) spills out into the cell medium almost as quickly as it is generated, thus not allowing sufficient time for its subsequent Cyp2a5-selective trans-3′-hydroxycotinine conversion. Thus, only when sufficient cotinine accumulates intracellularly with time (24 hours) is the trans-3′-hydroxycotinine conversion detectable (Fig. 8). This is also true of the other P450-dependent cotinine metabolites, whose relative increases were maximally detected at 24 hours. By contrast, the FMO3 metabolite nicotine-N-oxide was decreased, rather than increased, in gp78−/− hepatocytes, thereby revealing that FMO3 was, if at all, decreased rather than increased upon gp78 KO. The other issue is that nicotine itself may incur enhanced substrate competition by other P450s that not only are known to metabolize it (Zhou et al., 2010) but are also increased upon gp78 KO, which is not the case with the relatively more selective Cyp2a5 diagnostic probe. Nevertheless, these findings indicate that gp78 KO can statistically significantly increase the metabolism of pharmacologically active drugs by enhancing hepatic Cyp2a5 levels through protein stabilization.
Fig. 8.
Relative nicotine metabolism in cultured WT and gp78-KO mouse hepatocytes. PB-pretreated hepatocytes from WT and gp78-KO mice were cultured, and on day 4 incubated with nicotine for 0–24 hours. Aliquots of media were taken at the time points indicated and assayed as detailed in Materials and Methods. The insert in the cotinine production graph is a magnified view of the initial three time points demarcated by the blue square. 3-HC levels and 3-HC/cotinine ratios were determined at 24 hour. Values (mean ± S.D.) of three individual cultures. Statistical significance determined by the Student’s t test, *P < 0.05 vs. WT, **P < 0.01 vs. WT, ***P < 0.001 vs. WT.
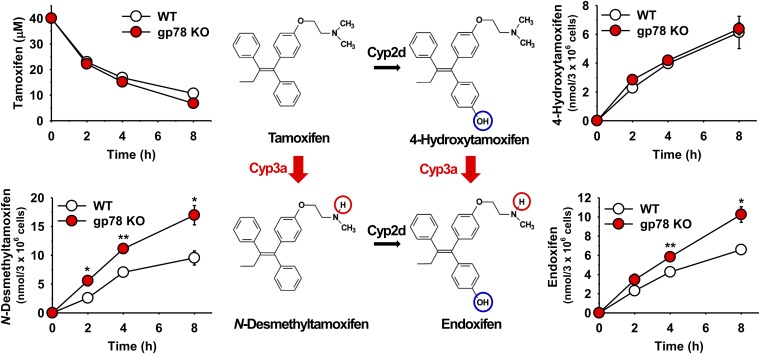
Pharmacologically relevant drugs often involve multiple hepatic P450s, each catalyzing a different step of their biotransformation pathway. Such biotransformation is relevant not only to their elimination, DDIs, and toxicity but also often to their pharmacological response through prodrug metabolic bioactivation. We therefore sought to determine the relevance of gp78 KO to the bioactivation of the estrogen receptor–positive breast cancer therapeutic prodrug tamoxifen. Tamoxifen bioactivation involves two concurrent alternate sequential metabolic routes (Fig. 9; Jacolot et al., 1991; Desta et al., 2004; Goetz et al., 2007): It can be first converted to 4-hydroxytamoxifen by Cyp2d, which is then N-demethylated by Cyps 3a to generate endoxifen. Alternatively, it could be first N-demethylated by Cyps 3a to N-desmethyltamoxifen, which is then 4-hydroxylated by Cyp2d to generate endoxifen (Jacolot et al., 1991; Desta et al., 2004). Both routes thus generate endoxifen, the ultimately desired bioactive chemotherapeutic entity. Our findings reveal that upon gp78 KO, although the Cyp2d-dependent 4-hydroxylation step remains unchanged, consistent with unaltered hepatic Cyp2d levels, the bioactivation of tamoxifen to endotoxifen is nevertheless statistically significantly increased consistent with functional Cyp3a stabilization, and would be therapeutically relevant (Fig. 9).
Fig. 9.
Relative tamoxifen metabolism in cultured WT and gp78-KO mouse hepatocytes. WT and gp78-KO mouse hepatocyte cultures were DEX-pretreated and on day 4 incubated with tamoxifen (40 μM) for 0–8 hours. Aliquots of cell media were taken at the time points indicated and assayed as detailed in Materials and Methods. Values (mean ± S.D.) of three individual cultures. Statistical significance determined by Student’s t test, *P < 0.05 or **P < 0.01 vs. WT.
In Vivo Relevance of the P450 Stabilization
Toxicological Implications.
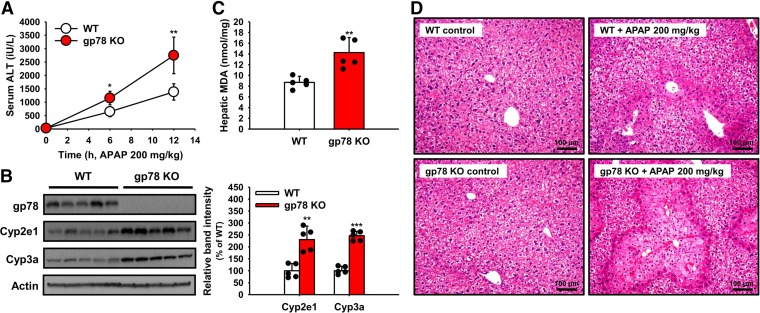
Our findings in cultured mouse hepatocytes upon genetic gp78 ablation, albeit providing proof of principle, may be equivocal and thus not entirely convincing of their plausible in vivo import. To specifically define their physiologic/toxicologic relevance in vivo, we employed the in vivo correlate of our APAP-treated cell culture model. Accordingly, male mice (8-week-old) treated with a mildly toxic APAP (200 mg/kg, i.p.) dose at time 0 documented progressively increased serum ALT levels at 6 and 12 hours after treatment that were statistically significantly higher in gp78-KO mice than those in correspondingly treated WT mice (Fig. 10A). Concurrently, as in Fig. 2, hepatic Cyp2e1 and Cyp3a content in these mice, not treated with any P450 inducers, was statistically significantly elevated in gp78-KO relative to WT mice 12 hours post-APAP (Fig. 10, B and C). Correspondingly, the relative increases in hepatic MDA-levels (Fig. 10C), a reflection of Cyp2e1/Cyp3a-mediated enhanced APAP metabolism to the redox-active NAPQI species and consequent ROS generation, were also more pronounced in the gp78-KO than in WT mice. Although the APAP (200 mg/kg) dose administered was mildly toxic, characteristic APAP-elicited hepatic injury was observed in all gp78-KO and WT mice at 12 hours postadministration (Mitchell et al., 1973; McGill et al., 2013; Bhushan et al., 2014). More importantly, the severity of the hepatic centrizonal gross necrosis elicited by the mildly toxic APAP dose was considerably greater in the gp78 KO than in the correspondingly treated WT mice—with pyknotic nuclei, karyorrhexis, karyolysis, and cytoplasmic eosinophilia of the necrotic hepatocytes bridging adjacent lobules; polymorphonuclear cell infiltration; and the congestion of the hepatic sinusoidal spaces in the centrilobular and midzonal regions. Furthermore, judging by the perimeter of the polymorphonuclear cell infiltration “collar” of these coalesced necrotic lobules, this APAP-elicited hepatic necrosis was much more extensive in the gp78-KO than in the WT mice. Collectively, these findings not only underscore the therapeutic/hepatotoxicological relevance of hepatic Cyp2e1 and Cyp3a stabilization upon gp78 KO but also validate similar findings in cultured hepatocytes (Fig. 7B).
Fig. 10.
In vivo APAP-elicited liver injury in WT and gp78-KO mice. After an overnight fast (12 hours), mice (N = 5 in each group) were treated with APAP and their serum ALT levels determined at 0, 6, and 12 hours (A) as detailed (Materials and Methods). Mice were killed at 12 hours and their livers used for Cyp2e1 and Cyp3a IB analyses (B) and hepatic MDA levels (C) as described (Materials and Methods). The five individual values used for the MDA (mean ± S.D.) values are included in the bar graph. Statistical significance determined by the Student’s t test, *P < 0.05 or **P < 0.01 vs. WT. Liver injury upon hematoxylin and eosin staining and microscopic histologic analyses of representative control and APAP-treated WT and gp78-KO mouse livers isolated at 12 hours is shown in the right panels (D). ***P < 0.001 vs. WT.
Discussion
We document herein for the first time that mouse hepatic Cyp2a5 and Cyps 2c, in addition to previously established Cyps 3a and Cyp2e1, are functionally stabilized upon liver-conditional gp78/AMFR KO. Furthermore, we provide concrete novel evidence that Cyp2a5 not only is primarily a ubiquitination target of gp78 E3-ligase, but also, despite the preferential gp78 mode of K48-linked rather than K63-linked ubiquitination (Li et al., 2007), Cyp2a5 is targeted to both UPD and ALD. On the basis of our findings regarding their human CYP2C9 ortholog, Cyps 2c are quite probably also gp78-ubiquitination substrates (Kim et al., 2016b). Thus, the observed hepatic P450 stabilization upon disruption of their UPD through gp78 KO provides additional therapeutically relevant examples of eukaryotic protein induction via protein stabilization (Schimke and Doyle, 1970).
The mouse hepatic P450s stabilized upon gp78 KO have corresponding human orthologs that are involved not just in the metabolism and elimination of many clinically prescribed drugs, dietary agents, and environmental xenobiotics but also in their biotransformation either to more pharmacologically active or even more toxic/carcinogenic/mutagenic species (Guengerich, 2015; Correia, 2018). Employing pharmacological agents with therapeutic and/or hepatotoxigenic potential, we document herein that such hepatic P450 functional stabilization, if occurring in humans likewise, would be of clinical relevance. Accordingly, most therapeutic agents identified as specific substrates of a given hepatic P450 that is functionally stabilized upon gp78 KO would experience enhanced clearance and accelerated termination of their pharmacological action. Their therapeutic responses consequently would be curtailed, much akin to the impaired drug responses observed upon co-ingestion of therapeutic or dietary P450 inducers. For drugs such as nicotine, metabolized predominantly by human CYP2A6, its functional stabilization, while accelerating nicotine clearance, may also lead to enhanced nicotine craving in chronic cigarette smokers, and thus to an altered behavioral response. Moreover, given the major CYP2A6 role in the bioactivation of tobacco nitrosamines, including the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and other toxic nitrosamines (Zhou et al., 2012; Zhu et al., 2013; Yuan et al., 2017), their carcinogenicity would also be enhanced.
Importantly, because many of these P450s are also involved in the metabolism of drugs (i.e., cancer chemotherapeutic agents) with narrow therapeutic indices, even minor oscillations in hepatic P450 content may translate into therapeutically undesirable losses of their pharmacological efficacy on the one hand, and toxicity on the other, as underscored previously (Peer et al., 2011) in support of the clinical relevance of our CHIP and gp78-shRNAi findings (Kim et al., 2010). Additionally, such enhanced hepatic P450 protein stability can also augment drug-elicited toxicity. Thus, the functional stabilization of hepatic Cyp2e1, and to a lesser extent that of Cyp3a, rendered the mildly toxic concentrations of APAP, a popular over-the-counter analgesic and a Cyp2e1/Cyp3a substrate, statistically significantly more hepatotoxic to gp78−/− hepatocytes than to corresponding WT both in culture (Fig. 8) and live mice (Fig. 10). Although Cyp3a levels are low without concurrent Cyp3a induction in cultured INH-pretreated gp78−/− hepatocytes, gp78 ablation would substantially synergize such toxicity in vivo owing to the functional stabilization of both hepatic Cyp2e1 and Cyps 3a (Figs. 4 and 10).
However, such a functional P450 protein stabilization can also be beneficial. Thus, our finding of the enhanced bioactivation of the breast cancer chemotherapeutic prodrug tamoxifen in gp78−/− relative to WT hepatocytes is particularly relevant and noteworthy. The E3 Ub-ligase gp78/AMFR was originally identified as the cell surface receptor for the prometastatic factor autocrine motility factor (Nabi et al., 1990). Later, it was found to function as the E3 Ub-ligase in the ERAD of several substrates, including the tumor metastatic suppressor KAI1 (Fang et al., 2001; Tsai et al., 2007; Chen et al., 2012). Indeed, gp78 levels are highly upregulated within various human tumors (including lung, breast, and colon) and their microenvironments, but not in adjacent normal tissue (Chiu et al., 2008; Fairbank et al., 2009). Importantly, this gp78 upregulation correlates with their invasiveness, the severity of their metastatic progression, and poor prognosis (Chiu et al., 2008; Fairbank et al., 2009). Thus, African-American breast cancer patients, despite their lower breast cancer incidence, exhibit markedly aggressive breast cancer forms and higher mortality rates than their European-American counterparts with lower mortality rates, albeit much higher breast cancer incidence (Chiu et al., 2008; Martin et al., 2009). Gene profiling via mRNA expression analyses revealed that gp78/AMFR is one of the two genes differentially elevated in the breast tumor epithelia and/or stroma of African-American patients versus those of their European-American cohorts. IB analyses documented a correspondingly increased gp78 protein content in these tissues (Martin et al., 2009). Likewise, a transgenic mouse mammary gland gp78-overexpression model also exhibited a hyperplastic phenotype, increased ductal branching, and dense alveolar lobule formation that correlated with its downregulation of the KAI1 metastatic suppressor (Joshi et al., 2010). Thus, given our findings of enhanced Cyp3a-mediated tamoxifen bioactivation upon gp78 KO (Fig. 9), pharmacological attenuation of gp78 function may be therapeutically desirable as an adjuvant to concurrent breast cancer chemotherapy with prodrugs (such as tamoxifen) that are bioactivated by P450s. Plausible adjuvants include aspirin, salicylates, and APAP that activate p38 MAPK-elicited gp78-degradation, resulting in CYP3A23 stabilization (Ohtsuki et al., 2019). However, although such a short-term therapeutic gp78-inhibition strategy warrants consideration, any long-term strategy may be contraindicated given that aged gp78-null mice reportedly develop NASH and human hepatocarcinoma (HHC) (Zhang et al., 2015a).
Over 600 known mammalian E3 Ub-ligases exist that are functionally redundant in substrate recognition and targeting (Metzger et al., 2014), and capable of serving in both E3 and E4 (poly-Ub chain elongation of existing mono-ubiquitinated residues) complementary roles (Morito et al., 2008; Wang et al., 2014; Zhang et al., 2015b; Menzies et al., 2018). Remarkably, despite these well recognized E3 features, the genetic ablation of just a single hepatic E3, gp78/AMFR, is sufficient to stabilize its target substrates, i.e., P450s, with relevant pharmacological/toxic consequences.
Indeed, we recently documented that upon genetic CHIP ablation in mice, hepatic Cyp2e1 was functionally stabilized irrespective of the presence of gp78 and possibly other redundant/complementary E3 Ub-ligases. This underscored the principal role of UbcH5a/CHIP/Hsp70 as the relevant E3 complex in Cyp2e1 proteolytic turnover (Kim et al., 2016a). This functional Cyp2e1 stabilization in vivo led to progressively increased hepatic lipid peroxidation and oxidative stress, JNK activation, and initial hepatic microvesicular steatosis that upon aging developed into macrovesicular steatosis, ushering a full-blown NASH syndrome over a 1-year period (Kim et al., 2016a). These findings revealed that abrogation of CHIP-mediated Cyp2e1 ubiquitination not only has acute therapeutic/pharmacological consequences but also severe, long-term pathophysiologic sequelae in mice.
Surprisingly, comparable Cyp2e1-functional stabilization upon liver-conditional gp78 KO, although toxicologically relevant, did not result in a similar hepatic microvesicular steatosis (D. Kwon and M. A. Correia, preliminary findings). In fact, fructose-induced hepatic triglyceride (TG) and free fatty acid content was statistically significantly lower in gp78−/− relative to gp78+/+ hepatocytes. This may stem from two plausible synergistic scenarios: 1) Concurrent stabilization of hepatic Insigs 1 and 2 upon gp78 KO suppresses SREB-elicited hepatic lipid biosynthesis as reported in a similar liver-conditional gp78-KO mouse model fed an obesity-inducing diet (Liu et al., 2012); and/or 2) the concurrent stabilization upon gp78 KO of apolipoprotein B100, an established gp78 substrate (Liang et al., 2003) critically involved in hepatic VLDL/LDL-packaging and export, accelerates extrahepatic TG export, thereby minimizing the potential for hepatic steatosis. Our observations and those of others (Liu et al., 2012) reveal that upon their liver-conditional gp78 deletion, the mice are leaner and healthier, whereas global genetic gp78 deletion enhances their risk of hepatic steatosis, NASH, and HCC (Zhang et al., 2015a).
Although the gp78/AMFR upregulation in cancerous tissues is widely documented and unequivocal (Chiu et al., 2008; Martin et al., 2009), less clear is whether genetic gp78 polymorphisms normally exist in the general population, engendering correspondingly high and low (defective) gp78 functional phenotypes. The PubMed online human AMFR Gene/dbSNP Variant/Clin Variant databases list various gp78/AMFR allelic variants, and classify some as benign and others as pathologic. Arg/Arg mutations to Arg/Gly or Gly/Gly at “locus 125/145” are claimed to correlate with a higher expression of gp78 mRNA/protein, and correspondingly increased incidence of coronary atherosclerotic disease in a Chinese population (Wang et al., 2018b). The authors indicate that the Arg/Arg residues at “locus” 145 or 125 were affected by the polymorphism. However, the corresponding residues in the human gp78/AMFR primary sequence deposited in PubMed are Val145 and Phe125. The higher incidence of neuroglioblastoma multiformis in an Iranian population has also been associated with an AMFR genetic polymorphism (Eishi Oskouei et al., 2018). The validity of these claims remains however to be verified more extensively. Although the gp78 SNPs listed in the PubMed Database are present along its entire 643 amino acid length, intriguingly, a few pertaining to basic (K, R) residues in the functional gp78 cytoplasmic domain target the very same positively charged residues that we documented through chemical crosslinking/proteomic analyses and site-directed mutagenesis to be involved in its molecular recognition of CYP3A4 and CYP2E1 acidic phosphodegrons (Wang et al., 2015). Whether any of these listed SNPs truly contribute to genetic gp78-polymorphisms in human populations, and/or correspondingly alter hepatic P450 content and function that trigger DDIs, remains to be elucidated.
In summary, our findings document that the liver-specific genetic ablation of the E3 Ub-ligase gp78/AMFR can result through ERAD disruption in concurrent stabilization of several therapeutically relevant P450s with potentially serious, unwarranted therapeutic and/or toxic consequences. Such outcomes could be further aggravated in patients requiring multiple drug therapies for their specific conditions. Whereas for most drugs, accelerated elimination and consequently shortened pharmacological response upon such P450-stabilization may be largely undesirable, this feature could also be ingeniously exploited as a therapeutic strategy in certain instances to enhance the metabolic activation of chemotherapeutic prodrugs with low therapeutic indices.
Acknowledgments
We thank Chris Her for liver cell isolation at the UCSF Liver Center Core on Cell & Tissue Biology, and Kevin Siao for histological assistance at the UCSF Liver Center Pathology Core supported by NIDDK Center Grant DK26743. We thank Dr. Alexandra Ioanoviciu for the training in the use of LC-MS system used in the tamoxifen metabolite analyses. We greatly appreciate the contributions of Lisa Yu and Trisha Mao for LC-MS/MS analyses of nicotine metabolites in cell-culture samples, and funding from NIDA P30 DA012393 (to N. Benowitz, UCSF) for supporting the laboratory resources used for these analyses.
Abbreviations
- ALD
autophagy-lysosomal degradation
- ALT
alanine aminotransferase
- AMF
autocrine motility factor
- AMFR
AMF-receptor
- APAP
acetaminophen
- CEC
3-cyano-7-ethoxycoumarin
- cgt
conditional gene trap
- CHIP
carboxy-terminus of Hsc70-interacting protein
- Cre
Cre-recombinase
- Ct
threshold cycle
- DDIs
drug-drug interactions
- DEX
dexamethasone
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- flp
floxed
- Flpe
flippase
- FMO3
flavin-monooxygenase
- HPLC-MS
high-performance liquid chromatography–mass spectrometry
- IB
immunoblotting
- INH
isoniazid
- KO
knockout
- 3-MA
3-methyladenine
- MDA
malondialdehyde
- MFC
7-methoxy-4-(trifluoromethyl)-coumarin
- NAPQI
N-acetylparaquinoneimine
- NASH
nonalcoholic steatohepatitis
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- P450
cytochrome P450
- PB
phenobarbital
- PCR
polymerase chain reaction
- shRNAi
short-hairpin RNA interference
- Ub
ubiquitin
- UPD
Ub-dependent proteasomal degradation
- WT
wild type
Authorship Contributions
Participated in research design: Kwon, Kim, Jacob, Correia.
Conducted experiments: Kwon, Kim, Liu.
Contributed new reagents or analytic tools: Kim, Jacob.
Performed data analysis: Kwon, Kim, Jacob.
Wrote or contributed to the writing of the manuscript: Kwon, Jacob, Liu, Correia.
Footnotes
This work was supported by the National Institutes of Health (Grants GM44037 and DK26506 to M.A.C.).
References
- Acharya P, Liao M, Engel JC, Correia MA. (2011) Liver cytochrome P450 3A endoplasmic reticulum-associated degradation: a major role for the p97 AAA ATPase in cytochrome P450 3A extraction into the cytosol. J Biol Chem 286:3815–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajt ML, Knight TR, Lemasters JJ, Jaeschke H. (2004) Acetaminophen-induced oxidant stress and cell injury in cultured mouse hepatocytes: protection by N-acetyl cysteine. Toxicol Sci 80:343–349. [DOI] [PubMed] [Google Scholar]
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol 19:4535–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz NL, Dains KM, Dempsey D, Yu L, Jacob P., III (2010) Estimation of nicotine dose after low-level exposure using plasma and urine nicotine metabolites. Cancer Epidemiol Biomarkers Prev 19:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan B, Walesky C, Manley M, Gallagher T, Borude P, Edwards G, Monga SP, Apte U. (2014) Pro-regenerative signaling after acetaminophen-induced acute liver injury in mice identified using a novel incremental dose model. Am J Pathol 184:3013–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binkhorst L, Mathijssen RH, Ghobadi Moghaddam-Helmantel IM, de Bruijn P, van Gelder T, Wiemer EA, Loos WJ. (2011) Quantification of tamoxifen and three of its phase-I metabolites in human plasma by liquid chromatography/triple-quadrupole mass spectrometry. J Pharm Biomed Anal 56:1016–1023. [DOI] [PubMed] [Google Scholar]
- Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, Bottomley J, Brown SD, Bürger A, Bult CJ, et al. (2012) The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome 23:580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham PC, Harman AW. (1991) Acetaminophen toxicity results in site-specific mitochondrial damage in isolated mouse hepatocytes. J Biol Chem 266:5049–5054. [PubMed] [Google Scholar]
- Cashman JR, Park SB, Yang ZC, Wrighton SA, Jacob P, III, Benowitz NL. (1992) Metabolism of nicotine by human liver microsomes: stereoselective formation of trans-nicotine N′-oxide. Chem Res Toxicol 5:639–646. [DOI] [PubMed] [Google Scholar]
- Chang TKH, Waxman DJ. (1996) The CYP2A subfamily, in Cytochromes P450: Metabolic and Toxicological Aspects (Ionnides C. ed) pp 99–134, CRC Press, Boca Raton, FL. [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. (2006) The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA 103:341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Du S, Fang S. (2012) gp78: a multifaceted ubiquitin ligase that integrates a unique protein degradation pathway from the endoplasmic reticulum. Curr Protein Pept Sci 13:414–424. [DOI] [PubMed] [Google Scholar]
- Chiu CG, St-Pierre P, Nabi IR, Wiseman SM. (2008) Autocrine motility factor receptor: a clinical review. Expert Rev Anticancer Ther 8:207–217. [DOI] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. (2014) Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol 21:325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Höhfeld J, Patterson C. (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol 3:93–96. [DOI] [PubMed] [Google Scholar]
- Correia MA. (2018) Drug biotransformation, in Basic and Clinical Pharmacology (Katzung BG. ed) pp 56–73, McGraw Hill & Lange. [Google Scholar]
- Correia MA, Davoll SH, Wrighton SA, Thomas PE. (1992) Degradation of rat liver cytochromes P450 3A after their inactivation by 3,5-dicarbethoxy-2,6-dimethyl-4-ethyl-1,4-dihydropyridine: characterization of the proteolytic system. Arch Biochem Biophys 297:228–238. [DOI] [PubMed] [Google Scholar]
- Dahmane E, Mercier T, Zanolari B, Cruchon S, Guignard N, Buclin T, Leyvraz S, Zaman K, Csajka C, Decosterd LA. (2010) An ultra performance liquid chromatography-tandem MS assay for tamoxifen metabolites profiling in plasma: first evidence of 4′-hydroxylated metabolites in breast cancer patients. J Chromatogr B Analyt Technol Biomed Life Sci 878:3402–3414. [DOI] [PubMed] [Google Scholar]
- Desta Z, Ward BA, Soukhova NV, Flockhart DA. (2004) Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 310:1062–1075. [DOI] [PubMed] [Google Scholar]
- Donato MT, Jiménez N, Castell JV, Gómez-Lechón MJ. (2004) Fluorescence-based assays for screening nine cytochrome P450 (P450) activities in intact cells expressing individual human P450 enzymes. Drug Metab Dispos 32:699–706. [DOI] [PubMed] [Google Scholar]
- Eishi Oskouei A, Rafiee L, Mahzouni P, Gharipour M, Javanmard SH. (2018) Association between autocrine motility factor receptor gene polymorphism (rs2440472, rs373191257) and glioblastoma multiform in a representative Iranian population. J Res Med Sci 23:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson E, Mkrtchian S, Halpert JR, Ingelman-Sundberg M. (1994) Substrate-regulated, cAMP-dependent phosphorylation, denaturation, and degradation of glucocorticoid-inducible rat liver cytochrome P450 3A1. J Biol Chem 269:18378–18383. [PubMed] [Google Scholar]
- Eliasson E, Mkrtchian S, Ingelman-Sundberg M. (1992) Hormone- and substrate-regulated intracellular degradation of cytochrome P450 (2E1) involving MgATP-activated rapid proteolysis in the endoplasmic reticulum membranes. J Biol Chem 267:15765–15769. [PubMed] [Google Scholar]
- Fairbank M, St-Pierre P, Nabi IR. (2009) The complex biology of autocrine motility factor/phosphoglucose isomerase (AMF/PGI) and its receptor, the gp78/AMFR E3 ubiquitin ligase. Mol Biosyst 5:793–801. [DOI] [PubMed] [Google Scholar]
- Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA 98:14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faouzi S, Medzihradszky KF, Hefner C, Maher JJ, Correia MA. (2007) Characterization of the physiological turnover of native and inactivated cytochromes P450 3A in cultured rat hepatocytes: a role for the cytosolic AAA ATPase p97? Biochemistry 46:7793–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette JR, Nelson SD, Mulder GJ, Jollow DJ, Mitchell JR, Pohl LR, Hinson JA. (1981) Formation of chemically reactive metabolites of phenacetin and acetaminophen. Adv Exp Med Biol 136 (Pt B):931–950. [PubMed] [Google Scholar]
- Girre C, Lucas D, Hispard E, Menez C, Dally S, Menez JF. (1994) Assessment of cytochrome P4502E1 induction in alcoholic patients by chlorzoxazone pharmacokinetics. Biochem Pharmacol 47:1503–1508. [DOI] [PubMed] [Google Scholar]
- Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, et al. (2007) The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat 101:113–121. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2015) Human cytochrome P450 enzymes, in Cytochrome P450: Structure, Mechanism and Biochemistry (Ortiz de Montellano P. ed) pp 523–785, Springer International publishing, New York. [Google Scholar]
- Han XM, Lee G, Hefner C, Maher JJ, Correia MA. (2005) Heme-reversible impairment of CYP2B1/2 induction in heme-depleted rat hepatocytes in primary culture: translational control by a hepatic alpha-subunit of the eukaryotic initiation factor kinase? J Pharmacol Exp Ther 314:128–138. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Jacob P, III, Benowitz NL. (2005) Metabolism and disposition kinetics of nicotine. Pharmacol Rev 57:79–115. [DOI] [PubMed] [Google Scholar]
- Jacob P, III, Yu L, Duan M, Ramos L, Yturralde O, Benowitz NL. (2011) Determination of the nicotine metabolites cotinine and trans-3′-hydroxycotinine in biologic fluids of smokers and non-smokers using liquid chromatography-tandem mass spectrometry: biomarkers for tobacco smoke exposure and for phenotyping cytochrome P450 2A6 activity. J Chromatogr B Analyt Technol Biomed Life Sci 879:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacolot F, Simon I, Dreano Y, Beaune P, Riche C, Berthou F. (1991) Identification of the cytochrome P450 IIIA family as the enzymes involved in the N-demethylation of tamoxifen in human liver microsomes. Biochem Pharmacol 41:1911–1919. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Höhfeld J, Patterson C. (2001) CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem 276:42938–42944. [DOI] [PubMed] [Google Scholar]
- Joshi B, Li L, Nabi IR. (2010) A role for KAI1 in promotion of cell proliferation and mammary gland hyperplasia by the gp78 ubiquitin ligase. J Biol Chem 285:8830–8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Upadhyay A, Kumar A, Mishra A. (2017) Gp78 E3 ubiquitin ligase: essential functions and contributions in proteostasis. Front Cell Neurosci 11:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Acharya P, Engel JC, Correia MA. (2010) Liver cytochrome P450 3A ubiquitination in vivo by gp78/autocrine motility factor receptor and C terminus of Hsp70-interacting protein (CHIP) E3 ubiquitin ligases: physiological and pharmacological relevance. J Biol Chem 285:35866–35877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Grenert JP, Patterson C, Correia MA. (2016a) CHIP(-/-)-mouse liver: adiponectin-AMPK-FOXO-activation overrides CYP2e1-elicited JNK1-activation, delaying onset of NASH: therapeutic implications. Sci Rep 6:29423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Wang Y, Nabavi N, Liu Y, Correia MA. (2016b) Hepatic cytochromes P450: structural degrons and barcodes, posttranslational modifications and cellular adapters in the ERAD-endgame. Drug Metab Rev 48:405–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Buters JT, Pineau T, Fernandez-Salguero P, Gonzalez FJ. (1996) Role of CYP2E1 in the hepatotoxicity of acetaminophen. J Biol Chem 271:12063–12067. [DOI] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y. (2007) A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446:333–337. [DOI] [PubMed] [Google Scholar]
- Liang JS, Kim T, Fang S, Yamaguchi J, Weissman AM, Fisher EA, Ginsberg HN. (2003) Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 cells. J Biol Chem 278:23984–23988. [DOI] [PubMed] [Google Scholar]
- Liao M, Faouzi S, Karyakin A, Correia MA. (2006) Endoplasmic reticulum-associated degradation of cytochrome P450 CYP3A4 in Saccharomyces cerevisiae: further characterization of cellular participants and structural determinants. Mol Pharmacol 69:1897–1904. [DOI] [PubMed] [Google Scholar]
- Liu TF, Tang JJ, Li PS, Shen Y, Li JG, Miao HH, Li BL, Song BL. (2012) Ablation of gp78 in liver improves hyperlipidemia and insulin resistance by inhibiting SREBP to decrease lipid biosynthesis. Cell Metab 16:213–225. [DOI] [PubMed] [Google Scholar]
- Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, Yfantis HG, Tsai YC, Williams EH, Lee DH, Stephens RM, et al. (2009) Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One 4:e4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Lebofsky M, Norris HR, Slawson MH, Bajt ML, Xie Y, Williams CD, Wilkins DG, Rollins DE, Jaeschke H. (2013) Plasma and liver acetaminophen-protein adduct levels in mice after acetaminophen treatment: dose-response, mechanisms, and clinical implications. Toxicol Appl Pharmacol 269:240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies SA, Volkmar N, van den Boomen DJ, Timms RT, Dickson AS, Nathan JA, Lehner PJ. (2018) The sterol-responsive RNF145 E3 ubiquitin ligase mediates the degradation of HMG-CoA reductase together with gp78 and Hrd1. eLife 7:e40009, doi: 10.7554/eLife.40009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta 1843:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Jollow DJ, Potter WZ, Davis DC, Gillette JR, Brodie BB. (1973) Acetaminophen-induced hepatic necrosis. I. Role of drug metabolism. J Pharmacol Exp Ther 187:185–194. [PubMed] [Google Scholar]
- Morishima Y, Peng HM, Lin HL, Hollenberg PF, Sunahara RK, Osawa Y, Pratt WB. (2005) Regulation of cytochrome P450 2E1 by heat shock protein 90-dependent stabilization and CHIP-dependent proteasomal degradation. Biochemistry 44:16333–16340. [DOI] [PubMed] [Google Scholar]
- Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, Tanaka K, Iwai K, Nagata K. (2008) Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRDeltaF508. Mol Biol Cell 19:1328–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Minami Y, Minami M, Chiba T, Tanaka K. (2001) CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2:1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabi IR, Watanabe H, Raz A. (1990) Identification of B16-F1 melanoma autocrine motility-like factor receptor. Cancer Res 50:409–414. [PubMed] [Google Scholar]
- Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, Funae Y, Shimada N, Kamataki T, Kuroiwa Y. (1996) Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos 24:1212–1217. [PubMed] [Google Scholar]
- Needham PG, Guerriero CJ, Brodsky JL. (2019) Chaperoning endoplasmic reticulum-associated degradation (ERAD) and protein conformational diseases. Cold Spring Harb Perspect Biol 11:a033928, doi: 10.1101/cshperspect.a033928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SD. (1990) Molecular mechanisms of the hepatotoxicity caused by acetaminophen. Semin Liver Dis 10:267–278. [DOI] [PubMed] [Google Scholar]
- Ohtsuki Y, Sanoh S, Santoh M, Ejiri Y, Ohta S, Kotake Y. (2019) Inhibition of cytochrome P450 3A protein degradation and subsequent increase in enzymatic activity through p38 MAPK activation by acetaminophen and salicylate derivatives. Biochem Biophys Res Commun 509:287–293. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Kopito RR, Christianson JC. (2013) The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol 5:a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabarcus MK, Hoe N, Sadeghi S, Patterson C, Wiertz E, Correia MA. (2009) CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases. Arch Biochem Biophys 483:66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer CJ, Sissung TM, Figg WD. (2011) CHIP and gp78-mediated ubiquitination of CYP3A4: implications for the pharmacology of anticancer agents. Cancer Biol Ther 11:549–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Magnuson MA. (2000) DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26:149–150. [DOI] [PubMed] [Google Scholar]
- Preston GM, Brodsky JL. (2017) The evolving role of ubiquitin modification in endoplasmic reticulum-associated degradation. Biochem J 474:445–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke RT, Doyle D. (1970) Control of enzyme levels in animal tissues. Annu Rev Biochem 39:929–976. [DOI] [PubMed] [Google Scholar]
- Siu EC, Wildenauer DB, Tyndale RF. (2006) Nicotine self-administration in mice is associated with rates of nicotine inactivation by CYP2A5. Psychopharmacology (Berl) 184:401–408. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, Veenstra T, Hewitt SM, Helman LJ, Khanna C, et al. (2007) The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med 13:1504–1509. [DOI] [PubMed] [Google Scholar]
- Wang HF, Figueiredo Pereira ME, Correia MA. (1999) Cytochrome P450 3A degradation in isolated rat hepatocytes: 26S proteasome inhibitors as probes. Arch Biochem Biophys 365:45–53. [DOI] [PubMed] [Google Scholar]
- Wang L, Bernert JT, Benowitz NL, Feng J, Jacob P, III, McGahee E, Caudill SP, Scherer G, Scherer M, Pluym N, et al. (2018a) Collaborative method performance study of the measurement of nicotine, its metabolites, and total nicotine equivalents in human urine. Cancer Epidemiol Biomarkers Prev 27:1083–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li HC, Zhou J, Yang B, Qiao CD, Liu YM, Zhang Z. (2018b) Correlation between the GP78 gene polymorphism and coronary atherosclerotic heart disease. Hellenic J Cardiol 59:8–13. [DOI] [PubMed] [Google Scholar]
- Wang X, Medzihradszky KF, Maltby D, Correia MA. (2001) Phosphorylation of native and heme-modified CYP3A4 by protein kinase C: a mass spectrometric characterization of the phosphorylated peptides. Biochemistry 40:11318–11326. [DOI] [PubMed] [Google Scholar]
- Wang Y, Guan S, Acharya P, Koop DR, Liu Y, Liao M, Burlingame AL, Correia MA. (2011) Ubiquitin-dependent proteasomal degradation of human liver cytochrome P450 2E1: identification of sites targeted for phosphorylation and ubiquitination. J Biol Chem 286:9443–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Guan S, Acharya P, Liu Y, Thirumaran RK, Brandman R, Schuetz EG, Burlingame AL, Correia MA. (2012) Multisite phosphorylation of human liver cytochrome P450 3A4 enhances Its gp78- and CHIP-mediated ubiquitination: a pivotal role of its Ser-478 residue in the gp78-catalyzed reaction. Mol Cell Proteomics 11:M111.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ha SW, Zhang T, Kho DH, Raz A, Xie Y. (2014) Polyubiquitylation of AMF requires cooperation between the gp78 and TRIM25 ubiquitin ligases. Oncotarget 5:2044–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim SM, Trnka MJ, Liu Y, Burlingame AL, Correia MA. (2015) Human liver cytochrome P450 3A4 ubiquitination: molecular recognition by UBC7-gp78 autocrine motility factor receptor and UbcH5a-CHIP-Hsc70-Hsp40 E2-E3 ubiquitin ligase complexes. J Biol Chem 290:3308–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liao M, Hoe N, Acharya P, Deng C, Krutchinsky AN, Correia MA. (2009) A role for protein phosphorylation in cytochrome P450 3A4 ubiquitin-dependent proteasomal degradation. J Biol Chem 284:5671–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf KK, Wood SG, Allard JL, Hunt JA, Gorman N, Walton-Strong BW, Szakacs JG, Duan SX, Hao Q, Court MH, et al. (2007) Role of CYP3A and CYP2E1 in alcohol-mediated increases in acetaminophen hepatotoxicity: comparison of wild-type and Cyp2e1(-/-) mice. Drug Metab Dispos 35:1223–1231. [DOI] [PubMed] [Google Scholar]
- Yuan JM, Nelson HH, Carmella SG, Wang R, Kuriger-Laber J, Jin A, Adams-Haduch J, Hecht SS, Koh WP, Murphy SE. (2017) CYP2A6 genetic polymorphisms and biomarkers of tobacco smoke constituents in relation to risk of lung cancer in the Singapore Chinese Health Study. Carcinogenesis 38:411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kho DH, Wang Y, Harazono Y, Nakajima K, Xie Y, Raz A. (2015a) Gp78, an E3 ubiquitin ligase acts as a gatekeeper suppressing nonalcoholic steatohepatitis (NASH) and liver cancer. PLoS One 10:e0118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Xu Y, Liu Y, Ye Y. (2015b) gp78 functions downstream of Hrd1 to promote degradation of misfolded proteins of the endoplasmic reticulum. Mol Biol Cell 26:4438–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Shen Y, Ballar P, Apostolou A, Agami R, Fang S. (2004) AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. J Biol Chem 279:45676–45684. [DOI] [PubMed] [Google Scholar]
- Zhou X, D’Agostino J, Xie F, Ding X. (2012) Role of CYP2A5 in the bioactivation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in mice. J Pharmacol Exp Ther 341:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhuo X, Xie F, Kluetzman K, Shu YZ, Humphreys WG, Ding X. (2010) Role of CYP2A5 in the clearance of nicotine and cotinine: insights from studies on a Cyp2a5-null mouse model. J Pharmacol Exp Ther 332:578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AZ, Binnington MJ, Renner CC, Lanier AP, Hatsukami DK, Stepanov I, Watson CH, Sosnoff CS, Benowitz NL, Tyndale RF. (2013) Alaska native smokers and smokeless tobacco users with slower CYP2A6 activity have lower tobacco consumption, lower tobacco-specific nitrosamine exposure and lower tobacco-specific nitrosamine bioactivation. Carcinogenesis 34:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]