Pandey, S., Assmann, S.M. (2004). The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632.
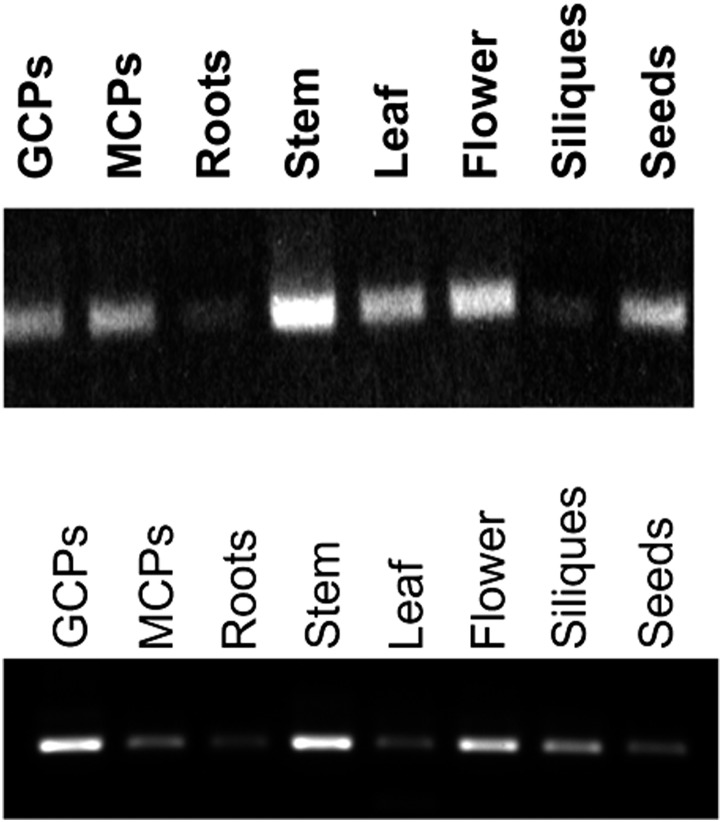
The original publication of this article included an error in Figure 5A. Specifically, the “MCPs” and “Roots” lanes were inappropriately reversed and duplicated as the “Siliques” and “Seeds” lanes. The authors apologize and accept responsibility for not detecting and correcting this issue prior to publication.
Figure 5A.
RT-PCR Profiling of GCR1 Expression.
RNA was extracted from Arabidopsis guard cell protoplasts (GCPs), mesophyll cell protoplasts (MCPs), mature roots, stems, mature rosette leaves, flowers, and siliques using the mirVana RNA isolation kit (Thermo) per the manufacturer’s instructions, with the optional Plant RNA Isolation Aid (Thermo) step included. Due to the abundance of polysaccharides in seeds, a different RNA extraction protocol designed to remove polysaccharides was used for seed RNA. Seeds were ground in liquid nitrogen, and the resultant powder was homogenized in 400 μL of extraction buffer (150 mM Tris-boric acid, pH 7.5, 2% SDS, and 50 mM EDTA), to which 100 μL of ethanol and then 44 μL of 5 M potassium acetate were sequentially added with mixing in between. After two rounds of phenol-chloroform extraction, nucleic acids were ethanol precipitated, then resuspended in DEPC-treated water, followed by RNA precipitation with 2 M lithium chloride. For all samples, 300 ng of total RNA was reverse transcribed using SuperScript III (Thermo), as per the manufacturer’s instructions, diluted 10×, and used as a template for amplification. PCR products were amplified using ExTaq polymerase (Takara) with 37 cycles of 95°C denaturation for 15 s, 54°C annealing for 15 s, and 72°C extension for 30 s, with the following primers: GCR1isp-RTF, 5′-CTACCTTGCTCTCTCTGATATGC-3′, and GCR1isp-RTR, 5′-CCATCTGTTCAACACCTTTAACTC-3′. Following amplification, products were separated on a 0.8% agarose gel in TBE buffer and imaged with a Gel Doc XR+ (Bio-Rad). PCR amplification was conducted three times with similar results observed each time.
The revised Figure 5A (lower panel) is the result of a new experiment that corrects the information in the original Figure 5A (upper panel). The figure legend provides methodological details of the new experiment. The original conclusion from this experiment, that expression “could be detected in all cell/tissue types assessed by RT-PCR, including stomatal guard cells (Figure 5A),” as stated in the main text of the article, is unaltered by this correction.
Both authors approve this correction.
Editors’ note: The corrected figure and accompanying text were reviewed by members of The Plant Cell editorial board. The authors are responsible for providing a complete listing and accurate explanations for all known errors or instances of inappropriate data handling or image manipulation associated with the original publication.
Footnotes
Articles can be viewed without a subscription.