FY is directly involved in poly(A) signal recognition and affects genome-wide poly(A) site usage through alternative polyadenylation.
Abstract
A crucial step for mRNA polyadenylation is poly(A) signal recognition by trans-acting factors. The mammalian cleavage and polyadenylation specificity factor (CPSF) complex components CPSF30 and WD repeat-containing protein33 (WDR33) recognize the canonical AAUAAA for polyadenylation. In Arabidopsis (Arabidopsis thaliana), the flowering time regulator FY is the homolog of WDR33. However, its role in mRNA polyadenylation is poorly understood. Using poly(A) tag sequencing, we found that >50% of alternative polyadenylation (APA) events are altered in fy single mutants or double mutants with oxt6 (a null mutant of AtCPSF30), but mutation of the FY WD40-repeat has a stronger effect than deletion of the plant-unique Pro-Pro-Leu-Pro-Pro (PPLPP) domain. fy mutations disrupt AAUAAA or AAUAAA-like poly(A) signal recognition. Notably, A-rich signal usage is suppressed in the WD40-repeat mutation but promoted in PPLPP-domain deficiency. However, fy mutations do not aggravate the altered signal usage in oxt6. Furthermore, the WD40-repeat mutation shows a preference for 3′ untranslated region shortening, but the PPLPP-domain deficiency shows a preference for lengthening. Interestingly, the WD40-repeat mutant exhibits shortened primary roots and late flowering with alteration of APA of related genes. Importantly, the long transcripts of two APA genes affected in fy are related to abiotic stress responses. These results reveal a conserved and specific role of FY in mRNA polyadenylation.
INTRODUCTION
Polyadenylation of eukaryotic mRNA is an essential posttranscriptional process achieved by poly(A) signal recognition, cleavage, and the addition of a poly(A) tail (Colgan and Manley, 1997; Elkon et al., 2013). Polyadenylation functions in mRNA stability, nuclear export, and translation initiation (Tian and Manley, 2017). At least 50% of genes in humans, animals, algae, and plants have more than one poly(A) site (Wu et al., 2011; Derti et al., 2012; Smibert et al., 2012; Ulitsky et al., 2012; Zhao et al., 2014; Fu et al., 2016). This common phenomenon is designated alternative polyadenylation (APA), which increases the complexity and diversity of transcriptomes and proteomes. In humans, APA affects immunity regulation, cancer formation, and cell reprogramming (Mayr and Bartel, 2009; Fu et al., 2011; Lin et al., 2012; Carpenter et al., 2014). In plants, APA functions in disease resistance, flowering time control, symbiosis, development, and reproduction (Bruggeman et al., 2014; Liu et al., 2014; Zhang et al., 2015; Cyrek et al., 2016; Pan et al., 2016; Lin et al., 2017; Zhou et al., 2019; Riester et al., 2019; Zeng et al., 2019).
During polyadenylation, poly(A) signals anchor the position of a cleavage site that eventually becomes the poly(A) site. In mammalian cells, four parts of the poly(A) signals are located around the pre-mRNA cleavage site. The predominant AAUAAA hexamer is located between 10 and 30 nucleotides upstream of the cleavage site. More than 50% of transcripts in humans preferentially use the AAUAAA poly(A) signal (Neve et al., 2016). The sequence elements at the cleavage site and its downstream element are mainly composed of the dinucleotide CA and U/GU-rich sequences, respectively. Some genes have an upstream element (USE, before the AAUAAA) with UGUA-containing hexamers or other similar repeats (Shi and Manley, 2015).
In plants, poly(A) signals consist of three major elements (Loke et al., 2005). Near upstream elements (NUEs) predominantly consist of AAUAAA. However, this canonical signal is less conserved in plants and embedded in only ∼10% of transcripts (Loke et al., 2005). The far UE (FUE) is a U-rich signal that is similar to USEs in humans. A cleavage element resides on both sides of the cleavage site and includes two U-rich regions, which is different from the elements at the cleavage site in human mRNAs. However, plant signals lack a downstream element (Loke et al., 2005). These similarities and differences indicate that the underlying mechanism of polyadenylation between mammals and plants is conserved but exhibits variability.
Poly(A) signals are recognized by trans-acting factors (Clerici et al., 2017, 2018; Sun et al., 2018). In mammals, biochemical studies have shown that pre-mRNA 3′ end processing requires four multi-unit protein complexes, cleavage and polyadenylation specificity factor (CPSF), cleavage stimulatory factor (CstF), cleavage factor I, and cleavage factor II, in addition to the single subunit poly(A) polymerase (Takagaki et al., 1989; Colgan and Manley, 1997). Among these, CPSF (assembled from CPSF160, WDR33, CPSF100, CPSF73, Fip1, and CPSF30), serves as the central complex for the recognition of the predominant AAUAAA signal and pre-mRNA cleavage (Shi et al., 2009; Michalski and Steiniger, 2015). CPSF30 and WDR33 directly bind to the AAUAAA signal (Chan et al., 2014; Schönemann et al., 2014). Recent studies have demonstrated that the CPSF160-WDR33-CPSF30 ternary complex has a high affinity for the AAUAAA signal and that CPSF160 functions as an essential scaffold that organizes CPSF30 and WDR33 to bind AAUAAA (Clerici et al., 2017; Sun et al., 2018). In addition, the A1 and A2 bases of AAUAAA are recognized specifically by the zinc finger 2 of CPSF30 and A4 and A5 bases are recognized specifically by the zinc finger 3 of CPSF30. WDR33 interacts with RNA at least in part via its N terminus, and the WD40-repeat of WDR33 contacts with the U3-A6 bases, indicating that the highly conserved WD40-repeat plays an important role in AAUAAA signal recognition (Schönemann et al., 2014; Sun et al., 2018).
Genetic and phylogenetic studies have revealed that polyadenylation trans-acting factors are evolutionarily conserved among eukaryotes (Hunt et al., 2008; Hunt et al., 2012). Notably, genetic evidence has shown that the plant CPSF30 is involved in NUE signal choice in Arabidopsis (Arabidopsis thaliana), where the knockout of AtCPSF30 (isolated as a mutant that enhances tolerance to oxidative stress, so it is named oxidative tolerant6 [oxt6]) leads to a shift from A-rich poly(A) signals to U-rich poly(A) signals (Thomas et al., 2012). However, the role of the WDR33 homolog, FY, in recognizing plant NUE signals remains unclear. FY is known as a flowering time regulator (Simpson et al., 2003) and a subunit of the CPSF complex with a special C terminus next to seven conserved WD40-repeats (Henderson et al., 2005). The C terminus harbors two plant-unique Pro-Pro-Leu-Pro-Pro (PPLPP) domains, which can interact with the Trp-Trp (WW) domain of the nuclear RNA binding protein FLOWERING CONTROL LOCUS A (FCA) and control plant flowering time (Simpson et al., 2003; Henderson et al., 2005). FCA/FY interaction suppresses FCA protein abundance by promoting the polyadenylation of FCA within intron 3 to generate a nonfunctional FCA-β transcript. The FCA/FY interaction is also important for properly positioning the polyadenylation site of the floral inhibitor gene FLOWERING LOCUS C (FLC) and controls flowering time (Henderson et al., 2005; Feng et al., 2011). In addition, FY influences seed dormancy by regulating the APA of DELAY OF GERMINATION1 (Cyrek et al., 2016).
In this study, we used a poly(A) tag sequencing (PAT-seq) approach to study the poly(A) profile in a set of defective fy mutants and fy oxt6 double mutants. We demonstrated that FY is indeed involved in AAUAAA signal recognition. Interestingly, we found that the WD40-repeat mutation of FY suppresses, whereas PPLPP-domain deficiency promotes, the A-rich signal recognition. Furthermore, the mutated WD40-repeat of FY shows a preference for using the proximal poly(A) site in 3′ untranslated regions (UTRs). However, the PPLPP-domain deficiency of FY results in a preference for using the distal poly(A) site in the 3′ UTRs. Importantly, we provided in vivo evidence that the long transcripts of ARMADILLO REPEAT KINESIN2 (ARK2) and a zinc ion binding protein affected by FY and AtCPSF30 play roles in salt and oxidative stress responses. Overall, our results reveal the role of FY in genome-wide mRNA polyadenylation.
RESULTS
fy Mutants and Poly(A) Profiling
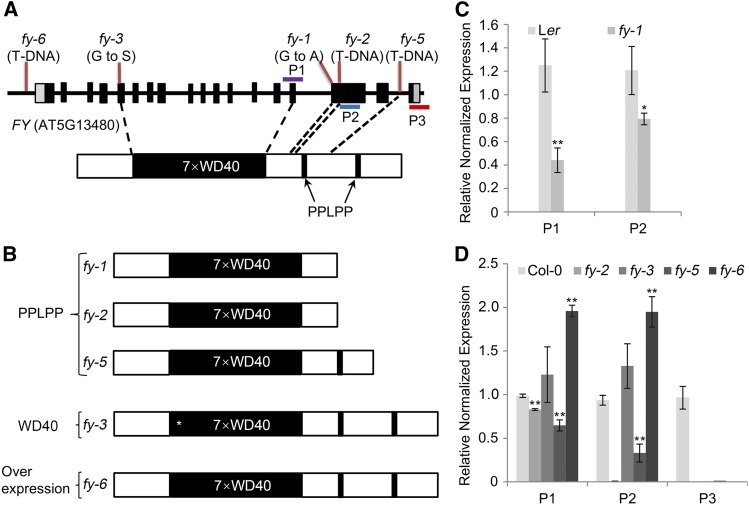
FY has seven WD40-repeats in the N terminus and two PPLPP-domains in the C terminus, and previous studies have reported a set of fy mutants with significant phenotypic outcomes (Henderson et al., 2005). Among these, fy-1 (G-A mutation at a splice-acceptor site results in premature termination), fy-2, and fy-5 are defective on the PPLPP-domain (Simpson et al., 2003; Henderson et al., 2005). Figure 1 shows a side-by-side comparison of various fy alleles using RT-qPCR. Both fy-1 and fy-2 have low transcript abundance and encode proteins that lack two PPLPP-domains, while the fy-5 mutant lacks the last PPLPP-domain (Figure 1). For uncertain reasons, the transcript levels of fy-5 were repeatedly quantified as a lower expression of FY than the wild type (Figure 1D), which is inconsistent with what was reported previously by Feng et al., (2011). The fy-3 allele introduces a Gly to Ser (G141S) change at the first WD40-repeat. The expression level of FY was not affected in fy-3, which is consistent with a previous report by Henderson et al., (2005). A T-DNA insertion on the promoter of FY resulted in its overexpression, and this mutant is designated the fy-6 line in this study to distinguish other FY WD40-repeat or PPLPP-domain mutants (Figure 1). These mutants were crossed with oxt6 (an AtCPSF30 knockout mutant) to generate double mutants.
Figure 1.
Schematic Representation of FY and Its Mutants, and the Transcriptional Level of FY.
(A) FY gene (top) and protein (bottom). The top of the gene shows the position of fy mutations. The black boxes and lines represent exons and introns, respectively. The FY protein contains seven WD40-repeats and two PPLPP-domains. The purple, blue, and red bars show the RT-qPCR amplicons located in 7×WD40-repeats (P1), the first PPLPP-domain (P2), and the second PPLPP-domain (P3) regions.
(B) Schema of FY in mutants. PPLPP-domain deficiency in fy-1, fy-2, and fy-5. The first WD40-repeat amino acid was changed in fy-3 and indicated by an asterisk (*). FY is overexpressed in fy-6 line.
(C) and (D) RT-qPCR quantification of FY transcription level in the mutants and wild types. The wild type of fy-1 is Ler, and wild type for the rest is Col-0. RT-qPCR quantification of FY expression levels in fy-1, fy-2, fy-3, fy-5, fy-6, and the wild type were done in the P1 and P2 regions, and only fy-5 was performed in the P3 region. Error bars represent sd from three biological replicates, and asterisks are indicative of statistically significant differences between the wild type and mutant using one-way ANOVA (*P-value < 0.05, **P-value < 0.01).
PAT-seq is an efficient method for genome-wide profiling of poly(A) site usage, mature transcripts abundance, and functional gene expression, as described in our previous publications (Fu et al., 2016; Lin et al., 2017; Hong et al., 2018). Thus, PAT-seq was used to uncover the poly(A) site usage and transcriptomic profiling of mutants described above (Supplemental Figure 1). A summary of the raw reads, mapped PATs, and PACs [poly(A) site clusters] for each library is provided in Table 1. In total, 48,457 PACs were identified, as shown in Supplemental Data Set 1. Principal component analysis showed replicates are repeated well (Supplemental Figure 2). Overall, these PACs mapped to 19,601 genes. Of these, 10,351 genes contain more than one PAC (defined as APA genes), reflecting ∼53% of APA genes (Supplemental Figure 3A) and 97.5% of the 19,601 genes are protein coding (Supplemental Figure 3B).
Table 1. Summary of PAT Mapping and poly(A) Site.
| Sample | Raw Read No. | PAT No. | PAC No. | Sample | Raw Read No. | PAT No. | PAC No. |
|---|---|---|---|---|---|---|---|
| Ler-rep1 | 20,492,374 | 13,402,960 | 37,477 | fy-6-rep1 | 6,729,599 | 4,230,734 | 33,644 |
| Ler-rep2 | 22,410,997 | 14,642,333 | 42,909 | fy-6-rep2 | 7,172,240 | 5,095,320 | 27,691 |
| Ler-rep3 | 23,726,124 | 15,800,072 | 43,454 | fy-6-rep3 | 15,213,057 | 7,498,700 | 37,900 |
| fy-1-rep1 | 20,289,695 | 13,194,738 | 43,544 | fy-2 oxt6-rep1 | 12,195,428 | 7,781,310 | 37,748 |
| fy-1-rep2 | 21,784,455 | 13,256,921 | 40,955 | fy-2 oxt6-rep2 | 10,590,120 | 6,098,745 | 35,439 |
| fy-1-rep3 | 21,389,641 | 14,460,622 | 43,889 | fy-2 oxt6-rep3 | 13,756,239 | 7,442,659 | 38,363 |
| Col-rep1 | 37,153,184 | 14,091,055 | 44,772 | fy-3 oxt6-rep1 | 17,320,310 | 8,939,522 | 40,325 |
| Col-rep2 | 31,094,544 | 10,583,248 | 42,190 | fy-3 oxt6-rep2 | 14,761,712 | 7,113,164 | 38,526 |
| Col-rep3 | 31,520,633 | 10,754,403 | 43,224 | fy-3 oxt6-rep3 | 11,275,339 | 5,462,609 | 38,468 |
| fy-2-rep1 | 17,334,713 | 8,520,124 | 42,288 | fy-6 oxt6-rep1 | 2,742,850 | 1,506,088 | 26,686 |
| fy-2-rep2 | 22,023,128 | 9,919,939 | 42,351 | fy-6 oxt6-rep2 | 4,060,464 | 1,511,615 | 26,496 |
| fy-2-rep3 | 20,640,430 | 9,105,296 | 41,932 | fy-6 oxt6-rep3 | 7,009,595 | 3,028,461 | 32,240 |
| fy-3-rep1 | 14,116,814 | 6,948,370 | 35,481 | oxt6-rep1 | 15,606,616 | 7,585,236 | 37,009 |
| fy-3-rep2 | 9,543,788 | 5,634,440 | 36,012 | oxt6-rep2 | 18,124,350 | 9,253,847 | 40,657 |
| fy-3-rep3 | 10,646,487 | 6,721,523 | 37,016 | oxt6-rep3 | 16,747,050 | 8,538,587 | 39,665 |
| fy-5-rep1 | 14,382,474 | 5,856,273 | 40,390 | fca-1-rep1 | 9,854,259 | 7,320,494 | 45,185 |
| fy-5-rep2 | 15,826,806 | 5,901,180 | 39,267 | fca-1-rep2 | 11,637,455 | 9,283,112 | 45,303 |
| fy-5-rep3 | 18,091,769 | 6,900,259 | 41,313 | fca-1-rep3 | 12,299,062 | 10,249,445 | 45,719 |
PAT No., numbers of individual tags after curation [to remove low-quality reads, invalid poly(T) reads and unmapped tags]; PAC No., numbers of PACs obtained after grouping poly(A) sites that lie within 24 nucleotides of adjacent sites; rep, repeat.
WD40-Repeat Mutation of FY Has a Stronger Effect on Poly(A) Site Usage Than PPLPP-Domain Deficiency
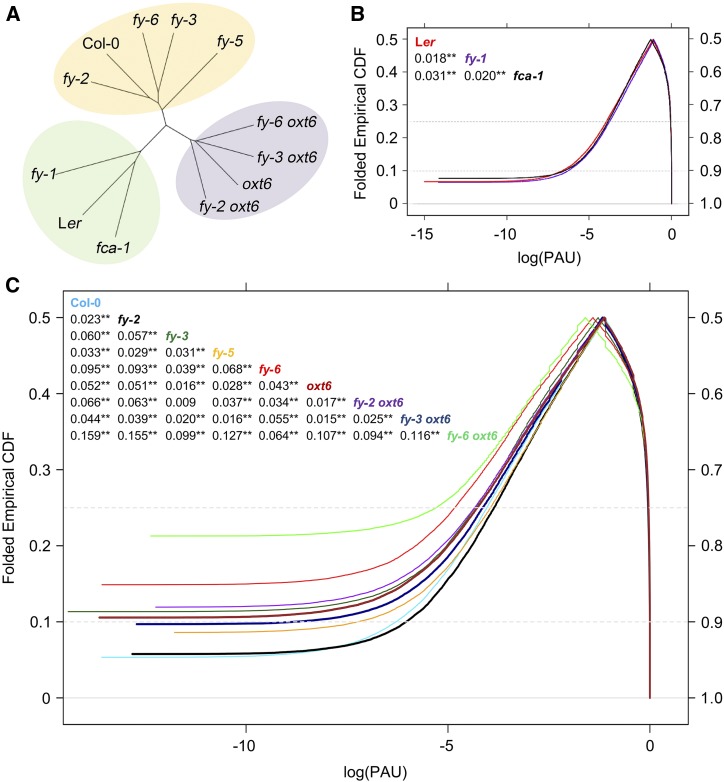
To assess the impact of different mutants on genome-wide poly(A) site usage, the fraction of each PAC within one gene was calculated to show the relative abundance of each isoform, which can be represented by poly(A) usage (PAU). Hierarchical cluster analysis based on PAU was used to distinguish distances among different samples. The results showed that fy-3 mutant and fy-6 line clustered together, as did wild-type Columbia-0 (Col-0), fy-2 and fy-5 were grouped (Figure 2A). This reflected that WD40-repeat mutation and the overexpression of FY may have a similar impact on the poly(A) profile. The profile of PPLPP-domain mutants (fy-2 and fy-5) was much closer to Col-0, indicating that they have less impact on global PAU than fy-3 mutant and fy-6 line. Double mutants of fy-2, fy-3, and fy-6 with oxt6 were grouped in a cluster with the oxt6 single mutant, and fy-2 oxt6 was distinguished from fy-3 oxt6 and fy-6 oxt6. Again, this indicates that the PPLPP-domain deficiency may be different from the WD40-repeat mutation in terms of affecting PAU. Mutants of fy-1 and fca-1 were grouped with a different ecotype, Landsberg erecta (Ler-0; Figure 2A). However, fy-1 was further away from ecotype Ler-0 than fca-1, suggesting that FY has a greater impact on polyadenylation than FCA in Arabidopsis.
Figure 2.
Analysis of Hierarchical Clustering and CDF Based on PAU.
PAU values were calculated as the ratio of its expression to the sum of the expression of all isoforms for each APA gene and are based on the average of three biological replicates.
(A) Hierarchical cluster analysis of PAU.
(B) and (C) Curves of CDF. The x axis is the log values of the ratio of poly(A) site in all isoforms of a single gene. The curve of CDF was based on a mountain plot to examine PAU distribution, and the mountain plot is formed by reflecting the two halves, folded at y = 50%. The Kolmogorov–Smirnov test is used to detect the differences in both location and shape of the empirical cumulative distribution functions between the mutant and its wild type. Compared with the wild type, the P-value of fy-3, fy-5, fy-6, oxt6, fy-2 oxt6, fy-3 oxt6, fy-6 oxt6, and fca-1 was less than 2.2e−16. fy-1 and fy-2 were 1.16e−06 and 1.88e−10, respectively. (B) and (C) were separated because of two different ecotype backgrounds. The numbers in the figure insert represent the maximum distance in paired comparison between two CDF curves. **P-value < 0.01.
The PAU values were plotted by cumulative distribution function (CDF) at the genomic level of individual mutants (Figures 2B and 2C). The PAU profiles of all mutants are significantly different from their wild types (Kolmogorov–Smirnov test, P-value < 0.001), reflecting the important role of FY in polyadenylation. In general, CDF curves merge between 0.5 and 0.6 on the right y axis, indicating that >50% of the PAU in mutants differed from that of the wild type (Figures 2B and 2C). Notably, the start site of CDF and the median (the point where the curves are folded) curves differed among samples, indicating a different sample unique poly(A) site usage and different PAU distribution profiles among those samples. The oxt6 mutant was reported to have a significantly different genome-wide poly(A) profile (Thomas et al., 2012). Accordingly, FY also coordinates genome-wide poly(A) site usage (Figures 2B and 2C).
WD40-repeat mutation and overexpression of FY shift PAU profiles more than PPLPP-domain deficiency (Figure 2C). The PPLPP-domains located in the C terminus of FY are specifically found in plants and not in human WDR33. These domains interact with FCA and function in flowering time control (Henderson et al., 2005). We found that the poly(A) profile of fca-1 is significantly different from that of the wild type (Ler-0; Figure 2B), suggesting that FCA also affects poly(A) site choice. Moreover, the poly(A) profiles of FY and CPSF30 double mutants were also different from both the wild type and single mutants with various alterations (Figure 2C). The CDF curve of fy-3 oxt6 is closer to Col-0 than their single mutants, whereas curves of fy-2 oxt6 and fy-6 oxt6 were much further away from Col-0 compared to their single mutants (Figure 2C).
Collectively, these lines of genetic evidence suggest that FY functions in the determination of poly(A) site usage, as does AtCPSF30. The WD40-repeat mutation of FY has a more significant influence than PPLPP-domain deficiency on poly(A) site choices. In addition, the overexpression of FY has the strongest impact and would be different with other single mutants for the PAU regulation. However, the interaction of plant-unique PPLPP-domains on FY and FCA results in a different and more complicated mechanism of polyadenylation than that in mammals.
fy Mutations Affect Poly(A) Signal Usage of NUE
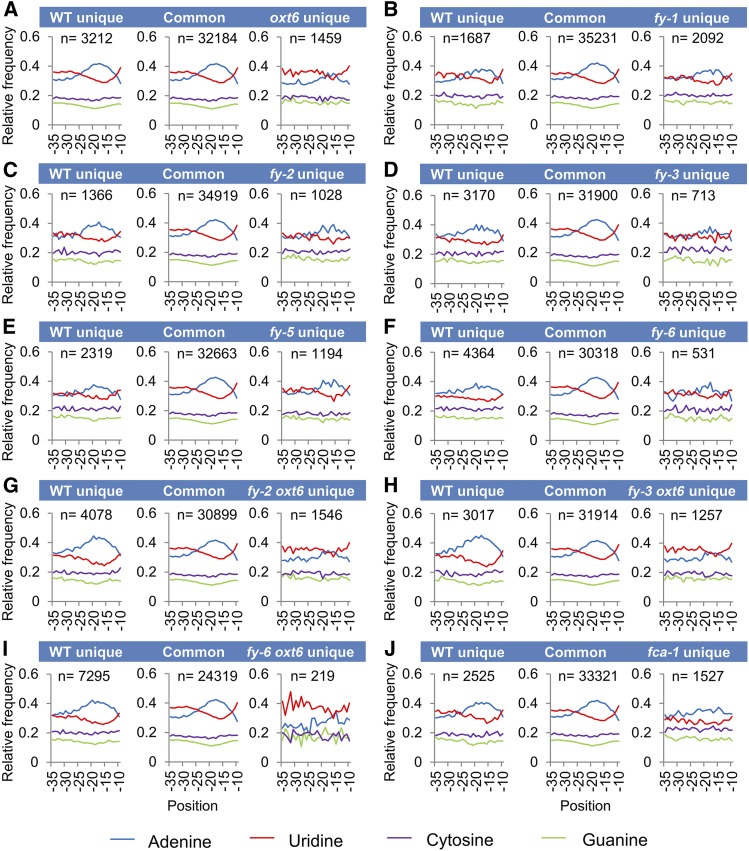
The cis-elements surrounding poly(A) sites, FUE, NUE, and cleavage element are important for plant polyadenylation (Loke et al., 2005). Previous studies revealed that mutations of one CPSF complex component (AtCPSF30 or AtCPSF100) resulted in an abnormal single nucleotide profile in NUE or FUE (Thomas et al., 2012; Lin et al., 2017). Thus, to elucidate the role of FY in poly(A) signal usage, poly(A) sites were grouped into three sets according to Thomas et al. (2012): those seen only in the wild type (wild-type unique PACs), those seen only in the mutants (mutant unique PACs), and those seen in both samples (common PACs), as shown in Supplemental Data Set 2.
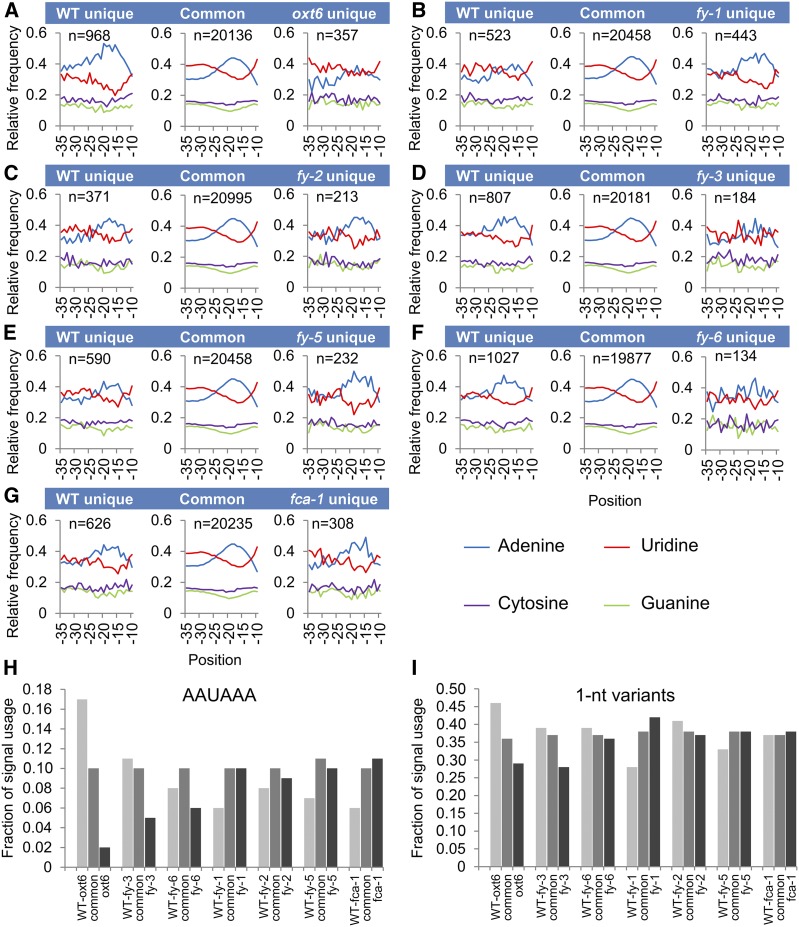
In order to identify canonical poly(A) signals, we focused on NUE regions between 10 and 35 bases upstream of poly(A) sites. As shown in Figure 3, an A-rich peak and low U content centered around 20 nucleotides upstream from the poly(A) site is shown in common PACs. A dramatic decrease in A usage is found in oxt6 unique PACs, as well as a dramatic increase in U usage (Figure 3A). The profile of the wild-type unique PACs is almost the same as that of common PACs (Figure 3A). These results are consistent with previous findings (Thomas et al., 2012). Generally, profiles of both the wild-type unique and mutant unique PACs of fy mutants are different from that of their common PACs (Figures 3B to 3F). The profiles of FY cryptic (mutant unique) PACs are consistent with that of AtCPSF30 cryptic PACs and have lower frequency of A usage than their common PACs. However, the profiles of FY authentic (wild-type unique) PACs have lower A usage than common PACs, which is different from that of AtCPSF30 authentic PACs. These indicate that FY function is not fully redundant with AtCPSF30 in poly(A) signal choice, but it is associated with A-rich NUE poly(A) signal usage. Moreover, in fy-3 mutant and fy-6 line, cryptic PACs appear to have a lower A usage than that in fy-1, fy-2, and fy-5, suggesting a different influence between the WD40-repeat mutation and the PPLPP-domain deficiency of FY. Moreover, the overexpression of both WD40-repeat and PPLPP-domain gives a similar phenotype to the WD40-repeat mutation, suggesting that the WD40-repeat may play a dominant role in poly(A) signal recognition. By coincidence, nucleotide composition profiles of cryptic PACs in fca-1 is similar to that of PPLPP-domain mutants, suggesting that FCA may interact with PPLPP-domain of FY to regulate APA (Figure 3J). Since oxt6 is an AtCPSF30 null mutant, whereas fy mutants are hypomorphic or overexpression plants, the single nucleotide profiles of double mutants with oxt6 appear to be similar to that of oxt6 (Figures 3G to 3I).
Figure 3.
Single Nucleotide Profiles of NUE of the Mutants.
(A) to (J) Nucleotide profiles of unique PACs in the different mutants, wild type (WT), and common PACs. oxt6 unique, sites seen only in the oxt6 mutant relative to WT; WT unique, sites seen only in WT relative to oxt6; Common, poly(A) sites seen in both the WT and oxt6. All other mutants are shown in the same way. n represents transcript number. The y axis indicates the fraction of nucleotide composition at x axis locations, for example, −10 indicates 10 nucleotides upstream of the poly(A) site.
Poly(A) Site Usage Pattern Coordinated by FY Is Associated with Canonical NUE Poly(A) Signals
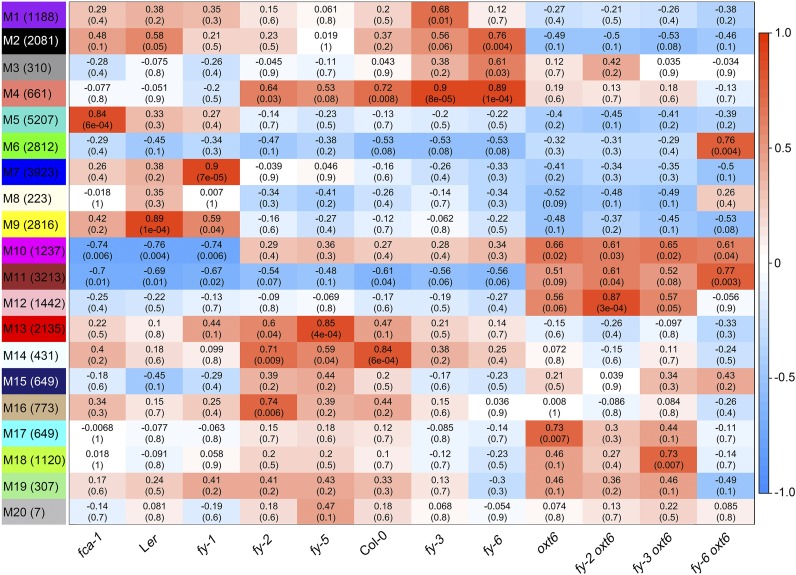
As described above, FY widely affects poly(A) site usage depending on its domains (Figure 2). Thus, to reduce the fuzziness of such a large amount of data, weighted gene coexpression network analysis (WGCNA) was used to cluster transcripts with similar poly(A) site usage patterns between samples. In total, 31,184 PAUs from APA gene transcripts were clustered into 19 module (M) types plus an M20, for which PAU patterns were not correlated well (Figure 4). Different modules had a different correlation with the samples. A higher correlation value (red) indicates that this module is positively associated with the sample, while a lower correlation value (blue) indicates that this module is negatively associated with the sample.
Figure 4.
A Heatmap of Module-Sample Associations.
(Left) The 20 modules (M1 to M20), with the number in parentheses representing the transcript number. Each row corresponds to a module, and each column corresponds to a mutant line or the wild type. The color scale on right shows module-mutant correlations from 1 (red) to −1 (blue). The color of each cell at the row–column intersection represents the correlation coefficient (top values) between the modules and samples. Red color indicates a high degree of positive correlation, and blue color indicates a high degree of negative correlation, between each module and the mutant or wild type. Each cell also contains the corresponding P-value (bottom values).
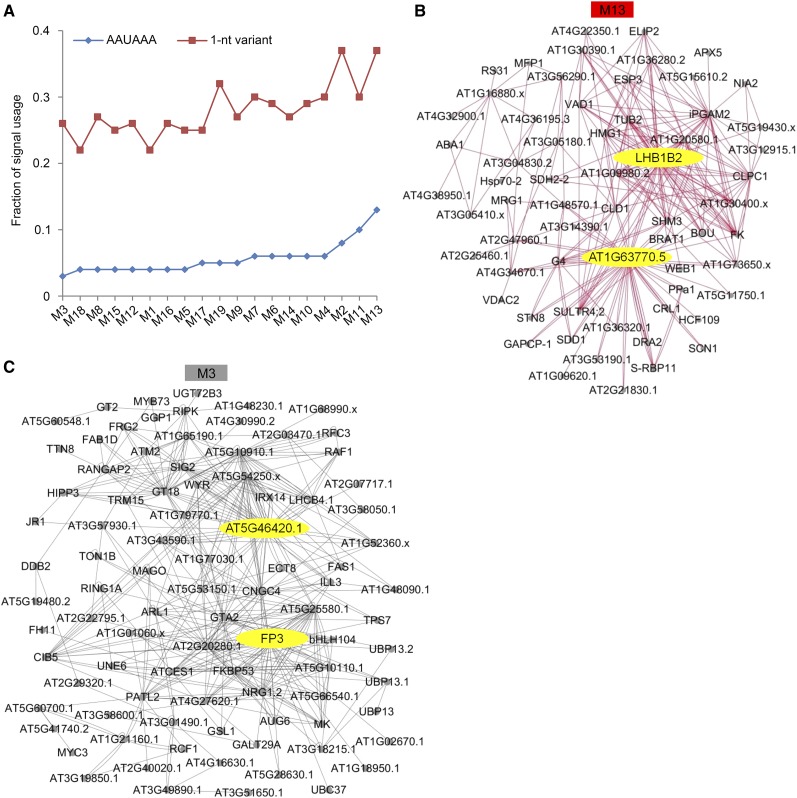
Such module clustering distinguishes each other by NUE poly(A) signal usage of transcripts, which shows the fractions of AAUAAA and 1-nucleotide variant of AAUAAA are different (Figure 5A). This analysis identified M13 as having a higher AAUAAA usage (∼15%) than others. However, M13 is negatively correlated with oxt6, indicating that AAUAAA usage in oxt6 is lower than Col-0. M13 has a higher correlation with PPLPP-domain mutants (fy-1, fy-2, and fy-5) than the wild type. However, M13 has a weaker correlation with fy-3 mutant and fy-6 line than Col-0. These results indicate that the PPLPP-domain deficiency and WD40-repeat mutation of FY oppositely affect polyadenylation. The coexpression network of M13 with 63 transcripts was visualized (Figure 5B). Two hub transcripts from AT2G34420 and AT1G63770 using AACAAA and AAUAAA signals, respectively, were identified. AT2G34420 (LHB1B2) is a chlorophyll a/b binding protein that is related to growth and seed dormancy (Li et al., 2007). AT1G63770 is a putative aminopeptidase involved in indole-3-acetic acid content, root development, and seed germination (Job et al., 2005; Sorin et al., 2006). Previous studies have shown that FY and AtCPSF30 play important roles in plant growth and seed germination (Jiang et al., 2012; Chakrabarti and Hunt, 2015). Thus, genes in this module may be mainly involved in plant growth and development and modulated by FY and AtCPSF30 through poly(A) signal choices.
Figure 5.
Coexpression Network Analysis of Specific Modules.
(A) AAUAAA signal and its one-nucleotide variant signal of the NUE region [between 10 and 35 bases upstream of poly(A) sites] are analyzed in each module. nt, nucleotide.
(B) and (C) Network of the 63 and 91 highly connected transcripts in M13 and M3, respectively. The networks were visualized using Cytoscape 3.6.0 software, and the protein name or gene ID (no protein name) are shown in the figure. Candidate hub genes in the module are shown in yellow.
Another module, M3, is positively correlated with fy-3, fy-6, and oxt6 but negatively correlated with fy-2 and fy-5 (both are PPLPP-domain mutants). This module contains the lowest AAUAAA usage, suggesting that noncanonical NUE poly(A) signal usages are overrepresented in the WD40-repeat–defective FY mutant (fy-3), overexpressed FY (fy-6 line), and AtCPSF30 knockout mutant (oxt6). Moreover, two hub transcripts from AT5G46420 and AT5G63530 were identified from M3 (Figure 5C); they used 1-nucleotide variant signal, AACAAA and UAUAAA, respectively. AT5G46420 and AT5G63530 encode 16S rRNA processing protein and farnesylated protein (FP3), respectively. The microarray result revealed that they are significantly reduced in CaLCuV-infected leaves, indicating that they play a key role in the defense response (Ascencio-Ibáñez et al., 2008). Previous research showed that AtCPSF30 is required for Pseudomonas syringae bacterial resistance (Bruggeman et al., 2014). Therefore, both FY and AtCPSF30 are involved in defense response by modulating poly(A) signal usage of related genes.
Taken together, FY and AtCPSF30 are both associated with AAUAAA poly(A) signal usage. However, FY function does not fully overlap with AtCPSF30. Interestingly, the PPLPP-domain deficiency of FY may act oppositely with its WD40-repeat mutation on canonical poly(A) signal usage.
FY Widely Affects APA and APA Gene Expression
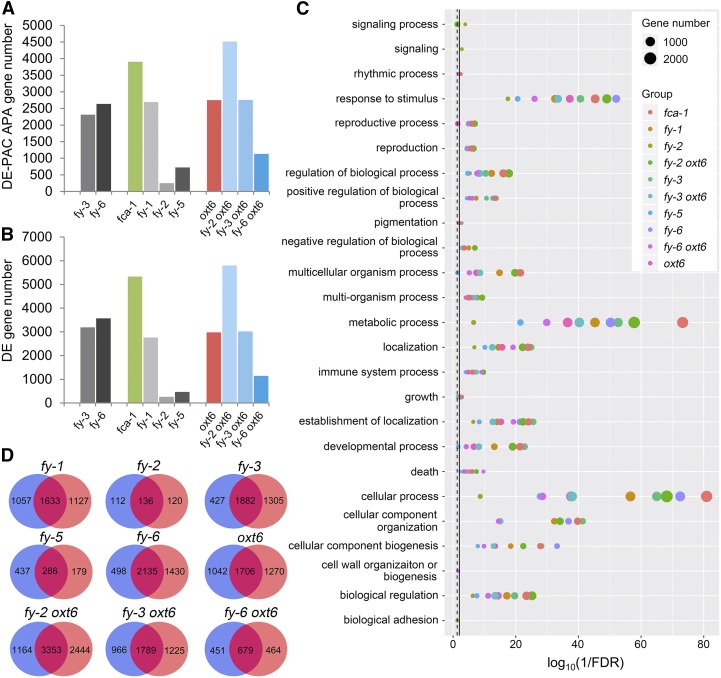
To further explore the role of FY in APA, the expression of each PAC was analyzed using the DESeq2 package. Different alleles result in a large variation of differentially expressed (DE) PAC APA genes and DE gene numbers (Figures 6A and 6B). These DE PAC APA genes were significantly enriched in plenty of biological processes, such as cellular process, response to stimulus, and developmental process (Figure 6C). Transcripts from >2000 APA genes were significantly differentially expressed in fy-3 (adjusted P-value < 0.05, Figure 6A). Moreover, total expression (all transcripts of one gene) of >3000 genes differed significantly in fy-3 (Figure 6B). These results indicate that the WD40-repeat mutation in FY widely impacts poly(A) site usage and gene expression. However, poly(A) site usage and gene expression were only significantly altered in several hundreds of genes in PPLPP-domain–related mutants, fy-2 and fy-5 (Figures 6A and 6B). This suggests that the PPLPP-domain deficiency has less impact on both poly(A) site usage and gene expression, which are consistent with the findings shown in Figure 2. Surprisingly, thousands of genes were affected in fy-1 and fca-1, which may be affected by the different genetic backgrounds. Two double mutants enhance the DE number (fy-2 oxt6 and fy-3 oxt6). However, fy-6 oxt6 contains fewer DE PAC and DE genes than single mutants only, indicating that overexpression of FY could partially rescue the expression variation induced by knocking out AtCPSF30 (Figures 6A and 6B). Detailed information of DE PAC is provided in Supplemental Data Set 3. Moreover, we found a large proportion of overlap (mostly >50%) between DE PAC APA genes and DE genes of each mutant (Figure 6D). These results suggest that DE genes in each mutant may be mainly contributed by APA of genes.
Figure 6.
Analysis of DE PAC APA Genes and DE Genes.
(A) and (B) Bar graph showing the number of DE-PAC APA genes and DE genes, which were analyzed by DESeq2 package. An adjusted P-value < 0.05 threshold was considered statistically significant.
(C) GO enrichment analysis of DE-PAC APA genes. All significant GO terms of biological process at the second level were shown. Solid line, FDR = 0.01; dashed line, FDR = 0.05.
(D) Venn diagram of DE-PAC APA genes and DE genes. The blue circle represents DE-PAC APA gene; the orange circle represents DE gene. The number in the circles show DE-PAC APA gene or DE gene count. The percentage of DE gene belong to DE-PAC APA gene is 58% in fy-1, 53% in fy-2, 59% in fy-3, 62% in fy-5, 60% in fy-6, 57% in oxt6, 58% in fy-2 oxt6, 59% in fy-3 oxt6, and 59% in fy-6 oxt6.
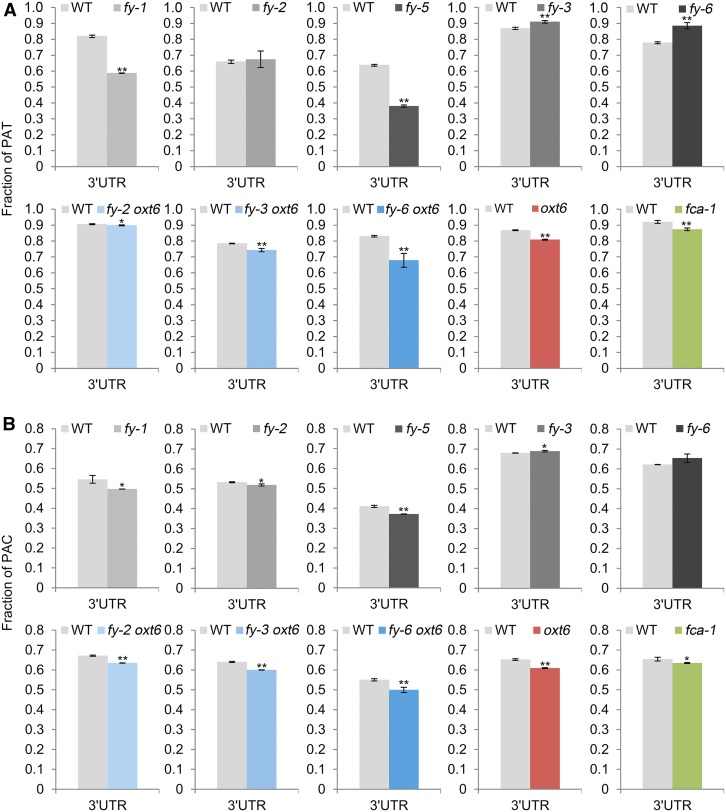
WD40-Repeat Mutation and PPLPP-Domain Deficiency in FY Antagonistically Affect NUE Poly(A) Signal Usage in 3′ UTRs
Single nucleotide profiles were different among genomic regions, and 3′ UTR polyadenylation was the most abundant event (Thomas et al., 2012; Lin et al., 2017). Thus, 3′ UTR PACs were extracted to study the NUE poly(A) signal. Generally, A usage of FY authentic PACs (wild-type unique) were less abundant than that in oxt6 (Figures 7A to 7F). Moreover, in the wild-type-fy-3 and wild-type-fy-6 comparisons, the wild-type unique PACs have a higher A usage than the wild-type unique PACs identified from the wild-type-fy-1, wild-type-fy-2, and wild-type-fy-5 pairwise comparisons. This trend was reversed in mutant unique PACs. For example, the profiles of the wild-type unique and fy-1 unique were opposite (Figure 7B), and this trend inverted in the wild-type-fy-3 and wild-type-fy-6 comparisons (Figures 7D and 7F). These results suggest that the WD40-repeat mutation and PPLPP-domain deficiency of FY may differently affect NUE poly(A) signal choice of 3′ UTR polyadenylation. Again, the WD40-repeat in FY may act an important role in poly(A) signal usage.
Figure 7.
Single Nucleotide Profiles of NUE Located in the 3′ UTR.
(A) to (I) Nucleotide profiles (see [A] to [G]), AAUAAA signal usage (H), and one-nucleotide variants of AAUAAA signal usage (I) of different mutant unique PACs, the wild-type (WT) unique PACs, and common PACs. oxt6 unique, sites seen only in the oxt6 mutant relative to WT; WT unique, sites seen only in WT relative to oxt6; common, poly(A) sites seen in both the WT and oxt6. All other mutants are shown in the same way. n represents transcript number. The y axis indicates the nucleotide composition at x axis locations. (H) and (I) WT-oxt6 indicates the control for oxt6 mutant. Others are labeled the same way.
Furthermore, the frequency of the canonical poly(A) signal, AAUAAA, and its 1-nucleotide variants was calculated (Figures 7H and 7I). By referring to WD40-repeat mutant (fy-3), the wild-type unique PACs have a higher AAUAAA frequency than fy-3 unique PACs (Figure 7H). Conversely, by referring to PPLPP-domain mutants, the wild-type unique PACs have a lower AAUAAA frequency than PPLPP-domain mutants’ unique PACs. Since both WD40-repeat and PPLPP-domain were overexpressed in fy-6 line, the frequency of AAUAAA in the wild-type unique PACs was not as high as that in fy-3 but still higher than fy-6 line unique PACs. These results indicate that WD40-repeat mutation and PPLPP-domain deficiency of FY antagonistically affect AAUAAA usage in 3′ UTR polyadenylation. The same trend (but weaker) was found in 1-nucleotide variants of AAUAAA (Figure 7I). Furthermore, FCA was reported to form a complex with FY and participated in polyadenylation (Simpson et al., 2003). The nucleotide composition and AAUAAA frequency of fca-1 appeared similar to PPLPP-domain deficiency mutants, rather than the WD40-repeat mutant or fy-6 line (Figures 7G and 7H). This confirmed that FCA engages in polyadenylation through interaction with the PPLPP-domain of FY.
From the above analysis, different domains mutation of FY inversely affect AAUAAA usage in 3′ UTR polyadenylation. In addition, it is clear that FY extensively impacts poly(A) site choices. Therefore, we speculated that mutations in FY would disrupt the distribution of 3′ UTR PATs and PACs at the genome level. To test this hypothesis, the genomic distributions of DE-PACs (|fold change| ≥ 2) and their PATs were determined. We found that the fraction of PAT reads and PACs in 3′ UTR were significantly increased in fy-3 mutant and fy-6 line (Figure 8), indicating that the WD40-repeat mutation in FY increases poly(A) site usage in 3′ UTR. However, compared with that in fy-3 mutant and fy-6 line, the distribution of PAT reads and PACs in 3′ UTR was opposite to that of fy-1 and fy-5, that is, the PPLPP-domain deficiency in FY decreases the poly(A) site usage in the 3′ UTR. Interestingly, oxt6 and its double mutants decrease the distribution of PAT reads and PACs in the 3′ UTR (Figure 8). These results show that the WD40-repeat mutation and PPLPP-domain deficiency in FY influence the expression of full-length transcripts differently.
Figure 8.
Distributions of PATs and PACs Selected from DE-PACs (|Fold Change| ≥ 2) in 3′ UTR.
(A) and (B) PATs distribution (A) and PACs distribution (B). Error bars represent sd from three biological replicates, with 10 plants in each repeat, and asterisks are indicative of statistically significant differences using one-way ANOVA (*P-value < 0.05; **P-value < 0.01).
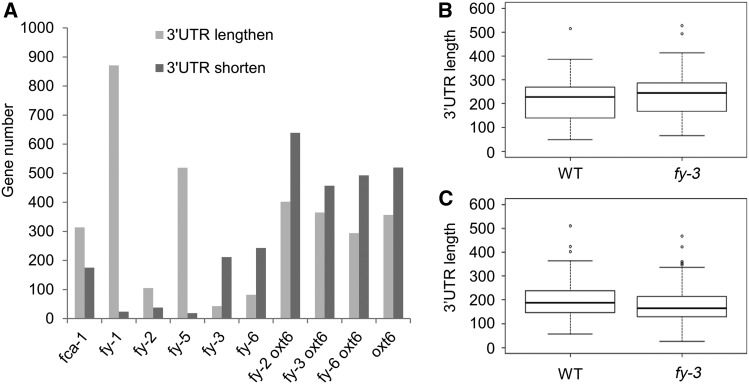
Importantly, we found that more genes used longer 3′ UTR in fy-1, fy-2, and fy-5 mutants (Figure 9A), indicating that the PPLPP-domain deficiency in FY results in a preference for using distal poly(A) sites rather than proximal sites in 3′ UTR. By contrast, more genes containing a shorter 3′ UTR were observed in the fy-3, fy-6, and oxt6 (Figure 9A). This tendency also occurred in the double mutants. The result implies that the WD40-repeat mutation in FY and the AtCPF30 knockout show a preference for using proximal poly(A) sites in 3′ UTRs. Again, the WD40-repeat mutation and PPLPP-domain deficiency in FY functional antagonistically affect poly(A) site usage in 3′ UTR. In 3′ UTR significantly lengthened genes, the average 3′ UTR length was increased by 25 nucleotides in fy-3 (Figure 9B); in 3′ UTR significantly shortened genes, the length was decreased by 22 nucleotides in fy-3 (Figure 9C). The variation of average 3′ UTR length was >20 nucleotides in other mutants.
Figure 9.
3′ UTR APA Analysis.
(A) Comparison of 3′ UTR significantly lengthen or shorten genes. For each gene, 3′ UTR average weighted length was defined as the sum of 3′ UTR length (the distance from each PAC location to the stop codon) of each PAC multiplied by its expression level (average value of three biological repeats normalized PATs) and then divided by the total expression level. A cutoff P-value of 0.05 was adopted for both significantly longer and shorter 3′ UTR between mutant and the wild type.
(B) and (C) Box plot was used to show the 3′ UTR average weighted length distribution of significantly lengthen or shorten in fy-3, respectively. WT, wild type.
WD40-Repeat Mutation of FY Affects the APA of Genes and Contributes to Phenotypic Outputs
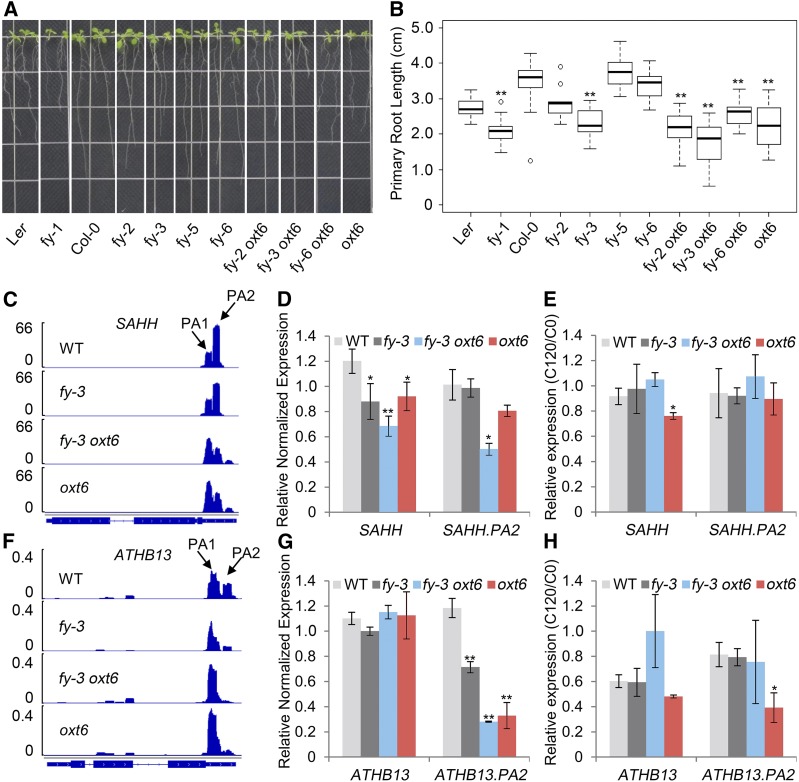
Phenotyping showed that primary root length varied between the wild type and mutants (including double mutants; Figures 10A and 10B). Among fy single mutants, primary root length had the most significant difference between fy-3 and Col-0. A similar trend was observed between oxt6, fy oxt6 double mutants, and the wild type (Figures 10A and 10B). Remarkably, the reduction of primary root length in fy-3 oxt6 was emphasized, potentially contributed by the combination effect of fy-3 and oxt6.
Figure 10.
Analysis of Primary Root Phenotype and Related Gene APA.
(A) Phenotype of root. Each line contained two seedlings.
(B) Root length was measured by ImageJ software. Box plots showing change in primary root length.
(C) and (F) Sequencing coverage of primary root–related gene SAHH and ATHB13 was visualized by Integrative Genomics Viewer software. WT, wild type.
(D) and (G) Distal transcript and total gene expression of primary root–related gene SAHH and ATHB13 were verified by RT-qPCR. Error bars represent sd from three biological replicates. WT, wild type.
(E) and (H) RNA stability assay. RT-qPCR analysis of distal transcript and total gene expression of primary root–related gene SAHH and ATHB13 in control and after 120 min of cordycepin conditions. Error bars represent sd from three biological replicates (pooling ∼10 plants per condition), and asterisks are indicative of statistically significant differences using one-way ANOVA (*P-value < 0.05; **P-value < 0.01). C0, control conditions; C120, mRNA after 120 min of cordycepin treatment; WT, wild type.
S-ADENOSYL-L-HOMOCYSTEINE HYDROLASE (SAHH), a gene encoding an S-adenosylhomocysteine hydrolase, was reported to control primary root length (Wu et al., 2009). We found that the poly(A) profiles among the 3′ UTR of SAHH were different among the wild type, fy-3, oxt6, and fy-3 oxt6, indicating that APA happened in mutants (Figure 10C). Normally, the distal poly(A) site (PA2) is preferred rather than the proximal site (PA1). By contrast, the preference usage was switched to PA1 in fy-3 oxt6, as well as in oxt6. Gene expression (total expression) of SAHH was quantified by RT-qPCR and showed that it was all significantly decreased in fy-3, oxt6, and fy-3 oxt6 by comparing to the wild type (Figure 10D, left). The PA2 abundance was also validated and appeared consistent with PAT-seq (Figure 10D, right). In fy-3, the distal site (PA2) was unchanged, whereas gene expression of SAHH significantly decreased, suggesting that PA1 usage decreased. Thus, in the case of SAHH, FY enhances PA1 usage, whereas AtCPSF30 promotes PA2 usage. To evaluate the impact of RNA turnover on gene expression, RNA stability assay was conducted. The results showed that the SAHH mRNA is stable in each mutant, even though it is a little bit less stable in oxt6 (Figure 10E). Thus, the variation of SAHH expression in fy-3, oxt6, and fy-3 oxt6 may be mainly contributed by APA.
Furthermore, we checked through another APA gene, ATHB13, which encodes a homeodomain leucine zipper class I (HD-Zip I) protein that regulates primary root development (Silva et al., 2016). The poly(A) profiles of ATHB13 were shifted to the proximal site in fy-3, oxt6, and fy-3 oxt6 (Figure 10F). PA2 of ATHB13 was mildly inhibited in fy-3 and strongly inhibited in oxt6 and fy-3 oxt6 (Figures 10F and 10G). This indicates that PA2 of ATHB13 may be affected by the combination of intact FY and AtCPSF30. However, total gene expression of ATHB13 was not changed in fy-3, fy-3 oxt6, and oxt6, which reflects the increase of PA1 that was compensated by the decrease of PA2. The RNA stability assay showed that ATHB13.PA2 was less stable in oxt6, reflecting that the APA of ATHB13 results in different isoform stability (Figure 10H).
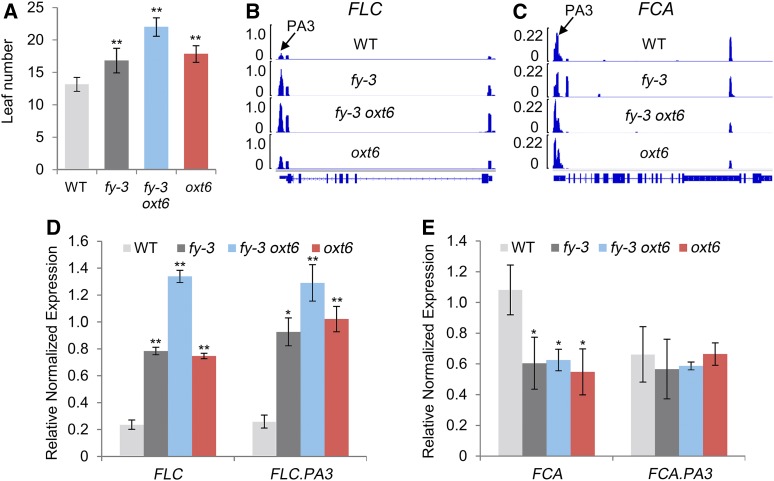
Both fy-3 and oxt6 exhibit significant late flowering, and fy-3 oxt6 double mutants flower much later than the others (Figure 11A). Thus, FY and AtCPSF30 synergistically affect flowering time. Indeed, the full-length transcript and total expression of FLC, which encodes an inhibitor of flowering, was overrepresented in fy-3 and oxt6 and dramatically overrepresented in fy-3 oxt6 (Figures 11B and 11D). Moreover, proximal poly(A) site usage within intron 3 and total expression levels of FCA decreased in fy-3, oxt6, and fy-3 oxt6 (Figures 11C and 11E). These are consistent with previous findings (Simpson et al., 2003), but now include the role of FY/CPSF30 in the APA of FCA and FLC.
Figure 11.
Analysis of Flowering Phenotype and Related Gene APA.
(A) Flowering time was measured by counting the number of rosette leaves at flowering under long-day photoperiods in the incubator. Each pool contained one plant. Each experiment comprised 18 pools, and three independent experiments were completed. WT, wild type.
(B) and (C) Sequencing coverage of FLC and FCA were visualized by Integrative Genomics Viewer software.
(D) and (E) Distal transcript and total gene expression of FLC and FCA were verified by RT-qPCR. Error bars represent sd from three biological replicates, and asterisks are indicative of statistically significant differences using one-way ANOVA (*P-value < 0.05; **P-value < 0.01).
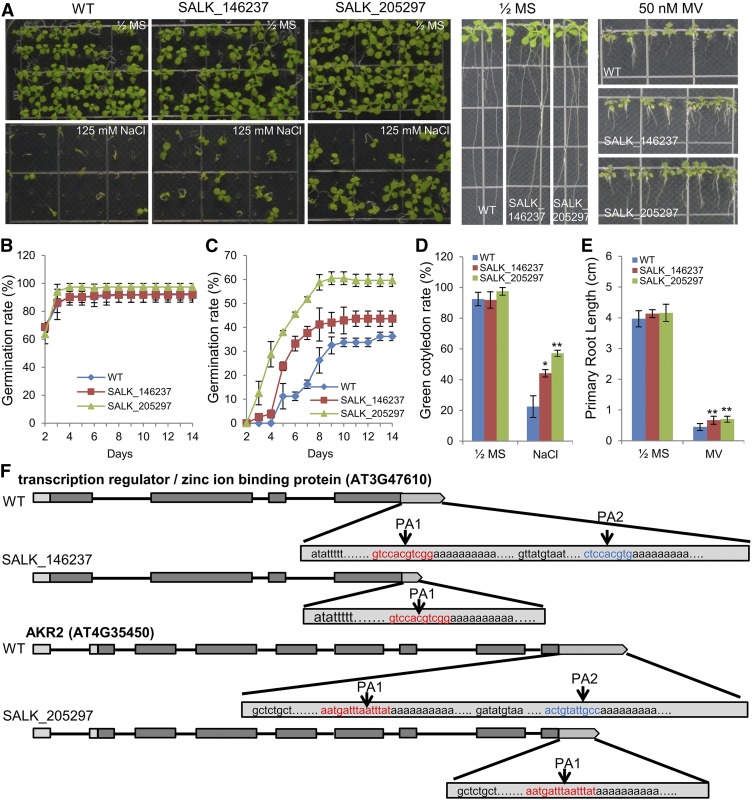
Mutation of FY and AtCPSF30 altered the APA of a transcriptional regulator (AT3G47610) and ANKYRIN REPEAT-CONTAINING PROTEIN2 (AKR2, AT4G35450; Supplemental Figures 4 and 5). Moreover, we found that the two mutants of these two genes carry T-DNA insertions between their APA sites, which may result in the loss of their full-length transcripts. Phenotypic studies showed that these two mutants have higher seed germination rates and green cotyledon rates than the wild type under salt stress (Figure 12), suggesting that these two mutants were less sensitive to salt stress. Moreover, the primary root length of the two mutants was longer than that of the wild type under oxidative stress induced by treatment with methyl viologen (MV), especially for the SALK_205297 mutant (Figure 12). Importantly, 3′ rapid amplification of cDNA ends (RACE) confirmed that the two mutants were long transcript deletions (Figure 12F), revealing that the above phenotypes indeed are related to the function of the long transcript. Collectively, we demonstrated in vivo that APA mediated by FY/CPSF30 can function in plant stress responses.
Figure 12.
Abiotic Stress Responses of the SALK_146237 and SALK_205297 and 3′ sRACE Analysis.
(A) Photographs of seedlings grown on one-half-strength MS medium or one-half-strength MS medium containing 125 mM NaCl at day 14 after the end of stratification and grown on one-half-strength MS or containing 50 nM MV at day 12. WT, wild type.
(B) and (C) Seed germination rates of the indicated genotypes grown on one-half-strength MS medium or one-half-strength MS medium containing 125 mM NaCl were quantified every day from the 2nd day to the 14th day after sowing. Three independent experiments were conducted. Forty seeds per genotype were measured in each replicate. Values are mean ± sd of three replications. WT, wild type.
(D) Cotyledon-greening percentages of the 14th day were recorded. WT, wild type.
(E) Root length was measured by ImageJ software. Error bars represent sd from three biological replicates, and asterisks are indicative of statistically significant differences using one-way ANOVA (*P-value < 0.05; **P-value < 0.01). WT, wild type.
(F) Illustration of gene constructs and 3′ RACE experiment results of AT3G47610 and AKR2. The red and blue region represent PA1 and PA2 region, respectively.
DISCUSSION
Role of FY-Mediated APA
FY is the Arabidopsis homolog of the polyadenylation factor Pfs2p in yeast and WDR33 in mammals (Simpson et al., 2003; Chan et al., 2014). However, the role of FY in poly(A) site choice at the genome level remained unclear. Our results herein demonstrate that FY is definitely involved in poly(A) site usage. Furthermore, the WD40-repeat mutation of FY has more influence on genome-wide poly(A) site usage than the PPLPP-domain deficiency (Figure 2). Interestingly, the WD40-repeat mutation in FY acts in an opposite manner as the PPLPP-domain deficiency in 3′ UTR APA, especially in the preference of single nucleotide usage and 3′ UTR length. Since the PPLPP-domain of FY is not found in WDR33 or Pfs2p (Henderson et al., 2005), this antagonistic effect of WD40-repeat and PPLPP-domain is plant unique. Moreover, FY was differentially expressed among tissues of Arabidopsis (Henderson et al., 2005), implying that FY may be involved in the differentiation of APA among tissues. Previous studies demonstrated that shortening 3′ UTR in mammalian cells resulted in the exception of microRNA (miRNA) targeting, leading to an increase of protein production (Sandberg et al., 2008; Bartel, 2009; Mayr and Bartel, 2009). However, we found that there is no obvious correlation between the 3′ UTR length switching and the gene expression in fy, oxt6, and their double mutants (data not shown). Moreover, our previous work also did not find an obvious negative correlation between 3′ UTR length variation and gene expressions in different rice (Oryza sativa) tissues (Zhou et al., 2019). It was reported that plant miRNAs predominately targeted to coding sequences (CDS; Carthew and Sontheimer, 2009). However, plant 3′ UTR were targeted by phased small interfering RNA, which is produced by miRNA targeted genes (Ma et al., 2018). Therefore, the relationship between 3′ UTR length and miRNA regulation in plants is complicated and remains to be further investigated.
Role of FY in the Recognition of Plant Poly(A) Signals
In this study, by using the PAT-seq approach, we provided genetic evidence that the mutation of WD40-repeat or PPLPP-domain deficiency in FY disrupt AAUAAA signal usage (Figures 3, 5, and 7). Importantly, WD40-repeat mutation and PPLPP-domain deficiency might also function antagonistically in polyadenylation (Figures 4 to 9). In addition, overexpression of both WD40-repeat and PPLPP-domain (fy-6 line) possess similar change with the WD40-repeat mutation (fy-3 mutant). These results indicate that the WD40-repeat in FY may play an important role in PAS recognition in the NUE region. By modeling in SWISS-MODEL (http://swissmodel.expasy.org), a protein three-dimensional structure of FY was found to be similar to WDR33 with 54.61% protein sequence identity and significant QMEAN Z-scores (−3.82). It was clear that the WD40-repeat of WDR33 can directly bind to AAUAAA signal (Schönemann et al., 2014; Clerici et al., 2017; Sun et al., 2018). Thus, WD40-repeat mutation of FY may directly affect AAUAAA signal recognition. However, further genetic and biochemical experiments should be performed to reveal the mechanism by which FY functions in plant polyadenylation.
Previous studies have shown that the PPLPP-domain (plant unique) of FY binds to the WW domain of FCA in vitro (Henderson et al., 2005). The FCA/FY interaction is well characterized in vitro and can be reproduced by using FCA/FY counterparts from other plant species (Lu et al., 2006). However, FY forms a stable complex with AtCPSF100 and AtCPSF160 in vivo, but not with FCA (Manzano et al., 2009). Thus, the FCA/FY interaction in vivo may be regulated or transient. Furthermore, FY/AtCPSF160-containing fractions and those containing FCA did not appear to overlap, suggesting that FY/AtCPSF and FCA/FY are two separate complexes. Importantly, it was found that FCA/FY interaction leads to an altered interaction in the FY/AtCPSF complexes (Manzano et al., 2009). Therefore, we speculate that FCA may compete with other CPSF factors to recruit FY in or out of the CPSF complex to affect poly(A) signal recognition and polyadenylation.
Since oxt6 is an AtCPSF30 null mutant and fy mutants are hypomorphic, fy-2 oxt6, fy-3 oxt6, and oxt6 have similar patterns for recognizing poly(A) signals (Figures 3 and 5). We also found fewer unique PACs in fy-6 oxt6 mutant compared with other double mutants (Figure 3). This result makes one speculate that FY overexpression may partially complement AtCPSF30 function in the recognition of poly(A) signals. Nevertheless, individual contributions of FY and AtCPSF30 to CPSF RNA binding specificity in plants remain to be determined by additional biochemical experiments. We also found that 2% to 4% AAUAAA signal still is used in pre-mRNA transcripts of fy oxt6 double mutants, suggesting that other polyadenylation factors may participate in the recognition of the AAUAAA signal in the absence of AtCPSF30 and FY. Previous studies showed that human Fip1 and CstF64 appeared to be able to crosslink with the AAUAAA signal (Martin et al., 2012), and Fip1 is in close association with the CPSF complex (Schönemann et al., 2014; Clerici et al., 2017; Sun et al., 2018). CstF64 and Fip1 are homologs of Arabidopsis CstF64 (AtCstF64) and FIPS (AtFIPS3 and AtFIPS5), respectively, and the three proteins can also interact with each other. In addition, AtFIPS can directly interact with AtCPSF30 (Hunt et al., 2008). The C-terminal of AtFIPS5 contains an RNA binding domain. AtFIPS5 may be the FUE recognition factor for polyadenylation in plants, suggesting that one or more of its interacting protein partners may be involved in the recognition of the NUE, FUE, and/or cleavage site (Forbes et al., 2006). AtCPSF100 resides at the center of the CPSF protein–protein interaction network (Hunt et al., 2008). However, recent studies have revealed that AtCPSF100 does not participate in NUE poly(A) signal selection but does affect the poly(A) signal recognition of the FUE (Lin et al., 2017). Hence, determining the full machinery of poly(A) signal recognition in plants requires additional research.
The Biological Functions of FY
APA regulation of gene expression participates in a subset of biological processes, including development, disease resistance, and abiotic stress tolerance in plants (Xu et al., 2006; Zhang et al., 2008; Xing and Li, 2011; Bruggeman et al., 2014; Ma et al., 2014). The results presented in this article demonstrate that FY comprehensively affects APA and gene expression and that these DE-PAC APA genes are involved in many biological processes, including cellular process, developmental process, and reproductive process, as determined by the gene ontology (GO) analysis. Indeed, our findings are consistent with previously known biological processes in which FY is involved, such as flowering time regulation. They are also consistent with previous reports where PPLPP-domain–deficient mutants (fy-1, fy-2, and fy-5) led to alterations in the poly(A) site usage of FCA and increased expression of FLC, especially in the fy-2 mutant (Supplemental Figures 6 and 7; Henderson et al., 2005; Feng et al., 2011). In addition, the Gly (G141) residue substitution occurred in the first WD40-repeat (fy-3) also demonstrates that intact WD40-repeats are required for mediating FLC expression (Supplemental Figure 7), as reported previously by Henderson et al., (2005). The amino acid substitution in fy-3 is predicted to affect a structural residue of the B-β-strand in the first propeller blade (Smith et al., 1999). Therefore, the G141S substitution may have a specific effect on FY-WD40-repeat interactions, resulting in an increase of FLC expression.
We also found that the WD40-repeat mutation (fy-3) affects primary root growth. This phenomenon could be related to the confirmed APA events of a couple of genes relate with root growth, SAHH and ATHB13. The RT-qPCR results showed that the gene expression level of SAHH was decreased in fy-3 (Figure 10). Previous reports have shown that sahh null mutants showed decreased primary root length (Wu et al., 2009). Therefore, downregulation of the SAHH gene may lead to shortened primary roots in fy-3. Knockout mutants, athb13, showed increased primary root length, suggesting that this transcription factor is a negative regulator of early root growth (Silva et al., 2016). Interestingly, the expression of distal poly(A) transcript from ATHB13 was decreased in fy-3, but gene expression of ATHB13 did not change, reflecting a switched usage of the poly(A) site. Moreover, SAHH and ATHB13 transcripts in WD40-repeat mutant are stable, further suggesting that FY regulates gene expression by mediating poly(A) site usage rather than directly modulating RNA stability. However, AtCPSF30 alters the stabilities of ATHB13 mRNA isoforms. Previous research showed that AtCPSF30 could localize in the cytoplasm by itself or colocalized with CPSF100 and is present in P-bodies (Rao et al., 2009), which are foci for mRNA surveillance and mRNA decay (Eulalio et al., 2007). Therefore, our results provided further evidence that AtCPSF30 plays a role in mRNA degradation.
METHODS
Plant Materials, Growth Conditions, and Phenotype Assays
The Arabidopsis (Arabidopsis thaliana) fy-1, fy-2, and fy-3 mutants were provided by Dr. Caroline Dean (John Innes Centre, UK). SALK_005697 (designated as fy-5), SALK_048649 (designated as fy-6 line), SALK_146237 (T-DNA insertion mutant of AT3G47610), and SALK_205297 (T-DNA insertion mutant of AT4G35450) were ordered from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). The fca-1 mutant (Arabidopsis Biological Resource Center stock: NS52) carries a point mutation at exon 13 introducing a premature termination codon, which contains both RRMs but lacks the WW domain (Macknight et al., 1997). A description of fy mutants and the PPLPP-domain and WD40-repeats are shown in Figure 1. The double mutants fy-2 oxt6, fy-3 oxt6, and fy-6 oxt6 were generated by crossing fy-2, fy-3, or fy-6 with oxt6, respectively. The fy-1 and fca-1 are in the Ler-0 genetic background. Other mutants are in the Col-0 background. Ler-0 and Col-0 are referred to as wild type in this study. Arabidopsis plants were grown under long-day conditions (16 h of illumination at 120 μmol m−2 s−1 of 400 to 700 nm continuous spectrum of composite white light and 8-h dark cycle) at a constant temperature of 22°C. Seeds for the following phenotypic analyses were collected at the same time.
For root length analyses, seeds were surface sterilized for 3 min, washed five times with sterilized distilled water, and then placed in the dark for 3 d at 4°C for synchronization, after which they were grown on 0.8% agar plates containing half-strength Murashige and Skoog (MS) medium or half-strength MS medium supplemented with 50 nM MV and 1% Suc for 11 d. At least six seeds of mutants and their wild type were sown on the same plates side by side. Three biological replicates were performed, and each replicate contained three plates. The root length was measured by ImageJ software. One-way ANOVA was applied to analyze statistically significant differences between the wild type and mutants. A P-value < 0.05 threshold was considered as statistical significance.
For flowering time tests, seeds were synchronized and then planted in soil. Each 6 × 6-cm pot contained one plant. Each experiment comprised of 18 pools, and three independent experiments were completed. Plants were grown in a controlled environment under long-day photoperiods in a growth chamber. Flowering time was measured by counting the number of rosette leaves at flowering as described previously by Macknight et al., (2002). For seed germination assays, the sterilized seeds were placed in the dark for 3 d at 4°C for synchronization, after which they were grown on half-strength MS medium (0.8% agar and 1% Suc) or half-strength MS medium supplemented with 125 mM NaCl. Three biological replicates were performed, and each replicate contained 40 seeds for each line on the same plate. Germination (emergence of radicles) and post-germination growth (green cotyledon appearance) were scored at the indicated time points.
For PAT-seq, seeds were synchronized and planted in soil for 14 d. At least 10 seedling shoots were collected for one replicate. Three biological replicates from different shoots and independent pools were accomplished for PAT-seq.
PAT-Seq Library Preparation and Sequencing
For PAT-seq libraries construction, samples of mutants and the wild type were prepared from three independent biological replicates. Total RNAs were isolated using the TRIzol reagent (Invitrogen), and their DNA was removed by using DNase I (Takara) following a column-based RNA purification. PAT-seq libraries were prepared from 2 µg of total RNA as described previously (de Lorenzo et al., 2017), with modifications. Briefly, RNA was fragmented in 5× first-strand buffer (Invitrogen) at 94°C for 4 min. RNA fragments with poly(A) tails were enrichened via oligo(dT)25 magnetic beads (New England Biolabs). RT was performed using barcoded oligo(dT)18 primers with SMARTSCRIBE enzyme (Clontech) for 2 h, and then 5′ adaptor for template switching was added. The last nucleotide of the 5′ adaptor was modified by locked nucleic acid modification. The generated cDNA was purified by AMPURE XP beads (Beckman), following by 18 PCR cycles with Phire II (Thermo Fisher Scientific) to produce PAT-seq libraries. The library was run on a 2% agarose gel, and 300- to 500-bp library fragments were purified. Libraries were qualified and quantified by Agilent Bioanalyzer 2100, Qubit 2.0 and qPCR. Finally, libraries were sequenced on the Illumina HiSeq 2500 platform at the facility located in the College of the Environment and Ecology, Xiamen University.
PAT and PAC Generation
The sequencing data were processed using previously described methods (Wu et al., 2011; Fu et al., 2016). Briefly, low-quality raw data were filtered out using FASTX-Toolkit (version 0.0.14, parameters “-q 10 -p 50 -v -Q 33”), and barcodes and poly(T) stretches of raw reads were trimmed. The remaining reads were mapped to the Arabidopsis reference genome (The Arabidopsis Information Resource 10; www.Arabidopsis.org) by Bowtie2 software (version 2.1.0, parameters “-L 25 -N 1 -i S,1,1.15–no-unal”). Potential internal priming reads were filtered out (Loke et al., 2005). As poly(A) site microheterogeneity is pervasive in plants, the mapped PATs within 24 nucleotides were grouped into one poly(A) cluster (PAC), which represents a cleavage site [known as a poly(A) site; Wu et al., 2011]. To facilitate the assignments of PACs to annotated genome, genes with annotated 3′ UTRs were extended for 120 nucleotides, and genes without annotated 3′ UTRs were extended by 338 nucleotides (Wu et al., 2011). To avoid uncertainty from low read counts, total reads of a PAC among all samples with <20 were discarded.
PAU Analysis
Filtered PACs were used for calculating PAU. PAU represents the ratio of reads in one PAC relative to total reads of the gene (Ha et al., 2018). Average PAUs among three biological replicates were used for calculating CDF and plotted by mountainplot package in R (Monti, 1995). By mountainplot, CDF was folded at 50% frequency to show the median of genome-wide PAU profile. Kolmogorov–Smirnov test was applied to judge the significant difference between two CDFs (Haslinger et al., 2010). A P-value < 0.05 threshold was considered as statistical significance.
Poly(A) Signal Analyses
The sequences 300 nucleotides upstream and 100 nucleotides downstream of unique and common poly(A) sites were extracted for single nucleotide profile analysis, as reported previously by Loke et al., (2005). In order to identify poly(A) signals, we focused on NUE regions between 10 and 35 bases upstream of poly(A) sites. The canonical AAUAAA signal and its 1-nucleotide variants were analyzed across all unique and common poly(A) sites as described previously by Loke et al., 2005). Sample unique PACs: PACs only expressed in a mutant (the sum of all PATs in three biological repeats was greater than 3), but not in the wild type (PAT of each biological repeat was equal to 0); or PACs only expressed in the wild type, but not in the mutant. Sample-common PACs: PACs expressed simultaneously in both the wild type and mutant. One-nucleotide variants of AAUAAA contained 18 hexamers (UAUAAA, CAUAAA, GAUAAA, AUUAAA, ACUAAA, AGUAAA, AAAAAA, AACAAA, AAGAAA, AAUUAA, AAUCAA, AAUGAA, AAUAUA, AAUACA, AAUAGA, AAUAAU, AAUAAC, and AAUAAG).
Transcript Coexpression Analysis
The WGCNA R package (Langfelder and Horvath, 2008; Zhan et al., 2015) was used to assess PAU profiles of APA genes across different mutants and the wild type. The average PAU values of 31,184 transcripts from three biological replicates were used for WGCNA. To calculate the adjacency matrix, we first calculated the Pearson correlation coefficients between every two transcripts across different mutants and the wild type. A soft threshold value of 7 was used to transform the adjacency matrix that was then transformed into a topological overlap matrix by the TOM similarity algorithm. Transcripts were hierarchically clustered based on topological overlap similarity. The Dynamic Tree Cut algorithm was used to detect clusters; the mergeCutHeight was 0.4. The modules were defined as branches from the tree cutting, and the minModuleSize was 30. These transcripts were clustered into 19 modules. The networks of M3 and M13 were filtered at adjacency thresholds of 0.1 and 0.3, respectively, and visualized in Cytoscape 3.6.0 software (Shannon et al., 2003). Intramodular connectivity was also calculated. Transcripts with high intramodular connectivity were considered as intramodular hub transcripts. The hub transcripts were obtained with >40 connectivity degree and were shown in yellow in network maps.
DE-PACs and DE Gene Analysis
DESeq2 package (version 1.14.1) was used to normalize read counts and process differential expression PACs (Anders and Huber, 2010). DE-PACs were calculated to uncover the poly(A) profile shift and to estimate the variance of expression levels for a set of genomic regions (5′ UTR, 3′ UTR, introns, CDS, and intergenic regions) based on read number within each feature. All PATs of the genes were summed for representing gene expression levels. Similarly, DE genes were calculated by DESeq2 package. An adjusted P-value was corrected using Benjamini–Hochberg method. An adjusted P-value < 0.05 threshold was considered statistically significant. DE-PAC APA-associated GO enrichment was performed using agriGO with The Arabidopsis Information Resource 10 annotation as the background (Du et al., 2010). False discovery rate (FDR)–corrected P-values < 0.05 were selected as statistically significant.
Identification of 3′ UTR Length
For 3′ UTR APA analysis, the average weighted length of each 3′ UTR of a gene was calculated as described previously by Fu et al., (2016). The 3′ UTR length of each PAC is the distance from each PAC location to the stop codon. For each gene, 3′ UTR average weighted length was defined as the sum of 3′ UTR length of each PAC multiplied by its expression level (average of three biological repeats normalized PATs) and then divided by the total expression level. A cutoff P-value of 0.05 was adopted for both significantly longer and shorter 3′ UTR. The box plot was used to show the length distribution.
RT-qPCR Analysis of Poly(A) Sites
Approximately 2 µg of high-quality total RNA free of DNA contamination was reverse transcribed with oligo(dT)18 primer by SMARTScribe reverse transcriptase (Clontech). RT-qPCR assays were performed using the CFX96TM real-time PCR detection system (Bio-Rad) with SYBR Premix Ex TaqII fluorescent dye (Roche). The relative expression values were determined by using UBQ10 as a housekeeping gene (Wang and Auwerx, 2017). Three biological replicates were performed for all experiments. Moreover, each replicate comprised of three technical repetitions. One-way ANOVA was used to analyze statistically significant differences between the wild type and mutants. A P-value < 0.05 threshold was considered as statistical significance. All primers used herein are listed in Supplemental Table 1.
mRNA Stability Assay
RNA stability assay was performed by using cordycepin to inhibit transcription (de Lorenzo et al., 2017). Briefly, 2-week-old seedlings were harvested, the soil attached on root surface was gently washed away, and whole plants were then transferred to a flask containing incubation buffer (15 mM Suc, 1 mM KCl, 1 mM PIPES, and 1 mM sodium citrate, pH 6.5). Cordycepin (Sigma-Aldrich) was dissolved in 50% ethanol. After 30 min of incubation (time 0), cordycepin solution was added to a final concentration of 200 mM. Seedlings were collected after 120 min and frozen in liquid nitrogen. Triplicate biological replicates were conducted with a pooling of ∼10 plants for each replicate. RNA extraction and RT-qPCR analysis were performed as described above. EIF4A was used as a reference gene (Fedak et al., 2016).
3′ RACE Analysis
3′ RACE was performed using SMART RACE cDNA Amplification Protocol (Clontech) according to the manufacturer’s instructions. One microgram of DNA-free total RNA was used to reverse to cDNA with oligo(dT)30 3′ RACE CDS primer A. The first PCR was amplified using Universal Primer A Mix (UPM; UPM-long and UPM-short mix) and gene special primer GSP1 with Phusion High-Fidelity DNA Polymerase (Life Technology). The second PCR was amplified using Nested Universal Primer A and GSP2. Multiple PCR products were purified and sequenced. All primers used herein are listed in Supplemental Table 1. Sequencing results were mapped to target gene by DNAMAN, and single nucleotide peaks were visualized by SeqMan.
Statistical Analysis
P-values were calculated with one-way ANOVA. See Supplemental Data Set 4 for detailed statistical results.
Accession Numbers
All PAT-seq raw data for this study are available at the National Center for Biotechnology Information website under accession number SRP145554.
Supplemental Data
Supplemental Figure 1. The general experimental process.
Supplemental Figure 2. Principal Component Analysis (PCA) of repeatability of three biological replications.
Supplemental Figure 3. APA gene number and gene type analysis.
Supplemental Figure 4. Sequencing coverage of AT3G47610 gene among WT, fy mutants and double mutants.
Supplemental Figure 5. Sequencing coverage of AKR2 gene among WT, fy mutants and double mutants.
Supplemental Figure 6. Sequencing coverage of FCA among WT, fy mutants and double mutants.
Supplemental Figure 7. Sequencing coverage of FLC among WT, fy mutants and double mutants.
Supplemental Table 1. RT-qPCR and 3′RACE primers used in this study.
Supplemental Data Set 1. List of poly(A) site clusters (PACs).
Supplemental Data Set 2. List of unique PACs and common PACs.
Supplemental Data Set 3. List of DE-PAC between mutant and wild type.
Supplemental Data Set 4. Statistical test results for one-way ANOVA.
Acknowledgments
We thank Caroline Dean for providing fy-1, fy-2, and fy-3 mutant seeds. We thank Haidong Qu, Xiuxiu Wang, Xiaoxuan Zhou, and Wenjia Lu for technical assistance, the American Journal Experts, and Taylor Li for language editing. This work was supported in part by the National Key R&D Project of China (grant 2016YFE0108800 to Q.Q.L.); the National Science Foundation http://dx.doi.org/10.13039/ (grant IOS-154173 to Q.Q.L.); and the China Postdoctoral Science Foundation http://dx.doi.org/10.13039/ (grants 2017M620274 and 2018T110649 to J.L.).
AUTHOR CONTRIBUTIONS
Q.Q.L. and Z.Y. designed the research; Z.Y. performed experiments; Z.Y., Q.Q.L., and J.L. contributed to data analysis and wrote and revised the article.
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio-Ibáñez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148: 436–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q., Garmier M., de Bont L., Soubigou-Taconnat L., Mazubert C., Benhamed M., Raynaud C., Bergounioux C., Delarue M. (2014). The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: A key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 165: 732–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S., Ricci E.P., Mercier B.C., Moore M.J., Fitzgerald K.A. (2014). Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 14: 361–376. [DOI] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M., Hunt A.G. (2015). CPSF30 at the interface of alternative polyadenylation and cellular signaling in plants. Biomolecules 5: 1151–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.L., Huppertz I., Yao C., Weng L., Moresco J.J., Yates J.R. III, Ule J., Manley J.L., Shi Y. (2014). CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 28: 2370–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Faini M., Aebersold R., Jinek M. (2017). Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex. eLife 6: e33111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Faini M., Muckenfuss L.M., Aebersold R., Jinek M. (2018). Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nat. Struct. Mol. Biol. 25: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan D.F., Manley J.L. (1997). Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11: 2755–2766. [DOI] [PubMed] [Google Scholar]
- Cyrek M., Fedak H., Ciesielski A., Guo Y., Sliwa A., Brzezniak L., Krzyczmonik K., Pietras Z., Kaczanowski S., Liu F., Swiezewski S. (2016). Seed dormancy in Arabidopsis thaliana is controlled by alternative polyadenylation of DOG1. Plant Physiol. 170: 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lorenzo L., Sorenson R., Bailey-Serres J., Hunt A.G. (2017). Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. Plant Cell 29: 1262–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A., Garrett-Engele P., Macisaac K.D., Stevens R.C., Sriram S., Chen R., Rohl C.A., Johnson J.M., Babak T. (2012). A quantitative atlas of polyadenylation in five mammals. Genome Res. 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R., Ugalde A.P., Agami R. (2013). Alternative cleavage and polyadenylation: Extent, regulation and function. Nat. Rev. Genet. 14: 496–506. [DOI] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Izaurralde E. (2007). P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8: 9–22. [DOI] [PubMed] [Google Scholar]
- Fedak H., Palusinska M., Krzyczmonik K., Brzezniak L., Yatusevich R., Pietras Z., Kaczanowski S., Swiezewski S. (2016). Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. USA 113: E7846–E7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Jacob Y., Veley K.M., Ding L., Yu X., Choe G., Michaels S.D. (2011). Hypomorphic alleles reveal FCA-independent roles for FY in the regulation of FLOWERING LOCUS C. Plant Physiol. 155: 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes K.P., Addepalli B., Hunt A.G. (2006). An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 281: 176–186. [DOI] [PubMed] [Google Scholar]
- Fu H., Yang D., Su W., Ma L., Shen Y., Ji G., Ye X., Wu X., Li Q.Q. (2016). Genome-wide dynamics of alternative polyadenylation in rice. Genome Res. 26: 1753–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sun Y., Li Y., Li J., Rao X., Chen C., Xu A. (2011). Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 21: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K.C.H., Blencowe B.J., Morris Q. (2018). QAPA: A new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biol. 19: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslinger R., Pipa G., Brown E. (2010). Discrete time rescaling theorem: Determining goodness of fit for discrete time statistical models of neural spiking. Neural Comput. 22: 2477–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Liu F., Drea S., Simpson G.G., Dean C. (2005). An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132: 3597–3607. [DOI] [PubMed] [Google Scholar]
- Hong L., Ye C., Lin J., Fu H., Wu X., Li Q.Q. (2018). Alternative polyadenylation is involved in auxin-based plant growth and development. Plant J. 93: 246–258. [DOI] [PubMed] [Google Scholar]
- Hunt A.G., Xing D., Li Q.Q. (2012). Plant polyadenylation factors: Conservation and variety in the polyadenylation complex in plants. BMC Genomics 13: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A.G., et al. (2008). Arabidopsis mRNA polyadenylation machinery: Comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 9: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Kumar S., Eu Y.J., Jami S.K., Stasolla C., Hill R.D. (2012). The Arabidopsis mutant, fy-1, has an ABA-insensitive germination phenotype. J. Exp. Bot. 63: 2693–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C., Rajjou L., Lovigny Y., Belghazi M., Job D. (2005). Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiol. 138: 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Asami T., Wu X., Tsang E.W., Cutler A.J. (2007). A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 145: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Xu R., Wu X., Shen Y., Li Q.Q. (2017). Role of cleavage and polyadenylation specificity factor 100: anchoring poly(A) sites and modulating transcription termination. Plant J. 91: 829–839. [DOI] [PubMed] [Google Scholar]
- Lin Y., Li Z., Ozsolak F., Kim S.W., Arango-Argoty G., Liu T.T., Tenenbaum S.A., Bailey T., Monaghan A.P., Milos P.M., John B. (2012). An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 40: 8460–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Xu R., Merrill C., Hong L., Von Lanken C., Hunt A.G., Li Q.Q. (2014). Integration of developmental and environmental signals via a polyadenylation factor in Arabidopsis. PLoS One 9: e115779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke J.C., Stahlberg E.A., Strenski D.G., Haas B.J., Wood P.C., Li Q.Q. (2005). Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 138: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Xu Z.K., Song R.T. (2006). OsFY, a homolog of AtFY, encodes a protein that can interact with OsFCA-gamma in rice (Oryza sativa L.). Acta Biochim. Biophys. Sin. (Shanghai) 38: 492–499. [DOI] [PubMed] [Google Scholar]
- Ma L., Guo C., Li Q.Q. (2014). Role of alternative polyadenylation in epigenetic silencing and antisilencing. Proc. Natl. Acad. Sci. USA 111: 9–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Chen C., Liu Y., Zeng M., Meyers B.C., Li J., Xia R. (2018). Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol. 217: 1535–1550. [DOI] [PubMed] [Google Scholar]
- Macknight R., Bancroft I., Page T., Lister C., Schmidt R., Love K., Westphal L., Murphy G., Sherson S., Cobbett C., Dean C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89: 737–745. [DOI] [PubMed] [Google Scholar]
- Macknight R., Duroux M., Laurie R., Dijkwel P., Simpson G., Dean C. (2002). Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell 14: 877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano D., Marquardt S., Jones A.M.E., Bäurle I., Liu F., Dean C. (2009). Altered interactions within FY/AtCPSF complexes required for Arabidopsis FCA-mediated chromatin silencing. Proc. Natl. Acad. Sci. USA 106: 8772–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Gruber A.R., Keller W., Zavolan M. (2012). Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports 1: 753–763. [DOI] [PubMed] [Google Scholar]
- Mayr C., Bartel D.P. (2009). Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski D., Steiniger M. (2015). In vivo characterization of the Drosophila mRNA 3′ end processing core cleavage complex. RNA 21: 1404–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti K.L. (1995). Folded empirical distribution function curves-mountain plots. Am. Stat. 49: 342–345. [Google Scholar]
- Neve J., Burger K., Li W., Hoque M., Patel R., Tian B., Gullerova M., Furger A. (2016). Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 26: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Oztas O., Zhang X., Wu X., Stonoha C., Wang E., Wang B., Wang D. (2016). A symbiotic SNARE protein generated by alternative termination of transcription. Nat. Plants 2: 15197–15201. [DOI] [PubMed] [Google Scholar]
- Rao S., Dinkins R.D., Hunt A.G. (2009). Distinctive interactions of the Arabidopsis homolog of the 30 kD subunit of the cleavage and polyadenylation specificity factor (AtCPSF30) with other polyadenylation factor subunits. BMC Cell Biol. 10: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riester L., Köster-Hofmann S., Doll J., Berendzen K.W., Zentgraf U. (2019). Impact of alternatively polyadenylated isoforms of ETHYLENE RESPONSE FACTOR4 with activator and repressor function on senescence in Arabidopsis thaliana L. Genes (Basel) 10: e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönemann L., Kühn U., Martin G., Schäfer P., Gruber A.R., Keller W., Zavolan M., Wahle E. (2014). Reconstitution of CPSF active in polyadenylation: Recognition of the polyadenylation signal by WDR33. Genes Dev. 28: 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Di Giammartino D.C., Taylor D., Sarkeshik A., Rice W.J., Yates J.R. III, Frank J., Manley J.L. (2009). Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Manley J.L. (2015). The end of the message: Multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 29: 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.T., Ribone P.A., Chan R.L., Ligterink W., Hilhorst H.W. (2016). A predictive coexpression network identifies novel genes controlling the seed-to-seedling phase transition in Arabidopsis thaliana. Plant Physiol. 170: 2218–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G., Dijkwel P.P., Quesada V., Henderson I., Dean C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787. [DOI] [PubMed] [Google Scholar]
- Smibert P., et al. (2012). Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Reports 1: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. (1999). The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Sorin C., Negroni L., Balliau T., Corti H., Jacquemot M.P., Davanture M., Sandberg G., Zivy M., Bellini C. (2006). Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 140: 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhang Y., Hamilton K., Manley J.L., Shi Y., Walz T., Tong L. (2018). Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc. Natl. Acad. Sci. USA 115: E1419–E1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y., Ryner L.C., Manley J.L. (1989). Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 3: 1711–1724. [DOI] [PubMed] [Google Scholar]
- Thomas P.E., Wu X., Liu M., Gaffney B., Ji G., Li Q.Q., Hunt A.G. (2012). Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24: 4376–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Manley J.L. (2017). Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I., Shkumatava A., Jan C.H., Subtelny A.O., Koppstein D., Bell G.W., Sive H., Bartel D.P. (2012). Extensive alternative polyadenylation during zebrafish development. Genome Res. 22: 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Auwerx J. (2017). Systems phytohormone responses to mitochondrial proteotoxic stress. Mol. Cell 68: 540–551.e5. [DOI] [PubMed] [Google Scholar]
- Wu X., Liu M., Downie B., Liang C., Ji G., Li Q.Q., Hunt A.G. (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 108: 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.Z., Li F.L., Kolenovsky A., Caplan A., Cui Y.H., Cutler A., Tsang E.W.T., Peterson R.L., Shelp B.J. (2009). A mutant deficient in S-adenosylhomocysteine hydrolase in Arabidopsis shows defects in root-hair development. Botany 87: 571–584. [Google Scholar]
- Xing D., Li Q.Q. (2011). Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip. Rev. RNA 2: 445–458. [DOI] [PubMed] [Google Scholar]
- Xu R., Zhao H., Dinkins R.D., Cheng X., Carberry G., Li Q.Q. (2006). The 73 kD subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol. Biol. 61: 799–815. [DOI] [PubMed] [Google Scholar]
- Zeng W., Dai X., Sun J., Hou Y., Ma X., Cao X., Zhao Y., Cheng Y. (2019). Modulation of auxin signaling and development by polyadenylation machinery. Plant Physiol. 179: 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J., Thakare D., Ma C., Lloyd A., Nixon N.M., Arakaki A.M., Burnett W.J., Logan K.O., Wang D., Wang X., Drews G.N., Yadegari R. (2015). RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27: 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Addepalli B., Yun K.Y., Hunt A.G., Xu R., Rao S., Li Q.Q., Falcone D.L. (2008). A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS One 3: e2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gu L., Hou Y., Wang L., Deng X., Hang R., Chen D., Zhang X., Zhang Y., Liu C., Cao X. (2015). Integrative genome-wide analysis reveals HLP1, a novel RNA-binding protein, regulates plant flowering by targeting alternative polyadenylation. Cell Res. 25: 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Wu X., Kumar P.K.R., Dong M., Ji G., Li Q.Q., Liang C. (2014). Bioinformatics analysis of alternative polyadenylation in green alga Chlamydomonas reinhardtii using transcriptome sequences from three different sequencing platforms. G3 (Bethesda) 4: 871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Fu H., Yang D., Ye C., Zhu S., Lin J., Ye W., Ji G., Ye X., Wu X., Li Q.Q. (2019). Differential alternative polyadenylation contributes to the developmental divergence between two rice subspecies, japonica and indica. Plant J. 98: 260–276. [DOI] [PubMed] [Google Scholar]