Circadian clock proteins REVEILLE4/8 regulate the expression of early heat shock–induced genes, including transcription factor genes, to prepare plants for exposure to high temperature during the day.
Abstract
Although much is known about plant responses to heat shock (HS), how plants sense high temperature and the primary HS signal transduction pathway leading to HS-regulated gene expression are still poorly understood. To identify primary transcription factors that mediate HS-regulated gene expression and their target genes, RNA sequencing was performed to detect genes whose expression is rapidly altered by HS in Arabidopsis (Arabidopsis thaliana). The results showed several genes were induced after only 5 min of HS treatment, suggesting that HS signaling occurs very rapidly. Analysis of the cis-elements in the promoters of genes upregulated by 10 min of HS treatment identified HEAT SHOCK FACTOR A1s (HSFA1s) and circadian clock proteins REVEILLE4 (RVE4) and RVE8 as essential transcription factors that independently mediate early HS-induced gene expression. Using hsfa1a/b/d/e and rve4/8 mutants, we identified subsets of HSFA1s- or RVE4/8-dependent early HS-induced genes and showed RVE4/8 regulate plant thermotolerance partially by regulating the expression of downstream transcription factors ETHYLENE RESPONSIVE FACTOR53 (ERF53) and ERF54, specifically around noon. These findings reveal a potential transcriptional regulatory hierarchy governing the first wave of HS-induced gene expression. They also provided important insight into the mechanism by which the circadian clock gates thermotolerance and prepares plants for exposure to high temperatures during the day.
INTRODUCTION
Temperature is one of the most important environmental factors affecting the diurnal and seasonal growth of plants, as well as their geographical distribution. Moderate increases in temperature above the optimum for plant growth (∼28°C for Arabidopsis [Arabidopsis thaliana]) stimulate thermomorphogenesis that is characterized by hypocotyl and petiole elongation, hyponastic leaf growth, and reduced leaf thickness (Koini et al., 2009; Quint et al., 2016). When the environmental temperature rises above the thermomorphogenesis-inducing temperature for extended periods, plants experience heat shock (HS) stress (Liu et al., 2015). HS stress has a profound impact on the plant life cycle, including suppressed seed germination (Hasanuzzaman et al., 2013), reduced fertility (Zinn et al., 2010), decreased photosynthetic efficiency (Bita and Gerats, 2013), inhibited grain filling (Wahid et al., 2007), and, in extreme cases, cell or plant death (Balk et al., 1999). HS significantly reduces global crop yields (Lobell et al., 2011). Therefore, understanding the molecular mechanisms by which plants sense and respond to HS stress will profoundly affect the future security of global agricultural productivity.
Pre-exposure to nonlethal high temperatures (generally ∼37°C for Arabidopsis) for certain periods can increase plant tolerance to lethal high temperatures (generally 42°C or higher), a process known as acquired thermotolerance (Mittler et al., 2012). During this process, exposure to nonlethal high temperature induces intracellular signaling pathways that alter gene expression, protein accumulation, and cellular metabolism to enhance plant tolerance to subsequent exposure to lethal high temperatures. In contrast to traditional chemical signals, which are kept outside the cell by the plasma membrane and usually require surface receptors to produce a cellular response, temperature is a physical signal; it reaches the cell surface and interior almost simultaneously. Therefore, cells can sense changes in environmental temperature at multiple sites, including the plasma membrane and intracellular compartments, and initiate cellular responses through a variety of mechanisms. Accordingly, several signaling pathways have been proposed to regulate cellular responses to elevated environmental temperatures, including a PHYTOCHROME B- and PHYTOCHROME-INTERACTING FACTOR4 (PIF4)–dependent thermomorphogenesis pathway (Jung et al., 2016; Legris et al., 2016), a cyclic nucleotide-gated calcium channel–calcium–calmodulin pathway (Liu et al., 2008; Saidi et al., 2009; Finka et al., 2012), a proteolytic cleavage-activated transcriptional activation pathway (Liu et al., 2007; Gao et al., 2008), a reactive oxygen species (ROS)–induced pathway (Mittler et al., 2011, 2012), and a H2A.Z-related pathway (Kumar and Wigge, 2010). Elevated temperatures also modulate cellular responses via phytohormone signaling and epigenetic mechanisms (Liu et al., 2015). However, because different studies used different high temperature and treatment durations, it is unclear whether high temperatures initiate these signaling pathways simultaneously, or whether there is a regulatory hierarchy in which a primary signaling pathway is activated by high temperature and subsequently affects other signaling pathways through crosstalk or transcriptional activation.
In general, signal transduction occurs very quickly. For example, plant exposure to brassinosteroids (BRs) for 5 min induced significant dephosphorylation of the transcription factor BRASSINAZOLE RESISTANT1 (BZR1; Wang et al., 2016). HS-induced cellular responses in plants can also occur quickly. For example, treatment at 37°C for 60 s increased the intracellular calcium concentration in wheat sheath cells; intracellular calcium concentration continued to increase with increasing HS period until it reached a plateau at ∼5 min (Liu et al., 2003). Similar results were obtained in Arabidopsis (Gao et al., 2012) and moss (Physcomitrella patens; Saidi et al., 2009). Using two-dimensional difference gel electrophoresis, it was found that 5 min of treatment at 37°C altered the abundance of at least 19 microsomal proteins in Arabidopsis seedlings (Wang et al., 2015). These HS-induced cellular responses occurred much more rapidly than those reported in many HS studies, in which plants were generally exposed to high temperatures for 30 min or longer. Meanwhile, 2 min of treatment at 37°C induced the formation of a chromatin puff in Drosophila larvae; the size of the puff peaked within 5 min of heat treatment (Belyaeva and Zhimulev, 1976). Moreover, HS treatment at 42°C for 2.5 min led to the up- or downregulation of numerous genes in cultured mouse cells (Mahat et al., 2016). These findings indicate that HS can induce rapid transcriptional responses in various organisms. Exploring how quickly heat treatment regulates the expression of HS-responsive genes (HSRs) in plants would help identify the major transcription factors that mediate the first wave of HS-regulated gene expression and the cellular responses that are directly regulated by the primary HS signal transduction pathway.
In this study, we systematically analyzed the transcriptional regulatory network in Arabidopsis plants after exposure to HS at 37°C using RNA sequencing (RNA-seq). We determined that HEAT SHOCK FACTOR A1 family proteins (HSFA1s), and the circadian clock proteins REVEILLE4 (RVE4) and RVE8, are among the primary transcription factors that mediate the first wave of HS-induced gene expression. In addition, we found that the mechanism by which RVE4/8 regulate thermotolerance in Arabidopsis around noon time involves the direct regulation of the expression of two downstream target transcription factors genes, ETHYLENE RESPONSIVE FACTOR53 (ERF53) and ERF54. Based on these results, a transcription regulatory hierarchy governing early HS-induced gene expression is proposed. Our study not only identified the transcription factors that regulate first wave of HS-regulated gene expression but also provide insight into how the circadian clock helps plants adapt to high temperatures during the day.
RESULTS
Identification of Rapidly Responding HS-Sensitive Genes by RNA-Seq
To determine how quickly HS treatment alters gene expression and to identify genes that are directly regulated by the primary HS signaling pathway, Arabidopsis seedlings (ecotype Columbia [Col-0]) were grown in agar plates for 1 week. With lids on to prevent withering of the seedlings due to sudden humidity and temperature changes, the seedlings were exposed to 37°C for 2.5, 5, 7.5, 10, 15, or 30 min and then snap frozen in liquid nitrogen for total RNA extraction and RNA-seq. As shown in Supplemental Figure 1, under our treatment conditions, the temperature inside the plates reached 28.7 ± 0.9, 31 ± 0.6, 32.7 ± 0.4, 33.9 ± 0.3, 35.5 ± 0.2, and 37.2 ± 0.4°C after HS treatment for 2.5, 5, 7.5, 10, 15, and 30 min, respectively.
Using a false discovery rate (FDR) < 0.05 and fold change of >2 or <0.5 as cutoffs, we identified 3259 early HS-regulated genes (eHSRs; Supplemental Data Set 1), including 938 upregulated and 2326 downregulated genes, whose expression was differentially affected by HS treatment during at least one exposure period. Five genes were initially downregulated by HS but subsequently upregulated as HS treatment time increased to 30 min. These genes are included in both HS downregulated and upregulated gene lists (Supplemental Data Set 1). Surprisingly, brief exposure to HS (2.5 min) reduced the expression of 877 genes but increased the expression of only 16 genes (Figure 1A; Supplemental Data Set 1). However, the upregulation of these genes was not detected in plants treated with HS for 5 min or longer. HS treatment for 5 min downregulated 722 genes and upregulated 6 genes. (Supplemental Data Set 1).
Figure 1.
RNA-Seq Analysis of Rapidly Responding HS-Regulated Genes.
(A) Number of differentially expressed genes following HS treatment at 37°C for the indicated time period.
(B) Gene expression paths within 30 min of HS treatment constructed using DREM. Each path represents a set of coexpressed genes. Split nodes (green dots) represent the divergence of a set of coexpressed genes, most likely due to the emergence of new regulatory events. A relatively small node implies that the expression levels of the genes going through that node are tightly centered around the node.
(C) GO classification of the genes in each DREM path (p1 to p13).
As the HS period increased, the number of HS-downregulated genes gradually decreased, while the number of HS-upregulated genes continued to increase (Figure 1A; Supplemental Data Set 1). HS treatment for 10 min appeared to be a threshold point; at 10 min, the number of HS-downregulated genes reached its lowest level, while the numbers of HS-downregulated and -upregulated genes increased rapidly beyond this point, possibly due to the activation of secondary or higher order transcriptional regulation. We then performed analysis using Dynamic Regulatory Events Miner (DREM), which summarizes the time-dependent emergence of new transcriptional regulatory events for groups of genes (Schulz et al., 2012). We detected 13 dynamic paths of differential gene expression within 30 min of HS treatment. Beginning at 10 min, the splitting of these nodes increased, most likely due to sequential transcriptional regulatory events (Figure 1B; Supplemental Data Set 1).
Gene Ontology (GO) enrichment analysis of genes in these paths showed that paths 1 to 5, which included HS-upregulated genes, were enriched for the categories related to abiotic stress responses, including responses to heat, temperature stimuli, high light, ROS, radiation, and protein folding. By contrast, paths 6 to 13, which included HS-downregulated genes, were enriched for categories involved in the regulation of plant growth and development, including cell surface receptor signaling, floral development, protein phosphorylation, plant organ morphogenesis, and so on (Figure 1C). These results suggest that HS induces the expression of stress-related genes to increase plant resistance to elevated environmental temperatures while suppressing the expression of growth- and development-related genes to ensure the appropriate energy and metabolite allocation, thereby increasing the likelihood of survival.
To further explore the potential transcriptional regulatory hierarchy that governs HS responses in plants, we constructed a gene coexpression network focusing on transcription factor–related eHSRs in DREM paths 1 to 5 as described previously (You et al., 2016). As observed in the network, as the HS treatment time increased, the number of gene clusters with similar HS-upregulated coexpression patterns increased, indicating that additional transcriptional events were activated (Supplemental Figure 2; Supplemental Data Set 3). Correspondingly, the number of HS-upregulated transcription factor genes increased with increasing treatment time. For example, MBF1c was the only transcriptional regulator among the eHSRs that was upregulated by 5 min of HS treatment, whereas at 7.5 min of HS treatment, the expression of MBF1c, HSFA2, HSFA7a, HSFB2b, and ZAT7 increased. When HS treatment was extended to 10 min, 16 transcription factor genes were identified among the HS-upregulated eHSRs (Supplemental Data Set 1). Taken together, these results indicate that increasing numbers of transcription factors are sequentially activated in response to longer HS treatment times.
Housekeeping genes are generally used as internal controls to ensure equal loading of samples in gene expression studies. To identify good internal control genes for future HS-regulated gene expression studies, we examined the expression patterns of ACTIN2 (AT3G18780), UBC30 (AT5G56150), TUBULIN4 (AT1G04820), PP2A A3 (AT1G13320), and UBQ5 (AT3G62250), which have been used as internal controls in previous studies (Franklin and Whitelam, 2007; Baron et al., 2012; Mu et al., 2013; Im et al., 2014; Zhang et al., 2016), based on our RNA-seq data. Although none of these genes were on our list of eHSRs, UBQ5 and PP2A A3 expression increased ≥1.5-fold following at least one HS exposure period, whereas the expression of UBC30, ACTIN2, and TUBULIN4 was relatively stable (Supplemental Figure 3). To validate these results, we measured the relative expression levels of UBC30, ACTIN2, and TUBULIN4 by RT-qPCR. As shown in Supplemental Figure 3, ACTIN2 and UBC30 expression was relatively stable in HS-treated plants, indicating that these genes could be used as internal controls in early HS-regulated gene expression studies in Arabidopsis. We chose UBC30 as an internal control for subsequent RT-qPCR.
The First Wave of HS-Upregulated Gene Expression Is Partially Mediated by HSFA1s
Transcription factors recognize the cis-regulatory elements in the promoter regions of their target genes to regulate their spatial and temporal expression patterns. To identify the primary transcription factors that mediate eHSR expression under HS conditions, we identified 1000-bp genomic sequences upstream of the translation start codons (ATG) of the 3259 eHSRs and subjected them to Multiple Em for Motif Elicitation (MEME) analysis. The cis-elements recognized by multiple transcription factor family proteins, including WRKY, MYB, and NAC transcription factors, were highly enriched among HS-downregulated eHSRs, suggesting that these transcription factors are involved in the suppression of early HS-regulated gene expression. In addition, the WRKY binding motif was the most highly enriched motif among genes downregulated by HS treatment for 2.5, 5, 7.5, and 15 min and the second most highly enriched motif among genes downregulated by 10 min of HS treatment. These findings suggest that WRKY family transcription factors might play critical roles in regulating the expression of early HS-downregulated genes (Supplemental Figure 4). However, it is also possible that transcriptional complexes containing WRKY, MYB, or NAC might be heat labile; thus, these complexes could be quickly inactivated by HS, resulting in the downregulation of their target genes. As the HS-induced expression of HSF and HEAT SHOCK PROTEIN (HSP) genes is generally considered a critical cellular response that increases plant thermotolerance, we focused our attention on the early HS-upregulated genes.
Few genes were upregulated following 2.5, 5, or 7.5 min of HS treatment (Figure 1A); therefore, our MEME analysis did not identify any enrichment of cis-elements from these groups of eHSRs. Interestingly, 59% of genes that were upregulated following 10 min of HS treatment contained the HSF binding cis-element heat shock element (HSE) in their promoters or 5′ untranslated regions (Figure 2A). As the HS period increased to 15 or 30 min, the number of HS-upregulated genes increased dramatically (Figure 1A), whereas the percentage of eHSRs containing HSEs decreased (Figure 2A), supporting our hypothesis that a secondary or higher order transcriptional regulatory mechanism emerged after 10 min of HS treatment. For eHSRs that were upregulated by 10 min of HS, many are likely the direct targets of the primary HS signal transduction pathway; these genes included 6 HSFs, 14 HSPs (most encoding small HSPs), and other genes involved in plant stress responses (e.g., heat stress, high light, wounding, salt stress), transcriptional regulation, protein stability, ROS responses, or hormonal signaling (Figure 2B; Supplemental Data Set 1).
Figure 2.
Rapid Upregulation of Genes by HS Is Partially Mediated by HSFs.
(A) Percentages of eHSRs upregulated by 10, 15, or 30 min of HS treatment containing cis-elements in their 1000-bp promoter regions recognized by the indicated transcription factors. C2H2dof, Dof-type zinc-finger DNA binding family protein; CPP, CXC domain-containing TSO1-like protein; Homeobox, homeodomain leucine zipper class I (HD-Zip I) protein; ND, AGAMOUS-like gene; NLP, plant regulator RWP-RK family protein; Orphan, paired amphipathic helix (PAH2) superfamily protein; PLATZ, PLATZ transcription factor; S1Falike, S1FA-like DNA binding protein.
(B) GO analysis of the eHSRs that were upregulated by 10 min of HS treatment.
HSFs are highly conserved transcription factors that regulate HS responses in eukaryotes (Scharf et al., 2012). The Arabidopsis genome contains 21 HSF genes, and HSFA1s are master transcription factors that regulate HS-induced gene expression (Liu et al., 2011; Yoshida et al., 2011). Based on RNA-seq, we identified several eHSRs that were regulated by 10 min of HS (including HSFA2, HSFA7a, HSFA7b, HSFB1, HSFB2a, and HSFB2b, MBF1c, and multiple small HSPs) that were previously shown to be directly regulated by HSFA1s in plants treated with HS for 60 min (Liu et al., 2011; Yoshida et al., 2011). These results suggest that HSFA1s are also master regulators of genes that are rapidly upregulated by 10 min of HS treatment. However, MEME analysis showed that ∼40% of the eHSRs that are upregulated by 10 min of HS treatment do not contain HSEs (Figure 2A), implying that additional primary transcription factors (other than HSFA1s) are involved in regulating HS-induced gene expression.
To verify this, we examined the basal and acquired thermotolerance of hsfa1-qk mutant, in which the expression of all four HSFA1 family transcription factors are knocked out by T-DNA insertions (Liu et al., 2011). Because the hsfa1-qk mutant was generated by crossing mutants produced in the Col-0 and Wassilewskija (Ws-2) backgrounds, we used both Col-0 and Ws-2 plants as controls (Liu et al., 2011). Similar to previous findings (Liu et al., 2011), the hsfa1-qk mutant is less resistant to HS treatment than the wild type. However, when the mutant was pretreated at 37°C for 30 min and subjected to the same HS conditions as in the basal thermotolerance assay, the survival rates of the seedlings greatly improved (Supplemental Figure 5). These results demonstrate that in the absence of HSFA1s, Arabidopsis plants can still sense elevated temperatures, regulate the expression of HS-related genes, and boost their tolerance to high environmental temperatures.
Identification of HSFA1-Dependent and -Independent eHSRs by RNA-Seq
To explore the HSFA1-dependent and -independent mechanisms that regulate early HS-induced gene expression, we compared the HS-regulated transcriptomes of wild-type versus hsfa1-qk plants. Meanwhile, because 1-week-old hsfa1-qk seedlings grown on agar plates were tiny and almost completely lacked expanded cotyledons (Supplemental Figure 6), we used 3-week-old, soil-grown seedlings as the difference in vegetative growth between the wild-type and hsfa1-qk plants was less dramatic (Supplemental Figure 6). We treated the seedlings at 37°C for 10 min before harvesting the leaves for RNA-seq. Using an FDR < 0.05 and fold change of >2 or <0.5 as cutoff values, we determined that 10 min of HS treatment differentially regulated the expression of 1790 genes (842 up- and 948 downregulated) in Col-0, 1049 genes (575 up- and 474 downregulated) in Ws-2, and 1495 genes (768 up- and 727 downregulated) in hsfa1-qk (Figure 3A; Supplemental Data Set 4). Therefore, many more HS-regulated genes were identified in soil-grown plants than in seedlings grown on agar plates, perhaps because the soil-grown plants were directly exposed to high temperature. Under this condition, the surfaces of the soil-grown plants reached the designated temperature much more rapidly than seedlings grown in agar plates (Supplemental Figure 1).
Figure 3.
Identification of HSFA1-Dependent and -Independent eHSRs.
(A) Venn diagram showing the number of eHSRs that were up- or downregulated in wild-type (Col-0 and Ws-2) and hsfa1-qk plants following HS treatment (at 37°C for 10 min; FDR < 0.05, fold change of >2 or <0.5).
(B) Heatmaps showing the expression patterns of HSF-dependent and -independent HS-regulated genes from (A). Log2(FPKM) values were used to generate the heatmap. FPKM, Fragments Per Kilobase per Million.
(C) GO classifications of the HSF-dependent and -independent HS-regulated genes shown in (B).
We identified 566 genes (331 up- and 235 downregulated) with similar changes in expression in response to HS in 3-week-old Col-0 and Ws-2 plants (Figure 3A). The small amount of overlap in eHSRs between Col-0 and Ws-2 indicates that different Arabidopsis ecotypes have different levels of sensitivity to HS or have evolved different mechanisms to adapt to increasing environmental temperatures. A comparison of these 566 genes with the eHSRs identified from 1-week-old seedlings listed in Supplemental Data Set 1 revealed that 40.46% (44.11% up- and 35.32% downregulated) of the genes were also identified as HS-regulated genes in 1-week-old Col-0 seedlings. This group of overlapping eHSRs might represent core HSRs with essential roles in early HS responses in Arabidopsis.
Of the 566 eHSRs that were similarly regulated by HS in Col-0 and Ws-2, 158 (47.73%) and 181 (77.02%) genes were up- or downregulated, respectively, to similar levels in hsfa1-qk, suggesting that these genes are regulated by HS in an HSFA1-independent manner. By contrast, 202 genes (155 up- and 47 downregulated, including HSFA2, HSFA7a, and HSFB2a) were not regulated by heat treatment in hsfa1-qk (or the fold change did not meet the selection criteria); these genes were considered to be HSFA1-dependent eHSRs. Additionally, 25 of the 566 eHSRs (18 up- and 7 downregulated) were up- or downregulated at least twofold (FDR < 0.05) in hsfa1-qk, but the changes in expression were much smaller in hsfa1-qk than in Col-0 or Ws-2. Thus, the HS-regulated expression of this small group of genes might be controlled by crosstalk between HSFA1s and (currently) unidentified transcription factors (Figure 3B; Supplemental Data Set 5).
GO enrichment analysis indicated that genes involved in plant growth and development were highly enriched among the HS-downregulated eHSRs identified in 3-week-old plants, regardless of whether these genes were regulated in an HSFA1-dependent or -independent manner (Figure 3C). Interestingly, the HSFA1-dependent HS-downregulated eHSRs from 3-week-old plants were more likely to be involved in plant growth processes (such as the response to auxin or gibberellin and organism development), whereas genes involved in cell differentiation processes (including the response to cytokinin, cell differentiation, and leaf morphogenesis) were more highly enriched among the HSFA1-independent HS-downregulated eHSRs from 3-week-old plants (Figure 3C).
Similar to the findings for 1-week-old seedlings, most HS-upregulated eHSRs from 3-week-old plants were related to plant stress responses. However, the HSFA1-dependent HS-upregulated eHSRs from 3-week-old plants were mainly involved in heat stress, chaperone binding, protein folding, and stress responses, whereas the HSFA1-independent HS-upregulated eHSRs from 3-week-old plants were more likely to be involved in drought, cold, salt, and abscisic acid responses (Figure 3C). Taken together, these results suggest that HSFA1s play essential roles in regulating the expression of genes that function in plant adaptation to HS, whereas other primary transcription factors involved in HS signaling mediate crosstalk with other abiotic stress signaling pathways.
RVE4 and RVE8 Regulate the Expression of HSFA1-Independent eHSRs
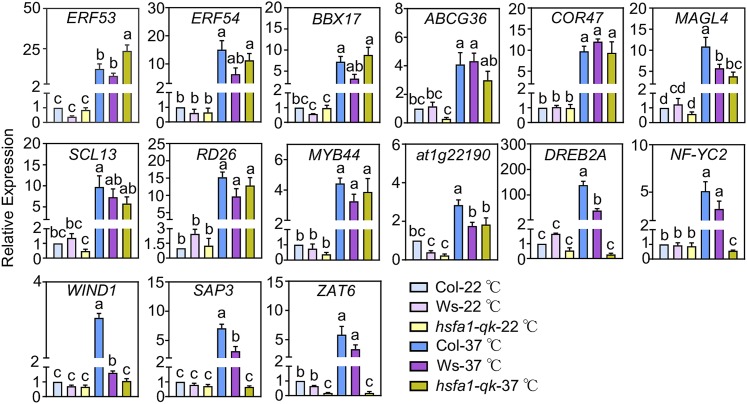
To validate our RNA-seq results, we performed RT-qPCR analysis of select eHSRs (mostly encoding transcription factors) from 3-week-old plants grown in soil under long-day (LD; 16-h-light/8-h-dark) conditions to investigate the effects of mutations in HSFA1s on their early HS-induced expression patterns. Consistent with our RNA-seq results, the HS-induced expression (after 10 min of treatment) of DREB2A, NF-YC2, WIND1, SAP3, and ZAT6 was abolished in hsfa1-qk, suggesting that the HS-induced expression of these genes is HSFA1 dependent. By contrast, ERF53, ERF54, BBX17, ABCG36, COR47, MAGL4, SCL13, RD26, MYB44, and At1g22190 were upregulated to similar levels under HS conditions in the hsfa1-qk and wild-type plants, suggesting that the HS-induced expression of these genes is HSFA1 independent. Together, these results indicate that our RNA-seq data are reliable (Figure 4).
Figure 4.
Validation of the HSFA1-Dependent and -Independent eHSRs by RT-qPCR.
Relative expression levels of the indicated genes in 3-week-old, HS-treated plants (at 37°C for 10 min). The gene expression levels are expressed as relative values compared with the value in non-heat-treated Col-0 seedlings and were analyzed by two-way ANOVA with Tukey’s significant difference test. Error bars indicate mean ± se of n ≥ 3, significant differences are indicated by different lowercase letters (P < 0.05).
To discover new primary transcription factors that mediate the transcriptional activation of HSFA1-independent eHSRs following HS exposure, we identified 1000 bp of genomic sequence located upstream of the ATG of the 331 HS-upregulated eHSRs shown in Supplemental Data Set 5 and subjected them to MEME analysis. The analysis suggested that these eHSRs might be regulated by HSF, basic leucine zipper (bZIP), NAC, basic helix-loop-helix (bHLH), BZR, and MYB transcription factors (Figure 5A). Not surprisingly, HSEs were highly enriched among the HSFA1-dependent eHSRs, but not among the HSFA1-independent eHSRs (Figures 5A to 5C). Interestingly, in contrast to bZIP, NAC, bHLH, and BZR transcription factors, which regulate both HSF-dependent and -independent eHSRs, a cis-element directly targeted by the MYB transcription factor RVE4 and its homologs RVE8, RVE7, and LHY was only enriched among HSFA1-independent eHSRs, suggesting that RVE4 family transcription factors specifically regulate HSFA1-independent eHSRs (Figures 5A to 5C).
Figure 5.
RVE4/8 Play an Important Role in Regulating Thermotolerance in Arabidopsis.
(A) Enrichment of the cis-elements recognized by various transcription factors in the 1000-bp promoter regions of HSFA1-dependent and -independent HS-upregulated genes.
(B) and (C) Top nine most highly enriched cis-elements for HSFA1-dependent (B) and -independent (C) HS-upregulated genes are shown.
(D) to (F) Comparison of basal and acquired thermotolerance in rve4, rve8, and rve4/8 plants. Seven-day-old seedlings grown at 22°C under continuous light were used. Seedlings recovered 7 d after HS were photographed (D), and the survival rates of the plants were calculated ([E] and [F]). Each dot represents the result from one biological replicate, error bars indicate mean ± se. Statistically significant differences are indicated by different lowercase letters (P < 0.05, two-way ANOVA with Tukey’s significant difference test).
RVE4 and RVE8 are components of the central oscillator of the circadian clock. RVE4 and RVE8 transcription is regulated by the circadian clock under both LD (Supplemental Figure 7) and short-day conditions (Hsu et al., 2013). RVE8 accumulates to its highest levels around Zeitgeber time (ZT) 3 to 6, that is, 3 to 6 h after turning on the light, under 12-h-light/12-h-dark growth conditions, when it binds to the evening elements (EEs) on the promoters of evening circadian clock genes, such as PSEUDO RESPONSE REGULATOR5 (PRR5), and activates their expression (Rawat et al., 2011; Hsu et al., 2013). RT-qPCR indicated that the expression of RVE4 and RVE8 was not significantly regulated by brief HS treatment (within 10 min; Supplemental Figure 8). Knocking out or knocking down the expression of RVE4, RVE8, or LHY alone slightly decreased the basal thermotolerance of the mutants, whereas simultaneously knocking out/down the expression of RVE4 and RVE8 significantly increased susceptibility of the rve4 rve8 mutant (rve4/8) to HS treatment (Supplemental Figure 9). To minimize the influence of the circadian phase changes of rve4/8 on its HS responses, we also grew the seedlings under 24 h of continuous light at 22°C for 7 d before subjecting them to HS treatment. Both basal and acquired thermotolerance were significantly reduced in rve4/8 versus the wild type (Figures 5D to 5F). Taken together, these results suggest that RVE4 and RVE8 play redundant, critical roles in regulating thermotolerance in Arabidopsis.
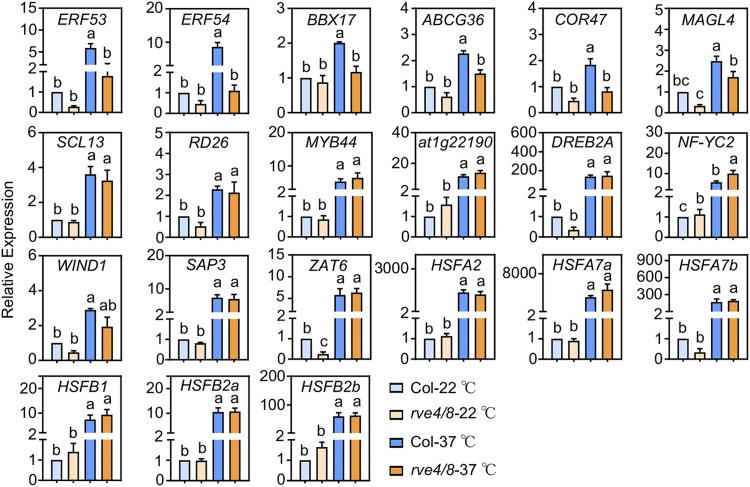
To investigate whether RVE4 and RVE8 are the primary transcription factors that mediate HS-induced, HSFA1-independent eHSRs expression, we performed RT-qPCR to test for the HS-induced expression of the HSFA1-independent genes listed in Figure 4 in rve4/8 double mutant plants. Under LD conditions, the HS-induced expression of ERF53, ERF54, ABCG36, BBX17, COR47, and MAGL4 was not significantly altered in rve4/8 plants after 10 min of heat exposure, demonstrating that the HS-induced expression of these eHSRs is dependent on RVE4/8 (Figure 6). By contrast, the expression of SCL13, RD26, MYB44, At1g22190, DREB2A, NF-YC2, WIND1, SAP3, ZAT6, HSFA2, HSFA7a, HSFA7b, HSFB1, HSFB2a, and HSFB2b in rve4/8 plants was induced by HS treatment to a level similar to that of the wild-type plants. These results suggest that other primary transcription factors (e.g., bZIP, NAC, and bHLH) are involved in mediating the HS-induced expression of SCL13, RD26, MYB44, and At1g22190.
Figure 6.
RT-qPCR Validation of the RVE4/8-Dependent and -Independent eHSRs.
Relative expression of the indicated genes in 3-week-old plants exposed to 37°C for 10 min. Gene expression levels are expressed relative to the value in non-heat-treated Col-0 seedlings. Error bars indicate mean ± se of n ≥ 3. Statistically significant differences are indicated by different lowercase letters (P < 0.05, two-way ANOVA with Tukey’s significant difference test).
To compare the contributions of RVE4/8 and HSFA1s to plant thermotolerance, we generated additional hsfa1a or hsfa1b mutations in the hsfa1d/1e T-DNA insertion double mutant using clustered regularly interspaced short palindromic repeat/associated protein 9 (CRISPR/Cas9). Among the mutants generated, hsfa1a-crispr contained an extra base in the coding sequence, while hsfa1b-crispr had a missing base in the coding sequence, resulting in a frameshift at codon 125 and codon 70 of HSFA1a and HSFA1b, respectively. These two CRISPR/Cas9 mutants were crossed with each other to generate the quadruple mutant (hsfa1-cq) for HSFA1s in the Col-0 ecotype background (Supplemental Figure 10).
The HS-regulated expression patterns of the eHSRs in the new hsfa1-cq mutant (Supplemental Figure 11) and the hsfa1-qk mutant (Figure 4) were similar. When hsfa1-cq plants were grown side by side with rve4/8 mutants and subjected to HS treatment, both basal and acquired thermotolerance were reduced in hsfa1-cq and rve4/8. However, hsfa1-cq was much more sensitive to HS treatment than rve4/8 (Supplemental Figure 12), suggesting that HSFA1s are more important than RVE4/8 for regulating thermotolerance in Arabidopsis.
ERF53 and ERF54 Are Direct Targets of RVE8
Among the RVE4/8-dependent HS-upregulated eHSRs were two homologous transcription factor genes: ERF53 and ERF54. Given that ERF53 overexpression increases thermotolerance in Arabidopsis (Hsieh et al., 2013) and that RVE4, RVE8, ERF53, and ERF54 are universally expressed genes (Supplemental Figure 13), we investigated whether the heat-induced expression of ERF53 and ERF54 is directly regulated by RVE4/8. We examined the promoter sequences of ERF53 and ERF54 and found two and three EE-like cis-elements located within 1000 bp upstream of the ATG of ERF53 and ERF54, respectively (Figure 7A). We therefore performed a chromatin immunoprecipitation (ChIP) assay using pRVE8:RVE8-YFP transgenic plants to test for the direct binding of RVE8 to the promoter regions of ERF53 and ERF54. Because we determined that RVE8 regulates ERF53 and ERF54 under HS condition in an HSFA1-independent manner, pHSFA1d:HSFA1d-GFP transgenic plants were used as an additional negative control. When expressed in rve4/8 or hsfa1-qk plants, the pRVE8:RVE8-YFP transgene recovered the HS-induced expression of ERF53 and ERF54 to the wild-type levels in rve4/8 (Supplemental Figure 14), and the pHSFA1d:HSFA1d-GFP construct rescued the dwarf phenotype of hsfa1-qk (Supplemental Figure 15), demonstrating that the RVE8-YFP and HSFA1d-GFP fusion proteins used in the ChIP assay were functional.
Figure 7.
RVE8 Regulates Plant Thermotolerance by Regulating the Expression of ERF53 and ERF54.
(A) Structural diagrams of the ERF53, ERF54, PRR5, and HSP70 promoters. The locations and sequences of putative EEs (a1-2, b1-3, c) in the ERF53, ERF54, and PRR5 promoters or HSE in the HSP70 promoters are shown.
(B) ChIP-qPCR assays of select EE- or HSE-containing regions from the promoters of ERF53, ERF54, PRR5, or HSP70 in pRVE8:RVE8-YFP or pHSFA1d:HSFA1d-GFP transgenic plants. GFP-trap agarose (GFP) or anti-myc agarose (control) beads were used for the pull-down assay.
(C) RVE8 activates the expression of ERF53 and ERF54 under HS conditions. The pERF53:luciferase or pERF54:luciferase expression vector was cotransformed with the p35S:YFP-RVE8 or p35S:CFP (control) expression vector into N. benthamiana leaves. Relative luciferase (LUC) activity represents the ratio between YFP-RVE8- and CFP-upregulated luciferase activity.
(D) and (E) Overexpression (OX) of ERF53 increases basal thermotolerance of both wild-type and rve4/8 plants. Each dot represents the result from one biological replicate. Error bars indicate mean ± se of n ≥ 3. Statistically significant differences are indicated by different lowercase letters (P < 0.05, two-way ANOVA with Tukey’s significant difference test).
No significant enrichment of EEs in the promoters of ERF53 and ERF54 was observed at room temperature, as determined by RT-qPCR. In plants treated at 37°C for 10 min, we detected a significant increase in the binding of RVE8-YFP to the EE motif located −169 to −161 bp upstream of the ATG in ERF53 and to the EE motifs located −479 to −471, −551 to −543, and −756 to −748 bp upstream of the ATG in ERF54 (Figure 7B). By contrast, RVE8-YFP bound strongly to the EE located at −389 to −381 bp in the PRR5 promoter; however, this binding was not regulated by HS treatment (Figures 7A and 7B). As a negative control, HSFA1d-GFP bound directly to the HSE motif in the HSP70 promoter (a direct target of HSFA1d) under HS conditions, where the binding of HSFA1d-GFP to the EE motifs in the ERF53 and ERF54 promoters was not detected (Figure 7B).
To obtain additional evidence that RVE8 is a transcriptional activator of ERF53 and ERF54, we performed a transient transcriptional reporter assay using Nicotiana benthamiana leaves. At room temperature, firefly luciferase activity in N. benthamiana leaves cotransformed with the p35S:YFP-RVE8 and pERF53:luciferase (or pERF54:luciferase) expression vector pair was similar to that of leaves cotransformed with p35S:Cyan fluorescent protein (p35S:CFP) and pERF53:luciferase (or pERF54:luciferase). A 10-min HS treatment significantly increased the luciferase signal in leaves cotransformed with p35S:YFP-RVE8 and pERF53:luciferase (or pERF54:luciferase) versus leaves cotransformed with p35S:CFP and pERF53:luciferase (or pERF54:luciferase; Figure 7C). These results help confirm the ChIP-qPCR data and suggest that RVE8 activates the expression of ERF53 and ERF54 only under HS conditions. Furthermore, overexpressing ERF53 increased the thermotolerance of both the wild-type and rve4/8 plants, which corresponded with the increased expression level of ERF53 (Figures 7D and 7E; Supplemental Figure 16). Taken together, these results demonstrate that ERF53 and ERF54 are direct targets of RVE8. In addition, HS stimulates the binding of RVE8 to the promoters of ERF53 and ERF54 and induces their expression to increase plant tolerance to HS treatment.
RVE8-Regulated Plant Thermotolerance Is Gated by the Circadian Clock
RVE8 protein accumulated to its highest level at ZT 3 to 6 (Supplemental Figure 17) under our growth (16-h-light/8-h-dark) conditions, and this is the time point at which we harvested seedlings for RNA-seq and HS treatment. To investigate whether RVE4/8 play a general role in regulating plant HS responses, or whether they regulate plant thermotolerance specifically around the time when their expression reaches its highest level, we subjected 7-d-old seedlings grown under LD conditions to 45°C HS treatment at ZT 0 to 1, ZT 6 to 7, ZT 15 to 16, and ZT 19 to 20. The thermotolerance of the wild-type plants increased at ZT 6 to 7 and ZT 15 to 16 but returned to a level similar to that at the beginning of the experiment (ZT 0 to 1) at ZT 19 to 20. By contrast, the thermotolerance of rve4/8 was similar to that of the wild type at ZT 0 to 1, ZT 15 to 16, and ZT 19 to 20 but significantly lower at ZT 6 to 7 (Figures 8A and 8B). Similarly, RT-qPCR analysis showed that HS-induced expression of ERF53, ERF54, and BBX17 (all RVE4/8-dependent eHSRs) was significantly lower in rve4/8 at ZT 6 to 7, but not at any other time point compared with the wild type (Figure 8C). By contrast, the HS-induced expression of HSFA2 was not altered in rve4/8 compared with the wild type at any time point (Figure 8D). Finally, the expression of the RVE8 target clock gene PRR5 was reduced in rve4/8 at ZT 6 to 7 compared with the wild type in both HS-treated and untreated conditions (Figure 8D). Taken together, these results demonstrate that the circadian clock plays important roles in regulating plant tolerance to high temperatures and that RVE4/8 regulate plant thermotolerance specifically around ZT 6 to 7 under LD conditions.
Figure 8.
Circadian Clock-Dependent Regulation of Plant Thermotolerance by RVE4/8.
(A) and (B) Basal thermotolerance analysis of Col-0 and rve4/8 plants at ZT 0 to 1, ZT 6 to 7, ZT 15 to 16, and ZT19 to 20. Seven-day-old seedlings grown at 22°C under LD conditions were transferred to a 45°C water bath. The plants were allowed to recover at 22°C for 7 d before being photographed (A) and assessed for survival (B). Each dot represents the result from one biological replicate.
(C) and (D) Effect of the circadian clock on the HS-induced upregulation of ERF53, ERF54, and BBX17 (C), HSFA2 (D). PRR5 was used as a negative control. Three-week-old, soil-grown Col-0 and rve4/8 plants were treated with 37°C for 10 min at different times (ZT). Error bars indicate mean ± se of n ≥ 3. Statistically significant differences are indicated by different lowercase letters (P < 0.05, two-way ANOVA with Tukey’s significant difference test).
DISCUSSION
In recent decades, significant progress has been made in elucidating the responses of plants to environmental HS at the molecular level. However, how plants sense HS and the primary HS signal transduction pathway leading to HS-regulated gene expression are not well understood. One reason for this situation is the lack of appropriate standards to determine whether the primary HS signal transduction pathway is affected in mutant or transgenic plants. Traditionally, differences in plant survival after HS treatment, and differences in the transcript levels of HSFs and HSPs between HS-treated and -untreated plants, have been used to evaluate whether specific genes play important roles in regulating plant HS responses. In these studies, the HS treatment time used was generally 30 min or longer. Under these conditions, genes whose expression is directly regulated via the primary HS signal transduction pathway are not usually separated from genes whose expression is indirectly regulated by a transcriptional cascade or signaling crosstalk. The cellular abundance of HSFs and HSPs in plants generally corresponds to their HS tolerance. Therefore, genes or signaling pathways that directly or indirectly affect HSF and/or HSP expression could alter plant thermotolerance (Swindell et al., 2007), which could lead to the incorrect conclusion that certain genes or pathways regulate HS signaling. However, this confusion could be avoided by reducing the HS treatment time to as short as possible.
In this study, we obtained a comprehensive view of how brief HS exposure affects the Arabidopsis transcriptome, including an assortment of eHSRs whose expression is rapidly regulated by HS. We determined that under HS conditions, transcriptional reprogramming occurs quickly; a 2.5-min HS treatment inhibited the expression of 877 genes (Figure 1A). This rapid heat-induced global gene suppression, which has also been reported previously (Duarte et al., 2016), could be caused by the rapid HS-induced dissociation of RNA polymerase II from DNA (Hieda et al., 2005). We also detected a second round of global HS-induced gene suppression, which occurred after 15 min of HS treatment. This second round of transcriptional suppression occurred after the HS-induced expression of a number of HSPs. HSPs are molecular chaperones that prevent high-temperature-induced protein aggregation by facilitating the correct folding of cellular proteins (Figures 1A and 2B). Thus, the molecular mechanism underlying the second round of HS-induced gene suppression is likely different from that during the first round.
RNA-seq showed that 5 min of HS treatment at 37°C upregulated the expression of MBF1c in 1-week-old seedlings grown on agar plates by at least twofold; the HS-induced expression of this gene continued to increase with prolongation of HS treatment time. Therefore, it appears that 5 min is the minimum amount of time required for the primary HS signal transduction pathway in plants, from HS sensing to alterations in nuclear gene expression. However, under our HS treatment conditions, the temperature inside the agar plates only reached an average of ∼28.7 ± 0.9°C after 2.5 min of HS treatment and 31 ± 0.6°C when HS treatment was extended to 5 min (Supplemental Figure 1), suggesting that plant HS signal transduction is accomplished even more rapidly and is evoked at a lower temperature than expected. In our RT-qPCR experiments, the twofold or higher HS-induced expression of a number of HSFs and many other eHSRs was stably reproduced in plants treated with HS for 10 min or longer. Therefore, we used 10 min of HS exposure in all subsequent early HS-induced gene expression experiments.
Increased HSF expression is generally used as an indicator of the turning on of plant HS responses. Activated HSFs bind to HSEs in the promoters of HSRs (e.g., HSPs and genes encoding ROS-producing and/or scavenging enzymes) and regulate their expression, thereby increasing plant thermotolerance. By analyzing the cis-elements present in the 1000-bp promoter region or 5′ untranslated region of each eHSR that was upregulated in response to 10 min of HS treatment, we found that HSEs, which are bound by HSFs, were highly enriched. Using hsfa1-qk plants, we demonstrated that HSFA1s are master regulators of rapid HS-induced gene expression; ∼50% of the eHSRs that are upregulated after 10 min of HS treatment in 3-week-old plants are regulated by HSFA1s (Figure 3). Interestingly, the expression of HSP90 and many other HSFA1-dependent HS-upregulated genes did not change within 10 min of HS treatment, but they were upregulated after 15 or 30 min of HS treatment (Supplemental Data Set 1). The HSFA1-dependent temporal regulation of HS-upregulated genes suggests that chromosome structure might be sequentially regulated by HS; for example, the HSFA1-dependent depletion of H2A.Z from nucleosomes is regulated by HS conditions (Cortijo et al., 2017). Alternatively, the binding of additional transcription factors might be responsible for upregulating the HSFA1-dependent eHSRs that require a longer HS treatment period.
The circadian clock regulates plant responses to daily temperature and light cycles. For example, in green and ripening grape (Vitis vinifera) fruit, HS triggers different transcriptomic responses depending on whether heat exposure occurs during the day or night, suggesting that the responses of plants to HS at different times of the day are not the same (Rienth et al., 2014). Plants also differ in their ability to withstand high temperatures at different times of day (Dickinson et al., 2018). The current results reinforce the notion that the thermotolerance of plants varies at different times of day (Figures 8A and 8B).
Recent studies have shown that various light receptors and the circadian clock protein TIMING OF CAB EXPRESSION1 (TOC1) may gate daily plant thermomorphogenesis (Zhu et al., 2016; Li et al., 2018); however, the molecular mechanism underlying circadian clock–dependent regulation of plant thermotolerance is largely unknown. Among the principal clock-regulating genes, the expression of RVE8 peaks 3 to 6 h after turning on the light (Supplemental Figure 19; Rawat et al., 2011), that is, around noon or in the early afternoon, when HS is most likely to occur. Temperature compensation of the circadian rhythm was disrupted in rve8- and RVE8-overexpressing plants (Rawat et al., 2011), suggesting that RVE8 plays an important role in integrating the temperature and circadian responses of plants.
In the current study, we provided several lines of evidence demonstrating that RVE8 and its close homolog RVE4 play important roles in regulating plant thermotolerance and the HS-induced expression of HSFA1-independent eHSRs. First, analysis of the cis-elements of the HS-upregulated eHSRs in 3-week-old plants revealed strong enrichment of the RVE4/8-recognizing EE motif only in HSFA1-independent eHSRs (Figures 5A to 5C). Second, knocking out/down the expression of RVE4 and/or RVE8 impaired both basal and acquired thermotolerance in plants grown under LD or continuous light conditions (Figures 5D to 5F; Supplemental Figure 9). Third, RT-qPCR showed that after 10 min of heat treatment, the HS-induced expression of ERF53, ERF54, ABCG36, BBX17, COR47, and MAGL4 was similar in the wild-type and hsfa1-qk plants but impaired in rve4/8 plants (Figures 4 to 6; Supplemental Figure 11). Of these RVE4/8-dependent HS-upregulated eHSRs, the overexpression of ERF53 increases plant resistance to HS (Figures 7D and 7E; Hsieh et al., 2013), suggesting that these RVE4/8-dependent eHSRs play critical roles in regulating plant thermoresponses. Fourth, ChIP-qPCR and transient transcription assays showed that RVE8 binds directly to the promoter regions of ERF53 and ERF54 and activates their expression only under HS conditions. Interestingly, the binding of RVE8 to the promoter of its circadian clock target gene PRR5 is not regulated by HS (Figures 7A to 7C). It was showed that overexpressing TOC1, another circadian clock gene directly targeted by RVE8, reduced acquired thermotolerance in plants, whereas the toc1 prr5 double mutant showed enhanced acquired thermotolerance (Rawat et al., 2011; Zhu et al., 2016). These findings, combined with the current results, suggest that the molecular mechanisms by which RVE8 regulates plant thermotolerance and circadian rhythms are not the same. Together, our results demonstrate RVE4/8 are primary transcription factors that function in parallel with HSFA1s to regulate the first wave of HS-induced gene expression and prepare plants for exposure to high temperatures during the day.
HSFA1s and RVE4/8 are constitutively expressed transcription factors and their expression is not significantly regulated by brief HS exposure, suggesting that HS signal transduction might activate HSFA1s and RVE4/8 posttranscriptionally. The DNA binding or transcriptional activity of HSFA1s can be altered by posttranslational modifications such as protein phosphorylation (Reindl et al., 1997; Liu et al., 2008), protein sumoylation (Miller et al., 2010), and/or oxidative damage caused by ROS (Jung et al., 2013). However, direct evidence that these posttranslational modifications are induced by HS in vivo is currently lacking. Additionally, HSFA1s are thought to be activated by the cytoplasmic protein response (Ohama et al., 2016). According to this model, HSP70 and HSP90 bind to HSFA1s under normal conditions and prevent their binding to DNA to influence gene expression. HS induces cytosolic protein misfolding and denaturation, and recruits molecular chaperones, including HSP70 and HSP90, to help refold the denatured proteins. As a result, HSFA1s are released and translocated into the nucleus where they regulate the expression of HS-related genes. Because the HS condition used in the current study was relatively mild (37°C) and the treatment time was relatively short (10 min or less), whether this treatment is sufficient to trigger the cytoplasmic protein response has yet to be determined.
By contrast, limited information is available about how RVE4/8 transcriptional activity is regulated. Given that RVE4/8 and HSFA1s might simultaneously regulate the first wave of HS-upregulated gene expression, it is likely that RVE4/8 and HSFA1s are activated via similar mechanisms under HS conditions. The identification of proteins that can bind to both RVE4/8 and HSFA1s under HS conditions would help decode the mechanisms leading to HS-induced gene expression.
SCL13, RD26, MYB44, and At1g22190 were induced to approximately the wild-type levels in hsfa1-qk and rve4/8 plants following 10 min of HS treatment (Figures 4 and 6), as revealed by RT-qPCR, suggesting that additional primary transcription factors regulate the first wave of HS-upregulated gene expression. Indeed, our MEME analysis showed that cis-elements that are recognized by bZIP, NAC, bHLH, and BZR transcription factors were highly enriched among eHSRs upregulated after 10 min of HS treatment (Figure 5A). Of these transcription factors, BZR transcription factors are key regulators of BR signaling. Exogenously applied BR increases plant tolerance to HS (Vardhini et al., 2010). In addition, BZR1 regulates thermotolerance in tomato by regulating the expression of its downstream target genes, the homologous receptor-like kinase genes FERONIA2 (FER2) and FER3 (Yin et al., 2018). The enrichment of BZR binding cis-elements in HS-upregulated eHSRs suggests that HS might rapidly modulate BR signaling to coordinate plant growth and stress responses.
In the current study, we identified numerous eHSRs, including many previously confirmed HS-regulated genes such as MBF1c, HSFA2, HSFA7a, HSFA7b, HSFB1, HSFB2a, HSFB2b, HSPs, GALACTINOL SYNTHASE1 (GolS1), GolS2, FKBP62, DREB2A, DREB2B, and bZIP28, genes related to superoxide production or inactivation, and so on. These findings suggest that the RNA-seq database generated in this study will be highly valuable for future in-depth data mining to uncover mechanisms that govern plant responses to HS. Through RT-qPCR, we demonstrated that the early upregulation of a select group of eHSRs (primarily encoding transcription factors) by HS is dependent on HSFA1s, RVE4/8, and currently unidentified transcription factors.
Based on these findings, we propose a model describing how a transcriptional regulatory hierarchy governs the first wave of HS-regulated gene expression (Figure 9). According to this model, increased environmental temperatures initiate a signal transduction pathway that activates primary transcription factors such as HSFA1s and RVE4/8 to initiate the first wave of HS-regulated gene expression and to upregulate the expression of genes encoding a number of transcription factors, including HSFA2, HSFA7a, HSFA7b, HSFB1, HSFB2a, HSFB2b, DREB2A, ZAT6, ERF53, ERF54, and so on. These transcription factors, in turn, activate the expression of their own set of downstream target genes to fine-tune cellular protein or metabolic dynamics to help plants adapt to increased environmental temperatures. However, it appears that RVE4/8 regulate plant thermotolerance specifically at approximately ZT 6 to 7, but not at ZT 15 to 16 (Figure 8). As the plants showed the highest thermotolerance at ZT 15 to 16 under our growth conditions, it would be interesting to investigate whether additional clock-related gene are activated at ZT 15 to 16 and increase the tolerance of Arabidopsis plants to HS. Finally, the 10-min HS-upregulated eHSRs identified in this study and the HS treatment conditions (10 min of HS treatment of plants grown in agar plates or soil) used in this study could be used as references for future HS signal transduction studies. Such studies could help uncover novel signaling mechanisms by which plants sense and adapt to increasing environmental temperatures.
Figure 9.
Proposed Model of the Transcriptional Regulatory Hierarchy Governing HS-Induced Gene Expression.
According to this model, HSFA1s and RVE4/8 serve as the primary transcription factors that are directly activated by HS signaling and initiate the expression of their downstream target genes, including a number of transcription factor genes, to regulate plant thermotolerance.
METHODS
Plant Growth and HS Treatment
Arabidopsis (Arabidopsis thaliana) seeds (40 to 50 per genotype) were surface sterilized and sown on glass plates containing 30 mL of growth medium (0.6% [w/v] Phytagel, 0.5× Murashige and Skoog basal salt mixture, and 1% [w/v] Suc, pH 5.7). The seeds were stratified at 4°C in the dark for 3 d and grown at 22°C under LD (16-h-light [∼100 μmol m−2 s−1]/8-h-dark) conditions or under constant light for 7 d in a growth chamber (Percival Scientific, cool-white fluorescent bulb). For the basal thermotolerance assay, at ZT 3 to 6, the plates were wrapped in plastic wrap, divided into two groups, and submerged directly in a water bath at 45°C (HS) or 22°C (control) for 12 to 13 min. For the acquired thermotolerance assay, at ZT 3 to 6, the plates were incubated in a water bath at 37°C for 30 min and transferred to a growth chamber at 22°C for 2 h of recovery before being submerged in a 45 or 22°C water bath for 15 to 16 min. For the diurnal thermotolerance assay, the plates were divided into five groups: HS-treated plates placed in a 45°C water bath for 13 min at ZT 0 to 1, ZT 6 to 7, ZT 15 to 16, and ZT 19 to 20, and the control. The heat-treated and control plants were allowed to recover at 22°C under LD conditions for 7 d before being assessed for survival. The temperature inside and outside the plates during the HS treatment was measured simultaneously using electronic thermometers with microprobe. The temperature was automatically recorded every 30 s.
Isolation of Arabidopsis Mutants
The T-DNA insertion mutants used in this study, SALK_051728 (rve4), SALK_053482 (rve8), SALK_031092 (lhy), SAIL_410_E01 (hsfa1d), and SALK_094943 (hsfa1e), were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). The confirmed mutants were backcrossed once with the corresponding wild-type plants to clean up the background. The rve4/8 double mutant was generated by crossing SALK_051728 with SALK_053482. F3 segregated homozygous single or double mutants were used in this study. The gene-specific primers used to genotype the mutants are listed in Supplemental Table 1.
To generate CRISPR/Cas9 mutants for HSFA1a and HSFA1b, guide RNA sequences were designed (CTAATGAAGGTTTCTTAAG for HSFA1a and AAGTATTTCAAGCACAACA for HSFA1b) and cloned into the CRISPR/Cas9 vector pHEC-401 as described by Xing et al. (2014). The CRISPR/Cas9 vectors were transformed into the hsfa1d hsfa1e T-DNA double homozygous mutant using the Agrobacterium tumefaciens strain GV3101–mediated floral dip method (Clough and Bent, 1998). T1 transgenic plants were selected on half strength Murashige and Skoog agar medium containing 50 µM hygromycin. The genomic DNA sequence containing the CRISPR/Cas9 target sites was amplified and sequenced to check for mutations. hsfa1a-cirspr and hsfa1b-cirspr T1 heterozygous plants were crossed with each other to generate the hsfa1-cq quadruple mutant, which was crossed once with hsfa1d hsfa1e to clean up potential background mutations. All primers used for construct generation and genotyping are listed in Supplemental Table 1.
RNA-Seq and Gene Expression Analysis
Seven-day-old seedlings grown on agar plates (covered with lids and sealed) or 3-week-old plants grown in soil at 22°C in a growth chamber under LD conditions were transferred to a new growth chamber with an inside temperature preset to 37°C for heat treatment for the designated time or kept in the same growth chamber without heat stress (control). Immediately after HS treatment (at ZT 3 to 6), the plates containing 7-d-old seedlings were snap frozen in liquid nitrogen, and the seedlings (40 to 50 individual plants) were harvested by scraping the agar surface with a spatula under liquid nitrogen. For 3-week-old plants, one to two rosette leaves each from five to six independent plants per genotype were harvested, pooled as one biological sample, and used for RNA-seq or RT-qPCR.
For RNA-seq, plant tissue was ground to a fine powder in liquid nitrogen, and total RNA was extracted from the tissue using an RNeasy Plant Mini Kit (Qiagen). Three biological replicates were used for RNA-seq and subsequent bioinformatic analyses. RNA-seq was performed on the HiSeq X platform (Illumina) using 150-bp double-ended reads. The reads were aligned to the Arabidopsis reference genome (version The Arabidopsis Information Resource 10 [TAIR10]) using TopHat (version 2.0.12; Trapnell et al., 2009; Kim et al., 2013). After filtering out low-quality reads with FASTX-Toolkit (version 0.0.13), uniquely aligned reads were counted for each annotated gene using the program htseq-count from the python package HTSeq (version 0.6.1; Anders et al., 2015). Differential gene expression was evaluated using the R package DESeq (Anders and Huber, 2010); P-values were adjusted using the Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995) to control the FDR. Differentially expressed genes were defined based on the following criteria: FDR < 0.05 and at least a twofold change in expression compared with the control samples.
For RT-qPCR, total RNA was extracted from the tissue using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was synthesized using a PrimeScript RT Reagent Kit with Genomic DNA Eraser (Takara Bio). HS-regulated gene expression was analyzed by RT-qPCR using SYBR Premix Ex Taq (Takara Bio) and a C1000 Touch Thermal Cycler CFX384 (Bio-Rad). The gene-specific primers used for gene amplification are listed in Supplemental Table 1. The relative abundance of the target transcripts was determined by the comparative threshold cycle method (Schmittgen and Livak, 2008), using UBC30 as an internal reference.
Bioinformatic Analysis
RNA expression DREM paths were constructed using DREM (version 2.0) as described by Ernst et al. (2007). The log2(fold change) values of the eHSRs for each HS treatment period were used for analysis. GO enrichment of genes in each DREM path was calculated using DREM (version 2.0) as follows: genes enriched = genes assigned − genes expected (Schulz et al., 2012). The remaining GO enrichment analysis of the eHSRs was performed using the online tool Database for Annotation, Visualization and Integrated Discovery (https://david.ncifcrf.gov/; Huang et al., 2009), using -log2(P-value) to evaluate enrichment levels. Heatmaps of the differentially expressed genes were generated using HemI (version 1.0; Deng et al., 2014). Enrichment of cis-elements in the −1000 to −1 promoter sequences of the eHSRs (downloaded from TAIR10; http://www.arabidopsis.org/) was performed using the online tool MEME (version 4.12.0; http://meme-suite.org/tools/centrimo; Bailey et al., 2009, 2015; Bailey and Machanick, 2012; O’Malley et al., 2016). Transcription factors were identified according to the Arabidopsis transcription factor database (http://planttfdb.cbi.pku.edu.cn/). eHSRs in DREM paths 1 to 5 were used to generate a gene coexpression network as described by You et al. (2016). The network was visualized using Cytoscape (version 3.7.1; Shannon et al., 2003).
Cloning and Generation of Transgenic Plants
The 1508-bp (ERF53) and 1502-bp (ERF54) promoter sequences (relative to the translation start site), the coding sequences of ERF53 and RVE8, and 2754-bp (RVE8), and 4547-bp (HSFA1d) genomic DNA sequences upstream of the stop codon (stop codon not included) were cloned into the pENTR/SD/D-TOPO vector (Invitrogen) and subcloned into Gateway-compatible binary vectors pHGWL7, pCAMBIA1390, pEarleyGate 104, pEarleyGate TW1 (a modified pEarleyGate 101 vector), and pGWB4 to generate the pHGWL-ERF53 (pERF53:luciferase), pHGWL-ERF54 (pERF54:luciferase), p1390-ERF53 (p35S:ERF53-myc), p104-RVE8 (p35S:YFP-RVE8), pTW1-RVE8 (pRVE8:RVE8-YFP), and pGWB4-HSFA1d (pHSFA1d:HSFA1d-GFP) expression vectors by LR cloning (Invitrogen). The expression vectors were incorporated into A. tumefaciens strain GV3101. Transgenic Arabidopsis plants were generated using the floral dip method. T3 transgenic homozygous plants were used for functional analysis unless otherwise indicated. Levels of the proteins in the transgenic plants were analyzed by immunoblots using anti-GFP antibody (dissolved in 500 µL of double-distilled water to create the stock, used as 1:1000 dilution; catalog no. 11814460001, Roche) or anti-myc antibody (1:5000 dilution; catalog no. M4439, Millipore); anti-tubulin antibody (1:5000 dilution; catalog no. T5168, Sigma-Aldrich) was used for equal loading control.
ChIP-qPCR
Seedlings expressing pHSFA1d:HSFA1d-GFP (hsfa1d background) and pRVE8:RVE8-YFP (Col-0 background) were grown in soil at 22°C under LD conditions for 3 weeks. At ZT 3 to 6, the seedlings were exposed to 37°C (HS) or kept at 22°C (control) for 10 min. Freshly harvested plant tissues were immediately cross-linked by vacuum infiltration with 1% (v/v) formaldehyde for 5 min and held for 5 min after releasing the vacuum. Cross-linking was quenched by submerging the tissues in 0.25 M Gly for 10 min. The tissues were ground into a fine powder in liquid nitrogen. ChIP was performed as described by Zhu et al. (2012). In brief, the isolated chromatin was fragmented (average length, 0.5 to 1 kb) by sonication at 4°C and immunoprecipitated using 10 μL of GFP-Trap agarose (ChromoTek) or anti-myc agarose beads for the control (catalog no. A7470, Millipore). The primers used for ChIP-qPCR are listed in Supplemental Table 1. Relative enrichment was calculated based on the fold change value compared with the UBC30 control using the comparative threshold cycle method (Schmittgen and Livak, 2008).
Transient Transcriptional Activation Assay
The A. tumefaciens strain GV3101 cells harboring the p35S:CFP, p104:RVE8, pHGWL-ERF53, and pHGWL-ERF54 expression vectors were mixed in pairs and co-infiltrated into leaf epidermal cells of 4-week-old Nicotiana benthamiana grown in the soil at 22°C under LD white light (at ∼100 μmol m−2 s−1, cool-white fluorescent bulb) in a growth room. Thirty-six hours after infiltration, the leaves were sprayed with firefly luciferase substrate and incubated in complete darkness for 5 min. Images were then captured using FUSION FX5 (Vilber Lourmat). After recording the bioluminescence signal from a leaf, the leaf was surface dried with Kimwipes (Sigma-Aldrich) and incubated in a growth chamber with the internal temperature preset to 37°C for 10 min. The leaves were sprayed with new luciferase substrate and incubated in the dark for 5 min. The images were recorded using the same settings as before HS treatment. ImageJ software was used to quantify the bioluminescence signal. Relative luciferase activity was determined based on the ratio between YFP-RVE8- and CFP-upregulated luciferase activity.
Experimental Replicates and Statistical Analysis
All RT-qPCR and thermoresponsive assay results in this article represent the average value from at least three biological replicates, and error bars represent ±se. The term “different biological replicates” refers to plants grown and HS treated on different days and harvested for analysis. Unless otherwise indicated, two-way ANOVA with Tukey’s honestly significant differences test was used to evaluate the differences across multiple data sets, and statistically significant differences are indicated by different lowercase letters, P < 0.05. ANOVA results are listed in Supplemental Data Set 6.
Data Availability
The RNA-seq data of this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) and were assigned with the following identification accession numbers: SRP187923 and SRP187905.
Accession Numbers
Sequence data from this article can be found in TAIR under the following accession numbers: UBC30 (AT5G56150), ACTIN2 (AT3G18780), TUBULIN4 (AT1G04820), UBQ5 (AT3G62250), PP2A A3 (AT1G13320), HSFA1a (AT4G17750), HSFA1b (AT5G16820), HSFA1d (AT1G32330), HSFA1e (AT3G02990), ERF53 (AT2G20880), ABCG36 (AT1G59870), BBX17 (AT1G49130), COR47 (AT1G20440), MAGL4 (AT1G73480), ERF54 (AT4G28140), SCL13 (AT4G17230), MYB44 (AT5G67300), AT1G22190, DREB2A (AT5G05410), NF-YC2 (AT1G56170), WIND1 (AT1G78080), SAP3 (AT2G27580), ZAT6 (AT5G04340), HSFA2 (AT2G26150), HSFA7a (AT3G51910), HSFA7b (AT3G63350), HSFB1 (AT4G36990), HSFB2a (AT5G62020), HSFB2b (AT4G11660), MBF1c (AT3G24500), RVE4 (AT5G02840), RVE8 (AT3G09600), LHY (AT1G01060), PRR5 (AT5G24470), HSP70 (AT3G12580), AGL95 (AT2G15660), HSFA4a (AT4G18880), bHLH157 (AT1G64625), RVE7 (AT1G18330), GBF6 (AT4G34590), bZIP68 (AT1G32150), ATAF1 (AT1G01720), GolS1 (AT2G47180), GolS2 (AT1G56600), FKBP62 (AT3G25230), DREB2B (AT3G11020), bZIP28 (AT3G10800), ZAT7 (AT3G46090).
Supplemental Data
Supplemental Figure 1. Time-dependent increases of the temperature inside or outside the plates during HS treatment at 37°C.
Supplemental Figure 2. A co-expression network of eHSRs.
Supplemental Figure 3. The expression of various reference genes in response to HS.
Supplemental Figure 4. Enrichment of cis-elements recognized by various transcription factors in HS down-regulated eHSRs.
Supplemental Figure 5. Acquired thermotolerance in hsfa1-qk.
Supplemental Figure 6. The vegetative growth of hsfa1-qk is slow.
Supplemental Figure 7. The circadian-dependent expression of RVE4 and RVE8 under long-day conditions.
Supplemental Figure 8. The expression of RVE4 and RVE8 is not regulated rapidly by HS.
Supplemental Figure 9. RVE4 and RVE8 redundantly regulate plant basal thermotolerance.
Supplemental Figure 10. Generation of the hsfa1-cq quadruple mutant.
Supplemental Figure 11. RT-qPCR validation of RVE4/8 and HSFA1s dependent and independent eHSRs using rve4/8 and hsfa1-cq mutants.
Supplemental Figure 12. HSFA1s are more important than RVE4/8 in regulating plant thermotolerance.
Supplemental Figure 13. RT-qPCR analysis of tissue-specific expression patterns of RVE4, RVE8, ERF53 and ERF54.
Supplemental Figure 14. Recovery of the HS-induced expression of ERF53 and ERF54 in rve4/8 mutants by the pRVE8:RVE8-YFP transgene.
Supplemental Figure 15. Complementation of the growth phenotypes of hsfa1-qk by the pHSFA1d:HSFA1d-GFP transgene.
Supplemental Figure 16. The level of ERF53 protein in ERF53 overexpression transgenic plants.
Supplemental Figure 17. Circadian-dependent accumulation of RVE8 proteins under long-day growth conditions.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Rapid HS regulated genes identified by RNaseq.
Supplemental Data Set 2. Classification of eHSRs in different DREM paths.
Supplemental Data Set 3. Co-expression analysis of eHSRs.
Supplemental Data Set 4. eHSRs regulated by 10 min of HS treatment in three-week-old soil grown Col, Ws and hsfa1-qk.
Supplemental Data Set 5. List of HSF-dependent and independent eHSRs identified from three-week-old soil grown plants.
Supplemental Data Set 6. ANOVA tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yee-Yung Charng (National Taiwan University) and Kazuko Yamaguchi-Shinozaki (University of Tokyo) for providing the hsfa1-qk mutant and Xiaodong Xu (Hebei Normal University) for providing the rev4, rve8, lhy, and rve4/8 mutants. This study was supported by grants from the National Natural Science Foundation of China (31670265), the “one hundred talents project” of Hebei Province (E2011100004), the Science and Technology Department of Hebei Province (15966306D), the Education Department of Hebei Province (GCC2014002), and the graduate innovation program from Hebei Province (CXZZBS2017094).
AUTHOR CONTRIBUTIONS
W.T. and D.S. designed the research. B.L. performed research under the supervision of W.T. and D.S. Z.G. analyzed the RNA-seq data. B.L. and X.L. did the bioinformatics analysis. B.L. and W.T. wrote the article together.
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2015). HTSeq--A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Machanick P. (2012). Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. (2009). MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37: W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Johnson J., Grant C.E., Noble W.S. (2015). The MEME suite. Nucleic Acids Res. 43 (W1): W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Leaver C.J., McCabe P.F. (1999). Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463: 151–154. [DOI] [PubMed] [Google Scholar]
- Baron K.N., Schroeder D.F., Stasolla C. (2012). Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana. Plant Sci. 188-189: 48–59. [DOI] [PubMed] [Google Scholar]
- Belyaeva E.S., Zhimulev I.F. (1976). RNA synthesis in the Drosophila melanogaster puffs. Cell Differ. 4: 415–427. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- Bita C.E., Gerats T. (2013). Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cortijo S., Charoensawan V., Brestovitsky A., Buning R., Ravarani C., Rhodes D., van Noort J., Jaeger K.E., Wigge P.A. (2017). Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in Arabidopsis. Mol. Plant 10: 1258–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H., Xue Y. (2014). HemI: A toolkit for illustrating heatmaps. PLoS One 9: e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P.J., Kumar M., Martinho C., Yoo S.J., Lan H., Artavanis G., Charoensawan V., Schöttler M.A., Bock R., Jaeger K.E., Wigge P.A. (2018). Chloroplast signaling gates thermotolerance in Arabidopsis. Cell Reports 22: 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte F.M., Fuda N.J., Mahat D.B., Core L.J., Guertin M.J., Lis J.T. (2016). Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes Dev. 30: 1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Vainas O., Harbison C.T., Simon I., Bar-Joseph Z. (2007). Reconstructing dynamic regulatory maps. Mol. Syst. Biol. 3: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finka A., Cuendet A.F., Maathuis F.J., Saidi Y., Goloubinoff P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24: 3333–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Whitelam G.C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39: 1410–1413. [DOI] [PubMed] [Google Scholar]
- Gao F., Han X., Wu J., Zheng S., Shang Z., Sun D., Zhou R., Li B. (2012). A heat-activated calcium-permeable channel--Arabidopsis cyclic nucleotide-gated ion channel 6--is involved in heat shock responses. Plant J. 70: 1056–1069. [DOI] [PubMed] [Google Scholar]
- Gao H., Brandizzi F., Benning C., Larkin R.M. (2008). A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 16398–16403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M., Nahar K., Alam M.M., Roychowdhury R., Fujita M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14: 9643–9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieda M., Winstanley H., Maini P., Iborra F.J., Cook P.R. (2005). Different populations of RNA polymerase II in living mammalian cells. Chromosome Res. 13: 135–144. [DOI] [PubMed] [Google Scholar]
- Hsieh E.J., Cheng M.C., Lin T.P. (2013). Functional characterization of an abiotic stress-inducible transcription factor AtERF53 in Arabidopsis thaliana. Plant Mol. Biol. 82: 223–237. [DOI] [PubMed] [Google Scholar]
- Hsu P.Y., Devisetty U.K., Harmer S.L. (2013). Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Im J.H., Cho Y.H., Kim G.D., Kang G.H., Hong J.W., Yoo S.D. (2014). Inverse modulation of the energy sensor Snf1-related protein kinase 1 on hypoxia adaptation and salt stress tolerance in Arabidopsis thaliana. Plant Cell Environ. 37: 2303–2312. [DOI] [PubMed] [Google Scholar]
- Jung J.H., et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889. [DOI] [PubMed] [Google Scholar]
- Jung H.S., Crisp P.A., Estavillo G.M., Cole B., Hong F., Mockler T.C., Pogson B.J., Chory J. (2013). Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc. Natl. Acad. Sci. USA 110: 14474–14479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19: 408–413. [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Legris M., Klose C., Burgie E.S., Rojas C.C., Neme M., Hiltbrunner A., Wigge P.A., Schäfer E., Vierstra R.D., Casal J.J. (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900. [DOI] [PubMed] [Google Scholar]
- Li B., Gao K., Ren H., Tang W. (2018). Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 60: 757–779. [DOI] [PubMed] [Google Scholar]
- Liu H.T., Li B., Shang Z.L., Li X.Z., Mu R.L., Sun D.Y., Zhou R.G. (2003). Calmodulin is involved in heat shock signal transduction in wheat. Plant Physiol. 132: 1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.T., Gao F., Li G.L., Han J.L., Liu D.L., Sun D.Y., Zhou R.G. (2008). The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 55: 760–773. [DOI] [PubMed] [Google Scholar]
- Liu H.C., Liao H.T., Charng Y.Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34: 738–751. [DOI] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Feng L., Li J., He Z. (2015). Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 6: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell D.B., Schlenker W., Costa-Roberts J. (2011). Climate trends and global crop production since 1980. Science 333: 616–620. [DOI] [PubMed] [Google Scholar]
- Mahat D.B., Salamanca H.H., Duarte F.M., Danko C.G., Lis J.T. (2016). Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell 62: 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V.B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011). ROS signaling: The new wave? Trends Plant Sci. 16: 300–309. [DOI] [PubMed] [Google Scholar]
- Mittler R., Finka A., Goloubinoff P. (2012). How do plants feel the heat? Trends Biochem. Sci. 37: 118–125. [DOI] [PubMed] [Google Scholar]
- Mu J., Tan H., Hong S., Liang Y., Zuo J. (2013). Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol. Plant 6: 188–201. [DOI] [PubMed] [Google Scholar]
- O’Malley R.C., Huang S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. [DOI] [PubMed] [Google Scholar]
- Ohama N., Kusakabe K., Mizoi J., Zhao H., Kidokoro S., Koizumi S., Takahashi F., Ishida T., Yanagisawa S., Shinozaki K., Yamaguchi-Shinozaki K. (2016). The transcriptional cascade in the heat stress response of Arabidopsis is strictly regulated at the level of transcription factor expression. Plant Cell 28: 181–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M., Delker C., Franklin K.A., Wigge P.A., Halliday K.J., van Zanten M. (2016). Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Rawat R., Takahashi N., Hsu P.Y., Jones M.A., Schwartz J., Salemi M.R., Phinney B.S., Harmer S.L. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindl A., Schöffl F., Schell J., Koncz C., Bakó L. (1997). Phosphorylation by a cyclin-dependent kinase modulates DNA binding of the Arabidopsis heat-shock transcription factor HSF1 in vitro. Plant Physiol. 115: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rienth M., Torregrosa L., Luchaire N., Chatbanyong R., Lecourieux D., Kelly M.T., Romieu C. (2014). Day and night heat stress trigger different transcriptomic responses in green and ripening grapevine (Vitis vinifera) fruit. BMC Plant Biol. 14: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi Y., Finka A., Muriset M., Bromberg Z., Weiss Y.G., Maathuis F.J., Goloubinoff P. (2009). The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 21: 2829–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K.D., Berberich T., Ebersberger I., Nover L. (2012). The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 1819: 104–119. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schulz M.H., Devanny W.E., Gitter A., Zhong S., Ernst J., Bar-Joseph Z. (2012). DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data. BMC Syst. Biol. 6: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell W.R., Huebner M., Weber A.P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhini B.V., Anuradha S., Sujatha E., Rao S.S.R. (2010). Role of brassinosteroids in alleviating various abiotic and biotic stresses-a review. Plant Stress 4: 55–61. [Google Scholar]
- Wahid A., Gelani S., Ashraf M., Foolad M.R. (2007). Heat tolerance in plants: An overview. Environ. Exp. Bot. 61: 199–223. [Google Scholar]
- Wang R., Liu M., Yuan M., Oses-Prieto J.A., Cai X., Sun Y., Burlingame A.L., Wang Z.Y., Tang W. (2016). The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B’ subunits. Mol. Plant 9: 148–157. [DOI] [PubMed] [Google Scholar]
- Wang X., Ma X., Wang H., Li B., Clark G., Guo Y., Roux S., Sun D., Tang W. (2015). Proteomic study of microsomal proteins reveals a key role for Arabidopsis annexin 1 in mediating heat stress-induced increase in intracellular calcium levels. Mol. Cell. Proteomics 14: 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H.L., Dong L., Wang Z.P., Zhang H.Y., Han C.Y., Liu B., Wang X.C., Chen Q.J. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Qin K., Song X., Zhang Q., Zhou Y., Xia X., Yu J. (2018). BZR1 transcription factor regulates heat stress tolerance through FERONIA receptor-like kinase mediated reactive oxygen species signaling in tomato. Plant Cell Physiol. 59: 2239–2254. [DOI] [PubMed] [Google Scholar]
- Yoshida T., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286: 321–332. [DOI] [PubMed] [Google Scholar]
- You Q., Zhang L., Yi X., Zhang K., Yao D., Zhang X., Wang Q., Zhao X., Ling Y., Xu W., Li F., Su Z. (2016). Co-expression network analyses identify functional modules associated with development and stress response in Gossypium arboreum. Sci. Rep. 6: 38436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., et al. (2016). OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 170: 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Sun Y., Wang Z.Y. (2012). Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). Methods Mol. Biol. 876: 173–188. [DOI] [PubMed] [Google Scholar]
- Zhu J.Y., Oh E., Wang T., Wang Z.Y. (2016). TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7: 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn K.E., Tunc-Ozdemir M., Harper J.F. (2010). Temperature stress and plant sexual reproduction: Uncovering the weakest links. J. Exp. Bot. 61: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The RNA-seq data of this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) and were assigned with the following identification accession numbers: SRP187923 and SRP187905.