Abstract
The world’s aging population is growing rapidly. Incidences of multiple pathologies, such as abdominal obesity, cardiovascular and cerebrovascular diseases, type 2 diabetes, and malignant neoplasms, increase sharply with age. Aged individuals possess a significantly shifted composition of gut microbiota, which is suggested to play important roles in aging associated pathologies. Whether the existing shifted structural composition of microbiota in aged populations can be reverted non-pharmacologically has not been studied so far. Here, we show an intestinal flora imbalance in old C57BL/6J mice with a remarkable dominant proportion of microbes promoting lipid metabolism and inflammation. Intriguingly, short-term (2 months) dietary restriction was enough to significantly revert the imbalance of intestinal flora in aged mice toward a more balanced structural composition as shown in young mice. Our study provides the first evidence that short-term dietary restriction in old mice can restore the already dysfunctional aged gut microbiota. Our study provides the first evidence that short-term dietary restriction in old mice can restore the already dysfunctional aged gut microbiota, which may help ameliorate aging-related disorders plaguing the vast elderly population.
Keywords: Aging, Dietary restriction, Gut microbiota, Rejuvenate, Obesity, Inflammation
Introduction
The trend towards an aging population is increasing, with the 2017 United Nations World Population Prospects report revealing that people over 60 years old will make up more than 25% of a nation’s population in the near future in many countries. Biological aging involves molecular and physical changes that increase the probability of developing certain diseases such as osteoporosis, arthritis, type 2 diabetes, hypertension, heart disease, and cancer. While the underlying mechanisms remain incompletely understood, it is generally believed to involve a progressive decline in organ functionality and tissue homeostasis, which is suggested to be closely related to microbiota shifts (Claesson et al. 2012; Liu and Rando 2011; Zapata and Quagliarello 2015). The number of bacteria in the body is estimated to be as much as 10 trillion, with the vast majority of bacteria residing in the colon. Imbalance of the intestinal microbiota can lead to increased permeability of the intestinal wall and dysfunction of the intestinal mucosal barrier (Bischoff et al. 2014; Clark et al. 2015; Thevaranjan et al. 2017). Furthermore, the intestinal microbiota can regulate lipid uptake and storage by invading intestinal mucosal cells and altering their circadian rhythm, contributing to the development of metabolic diseases, such as obesity, diabetes, and nonalcoholic fatty liver (Amar et al. 2011; Boursier et al. 2016; Wang et al. 2017).
It has been shown that the composition of the gut microbiota shifts towards increasing proinflammatory commensals and decreasing beneficial commensals microbes with age in both humans and rodents (Biagi et al. 2016; Biragyn and Ferrucci 2018; Hopkins et al. 2001; Jeffery et al. 2016), which is accompanied by an impairment of the intestinal barrier (Clark et al. 2015). Furthermore, gut dysbiosis contributes to a chronic proinflammatory state and could serve as a potential link between cancer risk in aging (Biragyn and Ferrucci 2018). Interventions that shift the gut microbiota towards a younger state might therefore help ameliorate aging associated pathologies. Recent studies have found that high sugar and high fat diet-induced obesity leads to changes in the composition of the intestinal microflora (Carmody et al. 2015; He et al. 2018). Obesity in humans and mice has also been found to be positively correlated with the ratio of Firmicutes/Bacteroidetes (F/B) (He et al. 2018; Ley et al. 2005, 2006; Turnbaugh et al. 2006, 2009). Studies have shown that changes in the composition of the gut microbiota in the elderly are not only related to chronic diseases such as obesity and inflammation, but also have a significant relationship with diet (Claesson et al. 2012). Dietary intervention has a great impact on the intestinal microbiota (Cotillard et al. 2013), a wealth of evidence suggest that dietary restriction (DR) has wide-ranging benefits in increasing the body’s general health status and in providing a nonspecific resistance to chronic diseases and metabolic derangements (Colman et al. 2009; Fontana et al. 2004; Ribaric 2012; Roth et al. 2001; Walford et al. 2002). Previous studies have explored the role of life-long DR on gut microbiota. Life-long DR, in combination with low-fat diet, maintained a structurally balanced architecture of the gut microbiota and improved colonic health (Kok et al. 2018; Zhang et al. 2013). However, DR was already initiated from young age when the gut microbiota was still healthy in all the above-mentioned studies. Whether, the existing imbalanced structural composition of microbiota in aged populations can be reverted non-pharmacologically has yet to be studied. Exploring the possibilities of how the established gut microbiota in aged organisms might be reformed could benefit the health of the growing elderly population.
In this study, we performed 16S rRNA gene sequencing of bacterial DNA extracted from freshly collected faecal samples to examine the effects of DR on the microbiota of aging mice. In line with previous studies, we found that the composition of the intestinal flora in old mice (20–22 months old) mainly shifted towards a proinflammatory state and promotion of lipid metabolism. Intriguingly, short-term (2 months) DR (dietary restriction) performed in aged mice significantly reverted the intestinal flora imbalance towards a more balanced structural composition as shown in young mice. This structural imbalance shown in old mice was rejuvenated by reducing the dominance of Clostridia, Clostridiales, and Firmicutes which have all been found to contribute to obesity and inflammation. Our study provides the first evidence that short-term DR in old organisms can rejuvenate the imbalanced composition of gut microbiota. Therefore, we identified a non-pharmacological and efficient way to rejuvenate gut microbiota in aged mice, which may benefit health in the elderly population.
Results
Bacterial community shifts towards dominant lipid-promoting and pro-inflammatory bacteria in aging intestine
To study whether microbiota composition changes upon aging, we collected fecal samples from young (2 months old) and old (20–22 months old) female mice. Hypervariable regions were sequenced by 16S rRNA high-throughput sequencing platform Illumina Hiseq 2500. 1,127,979 sequence reads were generated from the 16S rRNA gene V4 amplicon, with an average reading of 32,228 (± 2024 SEM) per subject.
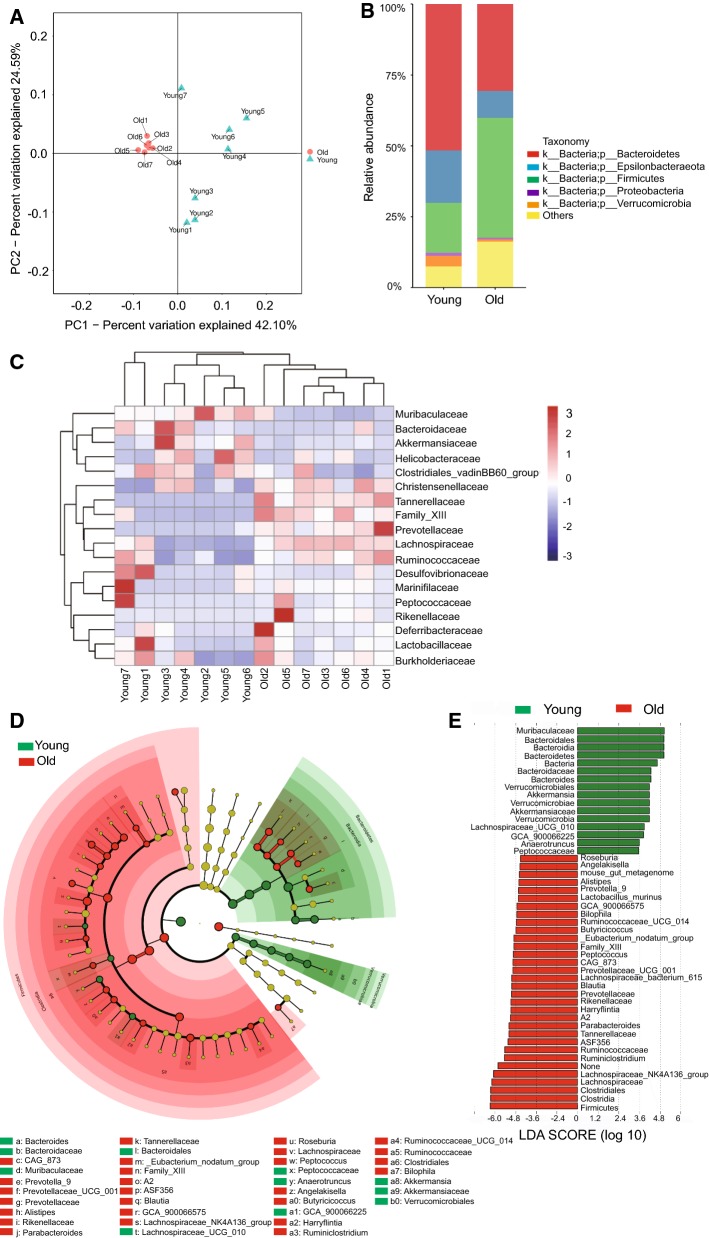
β-diversity analysis via the principal coordinate analysis (PCoA) analysis based on the Unweighted Unifrac distance indicated a clear separation between young intestinal flora and the old intestinal flora (Fig. 1a), indicating that the bacterial community was significantly changed upon aging. Analysis on the phyla level showed that Bacteroidetes was the most dominant phyla in young mice while it was significantly reduced in old mice (Fig. 1b). Epsilonbacteraeota and Firmicutes were the second most dominant phyla with an almost equal ratio in young mice, while Firmicutes increased to be the most abundant phyla in old mice (Fig. 1b). Bacterial taxa ranking at the top of the list in young mice all fell from the top positions in old mice; further analysis showed that families of Christensenellaceae, Family_XIII, Prevotellaceae, Lachnospiraceae, and Ruminococcaceae were all enriched in the gut microbiota of aged mice, while Bacteroidaceae and Muribaculaceae were enriched in the microbiota of young mice (Fig. 1c). To identify the most differently abundant taxa in aged mice, we used the linear discriminant analysis (LDA) effect size (LEfSe) method to assess the effect size of each taxon (Fig. 1e, f). In young mice, the intestinal flora was mainly enriched with Muribaculaceae, Bacteroidales, Bacteroidetes, Bacteroidaceae and Bacteroides, with all of these falling from the top-ranking positions in old mice. Old mice were comparatively enriched with Clostridia, Clostridiales and Firmicutes. The analysis further proved a significantly altered composition of gut microbiota in old mice compared to the young ones.
Fig. 1.
Alterations in the fecal microbial community structure of aging mice. Fecal samples of 2 months old (young) and 20–22 months old (old) mice were collected for analysis (n = 7 samples per group). a β-diversity analysis. The results of unweighted UniFrac PCoA were shown. b Relative abundance of bacteria at phylum level. The ratio of the average OTU for each group was shown. c Heatmap based on the relative abundance at family level. d Taxonomic cladogram from LEfSe showing differences in fecal taxa. Dot size is proportional to the abundance of the taxon. Letters correspond to the following taxa: a: Bacteroides, b: Bacteroidaceae, c: CAG_873, d: Muribaculaceae, e: Prevotella_9, f: Prevotellaceae_UCG_001, g: Prevotellaceae, h: Alistipes, i: Rikenellaceae, j: Parabacteroides, k: Tannerellaceae, l: Bacteroidales, m: _Eubacterium_nodatum_group, n: Family_XIII, o: A2, p: ASF356, q:Blautia, r: GCA_900066575, s: Lachnospiraceae_NK4A136_group, t: Lachnospiraceae_UCG_010, u: Roseburia, v: Lachnospiraceae, w: Peptococcus, x: Peptococcaceae, y: Anaerotruncus, z: Angelakisella, a0: Butyricicoccus, a1: GCA_900066225, a2: Harryflintia, a3: Ruminiclostridium, a4: Ruminococcaceae_UCG_014, a5: Ruminococcaceae, a6: Clostridiales, a7: Bilophila, a8: Akkermansia, a9: Akkermansiaceae, b0: Verrucomicrobiales. e LDA scores computed for differentially-abundant taxa in the fecal microbiomes of young and old mice. Length indicates effect size associated with a taxon. p = 0.05 for the Kruskal–Wallis test; LDA score > 2
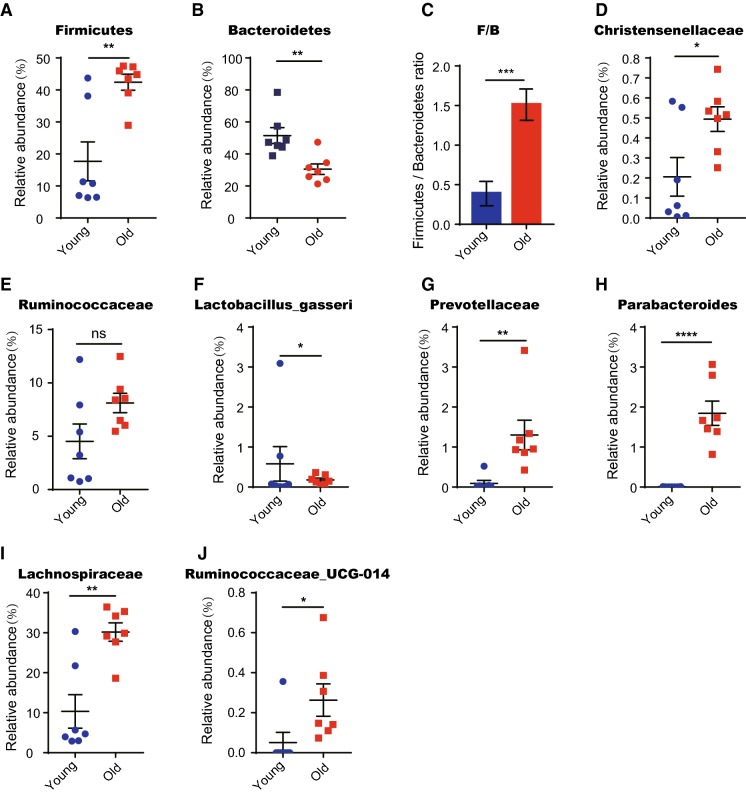
The Firmicutes/Bacteroidetes (F/B) ratio has been associated with obesity in humans and mice (He et al. 2018; Ley et al. 2005, 2006; Turnbaugh et al. 2006, 2009). Here we show that the proportion of Firmicutes was 18 ± 6.0% in young mice versus 42 ± 2.5% in old mice (Fig. 2a). Conversely, the proportion of Bacteroidetes decreased significantly from 52 ± 4.9% in young mice to 31 ± 3.3% in old mice (Fig. 2b). Therefore, the Firmicutes/Bacteroidetes ratio (F/B) in old mice was significantly higher than young mice (1.5 ± 0.2 in old mice vs. 0.4 ± 0.1 in young mice) (Fig. 2c). Interestingly, our study determined a significant increase of Ruminococcaceae and Christensenellaceae, and a decrease of Lactobacillus gasseri in old mice (Fig. 2d–f). These alterations in the structural microbiota composition in aged mice were reported to be positively related to fat accumulation (Bauer et al. 2018; Qin et al. 2018). Prevotellaceae, Parabacteroides, Oscillibacter, Lachnospiraceae, Ruminococcaceae-UCG-014 and Erysipelatoclostridium have all been reported to be increased in multiple inflammatory models, and whose related bacterial taxa were also enriched in aged mice (Fig. 2g–j) (Elinav et al. 2011; Peng et al. 2019; Qi et al. 2019). Taken together, the results indicate that aging leads to alterations in intestinal flora composition that have previously been positively correlated to lipid accumulation and inflammation.
Fig. 2.
Lipid-promoting and pro-inflammatory bacteria are enriched in aging mice. Fecal samples of 2 months old (young) and 20–22 months old (old) mice were collected for analysis (n = 7 samples per group). a, b, d–j Relative abundance based on OTUs of intestinal bacteria taxa which are significantly changed in old mice. Note that these taxa were all lipid-promoting or pro-inflammatory bacteria. c Ratio of Firmicutes/Bacteroidetes based on relative abundance of OTUs. Note a significant increase in the old mice compared to the young ones. Results were displayed as mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 by unpaired two-tailed Student’s t test
Short-term DR in old mice rejuvenates aging induced structural imbalance of gut microbiota
In line with previous studies, the above data indicate a clear aging phenotype involving compositional change of the gut microbiota. To investigate whether DR can rejuvenate the aged gut microbiota, we treated 20–22 months old female mice with 30% DR for 2 months and examined the composition of intestinal flora compared to identically aged female mice fed ad libitum (AL) as well as AL fed 2 months old mice.
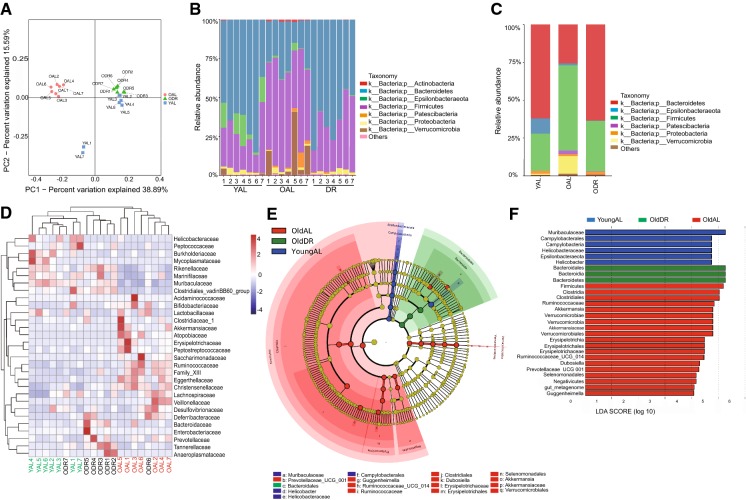
We first performed a diversity analysis among the three groups: young AL (2 months old mice fed with ad libitum), old AL (22–24 months old mice fed with ad libitum), and old DR (22–24 months old mice fed with DR for 2 months before analysis). The β-diversity analysis, PCoA analysis based on Unweighted Unifrac distance showed that old AL samples were clearly separated from the young AL samples, while the old DR samples were not. This finding demonstrates that the significantly altered composition and structure of an aged biological community can be reverted via short-term DR (Fig. 3a).
Fig. 3.
Short-term DR in old mice rejuvenates aging induced structural rearrangement of gut microbiota. 20–22 months old mice were treated with DR or AL diet for 2 months. Fecal samples of the following groups were collected for analysis: YAL (2 months old mice fed with ad libitum), OAL (22–24 months old mice fed with ad libitum), and ODR (22–24 months old mice pre-treated with DR for 2 months before sample collection) (n = 7 samples per group). a β-diversity analysis. The results of unweighted UniFrac PCoA of indicated groups were shown. b Relative abundance of bacteria at phylum level of individual sample based on OTUs. c The ratio of relative abundance at phylum level based on the average OTUs in each group. d Heatmap showing clustering of each sample at family level based on the relative abundance of OTUs. Note that hierarchical clustering shown that samples of ODR and YAL tend to cluster together. e Taxonomic cladogram from LEfSe showing differences in fecal taxa. Dot size is proportional to the abundance of the taxon. Letters correspond to the following taxa: a: Muribaculaceae, b: Prevotellaceae_UCG_001, c: Bacteroidales, d: Helicobacter, e: Helicobacteraceae, f: Campylobacterales, g: Guggenheimella, h: Ruminococcaceae_UCG_014, i: Ruminococcaceae, j: Clostridiales, k: Dubosiella, l: Erysipelotrichaceae, m: Erysipelotrichales, n: Selenomonadales, o: Akkermansia, p: Akkermansiaceae, q: Verrucomicrobiales. f LDA scores computed for differentially-abundant taxa in the fecal microbiomes of young (blue) old DR (green) and old AL (red). Length indicates effect size associated with a taxon. p = 0.05 for the Kruskal–Wallis test; LDA score > 2
Further analysis on the phyla level showed that the structure of the biological community of short-term DR mice converged with that of young AL mice, which was clearly different compared to old AL mice. In detail, Firmicutes was the most abundant phyla in the old AL mice, which was significantly decreased in young AL and old DR mice. The most abundant phyla in young AL and old DR mice was Bacteroidetes, which was clearly reduced in the old AL mice (Fig. 3b, c). At the family level, the composition and structure of the intestinal flora was similar in young AL and old DR mice, while it was distinct from the old AL mice (Fig. 3d). The LEfSe method was used to further assess the effect size of each taxon (Fig. 3e, f). The analysis showed that old DR mice had more Bacteroidales, Bacteroidia and Bacteroidia, with less Firmicutes, Clostridia and Ruminococcaceae, which is further evidence of microbiota rescue in aged individuals achieved by short-term DR (Fig. 3f).
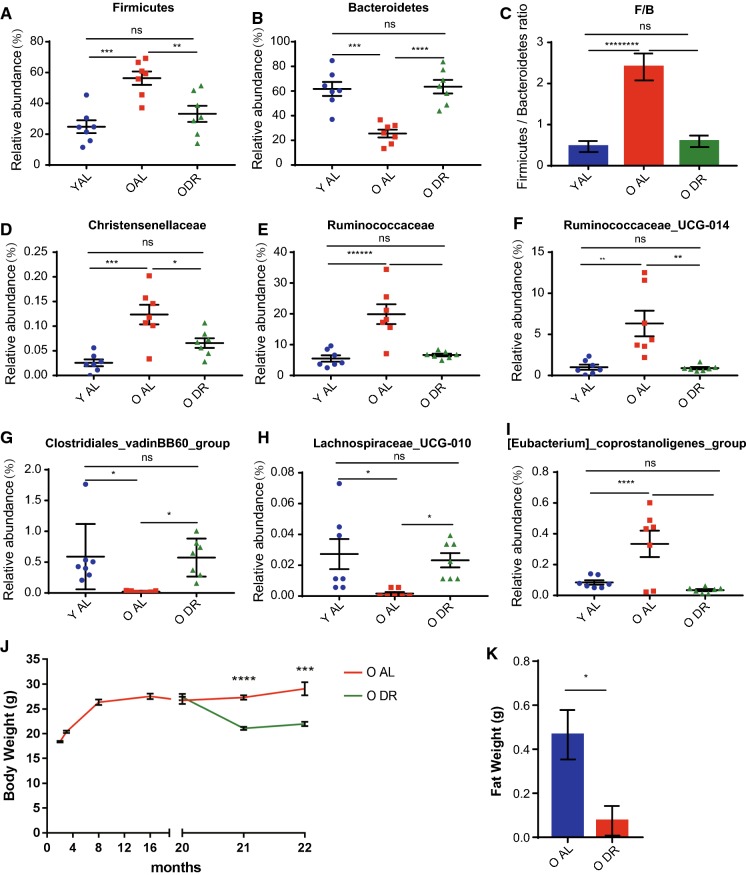
We further determined the effect of DR on bacterial taxa that were functionally relevant to obesity and inflammation. Firmicutes was the most abundant phyla in the old AL mice (56 ± 4.4%), while it was significantly less in young AL (25 ± 4.2%) and old DR mice (33 ± 5.3%) (Fig. 4a). The most abundant phyla in young AL and old DR mice was Bacteroidetes (62 ± 5.8% in young AL and 63 ± 5.5% in old DR), while it was clearly reduced in the old AL mice (26 ± 3.1%) (Fig. 4b). The F/B ratio in old AL mice was 2.4 ± 0.3, which was significantly reduced in the old DR mice to 0.6 ± 0.1, close to the level in the young mice at 0.5 ± 0.1 (Fig. 4c). Of note, short-term DR reverted almost all compositional changes of bacterial taxa associated with obesity and inflammation and brought their level close to that of young mice, including Bacteroidetes, Firmicutes, Christensenellaceae, Ruminococcaceae, and Ruminococcaceae_UCG_014 (Fig. 4a–f). Intriguingly, short-term DR additionally resulted in changes in more bacterial taxa that were reported to contribute to fat accumulation and inflammation, such as Clostridiales_vadinBB60_group, Lachnospiraceae_UCG-010, [Eubacterium]coprostanoligenes_group (Fig. 4g–i). These findings indicate that short-term DR was enough to induce changes in the composition of the commensal community towards a younger, healthier state which in turn may associate with less fat accumulation and decreased inflammation. We also note that short-term DR expectedly reduces the body weight and abdominal fat in old mice compared to old ad libitum fed mice (Fig. 4j, k).
Fig. 4.
Short-term DR reverted compositional alterations of bacterial taxa associated with obesity and inflammation in aging mice. 20–22 months old mice were treated with DR or AL diet for 2 months. Fecal samples of the following groups were collected for analysis: YAL (2 months old mice fed with ad libitum), OAL (22–24 months old mice fed with ad libitum), and ODR (22–24 months old mice pre-treated with DR for 2 months before sample collection) (n = 7 samples per group). a, b, d–i Relative abundance based on OTUs of intestinal bacteria taxa. c Ratio of Firmicutes/Bacteroidetes based on relative abundance of OTUs. Note that DR significantly rejuvenated all alterations of indicated taxa in aging mice. j, k Body and belly fat weight of indicated groups. Note a significant reduction upon DR. Results were displayed as mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 by unpaired two-tailed Student’s t test
Discussion
While lifelong DR has been proven to be a robust regimen to retard aging in many animal models, it is unpractical in humans (Colman et al. 2009; Goto 2006; Goto et al. 2007; Mattison et al. 2012; Weindruch 1996). Therefore, a number of studies considered the effects of DR initiated in later life and showed that late-onset DR could rejuvenate biological parameters that decline with age in rodents, such as improved protein and lipid metabolism and chromatin functions. These improvements in turn protect the functionalities of different organs, including the brain, skeletal muscle, and immune system (Goto 2006; Goto et al. 2007; Radak et al. 2002; Singh et al. 2012, 2015; Weindruch et al. 1982). However, the underlying mechanisms remains to be further elucidated.
The gut microbiota has been shown to play essential roles in multiple biological processes as well as pathologies. The current study described a significant structural rearrangement of gut microbiota in old mice (20–22 months old). The skewed intestinal flora has potential in promoting lipid accumulation and inflammation; therefore, it could be highly relevant to associated pathologies in aging. Whether and how the already aged microbiota could be rejuvenated has never been studied so far. Here, we provide the first experimental evidence that short-term (2 months) DR in old mice was enough to revert the already skewed gut microbiota. The equivalent change in humans would roughly be a 66–72 year old’s intestinal flora reverting back to a similar state to when they were around 23 years’ old (Dutta and Sengupta 2016). Our study showed that old mice receiving short-term DR exhibit a younger-shaped intestinal flora along with reduced body weight and abdominal fat. Assessing any potential contribution of a more balanced microbiome in old mice to weight loss will be an important topic of further research, especially given recent evidence demonstrating the rescue of DR-induced weight loss by fecal transplantation from AL mice (Wang et al. 2018).
Recent studies have found that obesity in humans and mice is primarily associated with changes in the relative abundance of Bacteroidetes and Firmicutes, and is positively correlated with the ratio of Firmicutes/Bacteroidetes (F/B) (He et al. 2018; Ley et al. 2005, 2006; Turnbaugh et al. 2006, 2009).Therefore, the elevated F/B ratio we observed in aged mice might serve as an underlying mechanism why fat accumulates with age in general. In line with previous studies, our study uncovered aging-associated changes of the gut microbiota, including increases of Firmicutes, Ruminococcaceae and Christensenellaceae, and decreases of Bacteroidetes and Lactobacillus gasseri. Interestingly, it was previously shown that Ruminococcaceae and Christensenellaceae are enriched in obese mice (Qin et al. 2018), and that Lactobacillus gasseri levels were reduced in high-fat diet mice (Bauer et al. 2018). Our study shows that short-term DR can reverse the imbalanced microbial community structure of aged mice and restore the proinflammatory and lipid metabolism promoting intestinal flora to a young level, which may reduce the incidence of age-related diseases.
Previous studies have shown that androgens influence gut microbiota which differs between males and females (Haro et al. 2016; Markle et al. 2013; Yurkovetskiy et al. 2013). While this study was performed on female mice for the benefit of co-housing before the dietary intervention, we would expect to observe the same rejuvenation effect of DR in old male mice. A recent assessment of calorie restriction (CR) noted that while its effect was invariably positive, the effect size could vary between mouse substrain, sex, and the extent of CR (Mitchell et al. 2016).
In this study we conclude that the existing proinflammatory and lipid-promoting microbiota in aged mice can be reverted non-pharmacologically to a young state using DR. This microbiota in old DR mice possesses a more balanced structural composition similar to that of young mice; the permanence and generalizability of this finding remains to be studied further.
Materials and methods
Animals and dietary intervention
C57BL/6 J female mice were obtained from Hunan SJA Laboratory Animal Co., Ltd. (Hunan, China) and maintained in the animal facilities of Nanchang Royo Biotech under pathogen-free conditions on a 12-h light/12-h dark cycle. 2 month old mice were used as a young cohort, and 20–22 month old mice used as an old cohort. Dietary interventions were performed according to a protocol from our previous publication (Tang et al. 2016). Briefly, bodyweight and age matched mice were randomly divided into either the AL-fed or DR-fed group. One week before the dietary intervention, mice were housed individually and daily food consumption was measured for every mouse to determine their AL-feeding rate. The average amount of food was determined after the 1-week measurement for every mouse. When initiating the feeding protocol, the AL mice were fed with unlimited access of food, while DR mice were fed with 70% the average amount of food according to the previous calculation. The calculated 70% food pellet was added to each cage daily at the same time, and was constant over the whole DR period. All mouse experiments were approved by the Animal Experimental Ethical Inspection of Nanchang Royo Biotech Co. Ltd (RYEI20170430-1).
Sample collection
Fecal pellets were directly collected from each mouse in 1.5 ml microtubes by positioning the microtube in the proximity of the anus of the mouse and collecting the pellets that were excreted. All samples stored at − 80 °C until DNA isolation.
Microbial DNA extraction, PCR amplification and Illumina Hiseq sequencing
DNA was extracted using DNA extraction kit (Minkagene Stool DNA kit) for the corresponding sample. The concentration and purity were measured using the NanoDrop One (Thermo Fisher Scientific, MA, USA). 16S rRNA genes of distinct region (V4) were amplified using specific primers (515F and 806R) with 12 bp barcodes. Primers were synthesized by Invitrogen (Carlsbad, CA, USA). PCR reactions, containing 25 μl 2 × Premix Taq (Takara Biotechnology, Dalian Co. Ltd., China), the PCR instrument was BioRad S1000 (Bio-Rad Laboratory, CA). The length and concentration of the PCR product were detected by 1% agarose gel electrophoresis. PCR products were mixed in equimolar ratios according to the GeneTools Analysis Software (Version4.03.05.0, SynGene). Then, the PCR mixture was purified with EZNA Gel Extraction Kit (Omega, USA). Then, sequencing libraries were generated using NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, USA) following the manufacturer’s recommendations and index codes were added. The library quality was assessed on the Qubit 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Lastly, the library was sequenced on an Illumina Hiseq 2500 platform and 250 bp paired-end reads were generated.
Bioinformatics and sequencing data analysis
Quality filtering on the paired-end raw reads were performed under specific filtering conditions to obtain the high-quality clean reads according to the Trimmomatic (V0.33, http://www.usadellab.org/cms/?page=trimmomatic) quality controlled process. At the same time, sequences were assigned to each sample based on their unique barcode and primer, after which the barcodes and primers were removed to get the paired-end clean reads. Paired-end clean reads were merged using FLASH (V1.2.11, https://ccb.jhu.edu/software/FLASH/) according to the relationship of the overlap between the paired-end reads, when at least 10 of the reads overlap the read generated from the opposite end of the same DNA fragment, the maximum allowable error ratio of the overlap region of 0.2, and the spliced sequences were called raw tags. Assigned Sequences to each sample based on their unique barcode and primer using Mothur software (V1.35.1, http://www.mothur.org), after which the barcodes and primers were removed to get the effective Clean Tags. Sequences analysis were performed by usearch software (V10, http://www.drive5.com/usearch/). Sequences with ≥ 97% similarity were assigned to the same OTU (Operational Taxonomic Units). An OTU is thought to possibly represent a species. The most frequently occurring sequence was extracted as representative sequence for each OTU and was screened for further annotation. For each representative sequence, the silva (for 16S, https://www.arb-silva.de/), database was used to annotate taxonomic information (set the confidence threshold to default to ≥ 0.5). The total number of otu sequences (No. of final seqs) and otu type (No. of OTUs) in otu_table were counted respectively. Based on the otu_table_subsampled, the annotation ratio on each classification level was calculated to obtain the sequence composition of each sample at each classification level. Based on the relative abundance of species at each classification level in otu_table, R software was used to draw the histogram, heat map and ternary phase diagram. Beta diversity analysis was used to evaluate differences of samples in species complexity. Beta diversity was determined using QIIME (Quantitative Insights Into Microbial Ecology) software. LDA Effect Size (LEfSe) analysis was used to find the biomarker of each group.
Statistics
All statistical analyses were performed using GraphPad Prism 7.0 software. The unpaired two-tailed Student’s t test and One-way ANOVA were used to calculate p values. Data are expressed as mean ± SEM.
Acknowledgements
This work was supported by the National Natural Science Fund of China (NSFC-81660520, NSFC-81660244 and NSFC-81860027), and the Jiangxi Provincial Nature Science Foundation (20171ACB21060, 20181BAB205060 and 2018ACB21034).
Author contributions
TZ, HC performed and analyzed majority of all experiments. DT participated in most of the experiments. ZT and LZ helped with sample collection and daily feeding for Figs. 1 and 2. YW and JW helped with sample collection and daily feeding for Figs. 3 and 4. GBG gave suggestions and helped with writing of the manuscript. DT and ST conceived and designed the experiments. The manuscript was written by HC, GBG and ST and commented on by all other authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ting Zeng and Hui Cui contributed equally to this work.
Change history
9/9/2019
In the original publication of the article, the alphabets in figures were published in upper case and mismatched with the figure legends. The corrected figures and figure legends are given in this Correction.
References
- Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez-Humaran LG, Smirnova N, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3:559–572. doi: 10.1002/emmm.201100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer PV, Duca FA, Waise TMZ, Dranse HJ, Rasmussen BA, Puri A, Rasti M, et al. Lactobacillus gasseri in the upper small intestine impacts an ACSL3-dependent fatty acid-sensing pathway regulating whole-body glucose homeostasis. Cell Metab. 2018;27:572–587. doi: 10.1016/j.cmet.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, Consolandi C, et al. Gut microbiota and extreme longevity. Curr Biol. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- Biragyn A, Ferrucci L. Gut dysbiosis: a potential link between increased cancer risk in ageing and inflammaging. Lancet Oncol. 2018;19:e295–e304. doi: 10.1016/S1470-2045(18)30095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff SC, Barbara G, Buurman W, Ockhuizen T, Schulzke JD, Serino M, Tilg H, et al. Intestinal permeability—a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. doi: 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, Rana A, et al. Distinct shifts in microbiota composition during drosophila aging impair intestinal function and drive mortality. Cell Rep. 2015;12:1656–1667. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- Dutta S, Sengupta P. Men and mice: relating their ages. Life Sci. 2016;152:244–248. doi: 10.1016/j.lfs.2015.10.025. [DOI] [PubMed] [Google Scholar]
- Elinav TSE, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome is a regulator of colonic microbial ecology and risk for colitis. Cell. 2011;145:745. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci USA. 2004;101:6659–6663. doi: 10.1073/pnas.0308291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S. Health span extension by later-life caloric or dietary restriction: a view based on rodent studies. Biogerontology. 2006;7:135–138. doi: 10.1007/s10522-006-9011-4. [DOI] [PubMed] [Google Scholar]
- Goto S, Takahashi R, Radak Z, Sharma R. Beneficial biochemical outcomes of late-onset dietary restriction in rodents. Ann N Y Acad Sci. 2007;1100:431–441. doi: 10.1196/annals.1395.048. [DOI] [PubMed] [Google Scholar]
- Haro C, Rangel-Zuniga OA, Alcala-Diaz JF, Gomez-Delgado F, Perez-Martinez P, Delgado-Lista J, Quintana-Navarro GM, et al. Intestinal microbiota is influenced by gender and body mass index. PLoS ONE. 2016;11:e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Cheng D, Peng C, Li Y, Zhu Y, Lu N. High-fat diet induces dysbiosis of gastric microbiota prior to gut microbiota in association with metabolic disorders in mice. Front Microbiol. 2018;9:639. doi: 10.3389/fmicb.2018.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut. 2001;48:198–205. doi: 10.1136/gut.48.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery IB, Lynch DB, O’Toole PW. Composition and temporal stability of the gut microbiota in older persons. ISME J. 2016;10:170–182. doi: 10.1038/ismej.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok DEG, Rusli F, van der Lugt B, Lute C, Laghi L, Salvioli S, Picone G, et al. Lifelong calorie restriction affects indicators of colonic health in aging C57Bl/6J mice. J Nutr Biochem. 2018;56:152–164. doi: 10.1016/j.jnutbio.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Liu L, Rando TA. Manifestations and mechanisms of stem cell aging. J Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, Gonzalez-Reyes JA, Cortassa S, et al. Effects of sex, strain, and energy intake on hallmarks of aging in mice. Cell Metab. 2016;23:1093–1112. doi: 10.1016/j.cmet.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yan Y, Wan P, Chen D, Ding Y, Ran L, Mi J, et al. Gut microbiota modulation and anti-inflammatory properties of anthocyanins from the fruits of Lycium ruthenicum Murray in dextran sodium sulfate-induced colitis in mice. Free Radic Biol Med. 2019;136:96–108. doi: 10.1016/j.freeradbiomed.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Qi Y, Chen L, Gao K, Shao Z, Huo X, Hua M, Liu S, et al. Effects of Schisandra chinensis polysaccharides on rats with antibiotic-associated diarrhea. Int J Biol Macromol. 2019;124:627–634. doi: 10.1016/j.ijbiomac.2018.11.250. [DOI] [PubMed] [Google Scholar]
- Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, Chrysovergis K, et al. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19:7. doi: 10.1186/s13059-018-1389-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radak Z, Takahashi R, Kumiyama A, Nakamoto H, Ohno H, Ookawara T, Goto S. Effect of aging and late onset dietary restriction on antioxidant enzymes and proteasome activities, and protein carbonylation of rat skeletal muscle and tendon. Exp Gerontol. 2002;37:1423–1430. doi: 10.1016/S0531-5565(02)00116-X. [DOI] [PubMed] [Google Scholar]
- Ribaric S. Diet and aging. Oxid Med Cell Longev. 2012;2012:741468. doi: 10.1155/2012/741468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth GS, Ingram DK, Lane MA. Caloric restriction in primates and relevance to humans. Ann N Y Acad Sci. 2001;928:305–315. doi: 10.1111/j.1749-6632.2001.tb05660.x. [DOI] [PubMed] [Google Scholar]
- Singh R, Lakhanpal D, Kumar S, Sharma S, Kataria H, Kaur M, Kaur G. Late-onset intermittent fasting dietary restriction as a potential intervention to retard age-associated brain function impairments in male rats. Age (Dordr) 2012;34:917–933. doi: 10.1007/s11357-011-9289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Manchanda S, Kaur T, Kumar S, Lakhanpal D, Lakhman SS, Kaur G. Middle age onset short-term intermittent fasting dietary restriction prevents brain function impairments in male Wistar rats. Biogerontology. 2015;16:775–788. doi: 10.1007/s10522-015-9603-y. [DOI] [PubMed] [Google Scholar]
- Tang D, Tao S, Chen Z, Koliesnik IO, Calmes PG, Hoerr V, Han B, et al. Dietary restriction improves repopulation but impairs lymphoid differentiation capacity of hematopoietic stem cells in early aging. J Exp Med. 2016;213:535–553. doi: 10.1084/jem.20151100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21:455–466. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford RL, Mock D, Verdery R, MacCallum T. Calorie restriction in biosphere 2: alterations in physiologic, hematologic, hormonal, and biochemical parameters in humans restricted for a 2-year period. J Gerontol A Biol Sci Med Sci. 2002;57:B211–B224. doi: 10.1093/gerona/57.6.B211. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357:912–916. doi: 10.1126/science.aan0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Huang M, You X, Zhao J, Chen L, Wang L, Luo Y, et al. Gut microbiota mediates the anti-obesity effect of calorie restriction in mice. Sci Rep. 2018;8:13037. doi: 10.1038/s41598-018-31353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R. Caloric restriction and aging. Sci Am. 1996;274:46–52. doi: 10.1038/scientificamerican0196-46. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Gottesman SR, Walford RL. Modification of age-related immune decline in mice dietarily restricted from or after midadulthood. Proc Natl Acad Sci USA. 1982;79:898–902. doi: 10.1073/pnas.79.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata HJ, Quagliarello VJ. The microbiota and microbiome in aging: potential implications in health and age-related diseases. J Am Geriatr Soc. 2015;63:776–781. doi: 10.1111/jgs.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li S, Yang L, Huang P, Li W, Wang S, Zhao G, et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun. 2013;4:2163. doi: 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]