Abstract
Indian rhesus macaque major histocompatibility complex (MHC) variation can influence the outcomes of transplantation and infectious disease studies. Frequently, rhesus macaques are MHC genotyped to identify variants that could account for unexpected results. Since the MHC is only one region in the genome where variation could impact experimental outcomes, strategies for simultaneously profiling variation in the macaque MHC and the remainder of the protein coding genome would be useful. Here we determine MHC class I and class II genotypes using target-capture probes enriched for MHC sequences, a method we term macaque exome sequence (MES) genotyping. For a cohort of 27 Indian rhesus macaques, we describe two methods for obtaining MHC genotypes from MES data and demonstrate that the MHC class I and class II genotyping results obtained with these methods are 98.1% and 98.7% concordant, respectively, with expected MHC genotypes. In contrast, conventional MHC genotyping results obtained by deep sequencing of short multiplex PCR amplicons were only 92.6% concordant with expectations for this cohort.
Keywords: Macaca mulatta, exome, major histocompatibility complex
Introduction
The major histocompatibility complex (MHC) is an intensively studied set of genes in macaques (Shiina et al. 2017; Wiseman et al. 2013). The genomic MHC region contains clusters of genes that encode the MHC class I complex and the MHC class II complex. Cells use MHC class I molecules to present intracellular peptides to immune cells like the CD8+ T cell or natural killer cells (Garcia and Adams 2005). MHC class I molecules accommodate intracellular peptides of varying specificity by having diverse amino acid sequences in the α1 and α2 subunits, which form the peptide-binding cleft (Silver and Watkins 2017; Loffredo et al. 2009). These α1 and α2 subunits correspond to exons 2 and 3, respectively, of an MHC class I gene (Malissen et al. 1982). Most of the polymorphisms that distinguish MHC class I alleles are concentrated in exons 2 and 3 (Williams 2001). Thousands of individual MHC allelic variants have been identified in the three most widely used macaque species for biomedical research: rhesus (Macaca mulatta), cynomolgus (Macaca fascicularis), and pig-tailed macaques (Macaca nemestrina) (Semler et al. 2018; Karl et al. 2017; Maccari et al. 2017).
The human MHC is termed the human leukocyte antigen complex (HLA). HLA class I has a single copy of the HLA-A, HLA-B, and HLA-C genes on each chromosome (Daza-Vamenta et al. 2004; Shiina et al. 2017). In contrast, macaques have a variable number of genes on each chromosome that encode MHC class I MHC-A and MHC-B proteins, and macaques lack an HLA-C orthologue (Daza-Vamenta et al. 2004; Shiina et al. 2017; Wiseman et al. 2013). Initial approaches to genotype macaque MHC class I relied on using sequence-specific PCR oligonucleotides to test for the presence or absence of individual alleles (Kaizu et al. 2007). More recently, deep sequencing genomic DNA or complementary DNA PCR amplicons spanning a highly variable region of exon 2 has become commonplace (Wiseman et al. 2009; Karl et al. 2013). Amplicon sequences can be used to genotype groups of closely related MHC class I alleles, which are denoted as lineages. For example, an amplicon deep sequence that corresponds to the rhesus macaque Mamu-A1*001 lineage demonstrates that an animal possesses Mamu-A1*001:01, Mamu-A1*001:02, or another closely related variant that has not yet been identified. This lineage-level reporting of MHC class I genotypes can be sufficient for designing experiments where specific MHC class I genotypes need to be matched between animals, balanced among experimental groups, or excluded entirely from a study (Loffredo et al. 2007; Loffredo et al. 2009; Muhl et al. 2002; Karl et al. 2013; Wiseman et al. 2009; Nomura et al. 2012; Mothe et al. 2003).
Amplicon deep sequencing can also be used for MHC class II genotyping. The human HLA class II genes DQA1, DQB1, DPA1, and DPB1 have direct orthologues in macaques (Otting et al. 2017). Both macaques and humans have a variable number of MHC class II DRB genes on a single chromosome, while the DRA gene is oligomorphic (Daza-Vamenta et al. 2004; Shiina et al. 2017), and typically is not used for genotyping purposes. MHC class II molecules are members of the immunoglobulin superfamily, but they differ from MHC class I in several ways. α and β subunits comprise the MHC class II heterodimer, with separate genes encoding α and β subunits (Brown et al. 1993). Highly polymorphic regions that are diagnostic for MHC class II allele lineages can be PCR amplified in a manner that is similar to the MHC class I genotyping. Exon 2 contains the most extensive polymorphism among MHC class II alleles (Williams 2001). Each of the DRB, DQA1, DQB1, DPA1, and DPB1 polymorphic MHC class II genes are sufficiently divergent that separate PCR amplicons are required for deep sequencing (Karl et al. 2014).
Comprehensive MHC class I and class II genotyping of macaques by amplicon deep sequencing requires preparation of six separate PCR amplicons for MHC class I and class II DRB, DQA1, DQB1, DPA1, and DPB1 (Karl et al. 2014; Karl et al. 2017). Despite this complexity, a major advantage to amplicon deep sequencing has been its cost effectiveness. The output from a single MiSeq sequencing run can be used to determine the MHC class I and MHC class II genotypes for up to 192 macaque samples. In recent years, improved sequencing hardware and software have prompted new approaches to MHC genotyping. Instead of utilizing PCR amplicons of variable gene regions to genotype samples, researchers can use human whole exome sequencing (WES) and whole genome sequencing (WGS) datasets to determine HLA genotypes (Xie et al. 2017; Kishikawa et al. 2019; Yang et al. 2014; Posey et al. 2016). Likewise, target capture approaches have been described for HLA genotyping with next-generation sequencing datasets (Cao et al. 2013; Wittig et al. 2015). In contrast, MHC genotyping of macaques using WGS (Xue et al. 2016; Bimber et al. 2017; de Manuel et al. 2018) or WES (Vallender 2011; Cornish et al. 2016) have not been reported to date. The macaque genome is being actively updated, but the reference database of known alleles is still incomplete (Zimin et al. 2014; Maccari et al. 2017). The unfinished state of the macaque reference database and the inherent intricacies that exist when attempting to map short sequence reads against complex, duplicated gene families both illustrate the significant challenges researchers can face with macaque genotyping.
In an era where the per-base cost of sequencing is dropping rapidly, we explored the feasibility of obtaining whole exome sequencing data while maintaining parity of results with the traditional MHC PCR amplicon approach. WES datasets provide investigators with the capability to examine sequence variants across the entire coding region of the genome. Although SIV researchers have traditionally focused on specific MHC genotypes that are associated with exceptional control of viral replication, there is also interest in evaluating variation in host restriction factor genes, such as TRIM5, Tetherin, and APOBEC3G receptor genes (Reynolds et al. 2018, Janaka et al. 2018, Krupp et al. 2013, Ericsen et al. 2014). Exome sequencing will also provide an opportunity for investigators in multiple fields to evaluate variants from a wide variety of pathways that are important for biomedical research (Haus et al. 2014). A key advantage of WES over WGS remains to be the cost per sample of generating sequence assays. Since WES only targets the coding regions, which are 1–2% of the genome, the amount of sequencing that is required is dramatically reduced. At our institutions, currently WES can be completed for less than 20% of the cost of a comparable WGS study. The ability to evaluate at least five times more animals for the same cost opens the possibility of characterizing much larger non-human primate cohorts, such as with breeding colonies of macaques, or complete retrospective experimental studies (de Manuel et al. 2018).
Here we introduce MHC genotyping via macaque exome sequencing (MES), which is an exome sequencing-based workflow for comprehensive MHC class I and class II genotyping in macaques. This workflow uses a commercially available human exome target-capture enrichment kit in conjunction with specialized spike-in target-capture probes to cope with the high copy number of macaque MHC genes. We show that accuracy of Indian rhesus macaque MHC class I and class II results from this workflow are comparable to conventional MiSeq genotyping when using exon 2 reference sequences.
Methods
Animals
Twenty-seven whole blood samples were collected from Indian rhesus macaques (Macaca mulatta). Five of these samples came from a breeding group of animals living at the Wisconsin National Primate Research Center (WNPRC). The remaining 22 samples were provided by Dr. Michele Di Mascio from the National Institutes of Health’s National Institutes of Allergy and Infectious Diseases. Blood sampling was performed under anesthesia and in accordance with the regulations and guidelines outlined in the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals, and the Weatherall report (Animal Welfare Act 1966; Weatherall 2006).
Data
Exome sequence datasets have been deposited in the sequence read archive (SRA) under BioProjects PRJNA527214 and PRJNA529708. Fasta reference sequences used for MHC genotyping, and sequence analysis scripts are available from https://go.wisc.edu/jb0926.
MHC class I and class II genotyping by amplicon deep sequencing
Genomic DNA was isolated from 250μL of whole blood using a Maxwell® 48 LEV Blood DNA Kit (Promega Corporation, Fitchburg, WI). Following isolation, DNA concentrations were determined with a Nanodrop 2000 and samples were normalized to 60 ng/ul. MHC class I and class II PCR amplicons were generated using exon 2-specific primers with adapters (CS1 and CS2) necessary for 4-primer amplicon tagging with the Fluidigm Access Array™ System (Fluidigm, San Francisco, CA, USA) by previously described methods (Karl et al. 2017; Karl et al. 2014). Pooled PCR products were purified using the AMPure XP beads (Agencourt Bioscience Corporation, Beverly, MA, USA) and quantified using the Quant-iT dsDNA HS Assay kit with a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s protocols. The MHC exon 2 genotyping amplicon pools were sequenced on an Illumina MiSeq instrument (San Diego, CA, USA) as previously described (Karl et al. 2017).
Analysis of the MiSeq exon 2 genotyping amplicon sequences was performed using a custom Python workflow. The workflow contained a step to remove oligonucleotide primers and sequencing adapters with bbduk, a step to merge reads using bbmerge, a step to identify unique sequences/remove chimeras with USEARCH, and finally mapping unique reads against a deduplicated reference database of rhesus macaque partial MHC class I and class II exon 2 sequences with bbmap (Bushnell et al. 2017; Edgar 2010). The IPD-MHC NHP database on the European Bioinformatics Institute website is continuously updated as new information from researchers is submitted to it, and we elected to download the release that was available on September 8, 2018 (Maccari et al. 2017). By doing so, we created a fixed snapshot of the reference sequences for our data analyses, and this ensured consistency for us when we tested our experimental hypotheses. For this publication, we define “IPD exon 2” as this database of reference sequences. SAM output files from bbmap were parsed with the Python package pandas to enumerate the reads from each animal that were identical to IPD exon 2 reference sequences. Mamu-A, -B, -DRB, -DQA1, -DQB1, -DPA1 and -DPB1 lineage-level haplotypes were inferred for each of the samples with a semi-automated custom workflow that identifies diagnostic alleles associated with previously defined rhesus macaque haplotypes (Karl et al. 2013; Otting et al. 2017).
MHC class I and class II genotyping by exome sequencing
Genomic DNA was isolated as described above and shipped to the Human Genome Sequencing Center at the Baylor College of Medicine. MHC and exon-containing genomic DNA fragments were selectively enriched using a custom target-capture probeset. Genome-wide exons were captured using SeqCap EZ HGSC VCRome2.1, an optimized human clinical exome probeset (Clark et al. 2013, Yang et al. 2013). SeqCap EZ HGSC VCRome2.1 contains probes designed to enrich 23,585 human genes and 189,028 non-overlapping exons. A low coverage audit was performed to identify rhesus macaque exons inferred from the reference genome rheMac2 that were not sufficiently enriched (<20× coverage) with these human probes, and an additional 22,884 rhesus macaque exons were incorporated into the genotyping probe design (Prall et al. 2017). Finally, and most importantly for MHC analyses, we modified the SeqCap EZ Design: Human MHC Design to selectively enrich MHC class I and class II sequences. This previous design was prepared by the Beijing Genome Institute in collaboration with Roche/Nimblegen and it targeted the complete 4.97Mb HLA region with non-redundant probes designed against 8 fully sequenced HLA haplotypes (Horton et al. 2008; Cao et al. 2013). For our macaque studies, we prepared a minimal MHC target capture design using a subset of these probes that are based on all functional HLA class I (HLA-A, -B, -C, and -E) and class II (HLA-DRA, -DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and -DPB1) genes. Probes were included to capture complete gene sequences (exons + introns + 3’ UTR) as well as ~1 kb of 5’ upstream flanking sequence. The BED file of rhesus rheMac2 target coordinates lifted over to rheMac8 was used to prepare this combined minimal MHC and supplemental rhesus spike-in probe design. Because derivation of MHC results from genotyping is paramount, we used a ratio of 2.5× spike-in probes to 1× VCRom2.1 probes. The supplemental probes for MHC and rhesus macaque were a single reagent, and the MHC-specific probes only constituted 609 kb of the 37.9 Mb probes (Prall et al. 2017).
An Illumina paired-end pre-capture library was constructed with 750 nanograms of DNA, as described by the Baylor College of Medicine Human Genome Sequencing Center. Pre-capture libraries were pooled into 10-plex library pools for target capture according to the manufacturer’s protocol. Samples were pooled in 10-plex sequence capture library pools for 151 bp paired-end sequencing in a single lane of an S4 flow cell on an Illumina NovaSeq 6000 at the Baylor College of Medicine Human Genome Sequencing Center.
Enumeration of MHC reads in exome data
The effective enrichment of MHC reads in the exome datasets was calculated by mapping the reads of each sample’s exome dataset against an individual genomic reference file for each individual locus: HLA-A exons 2–3 (NCBI Gene ID: 3105) and HLA-E exons 2–3 (Gene ID: 3133); HLA-DPA1 exons 2–4 (Gene ID: 3113); HLA-DPB1 exons 2–4 (Gene ID: 3115); HLA-DQA1 exons 2–4 (Gene ID: 3117); HLA-DQA2 exons 2–4 (Gene ID: 3118); HLA-DQB1 exons 2–4 (Gene ID: 3119); HLA-DQB2 exons 2–4 (Gene ID: 3120); HLA-DRB1 exons 2–4 (Gene ID: 3123), HLA-DRB3 exons 2–4 (Gene ID: 3125), HLA-DRB4 exons 2–4 (Gene ID: 3126) and HLA-DRB5 exons 2–4 (Gene ID: 3127). This mapping was done by using bbmap with default parameters, which corresponds to a minimum alignment identity of approximately 76% (Bushnell et al. 2017). Empirically, these mapping parameters are sufficient to map macaque MHC reads to their human orthologues. Mapped reads were written to a new fastq file using bbmap’s outm= parameter. To quantify the total number of reads in a sample and the number of reads extracted with our reference file, we created a custom Python script, which is available to download.
MHC genotyping from exome data
Two complementary data analysis strategies were employed to analyze the exome sequence data, and to verify reproducibility and confidence in the MHC genotyping results. For accuracy and quantification purposes, the expected MHC genotypes for each animal were established based on concordance among at least two out of the three described strategies and biological plausibility, e.g., no more than two alleles per Mamu-DQA1, -DQB1, -DPA1, or-DPB1 locus.
Strategy 1: MHC genotyping using Diagnostic Sub-Region (DSR)
The Diagnostic Sub-Region (DSR) was an intra-allelic region that encompassed polymorphisms, and these polymorphisms were distinguishable from alleles with similar sequences. Therefore, this method ensured the DSR was captured in at least one read for each called allele. MHC class I and class II reads initially were extracted from each animal by mapping the FASTQ reads to HLA class I and class II reference sequences containing exons 2–3 and exons 2–4, respectively, plus the intervening intron(s) as described above in ‘Enumeration of MHC reads in exome data’. Reads were mapped to these reference sequences using bbmap with default parameters and the parameter (qtrim=lr) (Bushnell et al. 2017).
Following extraction of MHC reads from the total exome sequences, the MHC reads were prepared for assembly using a modified version of a data pre-processing pipeline, which included tools from the BBTools package (Bushnell et al. 2017). Briefly, optical duplicates and reads from low-quality regions of the sequencing run were removed. Next, Illumina sequencing adapters were trimmed from the ends of sequencing reads. Any residual spike-in or PhiX sequences that inadvertently survived mapping to HLA class I and class II were then removed. Three rounds of error-correction and read merging were performed to create high-confidence merged reads that were well-supported by common kmers found in the extracted MHC reads. The error-corrected reads were not merged with a minimum overlap, but instead were separately mapped against the IPD exon 2 sequences using bbmapskimmer. The default settings for the software tool bbmapskimmer were used with the following modified parameters (semiperfectmode=t ambiguous=all ssa=t maxsites=50000 maxsites2=50000 expectedsites=50000) (Bushnell et al. 2017). The semiperfectmode setting accepted reads with perfect matches, as well as reads that extended off the end of contigs for no more than half of the length of the mapped read segment. The ambiguous setting and the ‘expectedsites’ setting reported the first 50,000 matched read segments that met the ‘semiperfectmode’ filtering. These settings were set exceedingly high in order to exhaustively map the reads to our IPD exon 2 reference file of approximately 874 alleles. Using semiperfect mode, these were the segment sequences with the longest matching length. This output file included all mapped reads to all IPD exon 2 reference allele sequences.
We then used samtools mpileup with the settings (-A -a --ff UNMAP -x -B -q 0 -Q 0) on the bbmapskimmer output to calculate a depth of coverage at each position for each IPD exon 2 sequence. Next, we removed any aligned reads to the IPD exon 2 database sequences that contained less than a minimum depth of coverage of two across the entire reference sequence. For ambiguously-mapped reads, alignments with the longest-matching region were selected for further analysis; ties among alignments were counted multiple times. To reduce the number of false positives, we used Python pandas to only report database sequence matches that had at least one unambiguously mapped read. Based on these mapping parameters, unambiguously-mapped reads must span the DSR. The MHC genotypes from this method were reported as the minimum depth of coverage for each IPD exon 2 database sequence per animal.
Strategy 2: De novo reconstruction of MHC sequences from exome reads
Most de novo sequence assemblers have been optimized for resolving long contigs, and are tolerant of small sequence mismatches that can otherwise fragment assemblies. In the case of MHC alleles, however, such closely related sequences were often biologically distinct. Because of the gene duplications in the macaque MHC, there were many valid MHC contigs that could be assembled from a single sample within the exome data. This is conceptually similar to the computational challenge of assembling viral haplotypes, where rapidly evolving viruses such as human immunodeficiency virus accumulate variants that frequently co-segregate as minor populations within an infected person (Baaijens et al. 2017). Therefore, we utilized the overlap assembly algorithm SAVAGE, originally designed to reconstruct viral haplotypes, to reconstruct MHC allele sequences from exome reads (Baaijens et al. 2017). Similar to Strategy 1 above, HLA-mapped reads were pre-processed using the BBTools package to remove low quality reads and optical duplicates. Next, adapters were trimmed, residual spike-in and PhiX sequences were removed, three steps of error-correction took place, and reads were merged. These merged reads, as well as high-confidence unmerged paired-end reads that could not be grouped into an overlapping merged read, were used for SAVAGE assembly. The following workflow was implemented in a reproducible snakemake workflow that fully documents parameter selection (Koster and Rahman 2012), and is available upon request.
SAVAGE is designed to construct individual haplotypes from the overlap graph of individual reads. We processed the totality of sequence data in a single patch in order to maximize the sensitivity of the contiguous reconstruction with parameter ‘--split 1’, as well as the parameter ‘--revcomp’ to handle reverse complement reads. The IPD exon 2 reference database that was used for MiSeq and DSR genotyping was also used to assess the quality of SAVAGE genotypes. The IPD sequences were mapped to contigs produced by SAVAGE using sensitive parameters in bbmap (minlength=100 vslow=t subfilter=0 indelfilter=0 lengthtag=t ignorefrequentkmers=t kfilter=100) designed to identify sequences that perfectly match SAVAGE contigs. A post-processing script refined these mappings, and only retained the mappings where the length of the mapped region was the same as the length of the IPD exon 2 sequence. These mappings indicated where the reference database sequence was fully and exactly contained within a SAVAGE contig.
Results
MHC reads are efficiently enriched using target-capture probes
We designed a custom target enrichment probeset that accounts for the extensive duplication of macaque MHC genes (Prall et al. 2017). A tripartite target capture system was used in this study. The first component is SeqCap EZ HGSC VCRome2.1, an optimized human clinical exome probeset. Since macaques are closely related to humans, SeqCap EZ HGSC VCRome2.1 can also be used in macaques, though some sequences that are most divergent between macaques and humans are not efficiently captured. The second component of the target capture system is an additional 22,884 rhesus macaque exon sequences that were not effectively captured with the SeqCap EZ HGSC VCRome2.1 reagent. The third component is a collection of probes designed to specifically enrich macaque MHC class I and class II sequences. These probes span the full length of HLA class I and class II genes including introns, 3’ UTRs, and approximately 1,000 bp of flanking 5’ sequence.
We obtained a median coverage of 100X for the target exon sequences across the genome with >20X coverage for 94.97% of bases that were targeted in this study. As illustrated in Table 1, an average of 70,549,789 Illumina sequence reads per sample were obtained for the 27 animals evaluated in this study. These reads were mapped against reference files containing representative genomic HLA exons 2–3 for HLA-A and HLA-E, and exons 2 – 4 for HLA-DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and -DPB1 sequences. We identified an average of 269,057 MHC class I and class II sequence reads per sample which corresponds to an average of 0.37% of the total sequence reads evaluated per sample (Table 1). In a previous study by Ericsen et al. (Ericsen et al. 2014), we found that MHC sequences only accounted for an average of 0.13% of the total Illumina sequence reads that were evaluated per animal when the standard HGSC VCRome2.1 panel was used alone for target capture (Supplementary Table 1). Thus, we achieved an almost three-fold increase of MHC genomic sequences after inclusion of the spike-in probes for target capture compared to use of the VCRome2.1 probeset alone.
Table 1.
Fraction of total exome sequence reads corresponding to exons 2 – 3 of MHC class I and exons 2 – 4 of MHC class II genes.
| Animal ID | SRA Accession | Total Raw Reads | MHC-I Reads Extracted | DRB1/3/4/5 Reads Extracted | DQA1 Reads Extracted | DQB1 Reads Extracted | DPA1 Reads Extracted | DPB1 Reads Extracted | MHC Reads Extracted | MHC % of total |
|---|---|---|---|---|---|---|---|---|---|---|
| r05029 | SAMN11131055 | 87,345,430 | 79,712 | 215,654 | 24,266 | 39,188 | 29,604 | 112,376 | 500,800 | 0.57 |
| r17099 | SAMN11131056 | 67,632,236 | 32,406 | 89,946 | 9,694 | 16,782 | 11,214 | 47,974 | 208,016 | 0.31 |
| r07010 | SAMN11173514 | 81,062,856 | 71,694 | 204,352 | 22,528 | 38,382 | 25,392 | 111,626 | 473,974 | 0.58 |
| r17041 | SAMN11131058 | 80,004,430 | 30,852 | 244,326 | 9,746 | 16,310 | 12,644 | 151,376 | 465,254 | 0.58 |
| r17061 | SAMN11131059 | 78,428,820 | 70,082 | 204,694 | 20,776 | 41,018 | 19,440 | 95,538 | 451,548 | 0.58 |
| J8R | SAMN11282383 | 80,868,782 | 45,164 | 87,864 | 11,878 | 19,736 | 13,810 | 57,146 | 235,598 | 0.29 |
| J01 | SAMN11282382 | 61,009,650 | 34,342 | 87,124 | 9,050 | 16,628 | 10,958 | 46,964 | 205,066 | 0.34 |
| ZC08 | SAMN11282384 | 60,693,394 | 31,168 | 81,586 | 10,392 | 15,660 | 10,172 | 45,748 | 194,726 | 0.32 |
| DGKG | SAMN11282378 | 80,530,772 | 48,658 | 130,256 | 12,788 | 22,682 | 13,372 | 62,990 | 290,746 | 0.36 |
| CF18 | SAMN11282368 | 68,593,710 | 37,792 | 96,842 | 10,864 | 18,226 | 13,118 | 51,556 | 228,398 | 0.33 |
| DE1AA | SAMN11282369 | 65,712,570 | 39,334 | 108,002 | 10,060 | 17,124 | 11,840 | 51,134 | 237,494 | 0.36 |
| DEG8 | SAMN11282370 | 68,490,344 | 35,566 | 116,824 | 10,404 | 19,278 | 12,294 | 55,086 | 249,452 | 0.36 |
| DEXX | SAMN11282371 | 67,451,626 | 33,900 | 128,128 | 10,442 | 18,898 | 9,778 | 53,258 | 254,404 | 0.38 |
| DF24 | SAMN11282372 | 67,768,724 | 41,362 | 96,172 | 10,606 | 18,176 | 10,976 | 50,612 | 227,904 | 0.34 |
| DF64 | SAMN11282373 | 65,762,818 | 37,426 | 107,974 | 11,222 | 15,632 | 11,446 | 51,214 | 234,914 | 0.36 |
| DF6T | SAMN11282374 | 71,472,222 | 41,160 | 101,102 | 11,052 | 18,794 | 13,492 | 51,626 | 237,226 | 0.33 |
| DFET | SAMN11282375 | 77,040,660 | 49,068 | 128,862 | 12,588 | 20,952 | 12,586 | 58,238 | 282,294 | 0.37 |
| DFV0 | SAMN11282377 | 73,877,184 | 41,924 | 98,866 | 11,630 | 17,690 | 13,952 | 52,394 | 236,456 | 0.32 |
| DJ94 | SAMN11282379 | 66,051,412 | 39,616 | 88,196 | 9,910 | 15,892 | 10,978 | 45,972 | 210,564 | 0.32 |
| HIH | SAMN11282380 | 76,491,578 | 41,308 | 85,732 | 11,148 | 18,366 | 14,102 | 50,746 | 221,402 | 0.29 |
| DFJT | SAMN11282376 | 62,249,064 | 37,728 | 78,860 | 9,666 | 15,268 | 11,426 | 43,086 | 196,034 | 0.31 |
| HLJ | SAMN11282381 | 68,288,558 | 41,748 | 94,570 | 9,960 | 17,240 | 12,756 | 48,356 | 224,630 | 0.33 |
| 37360 | SAMN11282363 | 62,310,824 | 38,200 | 97,452 | 9,866 | 15,024 | 10,320 | 46,140 | 217,002 | 0.35 |
| 0L7 | SAMN11282364 | 64,145,834 | 35,302 | 128,778 | 9,310 | 19,992 | 10,686 | 54,196 | 258,264 | 0.40 |
| 0R1 | SAMN11282365 | 64,789,208 | 36,462 | 86,772 | 10,656 | 17,258 | 11,982 | 46,124 | 209,254 | 0.32 |
| 0R8 | SAMN11282366 | 72,836,490 | 38,286 | 118,184 | 10,500 | 22,248 | 13,170 | 56,142 | 258,530 | 0.35 |
| 0TI | SAMN11282367 | 80,730,750 | 41,552 | 106,356 | 13,364 | 20,714 | 14,860 | 57,744 | 254,590 | 0.32 |
| Average | 70,549,789 | 42,660 | 119,018 | 12,014 | 20,487 | 13,569 | 61,310 | 269,057 | 0.37 |
MHC genotypes defined from target-enriched genomic DNA are comparable to those obtained by amplicon deep sequencing
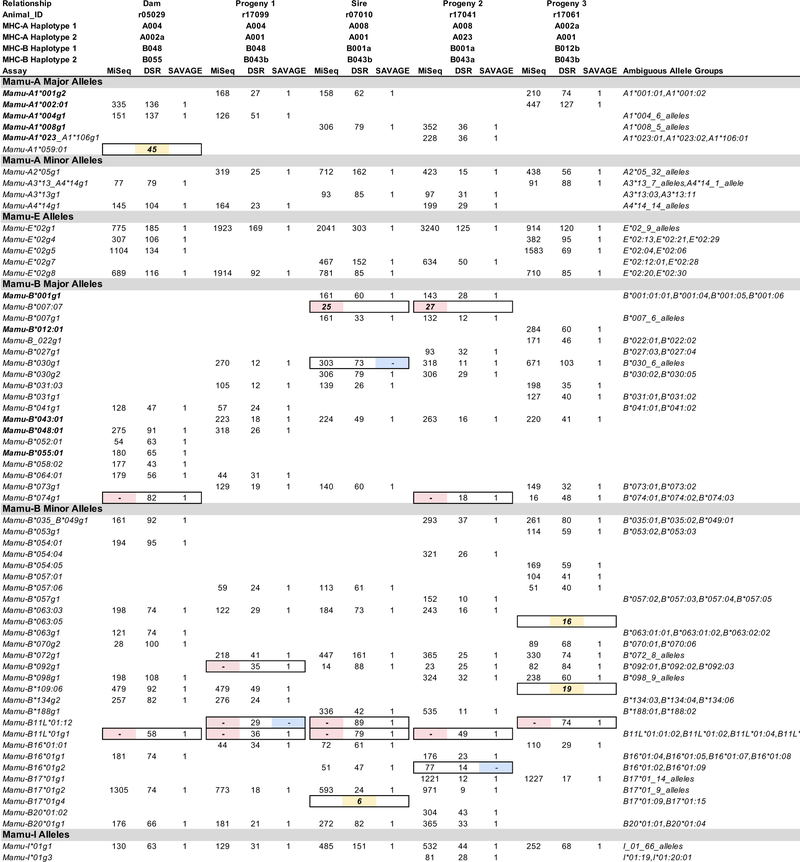
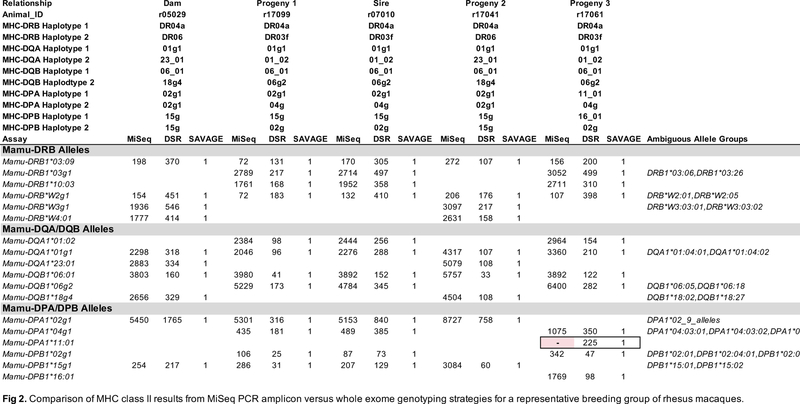
We hypothesized that MHC genotypes derived from target-enriched genomic sequence would be comparable in accuracy to MHC genotypes derived from conventional amplicon deep sequencing. In order to compare the accuracy of MHC genotyping results between the methods described here, the expected genotypes for each animal were defined by concordance among at least two out of the three described strategies, as well as biological plausibility. For example, no more than two alleles should be present for each of the MHC class II Mamu-DQA1, -DQB1, -DPA1, or -DPB1 loci. MHC class I and class II PCR amplicons were deep sequenced on an Illumina MiSeq from the same 27 animals that were evaluated by MES analysis. Representative MHC class I and class II genotypes supported by the amplicon data are shown in Figure 1 and Figure 2, respectively. These figures illustrate genotypes from a pedigreed family of Indian rhesus macaques: a sire, a dam, and three progeny that are paternal half-siblings. Although segregation data confirmed all expected genotypes that were shared among the directly related individuals in this breeding group, this was generally not possible, since pedigree information was unavailable for 22 of 27 rhesus macaques examined in this study. Supplementary Figures 1 and 2 show comprehensive genotyping results for all 27 animals.
Figure 1.
Comparison of MHC class I results from MiSeq PCR amplicon versus whole exome genotyping strategies for a representative breeding group of rhesus macaques. Results for each of the three methods are provided side-by-side in the columns for each macaque. Each row indicates the detection of a specific MHC class I allele or lineage group of closely related sequences that are ambiguous because they are identical over the IPD exon 2 database sequence. Values in the body of this figure indicate the number of sequence reads supporting each allele call for the MiSeq and DSR methods while alleles supported by a SAVAGE contig are reported with a “1”. Discrepancies between the MiSeq (pink), DSR (yellow) or SAVAGE (blue) methods are highlighted by filled cells with borders.
Figure 2.
Comparison of MHC class II results from MiSeq PCR amplicon versus whole exome genotyping strategies for a representative breeding group of rhesus macaques. Results for each of the three methods are provided side-by-side in the columns for these five related macaques. Each row indicates detection of a specific MHC class II allele or lineage group of closely related sequences that are ambiguous because they are identical over the IPD exon 2 database sequence. Values in the body of this figure indicate the number of sequence reads supporting each allele call for the MiSeq and DSR methods, while alleles supported by a SAVAGE contig are reported with a “1”. The Mamu-DPA1*11:01 allele missed by the MiSeq assay due to multiple mismatches versus the amplification primers is highlighted in pink.
The superset of alleles supported by at least two of the methods described in this manuscript (MiSeq amplicon genotyping, DSR genotyping of individual exome sequencing reads, or SAVAGE genotyping of contigs derived from exome data), was considered to be the expected MHC genotypes for these animals. A complication for this assertion was specific genotypes were difficult for MiSeq analysis to report correctly. As we have shown previously (Karl et al. 2017), the number of reads supporting each genotyping call was highly variable, ranging from tens of reads to thousands of reads per allele. A small subset of allelic variants contained nucleotide substitutions in their sequences within the binding sites of the PCR oligonucleotides that interfere with efficient PCR amplification. Members of the Mamu-B11L*01 allele lineage exemplify this issue; they have two nucleotide substitutions relative to the 5’ oligonucleotide that was used to generate MHC class I amplicons. These Mamu-B11L*01 sequences were routinely absent in MiSeq genotypes (Figure 1). Likewise, the Mamu-DPA1*11:01 allele has four nucleotide substitutions relative to the oligonucleotide pair used to generate DPA1 amplicons for MiSeq genotyping (Figure 2).
These nucleotide substitutions do not fully account for all differences in the abundance of sequence reads for each allele. Allele lineages, such as Mamu-B*074 and Mamu-B*098, exhibited significantly diminished PCR efficiency despite being perfectly matched with the oligonucleotides used for amplification (Figure 1). False positive genotyping calls were also noted in the MiSeq assay that resulted from intermolecular recombination during the PCR process (Ennis et al. 1990. Fichot et al. 2013, von Wintzingerode, et al. 1997). Artifacts from PCR amplification, dubbed ‘PCR chimeras’, arise when an incompletely extended PCR product serve as a primer with a partially mismatched template during subsequent cycles of PCR (Supplementary Figure 3). This was exemplified by read support for the presence of Mamu-B*007:07 in sire r07010, and progeny 2 r17041 (Figure 1). Mamu-B*007:07 only differs from the Mamu-B*007g1 allele group that was determined to be present in this pair of animals by a single nucleotide variant at the extreme 5’ end of the class I genotyping amplicon. Chimeric PCR products, equivalent to the Mamu-B*007:07 sequence, were formed between the 3’ portion of the Mamu-B*007g1 sequence and other allelic variants in these animals with this 5’ SNP such as Mamu-B20*01g1. These results illustrate that the MiSeq amplicon genotyping, while generally reflective of an animal’s MHC genotype, can yield both false positive and false negative results. When compared to the expected genotypes for this dataset, MiSeq amplicon genotyping has 90.4% MHC class I and 97.8% MHC class II concordance (Table 2).
Table 2.
Summary of discordant results for each MHC genotyping method by animal.
| MHC class I | MHC class II | |||||||
|---|---|---|---|---|---|---|---|---|
| Animal ID | Alleles | Discordance | Discordance | Discordance | Alleles | Discordance | Discordance | Discordance |
| r05029 | 28 | 2 | 1 | 10 | ||||
| r17099 | 25 | 3 | 1 | 12 | ||||
| r07010 | 26 | 3 | 1 | 1 | 12 | |||
| r17041 | 31 | 3 | 1 | 10 | ||||
| r17061 | 29 | 1 | 2 | 12 | 1 | |||
| J8R | 29 | 4 | 1 | 12 | 2 | |||
| J01 | 30 | 3 | 1 | 1 | 11 | 1 | ||
| ZC08 | 24 | 2 | 1 | 12 | ||||
| DGKG | 28 | 4 | 1 | 13 | ||||
| CF18 | 28 | 3 | 12 | |||||
| DE1AA | 30 | 5 | 1 | 13 | ||||
| DEG8 | 27 | 3 | 1 | 13 | ||||
| DEXX | 24 | 3 | 1 | 1 | 14 | |||
| DF24 | 32 | 4 | 1 | 12 | ||||
| DF64 | 30 | 3 | 1 | 1 | 13 | |||
| DF6T | 31 | 2 | 1 | 10 | ||||
| DFET | 30 | 3 | 1 | 1 | 13 | 1 | 1 | |
| DFV0 | 28 | 2 | 6 | |||||
| DJ94 | 34 | 3 | 2 | 1 | 12 | |||
| HIH | 30 | 3 | 10 | |||||
| DFJT | 28 | 2 | 3 | 11 | ||||
| HLJ | 29 | 2 | 1 | 1 | 11 | |||
| 37360 | 29 | 2 | 1 | 1 | 13 | 1 | ||
| 0L7 | 20 | 2 | 14 | |||||
| 0R1 | 17 | 1 | 12 | |||||
| 0R8 | 25 | 2 | 13 | 1 | ||||
| 0TI | 25 | 2 | 1 | 12 | ||||
| Total | 747 | 72 | 20 | 13 | 318 | 7 | 0 | 1 |
| Discordance (%) | 9.6 | 2.7 | 1.7 | 2.2 | 0.0 | 0.3 | ||
| Concordance (%) | 90.4 | 97.3 | 98.3 | 97.8 | 100.0 | 99.7 | ||
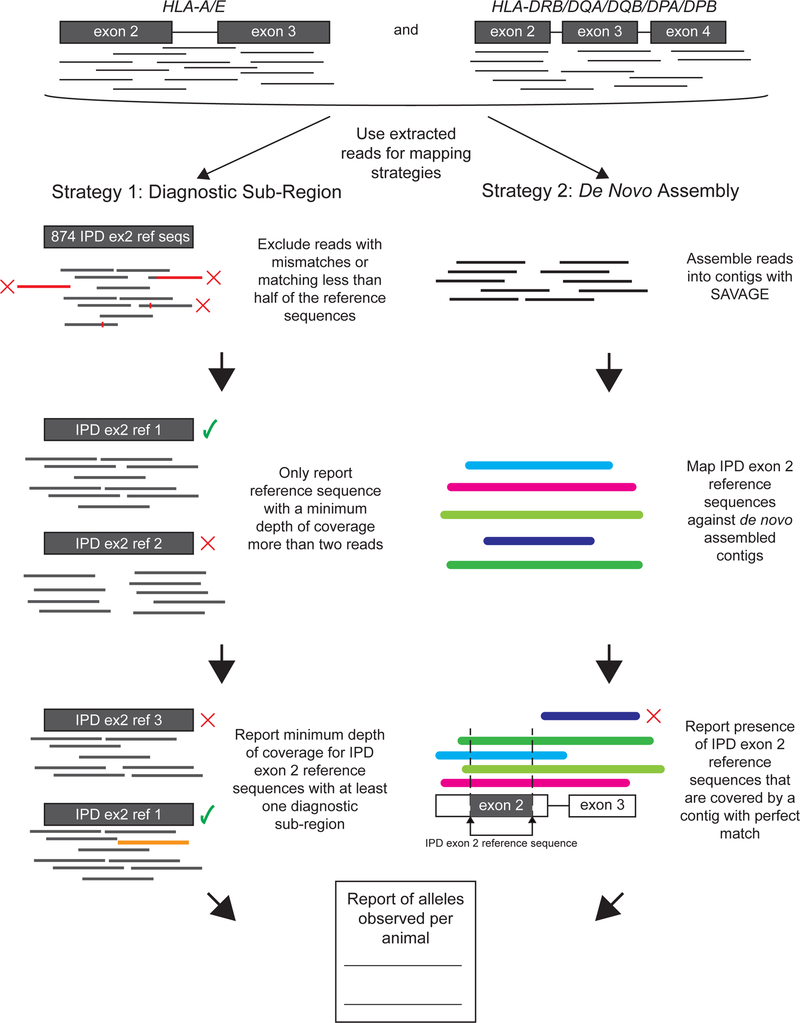
Two separate strategies were used to derive MHC genotypes from target-capture data using data from macaque exome sequencing. These strategies were outlined in Figure 3, and can be described as follows. The DSR analysis was a straightforward extension of the methodology used for deriving genotypes from MiSeq amplicons. MHC sequence reads were extracted from the total exome dataset. A simplified workflow that mapped those MHC reads to reference sequences, and then assigned the genotypes based on the presence of exome reads overlapping each position of the reference sequence, was problematic. This simplified workflow was extremely vulnerable to false positive genotypes, and the similarities among different MHC sequences often enabled those reads to map to multiple different alleles (Wiseman et al. 2013). Two or more reads each could partially match a portion of a sequence in the IPD exon 2 database, which could complement each other to provide support for an allele that was not biologically relevant. As discussed in the Methods, the DSR encompassed polymorphisms that discriminated among closely related alleles by requiring at least one mapped read to unambiguously map to the corresponding allele within the IPD exon 2 sequences. This strategy can identify specific polymorphisms of interest among the IPD exon 2 sequences to produce genotypes for MHC class I and MHC class II (Figures 1, 2 and Supplementary Figures 1 and 2). For the 27 animals evaluated in this study, DSR was 97.3% and 100% concordant with expected MHC class I and class II genotypes, respectively (Table 2).
Figure 3.
Description of strategies employed for MHC genotyping from macaque exome sequences. Exome sequencing reads are extracted by mapping to HLA-A/E exons 2–3 and HLA-DRB/DQA/DQB/DPA/DPB exons 2–4. These extracted sequences are then used for both analysis strategies. Strategy 1 maps the extracted reads to IPD exon 2 reference sequences. Only reads with perfect matches that extended off the end of contigs for no more than half of the length of the mapped read segment were retained. Next, each alignment of mapped sequences is checked to exclude those with a coverage that is less than 2 reads. The mapped alignments with this coverage are used to form a diagnostic sub-read, which is used to match alignments to the reference sequences. The minimum depth of coverage of the mapped alignments was reported in the output. Strategy 2 conducts a de novo assembly of the extracted reads using the computational tool SAVAGE. Then, the assembled contigs are mapped to IPD exon 2 reference sequences. To determine correct mapping, any IPD exon 2 reference sequences that fully cover and perfectly match a contig are reported in the output.
The DSR strategy did not consider sequences that were not among the IPD exon 2 sequences, and this lack of consideration contributes to overcalled alleles. The apparent Mamu-A1*059:01 allele in dam r05029 was erroneously derived from reads that were from Mamu-A1*004g1 and an unknown allele that was not among the sequences in the IPD exon 2 database. As a result of this unknown sequence not being among the known IPD exon 2 sequences, reads unambiguously mapped to Mamu-A1*059:01, which caused this allele to be overcalled. This type of overcalling will be mitigated with the discovery of additional allelic variants and their inclusion in future iterations of the IPD database.
The second approach for determining genotypes performed de novo assembly on MHC class I and class II reads from each sample. The resulting assembled contigs were then mapped against IPD exon 2 reference sequences to define perfectly matching contigs. Because most assemblers were not tuned for the challenge of assembling large numbers of contigs that differ from one another by as little as 1 bp, we relied on an assembler, SAVAGE, originally designed to reconstruct viral sequencing haplotypes. As shown in Figures 1, 2 and Supplementary Figure 1, contigs produced by SAVAGE matched the expected genotypes. Unlike the DSR method, SAVAGE assembled sequence contigs that may be useful for downstream analyses such as higher resolution MHC genotyping. False positives and false negatives among closely-related variants were mitigated by the inclusion of the filtering steps described under Strategy 2. Across all 27 samples, the SAVAGE contigs are 98.3% and 99.7% accurate with respect to the expected MHC genotypes (Table 2).
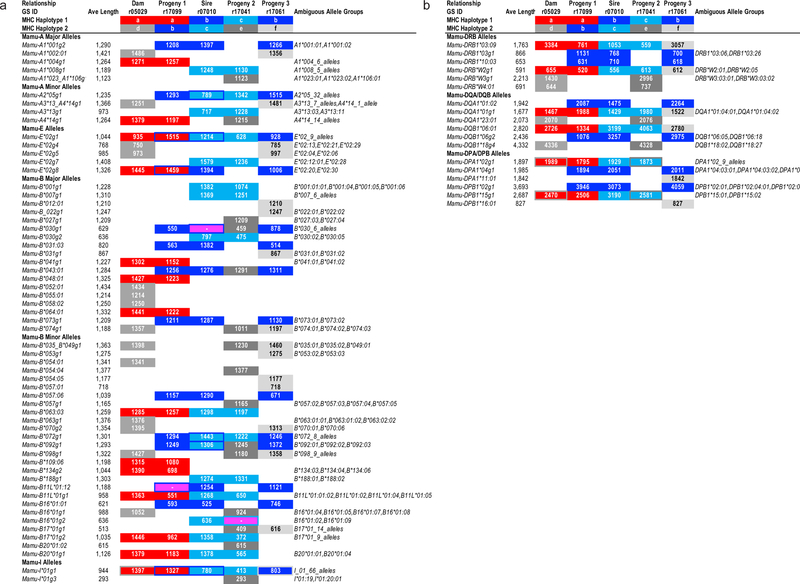
While this analysis focused on genotyping using the same exon 2 reference sequences that are commonly utilized for MiSeq amplicon analyses, the SAVAGE contigs are frequently much longer than these reference sequences (Figure 4). These extended contigs frequently contain exons 2 through 4, plus the intervening introns, and could be used to provide higher resolution genotyping than is possible using exon 2 sequence alone. Moreover, contigs that contain complete sequences for exons 2–3 of MHC class I and exon 2 of MHC class II alleles meet the minimum criteria for obtaining formal allele nomenclature for non-human primates from the IPD-MHC (Robinson et al. 2013).
Figure 4.
Lengths of MHC sequence contigs assembled by the SAVAGE method for a representative breeding group of rhesus macaques. Values in the body of this figure indicate contig lengths generated for these MHC sequences in each animal. (A) MHC class I genotyping results are illustrated for the five animals in this breeding group. Three false negative allele calls for SAVAGE versus the expected genotypes for these animals are highlighted in magenta, e.g., Mamu-B*030g1 in sire r07010. Sequences highlighted in red are associated with the maternal MHC Haplotype a that was inherited by progeny 1 from its dam. Sequences in progeny 1 and progeny 3 for their MHC Haplotype b that was inherited from sire r07010 are highlighted in dark blue while the light blue sequences represent the alternate paternal Haplotype c that was inherited by Progeny 2. Three unique extended MHC Haplotypes in this breeding group are indicated with shades of grey (Haplotypes d - f). MHC allele groups that are shared by both parental haplotypes are indicated by colored borders around filled cells. (B) MHC class II genotyping results are illustrated for this same breeding group.
Discussion
Here we describe MES genotyping of Indian rhesus macaques. In our estimation, this method will supersede MiSeq PCR amplicon genotyping as the most widely used macaque MHC genotyping assay in the future. This methodology has several compelling advantages. Most importantly, exome sequencing dramatically improves the overall quantity and quality of genomic information obtained from each sample. The same datasets used for MHC analyses may be used to evaluate protein-coding genetic variation throughout the genome. The loss of start and stop codons exome-wide can be obtained from the same datasets by modifying the workflows in silico, as opposed to requiring the sequencing of multiple new sets of target genes (Yang et al. 2013). Exome-wide datasets offer the promise of retrospectively identifying candidate DNA sequence variants that may be responsible for unexpected experimental outcomes in studies with macaques and other nonhuman primates. The availability of exome-wide sequences in conjunction with MHC genotypes may also increase the rigor of prospective macaque experiments by enabling more sophisticated balancing of experimental groups, and exclusion of animals whose genetics are likely to strongly bias experimental results (Loffredo et al. 2007; Reynolds et al. 2011; Haus et al. 2014).
These same exome datasets also offer the potential to improve the quality of MHC genotyping. Full-length long-read MHC transcript sequencing offers the highest resolution, but this technology can be labor intensive and difficult to scale (Karl et al. 2017; Semler et al. 2018). The MiSeq exon 2 amplicon approach is limited in its allelic resolution, but it is the current standard deep sequencing approach for high-throughput MHC genotyping in macaques. In this report, we compare two novel strategies for MHC genotyping from MES datasets to this standard MiSeq amplicon genotyping method. The results that we obtained with all three approaches show strong concordance with expected MHC genotypes. As illustrated in Figure 4a, MHC class I genomic contigs with an average length of approximately 1.1 kb can be assembled from sequence reads that were initially extracted from the whole exome datasets.
Following the initial enrichment step, MHC class II genomic contigs averaging approximately 1.9 kb in length could be assembled from whole exome sequence reads that mapped to HLA-DRB1, -DRB3, -DRB4, -DRB5, -DQA1, -DQB1, -DPA1 and -DPB1 reference sequences, which contained exons 2 – 4, and the pairs of introns (Figure 4b). The addition of HLA spike-in probes containing both exon and intron sequences for the target capture step, and the introduction of 151 bp paired end reads for the Illumina NovaSeq platform greatly facilitated our ability to assemble these extended MHC genomic contigs. Additional technological advances, including even longer sequence reads and more efficient assembly algorithms, will undoubtedly increase genomic contig lengths as well as MHC allelic resolution in future studies.
Both MHC genotyping approaches described here depend upon mapping exome sequence reads against a reference database of MHC class I and class II allele sequences. Currently, the IPD database mostly is restricted to coding regions for macaque MHC sequences, and many IPD entries are only partial transcript sequences that lack complete coding regions. It is also very challenging to correctly phase short Illumina sequence reads. The reads map to different exons of a specific allelic variant, and are separated by intronic sequences that span hundreds to thousands of base pairs in genomic DNA. Our results with the SAVAGE workflow (Figure 4) demonstrate that exome sequence reads that have been enriched with the enhanced MHC probe design described here can be assembled into genomic contigs that span multiple exons and introns. These contigs, therefore, could also be used to improve MHC reference databases, though this requires a major effort with more animals, and is beyond the scope of this manuscript.
The current cost of genotyping is relatively high compared to amplicon deep sequencing, primarily due to two expenses. First, the amount of sequence data needed for a single sample can be high. Compared to conventional MHC genotyping, where 192 macaques’ data can be collected on a single instrument run, exome data acquisition is much more expensive. On an Illumina NovaSeq instrument, which has a much higher run cost than the MiSeq, only 70 exome samples can be sequenced simultaneously per lane of a S4 flow cell. However, this cost of sequencing is rapidly decreasing, and as of early 2019, commercial providers have advertised sequencing for $9 USD per Gb of whole genome sequence data1. Second, a major expense in MHC genotyping is the production, validation, and use of target-capture arrays, as well as the development of in silico data analysis workflows. The approaches described here, in particular MHC genotyping from SAVAGE contigs produced by de novo assembly of exome reads, is flexible and should be adaptable as sequencing approaches evolve and improve.
A major goal for future studies will be to attempt to extend these contigs to encompass full length genomic MHC sequences using SAVAGE or other assembly software tools. The relatively compact genomic structure and consistent length of MHC class I genes increase the attainability of this goal. Establishment of comprehensive macaque MHC allele databases of extended genomic sequences will greatly facilitate mapping of exome sequence reads since they will be contiguous with the reference sequences instead of being interrupted by intervening sequences between each exon that are not included in current non-human primate IPD-MHC databases (Maccari et al. 2017).
These results demonstrate that MHC genotypes can be obtained by analyzing genomic DNA selectively enriched for MHC and protein-coding gene sequences. This represents an important advance for characterizing MHC genetics in macaques, and this suggests that analyses of whole exome and whole genome data will become the predominant method for studying macaque genetics in the coming decade.
Supplementary Material
Supplementary Figure 1. Comparison of MHC class I results from MiSeq PCR amplicon versus whole exome genotyping assays with the DSR and SAVAGE strategies for all 27 animals. Results for each of the three method are provided side-by-side in the columns for each macaque. Each row indicates the detection of a specific MHC class I allele or lineage group of closely related sequences that are ambiguous because they are identical over the IPD exon 2 database sequence. Values in the body of this figure indicate the number of sequence reads supporting each allele call for the MiSeq and DSR methods while alleles supported by a SAVAGE contig are reported with a “1”. Discrepancies between the MiSeq, DSR, or SAVAGE methods are highlighted by magenta-filled cells with borders. Abbreviated Mamu haplotype designations (Karl et al. 2013) for each individual are summarized below the Animal IDs. The Mamu class I and Mamu-DRB alleles that have been associated with each of these abbreviated haplotypes are listed in Supplementary Figure 2. Color coded cells indicate MHC sequences that are inferred to be associated with extended MHC haplotypes based on segregation in directly related individuals (first five animals) or by shared allele combinations in animals whose pedigree relationships are unknown (final 22 individuals). For example, the twenty MHC sequences (highlighted in red), associated with an extended haplotype that is shared by dam r05029 and her progeny r17099, can be summarized by the following string of abbreviated Mamu haplotype designations: Mamu-A004/B048/DR04a/DQA01g1/DQB06:01/DPA02g1/DPB15g.
Supplementary Figure 2. Abbreviated Mamu-A, -B and -DRB haplotype definitions for the animals evaluated in this study. The Mamu-A, -B and -DRB allele lineages that have been associated with each of the 43 abbreviated haplotypes observed in this study are listed here. These abbreviated haplotype designations were originally assigned based on the presence of a major “diagnostic” sequence that was typically the most abundant transcript associated with a haplotype in deep sequencing assays with cDNA templates (Karl et al. 2013). For example, the Mamu-A001 haplotype generally encodes a Mamu-A1*001g2 major transcript along with a minor Mamu-A2*05g1 transcript.
Supplementary Figure 3. Example of chimeric PCR amplification. The top panel shows the alignment of three IPD exon 2 reference sequences: Mamu-B20*01g1 (top, green), Mamu-B*007:07 (middle, blue), Mamu-B*007g1 (bottom, purple), and the location of the forward and reverse primers, SBT195F and SBT195R (top arrows, gray). The yellow boxes on the sequences represent SNPs between Mamu-B20*01g1 and Mamu-B*007:07 and between Mamu-B*007g1 and Mamu-B*007:07. The positions are relative to the end of the forward primer, with SNPs at position 7 and position 96 labeled accordingly. In this example, both Mamu-B20*01g1 and Mamu-B*007g1 are evident in the sample. During PCR, Mamu-B20*01g1 aborts amplification between positions 7 and 96. This aborted amplification becomes a “primer” in the next cycle and continues to amplify using Mamu-B*007g1 as a template. The resulting chimeric sequence is identical to Mamu-B*007:07 (Fichot et. al. 2013).
Supplementary Table 1. Fraction of total exome sequence reads corresponding to MHC class I and class II genes after target capture with the VCRome2.1 probe design alone. This exome sequence dataset was described previously by Ericsen and coworkers (Ericsen et al. 2014).
Acknowledgements
We gratefully acknowledge Michele Di Mascio and his group at the National Institute of Allergy and Infectious Diseases of the National Institutes of Health for providing rhesus macaque samples used in this study. We also gratefully thank Brian Bushnell for assistance with the BBTools software, and the WNPRC for providing samples from five related macaques.
This research was supported by contract HHSN272201600007C from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. This work was also supported in part by the Office of Research Infrastructure Programs/OD (P51OD011106) awarded to the Wisconsin National Primate Research Center at the University of Wisconsin-Madison. This research was also supported in part by grant R24-OD011173 from the National Institutes of Health. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grants RR15459–01 and RR020141–01.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Comment via Twitter 03/18/2019 @albertvilella https://twitter.com/AlbertVilella/status/1107524501645000705
References
- Animal Welfare Act. ‘The Animal Welfare Act - Public Law 89–544 Act of August 24, 1966. 1966. https://www.nal.usda.gov/awic/animal-welfare-act-public-law-89-544-act-august-24-1966. Accessed 05/01/2019
- Baaijens JA, Aabidine AZE, Rivals E, and Schonhuth A. 2017. De novo assembly of viral quasispecies using overlap graphs. Genome Res 27: 835–848. https://www.ncbi.nlm.nih.gov/pubmed/28396522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimber BN, Ramakrishnan R, Cervera-Juanes R, Madhira R, Peterson SM, Norgren RBJ, and Ferguson B. 2017. Whole genome sequencing predicts novel human disease models in rhesus macaques. Genomics 109: 214–220. https://www.ncbi.nlm.nih.gov/pubmed/28438488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, and Wiley DC. 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364: 33–39. https://www.ncbi.nlm.nih.gov/pubmed/8316295 [DOI] [PubMed] [Google Scholar]
- Bushnell B, Rood J, and Singer E. 2017. BBMerge - Accurate paired shotgun read merging via overlap. PLoS One 12: e0185056 https://www.ncbi.nlm.nih.gov/pubmed/29073143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Wu J, Wang Y, Jiang H, Zhang T, Liu X, Xu Y, Liang D, Gao P, Sun Y, Gifford B, D’Ascenzo M, Liu X, Tellier LC, Yang F, Tong X, Chen D, Zheng J, Li W, Richmond T, Xu X, Wang J, and Li Y. 2013. An integrated tool to study MHC region: accurate SNV detection and HLA genes typing in human MHC region using targeted high-throughput sequencing. PLoS One 8: e69388 https://www.ncbi.nlm.nih.gov/pubmed/23894464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Chen R, and Snyder M. 2013. Exome sequencing by targeted enrichment. Curr Protoc Mol Biol Chapter 7: Unit7.12 https://www.ncbi.nlm.nih.gov/pubmed/23547016 [DOI] [PubMed] [Google Scholar]
- Cornish AS, Gibbs RM, and Norgren RBJ. 2016. Exome screening to identify loss-of-function mutations in the rhesus macaque for development of preclinical models of human disease. BMC Genomics 17: 170 https://www.ncbi.nlm.nih.gov/pubmed/26935327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, and Geraghty DE. 2004. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 14: 1501–1515. https://www.ncbi.nlm.nih.gov/pubmed/15289473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Manuel M, Shiina T, Suzuki S, Dereuddre-Bosquet N, Garchon HJ, Tanaka M, Congy-Jolivet N, Aarnink A, Le Grand R, Marques-Bonet T, and Blancher A. 2018. Whole genome sequencing in the search for genes associated with the control of SIV infection in the Mauritian macaque model. Sci Rep 8: 7131 https://www.ncbi.nlm.nih.gov/pubmed/29739964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. https://www.ncbi.nlm.nih.gov/pubmed/20709691 [DOI] [PubMed] [Google Scholar]
- Ennis PD, Zemmour J, Salter RD, and Parham P. 1990. Rapid cloning of HLA-A,B cDNA by using the polymerase chain reaction: frequency and nature of errors produced in amplification. Proc Natl Acad Sci U S A 87: 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsen AJ, Starrett GJ, Greene JM, Lauck M, Raveendran M, Deiros DR, Mohns MS, Vince N, Cain BT, Pham NH, Weinfurter JT, Bailey AL, Budde ML, Wiseman RW, Gibbs R, Muzny D, Friedrich TC, Rogers J, and O’Connor DH. 2014. Whole genome sequencing of SIV-infected macaques identifies candidate loci that may contribute to host control of virus replication. Genome Biol 15: 478 https://www.ncbi.nlm.nih.gov/pubmed/25418588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichot Erin & Norman Robert. (2013). Microbial phylogenetic profiling with the Pacific Biosciences sequencing platform. Microbiome. 1 10.1186/2049-2618-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia KC, and Adams EJ. 2005. How the T cell receptor sees antigen--a structural view. Cell 122: 333–336. https://www.ncbi.nlm.nih.gov/pubmed/16096054 [DOI] [PubMed] [Google Scholar]
- Haus T, Ferguson B, Rogers J, Doxiadis G, Certa U, Rose NJ, Teepe R, Weinbauer GF, and Roos C. 2014. Genome typing of nonhuman primate models: implications for biomedical research. Trends Genet 30: 482–487. https://www.ncbi.nlm.nih.gov/pubmed/24954183 [DOI] [PubMed] [Google Scholar]
- Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, Hart E, Howe K, Jackson DK, Palmer S, Roberts AN, Sims S, Stewart CA, Traherne JA, Trevanion S, Wilming L, Rogers J, de Jong PJ, Elliott JF, Sawcer S, Todd JA, Trowsdale J, and Beck S. 2008. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 60: 1–18. https://www.ncbi.nlm.nih.gov/pubmed/18193213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janaka SK, Tavakoli-Tameh A, Neidermyer WJJ, Serra-Moreno R, Hoxie JA, Desrosiers RC, Johnson RP, Lifson JD, Wolinsky SM, and Evans DT. 2018. Polymorphisms in Rhesus Macaque Tetherin Are Associated with Differences in Acute Viremia in Simian Immunodeficiency Virus Deltanef-Infected Animals. J Virol 92 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6206476/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizu M, Borchardt GJ, Glidden CE, Fisk DL, Loffredo JT, Watkins DI, and Rehrauer WM. 2007. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics 59: 693–703. https://www.ncbi.nlm.nih.gov/pubmed/17641886 [DOI] [PubMed] [Google Scholar]
- Karl JA, Bohn PS, Wiseman RW, Nimityongskul FA, Lank SM, Starrett GJ, and O’Connor DH. 2013. Major histocompatibility complex class I haplotype diversity in Chinese rhesus macaques. G3 (Bethesda) 3: 1195–1201. https://www.ncbi.nlm.nih.gov/pubmed/23696100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl JA, Graham ME, Wiseman RW, Heimbruch KE, Gieger SM, Doxiadis GG, Bontrop RE, and O’Connor DH. 2017. Major histocompatibility complex haplotyping and long-amplicon allele discovery in cynomolgus macaques from Chinese breeding facilities. Immunogenetics 69: 211–229. https://www.ncbi.nlm.nih.gov/pubmed/28078358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl JA, Heimbruch KE, Vriezen CE, Mironczuk CJ, Dudley DM, Wiseman RW, and O’Connor DH. 2014. Survey of major histocompatibility complex class II diversity in pig-tailed macaques. Immunogenetics 66: 613–623. https://www.ncbi.nlm.nih.gov/pubmed/25129472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishikawa T, Momozawa Y, Ozeki T, Mushiroda T, Inohara H, Kamatani Y, Kubo M, and Okada Y. 2019. Empirical evaluation of variant calling accuracy using ultra-deep whole-genome sequencing data. Sci Rep 9: 1784 https://www.ncbi.nlm.nih.gov/pubmed/30741997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J, and Rahmann S. 2012. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. https://www.ncbi.nlm.nih.gov/pubmed/22908215 [DOI] [PubMed] [Google Scholar]
- Krupp A, McCarthy KR, Ooms M, Letko M, Morgan JS, Simon V, and Johnson WE. 2013. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog 9: e1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, and Watkins DI. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol 81: 8827–8832. https://www.ncbi.nlm.nih.gov/pubmed/17537848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, and Sette A. 2009. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol 182: 7763–7775. https://www.ncbi.nlm.nih.gov/pubmed/19494300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccari G, Robinson J, Ballingall K, Guethlein LA, Grimholt U, Kaufman J, Ho CS, de Groot NG, Flicek P, Bontrop RE, Hammond JA, and Marsh SG. 2017. IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res 45: D860–D864. https://www.ncbi.nlm.nih.gov/pubmed/27899604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissen M, Malissen B, and Jordan BR. 1982. Exon/intron organization and complete nucleotide sequence of an HLA gene. Proc Natl Acad Sci U S A 79: 893–897. https://www.ncbi.nlm.nih.gov/pubmed/6461010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, Allison DB, and Watkins DI. 2003. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol 77: 2736–2740. https://www.ncbi.nlm.nih.gov/pubmed/12552014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhl T, Krawczak M, Ten Haaft P, Hunsmann G, and Sauermann U. 2002. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J Immunol 169: 3438–3446. https://www.ncbi.nlm.nih.gov/pubmed/12218167 [DOI] [PubMed] [Google Scholar]
- Nomura T, Yamamoto H, Shiino T, Takahashi N, Nakane T, Iwamoto N, Ishii H, Tsukamoto T, Kawada M, Matsuoka S, Takeda A, Terahara K, Tsunetsugu-Yokota Y, Iwata-Yoshikawa N, Hasegawa H, Sata T, Naruse TK, Kimura A, and Matano T. 2012. Association of major histocompatibility complex class I haplotypes with disease progression after simian immunodeficiency virus challenge in burmese rhesus macaques. J Virol 86: 6481–6490. https://www.ncbi.nlm.nih.gov/pubmed/22491464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, van der Wiel MK, de Groot N, de Vos-Rouweler AJ, de Groot NG, Doxiadis GG, Wiseman RW, O’Connor DH, and Bontrop RE. 2017. The orthologs of HLA-DQ and -DP genes display abundant levels of variability in macaque species. Immunogenetics 69: 87–99. https://www.ncbi.nlm.nih.gov/pubmed/27771735 [DOI] [PubMed] [Google Scholar]
- Posey JE, Rosenfeld JA, James RA, Bainbridge M, Niu Z, Wang X, Dhar S, Wiszniewski W, Akdemir ZH, Gambin T, Xia F, Person RE, Walkiewicz M, Shaw CA, Sutton VR, Beaudet AL, Muzny D, Eng CM, Yang Y, Gibbs RA, Lupski JR, Boerwinkle E, and Plon SE. 2016. Molecular diagnostic experience of whole-exome sequencing in adult patients. Genet Med 18: 678–685. https://www.ncbi.nlm.nih.gov/pubmed/26633545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prall TM, Graham ME, Karl JA, Wiseman RW, Ericsen AJ, Raveendran M, Alan Harris R, Muzny DM, Gibbs RA, Rogers J, and O’Connor DH. 2017. Improved full-length killer cell immunoglobulin-like receptor transcript discovery in Mauritian cynomolgus macaques. Immunogenetics 69: 325–339. https://www.ncbi.nlm.nih.gov/pubmed/28343239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds MR, Sacha JB, Weiler AM, Borchardt GJ, Glidden CE, Sheppard NC, Norante FA, Castrovinci PA, Harris JJ, Robertson HT, Friedrich TC, McDermott AB, Wilson NA, Allison DB, Koff WC, Johnson WE, and Watkins DI. 2011. The TRIM5{alpha} genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J Virol 85: 9637–9640. https://www.ncbi.nlm.nih.gov/pubmed/21734037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Halliwell JA, McWilliam H, Lopez R, and Marsh SG. 2013. IPD--the Immuno Polymorphism Database. Nucleic Acids Res 41: D1234–40. https://www.ncbi.nlm.nih.gov/pubmed/23180793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler MR, Wiseman RW, Karl JA, Graham ME, Gieger SM, and O’Connor DH. 2018. Novel full-length major histocompatibility complex class I allele discovery and haplotype definition in pig-tailed macaques. Immunogenetics 70: 381–399. https://www.ncbi.nlm.nih.gov/pubmed/29134258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T, Blancher A, Inoko H, and Kulski JK. 2017. Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology 150: 127–138. https://www.ncbi.nlm.nih.gov/pubmed/27395034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver ZA, and Watkins DI. 2017. The role of MHC class I gene products in SIV infection of macaques. Immunogenetics 69: 511–519. https://www.ncbi.nlm.nih.gov/pubmed/28695289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender EJ 2011. Expanding whole exome resequencing into non-human primates. Genome Biol 12: R87 https://www.ncbi.nlm.nih.gov/pubmed/21917143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wintzingerode F, Gobel UB, and Stackebrandt E. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21: 213–229. [DOI] [PubMed] [Google Scholar]
- Weatherall D 2006. The use of non-human primates in research. 147 https://mrc.ukri.org/documents/pdf/the-use-of-non-human-primates-in-research/. Accessed 04/30/2019 [Google Scholar]
- Williams TM 2001. Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J Mol Diagn 3: 98–104. https://www.ncbi.nlm.nih.gov/pubmed/11486048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres EJ, Wright C, Harkins T, and O’Connor DH. 2009. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med 15: 1322–1326. https://www.ncbi.nlm.nih.gov/pubmed/19820716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bohn PS, Nimityongskul FA, Starrett GJ, and O’Connor DH. 2013. Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J 54: 196–210. https://www.ncbi.nlm.nih.gov/pubmed/24174442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig M, Anmarkrud JA, Kassens JC, Koch S, Forster M, Ellinghaus E, Hov JR, Sauer S, Schimmler M, Ziemann M, Gorg S, Jacob F, Karlsen TH, and Franke A. 2015. Development of a high-resolution NGS-based HLA-typing and analysis pipeline. Nucleic Acids Res 43: e70 https://www.ncbi.nlm.nih.gov/pubmed/25753671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Yeo ZX, Wong M, Piper J, Long T, Kirkness EF, Biggs WH, Bloom K, Spellman S, Vierra-Green C, Brady C, Scheuermann RH, Telenti A, Howard S, Brewerton S, Turpaz Y, and Venter JC. 2017. Fast and accurate HLA typing from short-read next-generation sequence data with xHLA. Proc Natl Acad Sci U S A 114: 8059–8064. https://www.ncbi.nlm.nih.gov/pubmed/28674023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Raveendran M, Harris RA, Fawcett GL, Liu X, White S, Dahdouli M, Rio Deiros D, Below JE, Salerno W, Cox L, Fan G, Ferguson B, Horvath J, Johnson Z, Kanthaswamy S, Kubisch HM, Liu D, Platt M, Smith DG, Sun B, Vallender EJ, Wang F, Wiseman RW, Chen R, Muzny DM, Gibbs RA, Yu F, and Rogers J. 2016. The population genomics of rhesus macaques (Macaca mulatta) based on whole-genome sequences. Genome Res 26: 1651–1662. https://www.ncbi.nlm.nih.gov/pubmed/27934697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, and Eng CM. 2013. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369: 1502–1511. https://www.ncbi.nlm.nih.gov/pubmed/24088041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, Veeraraghavan N, Hawes A, Chiang T, Leduc M, Beuten J, Zhang J, He W, Scull J, Willis A, Landsverk M, Craigen WJ, Bekheirnia MR, Stray-Pedersen A, Liu P, Wen S, Alcaraz W, Cui H, Walkiewicz M, Reid J, Bainbridge M, Patel A, Boerwinkle E, Beaudet AL, Lupski JR, Plon SE, Gibbs RA, and Eng CM. 2014. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA 312: 1870–1879. https://www.ncbi.nlm.nih.gov/pubmed/25326635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, Tharp GK, Marcais G, Roberts M, Ferguson B, Fox HS, Treangen T, Salzberg SL, Yorke JA, and Norgren RBJ. 2014. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct 9: 20 https://www.ncbi.nlm.nih.gov/pubmed/25319552 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Comparison of MHC class I results from MiSeq PCR amplicon versus whole exome genotyping assays with the DSR and SAVAGE strategies for all 27 animals. Results for each of the three method are provided side-by-side in the columns for each macaque. Each row indicates the detection of a specific MHC class I allele or lineage group of closely related sequences that are ambiguous because they are identical over the IPD exon 2 database sequence. Values in the body of this figure indicate the number of sequence reads supporting each allele call for the MiSeq and DSR methods while alleles supported by a SAVAGE contig are reported with a “1”. Discrepancies between the MiSeq, DSR, or SAVAGE methods are highlighted by magenta-filled cells with borders. Abbreviated Mamu haplotype designations (Karl et al. 2013) for each individual are summarized below the Animal IDs. The Mamu class I and Mamu-DRB alleles that have been associated with each of these abbreviated haplotypes are listed in Supplementary Figure 2. Color coded cells indicate MHC sequences that are inferred to be associated with extended MHC haplotypes based on segregation in directly related individuals (first five animals) or by shared allele combinations in animals whose pedigree relationships are unknown (final 22 individuals). For example, the twenty MHC sequences (highlighted in red), associated with an extended haplotype that is shared by dam r05029 and her progeny r17099, can be summarized by the following string of abbreviated Mamu haplotype designations: Mamu-A004/B048/DR04a/DQA01g1/DQB06:01/DPA02g1/DPB15g.
Supplementary Figure 2. Abbreviated Mamu-A, -B and -DRB haplotype definitions for the animals evaluated in this study. The Mamu-A, -B and -DRB allele lineages that have been associated with each of the 43 abbreviated haplotypes observed in this study are listed here. These abbreviated haplotype designations were originally assigned based on the presence of a major “diagnostic” sequence that was typically the most abundant transcript associated with a haplotype in deep sequencing assays with cDNA templates (Karl et al. 2013). For example, the Mamu-A001 haplotype generally encodes a Mamu-A1*001g2 major transcript along with a minor Mamu-A2*05g1 transcript.
Supplementary Figure 3. Example of chimeric PCR amplification. The top panel shows the alignment of three IPD exon 2 reference sequences: Mamu-B20*01g1 (top, green), Mamu-B*007:07 (middle, blue), Mamu-B*007g1 (bottom, purple), and the location of the forward and reverse primers, SBT195F and SBT195R (top arrows, gray). The yellow boxes on the sequences represent SNPs between Mamu-B20*01g1 and Mamu-B*007:07 and between Mamu-B*007g1 and Mamu-B*007:07. The positions are relative to the end of the forward primer, with SNPs at position 7 and position 96 labeled accordingly. In this example, both Mamu-B20*01g1 and Mamu-B*007g1 are evident in the sample. During PCR, Mamu-B20*01g1 aborts amplification between positions 7 and 96. This aborted amplification becomes a “primer” in the next cycle and continues to amplify using Mamu-B*007g1 as a template. The resulting chimeric sequence is identical to Mamu-B*007:07 (Fichot et. al. 2013).
Supplementary Table 1. Fraction of total exome sequence reads corresponding to MHC class I and class II genes after target capture with the VCRome2.1 probe design alone. This exome sequence dataset was described previously by Ericsen and coworkers (Ericsen et al. 2014).