Abstract
Metastasis remains the greatest challenge in the clinical management of cancer. Cell motility is a fundamental and ancient cellular behaviour that contributes to metastasis and is conserved in simple organisms. In this Review, we evaluate insights relevant to human cancer that are derived from the study of cell motility in non-mammalian model organisms. Dictyostelium discoideum, Caenorhabditis elegans, Drosophila melanogaster and Danio rerio permit direct observation of cells moving in complex native environments and lend themselves to large-scale genetic and pharmacological screening. We highlight insights derived from each of these organisms, including the detailed signalling network that governs chemotaxis towards chemokines; a novel mechanism of basement membrane invasion; the positive role of E-cadherin in collective direction-sensing; the identification and optimization of kinase inhibitors for metastatic thyroid cancer on the basis of work in flies; and the value of zebrafish for live imaging, especially of vascular remodelling and interactions between tumour cells and host tissues. While the motility of tumour cells and certain host cells promotes metastatic spread, the motility of tumour-reactive T cells likely increases their antitumour effects. Therefore, it is important to elucidate the mechanisms underlying all types of cell motility, with the ultimate goal of identifying combination therapies that will increase the motility of beneficial cells and block the spread of harmful cells.
In our quest to understand human cancer, why should we study amoebae, worms, flies and fish, and what can we learn from these simple organisms that is relevant to human disease? Even mammals such as mice differ considerably from people in myriad ways — some obvious (such as size and lifespan) and others less so (nocturnal versus diurnal)1. Thus, what can we learn of value from even simpler organisms?
The fact is that the more fundamental the biology, the better conserved it is. The genetic code is conserved from bacteria to humans, the basic architecture of the eukaryotic cell is similar across organisms and fundamental cell behaviours, such as motility, evolved early. Key molecular pathways, too, have been maintained over hundreds of millions of years of evolution.
It is not only the conserved features of model organisms that illuminate important biological mechanisms. Sometimes it is an unusual property of a particular organism that is most revealing. For example, genetic studies in the nematode Caenorhabditis elegans, an organism in which precisely the same 131 cells die during the development of every single animal, contributed key insights into the molecular mechanisms of apoptosis2. Early embryonic development of Drosophila melanogaster is also unusual, yet genetic screens for mutations that disrupt it yielded the discovery of the Hedgehog and WNT pathways3.
Few physicians or scientists would dispute the importance of these discoveries in simple organisms to human cancer. Nevertheless, it is not always obvious whether or how a particular experimental model will be relevant to cancer biology. Sometimes, relevance may become obvious only after years or decades, and all models have limitations. The direct study of human patient samples is also fraught with limitations and caveats, such as the inability to observe the process of metastasis as it unfolds, genetic heterogeneity of the patient population and constraints on sample sizes4. Nevertheless, DNA sequencing of thousands of human patient tumours has led to the identification of 54 oncogenes and 71 tumour suppressor genes that are repeatedly mutated in cancer5. The products of these genes fall into a handful of key molecular pathways that confer a selective growth advantage to both primary tumours and their metastatic colonies. Intriguingly, cancer cells co-opt these pathways from those that drive normal cell fate, proliferation and survival during embryonic and tissue development. We suggest that just as cancer cells exploit normal pathways to gain a growth advantage, they also hijack normal cellular processes and the molecular mechanisms that govern tissue morphogenesis to spread through the body in the process of metastasis (BOX 1). This idea leads to an important question. What are the cellular processes and molecular pathways that control normal morphogenesis in multicellular organisms?
Box 1 |. The elusive metastatic programme.
Many mutations that cause tumour growth and proliferation have been identified; however, metastasis genes have been more elusive225. While some metastasis genes have emerged226–229, our mechanistic understanding remains incomplete225,230–232. Why then has the metastatic programme escaped detection by the approaches that were successful in identifying tumour suppressors and oncogenes?
One answer might be that metastasis genes overlap substantially with those that drive growth and survival, making it impossible to tease out a separate programme. In fact, the signalling cascades that promote cancer growth and cell migration overlap extensively, perhaps because it is adaptive to migrate towards survival factors; this coupling can also eliminate errant cells. However, there is a key difference between the regulation of signalling pathways in chemotaxis compared with the regulation of pathways of growth, survival and proliferation. Chemotaxis requires spatially and temporally dynamic signalling, which is impaired by constitutive activation. For example, constitutively active RAS or PI3K signalling, which are frequently observed in cancer, should immobilize cells. Indeed, PTEN-mutant Dictyostelium discoideum are mostly paralysed233, as are Drosophila melanogaster border cells expressing RasG12V (REF 234). So how do cancer cells with activating mutations in these pathways spread? Perhaps motile immune cells, fibroblasts or cells lacking the mutation within the heterogeneous tumour serve as leader cells and guide or carry otherwise immobilized tumour cells97,219,235,236. Alternatively, different cells may employ distinct pathways for chemotaxis, so a given cell type may tolerate constitutively activating mutations in the PI3K pathway, for example, but not in RAS. In fact, Pten-mutant border cells are able to migrate normally (D.J.M., unpublished observation), as are some RasG12V-expressing tumour cells237. Such a mechanism might help explain why PTEN or RASG12V mutations are common in only particular types of cancer. A third possibility is that tumour cells adapt to higher than normal levels of signalling, as D. discoideum cells do while migrating up a chemoattractant gradient.
High-throughput sequencing has focused attention on mutations as the drivers of phenotypic diversity in tumours, whereas during embryonic development, the entire organism builds itself in the absence of meaningful mutations. Instead, changes in gene expression drive many varieties of cell movement, including epithelial to mesenchymal transition (EMT), mesenchymal to epithelial transition (MET) and colonization of new sites. Another possibility, then, is that epigenetic changes, rather than mutations, primarily drive metastasis.
In this Review, we focus specifically on cell migration and invasion, which are central to morphogenesis and to multiple aspects of tumour metastasis. We highlight examples of directed cell migration and invasion found in the normal development of non-mammalian organisms, the mechanistic insights gained from their study and their implications for understanding metastasis. Increasingly, non-mammalian models such as flies and fish are also used to model the abnormal spread of tumour cells throughout the body and to screen for drugs that block such behaviour. The advantages of model organisms include low cost, the ability to carry out large-scale genetic and pharmacological screens as well as biochemical analyses, and amenability to live, high-resolution fluorescence microscopy of cells interacting within native environments. These advantages have led to mechanistic insights into cell migration, invasion and metastasis that can be, and in some cases have been, tested for their importance to cancer invasion and metastasis in mammalian models and even clinical studies.
Origin and diversity of cell migration
Sensing and initiating directional movement in response to external cues is a fundamental property of biological systems — from individual cells to entire organisms. The ability to move towards food and other favourable environments and away from starvation and generally hostile conditions is essential for organismal survival. The first organisms to evolve these behaviours were unicellular. During the evolution of multicellular organisms, this primitive capacity of cells to move directionally has been adapted for essential processes including embryogenesis, adult tissue homeostasis and immune responses6–8. Thus, the molecular mechanisms of cell motility are fundamentally intertwined with cell survival, a connection that is likely relevant to metastasis. However, cell migration is not one single phenomenon. In disparate physiological contexts, distinct cell types exhibit a variety of morphologies, cell–cell interactions and types of movement. Indeed, cells can move in amoeboid, mesenchymal or epithelial modes, as individuals or in clusters, strands, streams, sheets or fluid-like masses and can even switch dynamically between different modes in response to changing environments9. This diversity in migratory dynamics is accomplished by differential regulation of forces in space and time9.
Key forces that are combined and tuned to different magnitudes and subcellular localizations to produce diverse cell migration behaviours include cell–substrate adhesion, cell–cell adhesion, cell cortex rigidity (which includes the plasma membrane and the underlying cortical cytoskeleton), actin polymerization-mediated protrusion and actomyosin contractility10,11. In cells that strongly adhere to surfaces coated with extracellular matrix (ECM) proteins via focal adhesion complexes and associated stress fibres, transient protrusions and retractions of the leading edge are driven by polymerizing and depolymerizing actin within the lamellipodium. Further back, in the lamellum, integrin-mediated adhesions couple to contractile filamentous (F)-actin stress fibres, engaging the ‘clutch’ of the cell. This engagement enables productive forward protrusion of the cell as new actin subunits are added to the fronts of anchored filament bundles. Young focal adhesions and stress fibres mature and become stronger, increasing forward protrusion12, whereas adhesions at the back loosen, resulting in anisotropic forces and forward movement. This type of behaviour, referred to as mesenchymal migration, is characteristic of fibroblasts migrating on rigid surfaces such as coverslips coated with ECM proteins and likely best models migration on a basement membrane in vivo9,13. By contrast, amoeboid migration refers to the movement of round or ellipsoid cells that do not strongly adhere to the ECM14. Amoeboid migration is either driven by high actomyosin activity that leads to rapid actin-rich front protrusions and back retractions or by actin-devoid protrusions known as blebs, which are driven by hydrostatic pressure and cytoplasmic flow15. In both cases, the rapid kinetics of protrusions and retractions, coupled with the weak and highly dynamic cell–substrate adhesions, result in fast and adaptable migration13,16. In contrast to single-cell mesenchymal or amoeboid migration, epithelial migration is characterized by migration of groups of cells that are inter-connected by cell–cell adhesions and move as clusters, sheets, strands or fluid-like masses17.
Additionally, the phenotype of migratory cells depends on the biochemical composition, stiffness and overall topography of the substrate. Conceptually, 1D, 2D or 3D environments can be distinguished18. An example of 1D migration is migration of cells along a single collagen fibre19. Cell migration on the endothelial lining of vessels or along a basement membrane surface is 2D migration. Finally, cells surrounded by matrix or other cells on all sides move through a 3D environment20,21.
While an early idea was that cells transition from one stable state, such as epithelial, mesenchymal or amoeboid, to another, we now appreciate that tumour cells are heterogeneous, plastic and adaptive. This is particularly important for understanding tumour cell migration, which can combine features of mesenchymal, epithelial and amoeboid modes to adapt to changing environments22,23.
Migration and metastasis
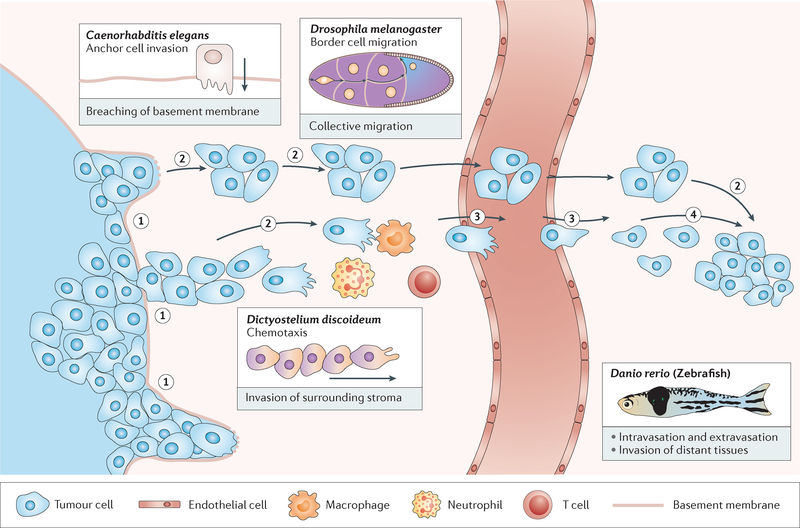
Tumour metastasis is a complex phenomenon that has been widely reviewed24–26. Key features of metastasis, specifically with respect to epithelial-derived carcinomas, include loss of epithelial polarity and breakdown of tissue architecture, breach of the basement membrane, intravasation of tumour cells into blood and/or lymphatic vessels, escape of tumour cells from vessels (extravasation), migration of tumour cells into a new tissue and expansion of the metastatic colony27 (FIG. 1).
Figure 1 |. Simple model organisms can be used to investigate aspects of tumour invasion and metastasis.
Metastasis is a multistep process during which tumour cells breach tissue borders (1); migrate in sheets, strands, streams and clusters (2); cross into and out of blood vessels (3) and form colonies at distant sites (4). Immune cells and endothelial cells in the tumour microenvironment also migrate, either increasing or inhibiting tumour growth and spread. Simple organisms provide models for individual steps or features of this complex process and have been successfully used to contribute key concepts and mechanistic insights to the field of cancer research.
A major challenge in understanding metastatic tumour spread in patients is that the process cannot be observed or manipulated directly. Although histological studies of human tumour samples have provided the clinically useful stage and grade classification system28,29, such approaches cannot reveal cellular or molecular dynamics of the metastatic process. In this regard, model organisms have much to offer. A key point is that simple model organisms need not recapitulate the entire metastatic programme. Rather, the goal is to dissect it into individual steps that can be studied in depth (FIG. 1). Here, we summarize some of the better-studied experimental models of both normal and abnormal cell behaviours that mimic features of tumour metastasis and highlight key insights relevant for understanding human cancer.
Non-mammalian model organisms
Dictyostelium discoideum and chemotaxis
The mechanisms by which tumour cells travel to metastatic sites are complex, involving the motility of tumour cells themselves as well as the hijacking of motile host cells (BOX 2). While the genetic basis of metastasis remains unclear (BOX 1), signalling via chemokines is known to stimulate tumour cell migration, invasion into the local environment, homing of tumour cells to lymphatic vessels and metastatic sites, and infiltration of immune cells into tumours30–33. The social amoeba Dictyostelium discoideum presents a simple model to study directed cell migration and chemokine signalling. The evolutionary conservation of chemokine signalling pathways, accessible genetics and amenability to live imaging make D. discoideum an important model to examine basic molecular mechanisms that govern chemokine-mediated chemotaxis34,35.
Box 2 |. Cell migration in metastasis.
The dominant view of the role of cell migration in metastasis has historically focused on the motility of tumour cells238,239 (see also BOX 3). The preference of specific cancers for particular metastatic sites has been attributed to two mechanisms. For example, when colon cancers metastasize to the liver240, cells appear to settle at the first opportunity, carried via the blood or lymphatics until they encounter capillary beds, where blood flow slows enough for them to attach to the vessel wall and migrate out241. Alternatively, tumour cells might disperse throughout the body, where metastases succeed only in permissive environments (known as the seed and soil hypothesis). This appears to be the case for the metastasis of breast cancer to bone242. In both models, tumour cells are passively distributed throughout the body, and the role of tumour cell motility is to enable them to reach the vasculature, exit from vessels and invade distant tissues. To this end, cells might specifically follow gradients of chemokines such as CXC-chemokine ligand 12 (CXCL12) to exit vessels and migrate towards permissive metastatic niches243,244. Alternatively, though, cancer cell clusters may settle passively as emboli, in which case motility of tumour cells themselves may not be essential for extravasation.
The motility of host cells is emerging as important for metastases. For example, as tumours grow and become hypoxic, they secrete angiogenic factors that cause blood vessels to migrate into the tumour, supplying it with oxygen and nutrients and bringing tumour cells close to vessels. Highly motile immune cells and cancer-associated fibroblasts likely also promote invasion and metastatic spread216,235,245,246. However, cell motility does not only promote metastasis. T cell migration into tumours improves antitumour immune responses247,248. In this case, therapeutic approaches that increase T cell migration into tumours may be helpful. Therefore, an effective therapeutic approach to preventing or treating metastasis on the basis of cell motility will likely require precise targeting of specific cell types rather than globally inhibiting migration.
During nutrient deprivation, D. discoideum cells enter a developmental programme leading to sporulation. During this process, cells chemotax towards secreted cAMP, leading to the formation of aggregates that differentiate into spore and stalk cells to form fruiting bodies36. Chemotactic migration is initiated when cAMP binds to the G protein-coupled receptor (GPCR) cAMP receptor 1 (cAR1). This leads to the dissociation of the G protein into Gα and Gβγ subunits, which then activate a variety of signalling cascades that converge to polarize the cell — a prerequisite for migration37. Cell polarization is accompanied by the redistribution of cytoskeletal components, with enrichment of dynamic F-actin and numerous actin-binding proteins at the front, leading edge and myosin II assembly on the sides and at the back, trailing edge (FIGS 1,2). These highly orchestrated events result in a type of amoeboid migration that allows D. discoideum to navigate complex environments at speeds that can exceed 20 μm/min (REF 34). Similarly, chemokine-mediated and growth factor-mediated chemotaxis has been observed in tumours. For example, the chemokine CXC-chemokine ligand 12 (CXCL12; also known as SDF1), which is involved in the chemoattraction of bone marrow-derived cells, can attract a variety of tumour cells, including those originating from breast, ovarian and colorectal tumours and melanoma38. In addition, immune cells, which chemotax to sites of inflammation and injury39, may be subverted by tumours to infiltrate tumour tissue and support tumour growth using similar chemotactic mechanisms40–42.
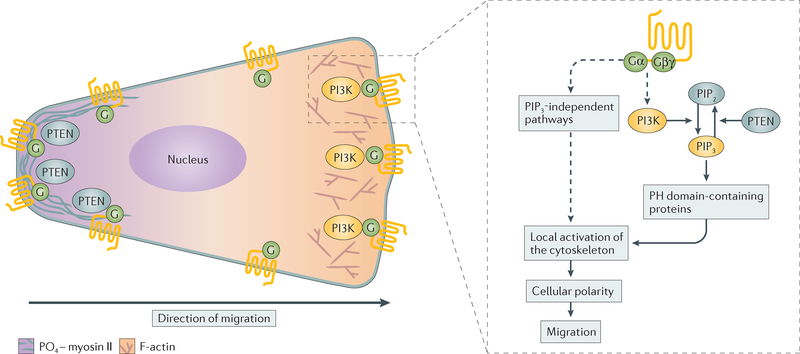
Figure 2 |. Regulation of chemoattraction in Dictyostelium discoideum.
A schematic demonstrating how chemotactic signals give rise to cell polarization — a prerequisite for cell migration — is presented. Following G protein-coupled receptor (GPCR) activation, PI3K and PTEN become spatially restricted to the front and back of Dictyostelium discoideum cells, respectively, giving rise to localized phosphatidylinositol-3,4,5-trisphosphate (PIP3) and the recruitment of pleckstrin homology (PH) domain-containing proteins at the front of cells. Together with PIP3-independent pathways, these events lead to polarized signals in which actin assembly occurs at the front and myosin phosphorylation (PO4–myosin II) occurs at the back of cells. F-actin, filamentous actin; PIP2, phosphatidylinositol-4,5-bisphosphate.
A central question in the field of chemotaxis has been how chemotactic signals initiate and maintain cell polarity. Uniform stimulation of D. discoideum cells with chemoattractants leads to transient increases in calcium influx, inositol triphosphate (IP3), cAMP and cGMP, as well as myosin II phosphorylation and actin polymerization. By contrast, when D. discoideum cells are exposed to gradients of chemoattractants, these responses are spatially restricted and persistent. This occurs because chemoattractant receptor activation elicits both stimulatory and inhibitory signals that allow cells to adapt to external signals and maintain chemotactic sensitivity to external gradients where the concentration of chemoattractant is increasing43,44. Work in D. discoideum provided the first evidence as to how this occurs. Use of green fluorescent protein (GFP) revealed that, whereas both cAR1 and its associated G protein remain uniformly distributed in chemotaxing cells, proteins harbouring pleckstrin homology (PH) domains that bind to phosphatidylinositol-3,4,5-trisphosphate (PIP3) specifically redistribute to the leading edge of chemotaxing cells45–47. This molecular underpinning of directed amoeboid migration was also observed in neutrophils48 and in fibroblasts in response to gradients of platelet-derived growth factor (PDGF)49. However, the role of localized PIP3 signals in mesenchymal and epithelial migration remains unclear50.
The asymmetrical distribution of PIP3 results from the spatial activation of PI3K at the front and PTEN at the sides and rear of cells51,52. These localized PIP3 signals therefore provide spatial orientation for PH domain-containing proteins that act as adaptors for specific downstream cascades, which, in the case of chemotaxis, spatially nucleate actin assembly53,54 (FIG. 2). In human cancers, mutations in the PI3K–PTEN–mTOR complex 1 (mTORC1) cascade are prevalent55,56, potentially dysregulating the PIP3 signals necessary for migration. In support of this, wortmannin-induced depletion of PIP3 is associated with an anti-migratory response to lysophosphatidic acid (LPA) in mouse B16 melanoma cells57, and PI3K–AKT signalling can increase invasion by upregulating matrix metalloproteinase 9 (MMP9)58,59. Additionally, PI3K is a key regulator of immune cell migration60 as well as cellular processes such as cell growth, survival and differentiation61,62. Thus, inhibitors of this pathway, which have been extensively studied to treat cancer62, likely exert pleiotropic effects on host cancer-associated cells, such as immune cells and fibroblasts, in addition to affecting the tumour cells directly.
Genetic screens are another powerful tool used to identify pathways controlling directed cell migration in D. discoideum. In this way, three independent pathways parallel to the PI3K–PTEN signalling cascade were identified in genetic screens for migration defective mutants: guanylyl cyclase (GC), phospholipase A2 (PLA2) and mTORC2 signalling43,63. However, only a few inhibitory mechanisms of the chemotactic response have been identified, including specific inhibitory Gα subunits (for example, Gα9) and phosphorylation of the cAMP receptor64. These inhibitory pathways confer adaptability of the chemokine signalling pathway, an essential feature of the chemotactic response that enables cells to detect increases in ligand concentrations over a large range of concentrations. Therefore, mutations in these pathways would also lead to defects in migration and metastasis.
The information gathered from D. discoideum has provided a blueprint to decipher chemotactic signalling in both leukocytes and cancer cells during inflammation, invasion and metastasis. Indeed, it has been proposed that in their most invasive form, metastatic cancer cells revert to the primitive mode of amoeboid migration that is shared by haematopoietic and D. discoideum cells65,66. Furthermore, GC, PLA2 and mTORC2 signalling have all been implicated in the regulation of migration of a variety of human cancer cells67–69. For example, PLA2 and its lipid mediators stimulate RHO-associated protein kinase (ROCK) signalling69,70, a key regulatory pathway of amoeboid migration66. Identifying the basic mechanisms that regulate chemotactic signalling may reveal how to increase the motility and invasion of tumour-reactive T cells into tumours. This is an important frontier in expanding the current success of immunotherapeutic agents, as a key limitation to the effectiveness of these treatments is the inability of T cells to infiltrate tumours in a large fraction of patients71.
Caenorhabditis elegans and anchor cell invasion
Tumours that form within epithelia have little potential to spread when they remain confined by the basement membrane. Basement membranes also surround blood vessels and thus present a barrier to intravasation and extravasation. Therefore, crossing basement membranes is a key step in metastasis72.
An elegant example of local basement membrane invasion is found in the nematode worm C. elegans73. During larval development, a single cell — the anchor cell — breaches two underlying basement membranes as part of normal morphogenesis of the vulva (FIGS 1,3). The power of this model derives from its simplicity, the reproducibility of this stereotyped event, the clarity of the live imaging and the ease of genetic screening74.
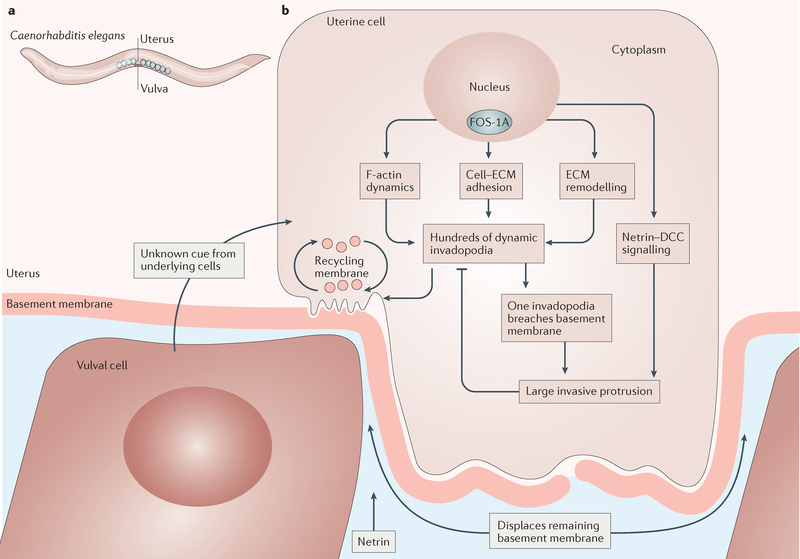
Figure 3 |. Caenorhabditis elegans anchor cell invasion.
a | During third larval instar development, the anchor cell breaches two basement membranes that separate this uterine cell from vulval cells, eventually connecting the uterus to the vulva. b | The molecular and cellular biological steps in anchor cell invasion begin in the nucleus, where activation of the transcription factor FOS-1A stimulates expression of many downstream target genes involved in the indicated processes, including the transcription factor egl-43 (orthologue of the EVI1 proto-oncogene), the protocadherin cdh-3, the extracellular matrix (ECM) protein him-4 (orthologue of hemicentin genes) and zmp-1 (orthologue of the matrix metalloproteinase (MMP) genes), suggesting a highly conserved programme for basement membrane invasion. DCC, deleted in colorectal carcinoma; F-actin, filamentous actin.
Specification of the anchor cell fate is the first known step in activating this physiological invasion programme. Cell cycle arrest in G1 follows and is essential for further progress75. Chromatin modifications ensue, followed by upregulation of a set of transcription factors, including FOS-1A, which is the C. elegans orthologue of proto-oncogene FOS76, a protein strongly associated with cancer cell invasion77. In vertebrates, FOS dimerizes with proto-oncogene JUN to form the transcription factor activator protein 1 (AP-1), which stimulates expression of MMPs. In C. elegans, downstream transcriptional targets of FOS-1A include genes that encode cell–matrix adhesion molecules such as hemicentin and proteins that weaken or degrade basement membranes such as zinc metalloprotease 1 (ZMP-1), a membrane-type MMP78.
An as-yet-unidentified cue from the underlying vulval cells activates the RHO-family GTPase CDC-42 within the anchor cell, causing the formation of a special invasive domain within the plasma membrane that is enriched in specific lipids, cell–matrix adhesion proteins, netrin–deleted in colorectal carcinoma (DCC) signalling, F-actin regulators such as RAC, the enabled/vasodilator-stimulated phosphoprotein (ENA/VASP) family of proteins, cofilin and matrix remodelling factors, including MMPs79 (FIG. 3). Within this invasive domain, hundreds of dynamic protrusions called invadopodia form. Eventually, one of them drills through the basement membrane, leading to the recruitment of more F-actin regulators and the formation of a large invasive protrusion, which feeds back to shut down further production of dynamic invadopodia. This large protrusion then pushes the remaining basement membrane aside, allowing the anchor cell to reach the underlying vulva cells to form the vulva80. Similarly, tumour cells extend actin-based invadopodia into the ECM and secrete MMPs78,81–83.
Virtually all the molecular components of this system, including FOS, RHO GTPases, MMPs and laminin (a major component of basement membranes), are conserved in mammals (including humans), and in many cases, they are clearly associated with invadopodia formation during tumour cell invasion81,84,85. Therefore, the exquisite precision in the mechanistic analysis of this system has yielded several striking observations that are likely to be relevant to cancer cell invasion across basement membranes. One example is the observation that an initially tiny hole in the basement membrane is enlarged by the formation of a massive protrusion that pushes the remaining basement membrane aside80. This shows that the basement membrane can be removed, at least in part, through such physical means. In support of the general idea that cells can facilitate breaching of the basement membrane through mechanical (non-enzymatic) means in mammals too, a recent paper has shown that cancer-associated fibroblasts (CAFs) can also perform such a function86. This alternative mechanism to generating tracks by matrix degradation may be one of many reasons why MMP inhibitors have not been clinically successful in treating tumours87,88. Thus, illuminating the mechanisms of anchor cell invasion may lead to the identification of drug targets that might be effective in combination with MMP inhibitors.
Another intriguing observation is that cell cycle arrest in G1 is a prerequisite for anchor cell invasion75. If tumour cells similarly cannot divide and migrate at the same time, in a phenomenon called the ‘go or grow’ hypothesis89,90, then they must either temporarily exit the cell cycle in order to invade, which is certainly possible given the timescale over which metastasis occurs relative to the cell cycle, or invade as a cooperative group of cells in which some cells continue to proliferate while others exit the cell cycle for invasion and/or migration, as has been observed in studies of tumour invasive fronts91. Indeed, a dichotomy between proliferation and migration has been observed in some neurological tumours such as astrocytomas89,90,92, and emerging evidence suggests that the ‘metastatic unit’ is commonly formed of polyclonal groups of cells rather than single cells93. Within these polyclonal groups, immobile tumour cells might be guided by highly motile tumour cells, immune cells or fibroblasts (known as the ‘piggy-back theory’) through physical or paracrine interactions94–96. In at least one example in H1299 non-small-cell lung cancer (NSCLC) cells growing in 3D spheroids, the highly migratory and invasive ‘leader’ cells also actively promote the survival and proliferation of the less mobile ‘followers’ (REF 97). A clear example of cooperative migration of two mutually dependent cell types that occurs during normal development is described in the next section.
Drosophila melanogaster border cells
During normal morphogenesis as well as during tumour invasion, cells often move in groups18,98, and at least in some animal models, cell clusters are more effective than individual cells at metastatic spread91,93,99. In colorectal cancer, and increasingly in additional cancers, detached cell clusters, or ‘buds’, are recognized as clinically important in biopsy samples and correlate with poor prognosis100. Cooperation appears to improve group survival and spreading. This may be owing to the shielding of inner cells of the group from the immune system or to the combination of individuals with specialized skills, such as combining highly proliferative cells with motile cells. An example of cooperative, collective cell migration during normal development can be found in the border cells of the D. melanogaster ovary101.
Migratory border cells originate within an epithelial monolayer of somatic cells in a structure called the egg chamber (FIGS 1,4). Border cells move as a group consisting of two distinct cell types, each of which depends upon the other. Two cells at the centre of the cluster, called polar cells, cannot move autonomously; instead, they activate motility in the neighbouring cells. These migratory border cells then carry the polar cells ~150 μm, squeezing between large cells called nurse cells and eventually arriving at their destination, the oocyte. If this migration fails, females are sterile102. Border cell migration, such as anchor cell invasion in C. elegans, begins with cell-fate specification and activation of a transcriptional programme. Polar cells secrete a cytokine, Unpaired (Upd1), which activates Janus kinase (Jak) in nearby cells, activating signal transducer and activator of transcription (Stat) and specifying them as the migratory population103 (FIG. 4). Whereas mammals possess dozens of cytokines, five JAKs and seven STATs, the simpler fly genome encodes just three Upds, one Jak and one Stat (encoded by Stat92E). The discovery that Jak–Stat signalling stimulates border cell migration was the first evidence for the role of this pathway in cell motility in vivo103–105. JAK–STAT signalling also promotes the motility of human cancer cells in vitro and in orthotopic xenografts106,107 and contributes to cell motility and metastasis of prostate108, pancreatic109, hepatocellular110, lung111 and other carcinomas in a variety of experimental models (reviewed in REF 112). Additionally, in another example of cooperative, collective cell migration, JAK–STAT signalling in CAFs promotes cancer cell invasion113.
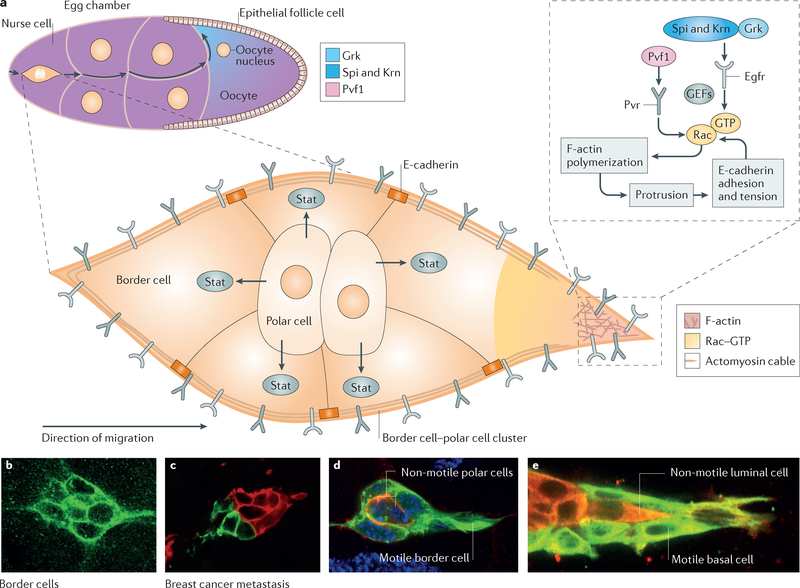
Figure 4 |. Border cell migration in Drosophila melanogaster.
A schematic drawing of a stage 9 egg chamber (upper left) showing the germline cells (nurse cells and oocyte) surrounded by the epithelial layer of follicle cells is presented (part a). The border cells squeeze between the nurse cells as they migrate towards the oocyte. The enlarged view of the migrating cluster shows two non-migratory polar cells, which secrete the cytokine Unpaired 1 (Upd1) to activate Jak–signal transducer and activator of transcription (Stat) signalling and motility in five neighbouring migratory border cells. Secreted platelet-derived growth factor (PDGF)- and vascular endothelial growth factor (VEGF)-related factor 1 (Pvf1) from the germ line activates PDGF- and VEGF-receptor-related (Pvr), while Spitz (Spi), Keren (Krn) and Gurken (Grk) activate epidermal growth factor receptor (Egfr). The receptor tyrosine kinases (RTKs) then activate Rac, which stimulates act in polymerization and protrusion. The border cells migrate in between nurse cells in the absence of detectable extracellular matrix (ECM) and thus use homophilic E-cadherin-mediated adhesion for tract ion on the nurse cells as well as for mechanical coupling between all cells of the cluster. Confocal images of border cell clusters (parts b and d) and breast cancer (parts c and e) that show similarities in forming clusters (parts b and c) and in heterotypic composition (parts d and e) are presented. F-actin, filamentous actin; GEF, guanine nucleotide exchange factor. Part c is reproduced with permission from REF. 93, Proceedings of the National Academy of Sciences. Part e is reproduced with permission from REF 91, Elsevier. Parts b and d are the authors’ own images.
Within border cells, Stat is a key node in a transcriptional network that activates the expression of hundreds of target genes104,105,114, many of which contribute to migration. One transcriptional target is the receptor tyrosine kinase (RTK) PDGF- and vascular endothelial growth factor (VEGF)-receptor related (Pvr), which serves as a chemoattractant receptor115. Direction-sensing by border cells is relatively complex, considering their straightforward trajectory. At least four chemoattractants secreted by the germline cells (15 nurse cells and one oocyte) activate two RTKs, Pvr and epidermal growth factor receptor (Egfr), expressed on border cell surfaces115–118 (FIG. 4). Ectopic expression of the ligands for these RTKs is sufficient to redirect the cells, demonstrating their role in providing directional information117. RTK signalling activates both the Mapk–Erk pathway and the Rho GTPase, Rac101. The role of Rac in promoting protrusion and migration in vivo was first demonstrated in the border cell system119. This protein is now known to be a critical node in the signalling and cytoskeletal networks that govern the motility of both normal and cancer cells120,121. Activation of Rac in one cell of the migrating border cell cluster is sufficient to redirect the whole group, showing that differential Rac activity within the group is key to collective guidance122. Interestingly, Rac activation requires Pvr through the action of multiple guanine nucleotide exchange factors (GEFs)115,123 and Rab5-dependent endocytosis124, consistent with work in HeLa cells125. Furthermore, Rab11-dependent activation of moesin is essential for the lead cell to inhibit Rac activation in the followers124, providing specific insight into the mechanisms that maintain asymmetric signalling within a cell cluster.
While parallels between border cell migration and ovarian cancer motility have been noted126, which may relate to their common origin from somatic cells of the female reproductive organ, border cells may also serve more generally as a model for dissemination of collectively moving, heterogeneous groups of migratory and non-migratory cells. Determining the contribution of this type of motility to tumour metastasis is currently a very active line of investigation in lung127, breast99, pancreatic128, prostate129 and other types of cancer26.
Another key transcriptional target of Stat in the border cells is E-cadherin. Border cells elevate E-cadherin expression during migration130, highlighting an important difference between collective cell migration and the epithelial to mesenchymal transition (EMT) (BOX 3). Moreover, E-cadherin promotes border cell migration by three distinct mechanisms131, suggesting that molecular programmes distinct from EMT exist to promote collective cell migration. First, high levels of E-cadherin at interfaces between polar cells and border cells bind the cluster together to ensure collective behaviour. Second, E-cadherin molecules on the border cells bind E-cadherin molecules on the nurse cells between which they migrate (in the absence of substantial ECM). At the leading edge, a positive feedback mechanism between Pvr and Egfr signalling, Rac-mediated actin polymerization and E-cadherin-mediated adhesion produces a large forward-directed protrusion131 (FIG. 4). Here, E-cadherin fulfils a function similar to that of integrins at the leading edge of a cell migrating through ECM21,132. Protrusions are more frequent, larger and persist longer in the leading cell(s) of the cluster. Third, E-cadherin-containing adherens junctions between individual border cells mechanically couple them so that the lead cell, pulling on the following cells, inhibits protrusions to the side and rear of the cluster and coordinates the motile forces of individual cells to promote collective forward movement of the group. A simple physical model predicts that in 3D but not 2D environments, larger cell clusters should migrate faster133. In border cells, this correlation between size and speed holds true until clusters become so large that viscous drag from the surrounding tissue impedes their movement133.
Box 3 |. The epithelial to mesenchymal transition controversy.
The epithelial to mesenchymal transition (EMT) is a programme used repeatedly during embryonic development249. EMT refers to characteristic morphological changes that enable cells to leave an epithelium and move to a new location, where they can undergo the reverse process (mesenchymal to epithelial transition (MET)) to form new epithelial organs. A conserved molecular programme that involves expression of transcription factors including SNAIL1, TWIST, ZEB1 and ZEB2 (REF. 250) drives EMT and inhibits apical–basal polarity components and cell–cell adhesion proteins such as E-cadherin. EMT confers increased motility, the ability to survive when detached from the basement membrane extracellular matrix (ECM) and from epithelial neighbours, as well as radioresistance and chemoresistance, properties that should provide a selective advantage to tumour cells250–252. A vast body of literature correlates EMT–MET with metastasis238,250. However, there are challenges to the simplest form of the EMT–MET hypothesis:
Full EMT is rarely observed in histological specimens253,254
Loss of E-cadherin does not always occur during tumour development138
EMT may be required not for the metastatic tumour spread of all cancer types but rather for acquisition of drug resistance251,252
Single tumour cells are rarely found in clinical tumour specimens254, although single circulating tumour cells can be found99
Indeed, circulating tumour cell clusters isolated from patient blood are 50 times more efficient at establishing metastases than single circulating tumour cells99, and artificially clustering breast cancer cells increase their metastatic efficiency by 100-fold93. Cell clustering could provide a number of advantages such as allowing mixtures of highly motile and highly proliferative cells to spread together and shielding the innermost cells from the immune system. Thus, two distinct conceptual frameworks have emerged. Collective cell motility programmes distinct from EMT may mediate metastasis. Alternatively, EMT proponents favour models in which cells adopt intermediate states that are most effective at metastasis. Resolution of this controversy is an exciting open area of research.
Interestingly, while many investigators have focused on the motility of individual tumour cells as the driving force for cancer metastasis, recent studies demonstrate that circulating tumour cell clusters are more efficient at seeding distant metastases and correlate with worse clinical outcomes than single circulating tumour cells99,134. E-Cadherin expression varies greatly among tumour cell lines and among tumour types135. Although a large body of work suggests that reduced or atypical E-cadherin expression is a sign of increased malignancy in tumours136, loss-of-function mutations in the gene encoding E-cadherin (CDH1) are prevalent only in gastric cancer (CbioPortal). E-cadherin may more commonly be dynamically regulated epigenetically in motile cells, enabling a greater variety of behaviours. For example, cells might undergo partial or transient downregulation of E-cadherin followed by its re-expression when they colonize distant sites137. Elevated E-cadherin is commonly found in circulating tumor cell clusters, established metastases and is characteristic of ovarian and inflammatory breast cancers138, both of which have poor prognoses. Like migrating border cells, collectively invading breast cancer cells are connected by homotypic E-cadherin junctions, which may similarly mechanically couple the cells139. Heterotypic E-cadherin–N-cadherin (also known as cadherin 2) junctions between tumour cells and CAFs also facilitate cooperative tumour cell movement and invasion96.
As cadherins promote collective, cooperative, cell-on-cell migration during border cell migration by multiple mechanisms, it may be that cadherins also promote metastasis by similar mechanisms during collective tumour cell dissemination. This raises the general notion that molecules with a negative effect on individual cell migration might actively promote collective motility. Border cells also require apical–basal polarity to coordnate their movements140, raising the possibility that apical–basal polarity might be required for the spread of cancer by cell collectives. In addition, border cells require the Src42A tyrosine kinase for their migration141, raising the possibility that SRC kinases could have distinct roles in collective versus individual cell migration. Although it has not been studied extensively, there is evidence for SRC promoting collective cancer cell motility in 3D142.
In addition to border cells, many diverse modes of cell motility are found throughout normal fly development. Additional models of individual and collective cell motility include primordial germ cells (PGCs), which exhibit individual, amoeboid motility similar to immune cells143–145; gastrulating mesoderm, which undergoes EMT mediated by the transcription factors Twist and Snail146; haemocytes, the macrophages of the fly147,148. caudal visceral mesoderm, which undergoes EMT followed by collective, mesenchymal migration149; dorsal closure, which represents an epithelial sheet movement150; tracheal and salivary gland morphogenesis, which serve as general models for the development of tubular organs such as blood vessels151; and larval histoblasts, another example of epithelial sheet movement152. Thus, D. melanogaster can serve as a model for studies of many types of cell migration, with the technical advantages of sophisticated genetics and in vivo live imaging.
Drosophila melanogaster and tumour metastasis
D. melanogaster can also serve as a more direct model for tumour metastasis. The first mutations that cause metastatic behaviour were characterized in D. melanogaster in the 1970s, before the tumour suppressor gene concept was firmly established in cancer biology153. Genes such as lethal (2) giant larvae (l(2)gl), discs large (dlg) and scribbled (scrib) are named for the phenotypes they cause in developing D. melanogaster larvae. Cells homozygous for these mutations grow excessively and exhibit increased MMP expression, cell migration and the ability to spread throughout the organism when transplanted into a wild-type host153. These genes encode highly conserved proteins (LLGL, DLG and SCRIB in humans, respectively) that form a complex in epithelial cells that is essential for establishing and maintaining apical–basal polarity in animals from flies to humans154. The SCRIB–DLG–LLGL complex normally establishes the basolateral domain of an epithelial cell by excluding apical proteins such as partitioning defective 3 (PAR3), PAR6 and atypical protein kinase C (aPKC) from that region155,156. Mutual antagonism between the PAR and SCRIB complexes ensures establishment and maintenance of two distinct membrane regions of differing compositions. This polarity promotes regular patterns of oriented cell division and the establishment of adherens and tight junctions and thus results in well-organized epithelial architecture157. Loss of apical–basal polarity and aberrant expression or localization of human homologues of polarity proteins such as SCRIB, DLG and LLGL or PAR3 and PAR6 are considered hallmarks of carcinomas. As such, these proteins have been suggested to be tumour suppressors in humans, highlighting the relevance of this model to human cancer154,158. However, it is intriguing that the human homologues of scrib, dlg and l(2)gl are rarely deleted or mutated in cancer; rather, these genes are frequently amplified and/or overexpressed (CbioPortal)159–161. These observations indicate that the roles of SCRIB, DLG and LLGL proteins in tumour progression are poorly understood.
One possible explanation for the observed overexpression of SCRIB, DLG and LLGL proteins in human cancers is that the overexpression causes mislocalization and a dominant-negative effect162. However, if this were true, then SCRIB, DLG and/or LLGL1 (the human gene encoding LLGL) should be mutated or deleted at least as often as they are amplified or overexpressed, which is not the case. Another possible explanation is that polarity proteins might suppress tumour initiation but promote tumour progression, similar to transforming growth factor-β (TGFβ) signalling, which is a tumour suppressor and metastasis promoter163. If this is true, these polarity proteins would presumably have to be suppressed by epigenetic mechanisms early in tumorigenesis and then re-expressed during metastasis, as proposed for E-cadherin in EMT and mesenchymal to epithelial transition (MET)137. Maintenance of apical–basal polarity is a key feature of collective epithelial motility140, so one possibility is that SCRIB, DLG and LLGL promote tumour progression by facilitating collective cell migration, though it is unclear how overexpression would enhance that behaviour. Further studies of the phenotypes caused by overexpression of SCRIB, DLG and LLGL are warranted.
The protein Scrib emerged independently in a systematic screen for metastatic cell behaviour in flies157. Clones of cells expressing active Ras (RasG12V) in larval tissues overgrow but do not spread157. Therefore, a screen was conducted to identify recessive mutations that together with RasG12V expression cause tumour cells to breach the basement membrane and spread throughout the fly; this identified scrib164,165. Clones of cells lacking scrib are normally eliminated by apoptosis; however, scrib−/− cooperates with RasG12V to cause massive overgrowth, EMT and MMP expression, leading to metastatic spread and colonization of distant organs157. Mutations in dlg and l(2)gl can substitute for scrib mutations in this context, thereby implicating loss of the basolateral complex as key to the metastatic phenotype157. Tissue injury or stress can also substitute for scrib−/−, suggesting that cells perceive loss of polarity as an injury or stress. Exactly how RasG12V contributes is not clear because stimulating growth and proliferation, by overexpression of Myc or Akt, or inhibiting apoptosis allows scrib−/− cells to survive but is insufficient to substitute for RasG12V in promoting spread throughout the organism164.
Remarkably, RasG12V and scrib mutations do not have to occur in the same cells to promote metastasis. RasG12V-expressing cells adjacent to scrib−/− cells overgrow and metastasize166. The scrib−/− cells secrete Upd cytokines, activating the Jak–Stat signalling that is essential for tumour growth and metastasis. Importantly, the cytokines promote tumour growth even after the initial scrib−/− cells are outcompeted by the RasG12V-expressing cells. This key insight from the fly could explain why SCRIB, LLGL1 and DLG mutations are rarely detected in human cancer. Such cells may transiently promote tumour progression but ultimately become outcompeted.
Screens for additional mutations that enhance or suppress the RasG12Vscnb−/− phenotype have uncovered many genes contributing to metastasis in the fly167. A summary of some salient features of the pathways are illustrated in FIG. 5. Jun N-terminal kinase (Jnk; also known as Mapk) is a stress-activated kinase that at high levels triggers apoptosis168. However, in scrib−/− cells, Jnk promotes proliferation and invasion169. Jnk signalling also stimulates cells to multiply in tissues that need repair (known as compensatory growth) and sustains tumour growth and invasion in flies170–172. Many of these findings are also true in human cancers and may well contribute to metastasis173. For example, activation of JNK is necessary for cellular transformation by oncogenic RAS, JNK stimulates cell proliferation by producing cytokines that activate the JAK–STAT pathway, and JNK promotes tumorigenesis in hepatocellular carcinoma, gastric cancer, and tobacco-induced lung cancer (reviewed in REF 173).
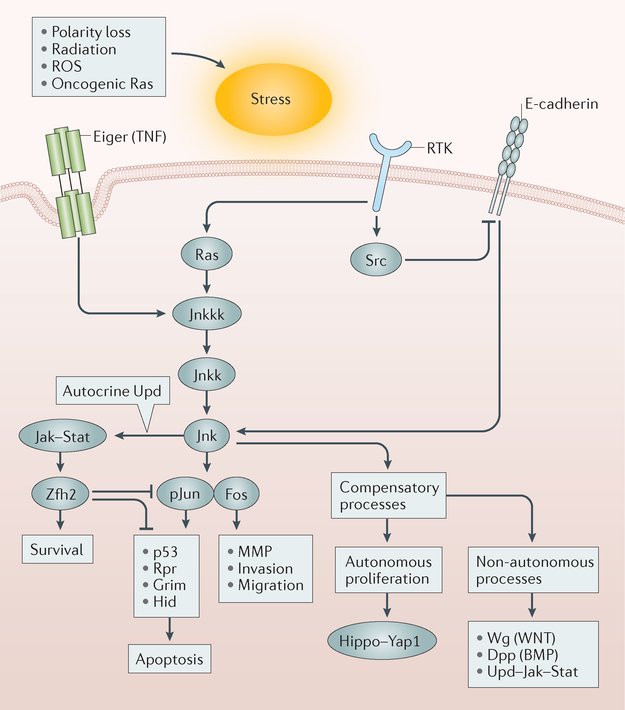
Figure 5 |. Jnk pathway signalling is a key part of metastatic spread in Drosophila melanogaster.
Metastasis models in Drosophila melanogaster have revealed that a variety of stresses activate the Jun N-terminal kinase (Jnk) pathway, which at high levels of signalling can lead to apoptosis. Alternatively, at lower levels or in the presence of oncogenic Ras, the Jnk pathway leads to autonomous and non-autonomous proliferation, expression of matrix metalloproteinases (MMPs), loss of E-cadherin and increased motility. Thus, the Jnk pathway is central to metastatic spread in the fly and likely also in human cancer173. BMP, bone morphogenetic protein; Dpp, decapentaplegic; Hid, Head involution defective; Jak, Janus kinase; Jnkk, Jnk kinase; Jnkkk, Jnkk kinase; pJun, phosphorylated Jun; ROS, reactive oxygen species; Rpr, Reaper; RTK, receptor tyrosine kinase; Stat, signal transducer and activator of transcription; TNF, tumour necrosis factor; Upd, unpaired; Wg, Wingless; Yap1, Yes-associated protein 1; Zfh2, zinc-finger homeodomain 2.
A genetic screen for mutations that enhance or suppress fly eye defects caused by overexpression of the oncogenic form of the RET RTK gene, which is the gene mutated in the human cancer syndrome multiple endocrine neoplasia (MEN), a disease that includes medullary thyroid carcinoma, identified mutations in the RAS, SRC, JNK and PI3K pathways174, illuminating critical downstream signalling pathways of oncogenic RET in vivo. In particular, the SRC pathway promotes cell invasion by altering E-cadherin-mediated adhesion, inhibiting apoptosis and activating MMP expression (reviewed in REF 175).
Remarkably, Dar et al.176 tested a library of kinase inhibitors in a RET-driven D. melanogaster model in which a constitutively active form of the fly homologue of RET was expressed in the eye. They identified one kinase inhibitor with improved efficacy and reduced toxicity compared with vandetanib, which is the approved treatment for RET-dependent thyroid cancer. The new drug also proved effective in mouse xenograft studies. For a detailed review of D. melanogaster as a model for identifying cancer drugs, see the publication by Sonoshita and Cagan177. Thus, despite the lack of blood or lymphatic vessels, D. melanogaster is a powerful model for identifying molecular pathways and cell–cell interactions that contribute to metastasis and systemic effects such as cachexia178,179 as well as promising drugs to treat cancer.
The zebrafish as a model for bleb-driven migration
Cell motility driven by membrane blebbing is part of the repertoire of motility behaviours that cancer cells use to reach distant sites180. Cellular blebs are simple structures thought to represent a primordial mode of migration181. Remarkably, cells such as normal human fibroblasts, plated in confined spaces with limited cell–substrate adhesion switch from a slow-moving mesenchymal phenotype to a fast-moving amoeboid migratory phenotype with the appearance of cellular blebs and high cellular contractility182.
In addition to being a powerful model system to study immune cell migration in vivo183, the zebrafish (Danio rerio) has been central in our understanding of the mechanisms that regulate bleb formation through the study of PGC migration184. PGCs acquire the capacity to migrate by undergoing a series of differentiation steps beginning 3 hours after fertilization. PGC differentiation depends on de novo transcription, the RNA-binding protein dead end protein homologue 1 and the downregulation of E-cadherin185. Once polarized and motile, PGCs chemotax towards dynamic patches of Cxcl12a to reach their final destination, the primordial gonad. Cxcl12a binds to the GPCR CXC-chemokine receptor 4b (Cxcr4b) and promotes the formation of cellular blebs that mediate PGC migration. Cxcr4b is uniformly distributed around the periphery of PGCs, but its activation is restricted to the side of the cells that is exposed to the highest concentration of Cxcl12a, leading to cell polarization and directional migration186. Interestingly, and in contrast to D. discoideum, neutrophils and fibroblasts47–49, PGCs exhibit a uniform distribution of PIP3 on their surfaces, and depletion of PIP3 from PGCs by expression of a dominant-negative form of the regulatory subunit of class 1A Pi3k does not alter their ability to migrate directionally187. Instead, chemokine receptor signalling triggers local calcium increases at the front of migrating PGCs. This results in a localized myosin activity that causes the blebs188. Furthermore, elevated pH at the front of migrating PGCs has recently been observed and proposed to be involved in their polarized migration189. While the advantages of bleb-based migration are not fully understood, the fast, energy-efficient formation of blebs appears to be one mechanism by which individual cells can effectively explore complex 3D environments15,190,191. Intriguingly, zebrafish lateral mesendoderm progenitors alternate migratory phenotypes using blebs and actin-rich protrusions to migrate in a manner characterized by ‘runs’ of high directional persistence and formation of directed actin-rich protrusions, whereas less oriented ‘tumbles’ occur when blebbing is increased192.
Similar switching between migratory phenotypes is also observed when tumour cells encounter changing environments. For example, experiments using micropatterns revealed that MDA-MB-231 breast cancer cells switch from a migratory phenotype dependent on integrin adhesion and actin polymerization in unconfined spaces to a tubulin-dependent migratory phenotype in confined spaces193,194. These findings suggest that the physical properties of the cellular microenvironment are one parameter that can induce switching between migratory phenotypes. However, it is not yet fully understood which cellular or microenvironmental parameters, such as matrix composition, matrix density or cytokine levels, trigger such switches or what the consequences of switching migratory phenotypes are. The zebrafish model, which offers an imageable, 3D model of cell migration, may help answer these questions.
The zebrafish as a vertebrate model for tumour progression
In the past decade, zebrafish have emerged as a useful vertebrate model system to study the metastatic cascade in vivo. Easy and cost-efficient to maintain relative to mammals, zebrafish can be genetically manipulated, have an innate and adaptive immune system, vasculature, and have many of the same organs as humans195–197. Zebrafish embryos are particularly suitable for in vivo, high-resolution, single-cell imaging, as they are transparent and develop outside the mother198–200. In addition to these advantages, zebrafish do not have a functional adaptive immune system until 14 days after fertilization and thus lend themselves to xenograft models201. Recently, optically clear and immunocompromised V(D)J recombination-activating protein 2 (Rag2)-mutant fish, which harbour a reduced number of T cells and B cells, became available for xenograft studies in embryos and adult fish202–204. Inoculation of immunocompromised zebrafish with patient-derived breast or neuroendocrine tumour tissue resulted in metastatic tumour spread that reflected or predicted the disease course in patients205,206. Thus, zebrafish represent a lower-cost model that may be used to predict metastatic tumour spread on the basis of patient material.
On a mechanistic level, the zebrafish model has been used to study intravasation and extravasation of tumour cells — critical steps during tumour spread that are still poorly understood, are difficult to observe in vivo and cannot be modelled in flies or worms. Injection of GFP-labelled human tumour cells into the abdominal cavity of embryonic zebrafish revealed that tumour cells secrete VEGF, which stimulates vascular remodelling and the formation of openings (called portholes) at the sites of remodelling. The cells then extend protrusions into the portholes in a RHOC-dependent manner, followed by tumour cell intravasation and dispersal207. Inprostate cancer cells, RHOC or ROCK1 and ROCK2 depletion by RNAi was also shown to reduce cancer cell adhesion to endothelial cells and transendothelial migration208. Furthermore, this vascular remodelling has been observed in extravasating tumour cells in mice using electron microscopy and has been shown to be increased by the presence of VEGF209. Similarly, using real-time intravital imaging, the Condeelis group has shown that VEGF signalling in macrophages causes the local loss of endothelial cell junctions and vascular permeability210, which could possibly also involve porthole-like structures.
Injection of GFP-labelled human cancer cell lines into the common cardinal vein of fish embryos revealed that extravasation of tumour cells is also an active process. Real-time intravital imaging in zebrafish revealed that at sites of extravasation, arrested tumour cells induce clustering of endothelial cells and alterations of cell–cell junctions211. Moreover, it was shown that while intravascular migration of tumour cells is dependent on β1 integrin adhesion to the blood vessel wall, the expression of TWIST, an EMT-related transcription factor, increased intravascular migration and extravasation in a β1 integrin-independent manner211. The zebrafish model thus extends and confirms the anchor cell model in C. elegans that identified a role for β1 integrin in anchor cell invasion212. More recently, β1 integrin has been shown to be required for tumour cell extravasation in a 3D model of human microvasculature, specifically mimicking invasion past the endothelial basement membrane213, and CDC42 has been reported to promote transendothelial migration of prostate cancer cells through β1 integrin214. Thus, β1 integrin is implicated in transendothelial and transepithelial migration in three different model systems, implying that this integrin has a conserved role in the migration of cells across basement membranes.
Interactions between tumour cells and other types of cells during tumour progression is another area that is highly relevant for our understanding of tumour disease, but difficult to observe. Of special interest are immune cells that have a complex role in tumour development215. For example, macrophages can support or limit tumour growth216. In mammals, interactions between tumour cells and macrophages through an EGF–macrophage colony-stimulating factor 1 (CSF1) paracrine loop enable the formation of a tripartite arrangement between tumour cells, macrophages and endothelial cells (referred to as the tumour microenvironment of metastasis (TMEM)) that is critical for intravasation216–220. Intravasation of tumour cells within the primary mouse mammary tumour occurs preferentially at the TMEM and is dependent on VEGF signalling from macrophages and subsequent loss of vascular junctions and transiently increased vascular permeability210. Interestingly, while inhibition of Vegf signalling in zebrafish inhibits primary tumour vascularization, it has been shown to increase the formation of micrometastases at distal sites by increasing neutrophil migration, which deforms the collagen matrix and supports tumour cell invasion221. In zebrafish, human tumour-associated macrophages (TAMs) such as activated M2 macrophages and TAMs isolated from breast, lung, colorectal and endometrial cancers, increase metastasis of murine T241 fibrosarcoma cells by interacting with tumour cells and facilitating intravasation222.
Thus, the zebrafish emerges as a non-mammalian model system that lends itself to cost-efficient studies of aspects of tumour-related disease. Furthermore, zebrafish can easily be exposed to carcinogens or drugs, making them a potential system for high-throughput screening studies and personalized treatments196,223.
Perspectives
Despite decades of study, metastasis remains the major cause of mortality in patients with cancer. In addition to traditional mouse models, non-mammalian model organisms will continue to contribute key insights into the basic mechanisms of both normal and pathological cell migration as well as the development of therapies for cancer and metastasis. These models offer advantages that include high-resolution live imaging, the ability to screen thousands of organisms relatively quickly and inexpensively, and genetic tractability. Studies of D. discoideum have uncovered the complex signalling networks that drive chemokine-directed cell motility, which is also a feature of tumour and immune cell migration. Work in C. elegans demonstrated that cells can push basement membranes aside in addition to enzymatically degrading them, offering a new mechanism to target for this key step in metastasis, assuming it proves to be conserved in human cancer. In D. melanogaster and zebrafish, diverse modes of collective and individual cell migration that occur during normal morphogenesis as well as in simple and relatively inexpensive models of tumour metastasis have been described. Border cells show that molecular mechanisms of cooperative, collective cell migration can differ in important ways from those of single-cell migration, which is exemplified by the positive role of E-cadherin in collective direction-sensing of the border cells. D. melanogaster metastasis models have revealed signalling networks within tumour cells and between tumour cells and their neighbours in addition to systemic factors that promote the growth and spread of abnormal cells. D. melanogaster is also a valuable model for cancer drug discovery and optimization. Zebrafish offer the simplest and least expensive model for directly observing intravasation and extravasation.
While the experimental advantages of these models have elucidated critical concepts as well as molecular pathways and candidate treatments, many open questions remain. It is important to appreciate that cell migration contributes both positively and negatively to tumour metastasis. For example, stromal cells such as tumour-associated fibroblasts and macrophages can enable tumour cell invasion, intravasation, extravasation and colonization of new sites217,220,224, whereas T cell migration into a solid tumour is essential for the success of state-of-the-art immune therapies71. Therefore, it will be crucial to reveal the molecular underpinnings of all types of motility and to identify differences between them that can be exploited therapeutically. As described in this Review, examples of different types of normal and abnormal cell migration abound in non-mammalian model organisms and are likely to yield key insights into how to promote beneficial antitumour T cell migration while inhibiting the motility of metastasis-promoting cells. Understanding how tumour cells switch between different modes may also present opportunities for therapeutic intervention.
All models have limitations. For example, D. discoideum, D. melanogaster and C. elegans lack blood and lymphatic vessels, which are recognized barriers to metastasizing cancer cells in mammals. However, time and again, we are surprised by the degree of conservation of fundamental cellular properties and their underlying molecular mechanisms, and many important insights into the molecular mechanisms driving cancer have come from the study of normal cell and developmental biology. Components of the WNT, Hedgehog, RTK, MAPK, Notch and apoptotic pathways, as well as telomerase and more, were elucidated from work on the normal biology of simple model organisms. Now, direct modelling of metastasis and testing of drugs in organisms ranging from D. melanogaster to zebrafish and mice also appear extremely promising. Breakthroughs frequently occur when work in simple animals and mammalian models converge.
Lamellipodium.
A quasi-two-dimensional structure localized at the leading edge of motile cells that contains a highly dynamic actin network.
Lamellum.
A structure containing stable actin filaments and mature adhesion sites localized just behind the lamellipodium.
Basement membrane.
A thin, fibrous membrane that separates epithelium, mesothelium or endothelium from the underlying stroma.
Chemokines.
A family of low molecular mass proteins that are secreted by various cells and regulate a variety of responses, including cell migration, morphogenesis and proliferation as well as angiogenesis by binding to G protein-coupled receptors.
Matrix metalloproteinase.
(MMP). Calcium-dependent, zinc-containing endopeptidases that degrade matrix proteins.
Mesendoderm.
An embryonic tissue layer that differentiates into mesoderm and endoderm.
Directional persistence.
A measure commonly defined as the ratio of displacement to trajectory length.
Acknowledgements
The authors thank their anonymous reviewers for helpful suggestions and careful reading of the manuscript. This work was funded by the Intramural Research Program, National Cancer Institute, National Institutes of Health, by internal funding from the University of Michigan to C.A.P and by NIH grants R01GM73164 and R01GM46425 to D.J.M.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Seok J et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl Acad. Sci. USA 110, 3507–3512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis HM & Horvitz HR Genetic control of programmed cell death in the nematode C. elegans. Cell 44, 817–829 (1986). [DOI] [PubMed] [Google Scholar]
- 3.Nüsslein-Volhard C & Wieschaus E Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 (1980). [DOI] [PubMed] [Google Scholar]
- 4.Klein CA Selection and adaptation during metastatic cancer progression. Nature 501, 365–372 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin T, Xu X & Hereld D Chemotaxis, chemokine receptors and human disease. Cytokine 44, 1–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charest PG & Firtel RA Big roles for small GTPases in the control of directed cell movement. Biochem. J 401, 377–390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarris M & Sixt M Navigating in tissue mazes: chemoattractant interpretation in complex environments. Curr. Opin. Cell Biol 36, 93–102 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Te Boekhorst V, Preziosi L & Friedl P Plasticity of cell migration in vivo and in silico. Annu. Rev. Cell Dev. Biol 32, 491–526 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Lauffenburger DA & Horwitz AF Cell migration: a physically integrated molecular process. Cell 84, 359–369 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Montell DJ Morphogenetic cell movements: diversity from modular mechanical properties. Science 322, 1502–1505 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Riveline D et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol 153, 1175–1186 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt S & Friedl P Interstitial cell migration: integrin-dependent and alternative adhesion mechanisms. Cell Tissue Res. 339, 83–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lämmermann T & Sixt M Mechanical modes of “amoeboid” cell migration. Curr. Opin. Cell Biol 21, 636–644 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Paluch EK & Raz E The role and regulation of blebs in cell migration. Curr. Opin. Cell Biol 25, 582–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lämmermann T et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 453, 51–55 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Friedl P & Wolf K Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3, 362–374 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Haeger A, Wolf K, Zegers MM & Friedl P Collective cell migration: guidance principles and hierarchies. Trends Cell Biol. 25, 556–566 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Doyle AD, Wang FW, Matsumoto K & Yamada KM One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol 184, 481–490 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver VM, Howlett AR, Langton-Webster B, Petersen OW & Bissell MJ The development of a functionally relevant cell culture model of progressive human breast cancer. Semin. Cancer Biol 6, 175–184 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Wolf K et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol 9, 893–904 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Friedl P & Wolf K Plasticity of cell migration: a multiscale tuning model. J. Cell Biol 188, 11–19 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedl P, Sahai E, Weiss S & Yamada KM New dimensions in cell migration. Nat. Rev. Mol. Cell Biol 13, 743–747 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Pickup MW, Mouw JK & Weaver VM The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 15, 1243–1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena M & Christofori G Rebuilding cancer metastasis in the mouse. Mol. Oncol 7, 283–296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert AW, Pattabiraman DR & Weinberg RA Emerging biological principles of metastasis. Cell 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Bloom HJG The value of histology in the prognosis and classification of breast cancer. Proc. R. Soc. Med 51, 122–126 (1957). [PMC free article] [PubMed] [Google Scholar]
- 29.Gordetsky J & Epstein J Grading of prostatic adenocarcinoma: current state and prognostic implications. Diagn. Pathol 11, 25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller A et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50–56 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Qian B-Z & Pollard JW Macrophage diversity enhances tumor progression and metastasis. Cell 141, 39–51 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shields JD et al. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell 11, 526–538 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Sceneay J, Smyth MJ & Möller A The premetastatic niche: finding common ground. Cancer MetastasisRev. 32, 449–464 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Artemenko Y, Lampert TJ & Devreotes PN Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell. Mol. Life Sci 71, 3711–3747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eichinger L et al. The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chisholm RL & Firtel RA Insights into morphogenesis from a simple developmental system. Nat. Rev. Mol. Cell Biol 5, 531–541 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Nichols JM, Veltman D & Kay RR Chemotaxis of a model organism: progress with Dictyostelium. Curr. Opin. Cell Biol 36, 7–12 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Roussos ET, Condeelis JS & Patsialou A Chemotaxis in cancer. Nat. Rev. Cancer 11, 573–587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolaczkowska E & Kubes P Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol 13, 159–175 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Wculek SK & Malanchi I Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 528, 413–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruffell B, Affara NI & Coussens LM Differential macrophage programming in the tumor microenvironment. Trends Immunol. 33, 119–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sica A, Schioppa T, Mantovani A & Allavena P Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur. J. Cancer 42, 717–727 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Swaney KF, Huang C-H & Devreotes PN Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys 39, 265–289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parent CA & Devreotes PN A cell’s sense of direction. Science 284, 765–770 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Xiao Z, Zhang N, Murphy DB & Devreotes PN Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J. Cell Biol 139, 365–374 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin T, Zhang N, Long Y, Parent CA & Devreotes PN Localization of the G protein betagamma complex in living cells during chemotaxis. Science 287, 1034–1036 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB & Devreotes PN G protein signaling events are activated at the leading edge of chemotactic cells. Cell 95, 81–91 (1998).This work establishes D. discoideum as a model for imaging key biochemical events underlying chemotaxis at the leading edge using a novel biosensor for G protein signalling.
- 48.Servant G et al. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287, 1037–1040 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider IC & Haugh JM Quantitative elucidation of a distinct spatial gradient-sensing mechanism in fibroblasts. J. Cell Biol 171, 883–892 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bear JE & Haugh JM Directed migration of mesenchymal cells: where signaling and the cytoskeleton meet. Curr. Opin. Cell Biol 30, 74–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iijima M & Devreotes P Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109, 599–610 (2002).This paper demonstrates the key role of the PTEN tumour suppressor in chemotaxis.
- 52.Funamoto S, Meili R, Lee S, Parry L & Firtel RA Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109, 611–623 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Bagorda A & Parent CA Eukaryotic chemotaxis at a glance. J. CellSci 121, 2621–2624 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Comer FI & Parent CA PI 3-kinases and PTEN: how opposites chemoattract. Cell 109, 541–544 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Mayer IA & Arteaga CL The PI3K/AKT pathway as a target for cancer treatment. Annu. Rev Med 67, 11–28 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Engelman JA Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer 9, 550–562 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Jongsma M, Matas-Rico E, Rzadkowski A, Jalink K & Moolenaar WH LPA is a chemorepellent for B16 melanoma cells: action through the cAMP-elevating LPA5 receptor. PLoS ONE 6, e29260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shukla S et al. Activation of PI3K-Akt signaling pathway promotes prostate cancer cell invasion. Int. J. Cancer 121, 1424–1432 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Kim D et al. Akt/PKB promotes cancer cell invasion via increased motility and metalloproteinase production. FASEB J. 15, 1953–1962 (2001). [DOI] [PubMed] [Google Scholar]
- 60.Hawkins PT & Stephens LR PI3K signalling in inflammation. Biochim. Biophys. Acta 1851, 882–897 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Zhang S & Yu D PI(3)king apart PTEN’s role in cancer. Clin. Cancer Res 16, 4325–4330 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Okkenhaug K, Graupera M & Vanhaesebroeck B Targeting PI3K in cancer: impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. CancerDiscov. 6, 1090–1105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rericha EC & Parent CA Steering in quadruplet: the complex signaling pathways directing chemotaxis. Sci. Signal 1, e26 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Brzostowski JA et al. Phosphorylation of chemoattractant receptors regulates chemotaxis, actin reorganization and signal relay. J. Cell Sci 126, 4614–4626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Zijl F, Krupitza G & Mikulits W Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat. Res 728, 23–34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yilmaz M & Christofori G Mechanisms of motility in metastasizing cells. Mol. Cancer Res 8, 629–642 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Arshad N & Visweswariah SS The multiple and enigmatic roles of guanylyl cyclase C in intestinal homeostasis. FEBS Lett. 586, 2835–2840 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Cybulski N & Hall MN TOR complex 2: a signaling pathway of its own. Trends Biochem. Sci 34, 620–627 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Park JB et al. Phospholipase signalling networks in cancer. Nat. Rev. Cancer 12, 782–792 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Choi JW et al. LPA receptors: subtypes and biological actions. Annu. Rev. Pharmacol. Toxicol 50, 157–186 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Motz GT & Coukos G Deciphering and reversing tumor immune suppression. Immunity 39, 61–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rowe RG & Weiss SJ Breaching the basement membrane: who, when and how? Trends Cell Biol. 18, 560–574 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Sherwood DR & Sternberg PW Anchor cell invasion into the vulval epithelium in C. elegans. Dev. Cell 5, 21–31 (2003).This work establishes C. elegans as a powerful model for cells crossing basement membranes owing to the exceptionally clear live imaging.
- 74.Sherwood DR Cell invasion through basement membranes: an anchor of understanding. Trends Cell Biol. 16, 250–256 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Matus DQ et al. Invasive cell fate requires G1 cell-cycle arrest and histone deacetylase-mediated changes in gene expression. Dev. Cell 35, 162–174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sherwood DR, Butler JA, Kramer JM & Sternberg PW FOS-1 promotes basementmembrane removal during anchor-cell invasion in C. elegans. Cell 121, 951–962 (2005).This work uses genetic screening and live imaging to dissect molecular pathways of basement membrane removal in C. elegans with unprecedented clarity and precision.
- 77.Ozanne BW, Spence HJ, McGarry LC & Hennigan RF Transcription factors control invasion: AP-1 the first among equals. Oncogene 26, 1–10 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Hastie EL & Sherwood DR A new front in cell invasion: The invadopodial membrane. Eur. J. Cell Biol 95, 441–448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lohmer LL et al. A sensitized screen for genes promoting invadopodia function in vivo: CDC-42 and Rab GDI-1 direct distinct aspects of invadopodia formation. PLoS Genet. 12, e1005786 (2016).This work exploits the power of C. elegans genetics to identify key genes required for invadopodia formation and function in vivo.
- 80.Hagedorn EJ et al. The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J. Cell Biol 201, 903–913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murphy DA & Courtneidge SA The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol 12, 413–426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Castro-Castro A et al. Cellular and molecular mechanisms of MT1-MMP-dependent cancer cell invasion. Annu. Rev. Cell Dev. Biol 32, 555–576 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Foxall E, Pipili A, Jones GE & Wells CM Significance of kinase activity in the dynamic invadosome. Eur. J. Cell Biol 95, 483–492 (2016). [DOI] [PubMed] [Google Scholar]
- 84.Pozzi A, Yurchenco PD & Iozzo RV The nature and biology of basement membranes. Matrix Biol. 57-58, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelley LC, Lohmer LL, Hagedorn EJ & Sherwood DR Traversing the basement membrane in vivo: a diversity of strategies. J. Cell Biol 204, 291–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Glentis A et al. Cancer-associated fibroblasts induce metalloprotease-independent cancer cell invasion of the basement membrane. Nat. Commun 8, 924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kessenbrock K, Wang C-Y & Werb Z Matrix metalloproteinases in stem cell regulation and cancer. Matrix Biol. 44-46, 184–190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kessenbrock K, Plaks V & Werb Z Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Corcoran A & Del Maestro RF Testing the “Go or Grow” hypothesis in human medulloblastoma cell lines in two and three dimensions. Neurosurgery 53, 174–184 (2003). [DOI] [PubMed] [Google Scholar]
- 90.Garay T et al. Cell migration or cytokinesis and proliferation? — revisiting the “go or grow” hypothesis in cancer cells in vitro. Exp. Cell Res. 319, 3094–3103 (2013). [DOI] [PubMed] [Google Scholar]
- 91.Cheung KJ, Gabrielson E, Werb Z & Ewald AJ Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 155, 1639–1651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giese A et al. Dichotomy of astrocytoma migration and proliferation. Int. J. Cancer 67, 275–282 (1996). [DOI] [PubMed] [Google Scholar]
- 93.Cheung KJ et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl Acad. Sci. USA 113, E854–E863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Man Y-G et al. Tumor-infiltrating immune cells promoting tumor invasion and metastasis: existing theories. J. Cancer 4, 84–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ewald AJ Pulling cells out of tumours. Nat. Cell Biol 19, 147–149 (2017). [DOI] [PubMed] [Google Scholar]
- 96.Labernadie A et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol 19, 224–237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Konen J et al. Image-guided genomics of phenotypically heterogeneous populations reveals vascular signalling during symbiotic collective cancer invasion. Nat. Commun 8, 15078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Theveneau E & Mayor R Collective cell migration of epithelial and mesenchymal cells. Cell. Mol. Life Sci 70, 3481–3492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aceto N et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158, 1110–1122 (2014).This work demonstrates that circulating breast tumour cell clusters are more effective than single cells at colonizing distant sites.
- 100.Grigore AD, Jolly MK, Jia D, Farach-Carson MC & Levine H Tumor budding: the name is EMT Partial EMT J. Clin. Med 5, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Montell DJ, Yoon WH & Starz-Gaiano M Group choreography: mechanisms orchestrating the collective movement of border cells. Nat. Rev. Mol. Cell Biol 13, 631–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Montell DJ, Rorth P & Spradling AC slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes Drosophila C/EBP. Cell 71, 51–62 (1992).This paper establishes D. melanogaster border cells as a genetic model for studying collective epithelial cell motility in vivo.
- 103.Silver DL & Montell DJ Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell 107, 831–841 (2001).This work exploits the power of genetic screening in mosaic clones to show for the first time that JAK–STAT signalling is necessary and sufficient to activate motility within normal epithelial cells in vivo.
- 104.Starz-Gaiano M, Melani M, Wang X, Meinhardt H & Montell DJ Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev. Cell 14, 726–738 (2008). [DOI] [PubMed] [Google Scholar]
- 105.Yoon WH, Meinhardt H & Montell DJ miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat. Cell Biol 13, 1062–1069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Silver DL, Naora H, Liu J, Cheng W & Montell DJ Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 64, 3550–3558 (2004).This work demonstrates the role of STAT3 in cancer cell motility in vitro.
- 107.Yue P et al. Hyperactive EGF receptor, Jaks and Stat3 signaling promote enhanced colony-forming ability, motility and migration of cisplatin-resistant ovarian cancer cells. Oncogene 31, 2309–2322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gu L et al. Stat5 promotes metastatic behavior of human prostate cancer cells in vitro and in vivo. Endocr. Relat. Cancer 17, 481–493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moser C et al. STAT5b as molecular target in pancreatic cancer — inhibition of tumor growth, angiogenesis, and metastases. Neoplasia 14, 915–925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Niwa Y et al. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 24, 6406–6417 (2005). [DOI] [PubMed] [Google Scholar]
- 111.Chuang C-H et al. Molecular definition of a metastatic lung cancer state reveals a targetable CD109-Janus kinase-Stat axis. Nat. Med 23, 291–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Teng Y, Ross JL & Cowell JK The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT 3, e28086 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuzet S-E & Gaggioli C Fibroblast activation in cancer: when seed fertilizes soil. Cell Tissue Res. 365, 607–619 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Wang X et al. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell 10, 483–495 (2006). [DOI] [PubMed] [Google Scholar]
- 115.Duchek P, Somogyi K, Jékely G, Beccari S & Rørth P Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107, 17–26 (2001). [DOI] [PubMed] [Google Scholar]
- 116.McDonald JA, Pinheiro EM, Kadlec L, Schupbach T & Montell DJ Multiple EGFR ligands participate in guiding migrating border cells. Dev. Biol 296, 94–103 (2006). [DOI] [PubMed] [Google Scholar]
- 117.McDonald JA, Pinheiro EM & Montell DJ PVF1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development 130, 3469–3478 (2003). [DOI] [PubMed] [Google Scholar]
- 118.Duchek P & Rørth P Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science 291, 131–133 (2001). [DOI] [PubMed] [Google Scholar]
- 119.Murphy AM & Montell DJ Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J. Cell Biol 133, 617–630 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ridley AJ Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol 36, 103–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zegers MM & Friedl P Rho GTPases in collective cell migration. Small GTPases 5, e28997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang X, He L, Wu YI, Hahn KM & Montell DJ Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol 12, 591–597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fernández-Espartero CH et al. GTP exchange factor Vav regulates guided cell migration by coupling guidance receptor signalling to local Rac activation. J. Cell Sci 126, 2285–2293 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Ramel D, Wang X, Laflamme C, Montell DJ & Emery G Rab11 regulates cell-cell communication during collective cell movements. Nat. Cell Biol 15, 317–324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Palamidessi A et al. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell 134, 135–147 (2008). [DOI] [PubMed] [Google Scholar]
- 126.Naora H & Montell DJ Ovarian cancer metastasis: integrating insights from disparate model organisms. Nat. Rev. Cancer 5, 355–366 (2005). [DOI] [PubMed] [Google Scholar]
- 127.Hou J-M et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol 30, 525–532 (2012). [DOI] [PubMed] [Google Scholar]
- 128.Maddipati R & Stanger BZ Pancreatic cancer metastases harbor evidence of polyclonality. CancerDiscov. 5, 1086–1097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gundem G et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Niewiadomska P, Godt D & Tepass U DE-Cadherin is required for intercellular motility during Drosophila oogenesis. J. Cell Biol 144, 533–547 (1999).This work demonstrates a surprising requirement for E-cadherin-mediated adhesion between border cells and nurse cells during border cell migration.
- 131.Cai D et al. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157, 1146–1159 (2014).This study uses sophisticated genetics and live imaging to show that E-cadherin serves three distinct positive roles in promoting collective direction-sensing during border cell migration.
- 132.Desgrosellier JS & Cheresh DA Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cai D et al. Modeling and analysis of collective cell migration in an in vivo three-dimensional environment. Proc. Natl Acad. Sci. USA 113, E2134–E2141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cheung KJ & Ewald AJ A collective route to metastasis: seeding by tumor cell clusters. Science 352, 167–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reinhold WC et al. Multifactorial regulation of E-cadherin expression: an integrative study. Mol. Cancer Ther 9, 1–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jeanes A, Gottardi CJ & Yap AS Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jolly MK, Ware KE, Gilja S, Somarelli JA & Levine H EMT and MET: necessary or permissive for metastasis? Mol. Oncol 11, 755–769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rodriguez FJ, Lewis-Tuffin LJ & Anastasiadis PZ E-Cadherin’s dark side: possible role in tumor progression. Biochim. Biophys. Acta 1826, 23–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shamir ER et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J. Cell Biol 204, 839–856 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pinheiro EM & Montell DJ Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development 131, 5243–5251 (2004). [DOI] [PubMed] [Google Scholar]
- 141.Sallak Y, Torres AY, Yin H & Montell D Src42A required for collective border cell migration in vivo. bioRxiv 10.1101/186049 (2017). [DOI] [Google Scholar]
- 142.Serrels A, Canel M, Brunton VG & Frame MC Src/FAK-mediated regulation of E-cadherin as a mechanism for controlling collective cell movement: insights from in vivo imaging. Cell Adh. Migr. 5, 360–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cai D & Montell DJ Diverse and dynamic sources and sinks in gradient formation and directed migration. Curr. Opin. Cell Biol 30, 91–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pocha SM & Montell DJ Cellular and molecular mechanisms of single and collective cell migrations in Drosophila: themes and variations. Annu. Rev. Genet 48, 295–318 (2014). [DOI] [PubMed] [Google Scholar]
- 145.Kunwar PS, Siekhaus DE & Lehmann R In vivo migration: a germ cell perspective. Annu. Rev. Cell Dev. Biol 22, 237–265 (2006). [DOI] [PubMed] [Google Scholar]
- 146.Bae Y-K, Trisnadi N, Kadam S & Stathopoulos A The role of FGF signaling in guiding coordinate movement of cell groups: guidance cue and cell adhesion regulator? Cell Adh. Migr 6, 397–403 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ratheesh A, Belyaeva V & Siekhaus DE Drosophila immune cell migration and adhesion during embryonic development and larval immune responses. Curr. Opin. Cell Biol 36, 71–79 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Evans IR & Wood W Drosophila blood cell chemotaxis. Curr. Opin. Cell Biol 30, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Trisnadi N & Stathopoulos A Ectopic expression screen identifies genes affecting Drosophila mesoderm development including the HSPG Trol. G3 5, 301–313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Heisenberg C-P Dorsal closure in Drosophila: cells cannot get out of the tight spot. Bioessays 31, 1284–1287 (2009). [DOI] [PubMed] [Google Scholar]
- 151.Andrew DJ & Ewald AJ Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev. Biol 341, 34–55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bischoff M Lamellipodia-based migrations of larval epithelial cells are required for normal closure of the adult epidermis of Drosophila. Dev. Biol 363, 179–190 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gateff E Malignant neoplasms of genetic origin in Drosophila melanogaster. Science 200, 1448–1459 (1978).This study establishes D. melanogaster as a tumour model and demonstrates that mutations can cause neoplasms in this organism.
- 154.Elsum I, Yates L, Humbert PO & Richardson HE The Scribble-Dlg-Lgl polarity module in development and cancer: from flies to man. Essays Biochem. 53, 141–168 (2012). [DOI] [PubMed] [Google Scholar]
- 155.Bilder D, Li M & Perrimon N Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113–116 (2000). [DOI] [PubMed] [Google Scholar]
- 156.Bilder D & Perrimon N Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403, 676–680 (2000). [DOI] [PubMed] [Google Scholar]
- 157.Campanale JP, Sun TY & Montell DJ Development and dynamics of cell polarity at a glance. J. Cell Sci 130, 1201–1207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Halaoui R & McCaffrey L Rewiring cell polarity signaling in cancer. Oncogene 34, 939–950 (2015). [DOI] [PubMed] [Google Scholar]
- 159.Feigin ME et al. Mislocalization of the cell polarity protein scribble promotes mammary tumorigenesis and is associated with basal breast cancer. Cancer Res. 74, 3180–3194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lin W-H, Asmann YW & Anastasiadis PZ Expression of polarity genes in human cancer. Cancer Inform. 14, 15–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vaira V et al. Aberrant overexpression of the cell polarity module scribble in human cancer. Am. J. Pathol 178, 2478–2483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhan L et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135, 865–878 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Roberts AB & Wakefield LM The two faces of transforming growth factor beta in carcinogenesis. Proc. Natl Acad. Sci. USA 100, 8621–8623 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pagliarini RA & Xu T A genetic screen in Drosophila for metastatic behavior. Science 302, 1227–1231 (2003). [DOI] [PubMed] [Google Scholar]
- 165.Brumby AM & Richardson HE scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 22, 5769–5779 (2003).References 164 and 165 establish D. melanogaster as a model for genetic screening for mutations that promote the spread of cells expressing oncogenic Ras or Notch throughout the fly larva.
- 166.Wu M, Pastor-Pareja JC & Xu T Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature 463, 545–548 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chi C et al. Disruption of lysosome function promotes tumor growth and metastasis in Drosophila. J. Biol. Chem 285, 21817–21823 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dhanasekaran DN & Reddy E P JNK signaling in apoptosis. Oncogene 27, 6245–6251 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Leong GR, Goulding KR, Amin N, Richardson HE & Brumby AM Scribble mutants promote aPKC and JNK-dependent epithelial neoplasia independently of Crumbs. BMC Biol. 7, 62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Igaki T, Pagliarini RA & Xu T Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr. Biol 16, 1139–1146 (2006). [DOI] [PubMed] [Google Scholar]
- 171.Sun G & Irvine KD Ajuba family proteins link JNK to Hippo signaling. Sci. Signal 6, ra81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rudrapatna VA, Bangi E & Cagan RL Caspase signalling in the absence of apoptosis drives Jnk-dependent invasion. EMBO Rep. 14, 172–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen F JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer Res. 72, 379–386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Read RD et al. A Drosophila model of multiple endocrine neoplasia type 2. Genetics 171, 1057–1081 (2005).This study establishes that D. melanogaster, improbably, serves as a model for multiple endocrine neoplasia type 2 (MEN2).
- 175.Das TK & Cagan RLA Drosophila approach to thyroid cancer therapeutics. Drug Discov. Today Technol 10, e65–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dar AC, Das TK, Shokat KM & Cagan RL Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486, 80–84 (2012).This study demonstrates the power of D. melanogaster to identify a kinase inhibitor that when used in combination with inhibitors of other targets renders it more effective and safer than another drug already in clinical use for MEN2.
- 177.Sonoshita M & Cagan RL Modeling human cancers in drosophila. Curr. Top. Dev. Biol 121, 287–309 (2017). [DOI] [PubMed] [Google Scholar]
- 178.Figueroa-Clarevega A & Bilder D Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev. Cell 33, 47–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kwon Y et al. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell 33, 36–46 (2015).References 178 and 179 show that D. melanogaster can model systemic features such as cachexia.
- 180.Petrie RJ & Yamada KM Multiple mechanisms of 3D migration: the origins of plasticity. Curr. Opin. Cell Biol 42, 7–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zatulovskiy E, Tyson R, Bretschneider T & Kay RR Bleb-driven chemotaxis of Dictyostelium cells. J. Cell Biol 204, 1027–1044 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liu Y-J et al. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 160, 659–672 (2015). [DOI] [PubMed] [Google Scholar]
- 183.Harvie EA & Huttenlocher A Neutrophils in host defense: new insights from zebrafish. J. Leukoc. Biol 98, 523–537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Paksa A & Raz E Zebrafish germ cells: motility and guided migration. Curr. Opin. Cell Biol 36, 80–85 (2015). [DOI] [PubMed] [Google Scholar]
- 185.Blaser H et al. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J. Cell Sci 118, 4027–4038 (2005). [DOI] [PubMed] [Google Scholar]
- 186.Meyen D et al. Dynamic filopodia are required for chemokine-dependent intracellular polarization during guided cell migration in vivo. elife 4, e05279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dumstrei K, Mennecke R & Raz E Signaling pathways controlling primordial germ cell migration in zebrafish. J. Cell Sci 117, 4787–4795 (2004). [DOI] [PubMed] [Google Scholar]
- 188.Blaser H et al. Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev. Cell 11, 613–627 (2006). [DOI] [PubMed] [Google Scholar]
- 189.Tarbashevich K, Reichman-Fried M, Grimaldi C & Raz E Chemokine-dependent pH elevation at the cell front sustains polarity in directionally migrating zebrafish germ cells. Curr. Biol 25, 1096–1103 (2015). [DOI] [PubMed] [Google Scholar]
- 190.Charras G & Paluch E Blebs lead the way: how to migrate without lamellipodia. Nat. Rev. Mol. Cell Biol 9, 730–736 (2008). [DOI] [PubMed] [Google Scholar]
- 191.Bereiter-Hahn J, Lück M, Miebach T, Stelzer HK & Vöth M Spreading of trypsinized cells: cytoskeletal dynamics and energy requirements. J. Cell Sci 96, 171–188 (1990). [DOI] [PubMed] [Google Scholar]
- 192.Diz-Muñoz A et al. Steering cell migration by alternating blebs and actin-rich protrusions. BMC Biol. 14, 74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Balzer EM et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 26, 4045–4056 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stroka KM & Konstantopoulos K Physical biology in cancer. 4. Physical cues guide tumor cell adhesion and migration. Am. J. Physiol, Cell Physiol 306, C98–C109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ignatius MS, Hayes M & Langenau DM In vivo imaging of cancer in zebrafish. Adv. Exp. Med. Biol 916, 219–237 (2016). [DOI] [PubMed] [Google Scholar]
- 196.Spaink HP et al. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods 62, 246–254 (2013). [DOI] [PubMed] [Google Scholar]
- 197.Taylor AM & Zon LI Zebrafish tumor assays: the state of transplantation. Zebrafish 6, 339–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhao S, Huang J & Ye J A fresh look at zebrafish from the perspective of cancer research. J. Exp. Clin. Cancer Res 34, 80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Starnes TW & Huttenlocher A Neutrophil reverse migration becomes transparent with zebrafish. Adv. Hematol 2012, 398640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.White RM et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189 (2008).This paper discusses the use of transparent zebrafish as a xenograft model that allows visualization of tumour spread.
- 201.White R, Rose K & Zon L Zebrafish cancer: the state of the art and the path forward. Nat. Rev. Cancer 13, 624–636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Moore JC et al. Single-cell imaging of normal and malignant cell engraftment into optically clear prkdc-null SCID zebrafish. J. Exp. Med 213, 2575–2589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tang Q et al. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat. Methods 11, 821–824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Drabsch Y, Snaar-Jagalska BE & Dijke P Fish tales: The use of zebrafish xenograft human cancer cell models. Histol. Histopathol 32, 673–686 (2017). [DOI] [PubMed] [Google Scholar]
- 205.Mercatali L et al. Development of a patient-derived xenograft (PDX) of breast cancer bone metastasis in a zebrafish model. Int. J. Mol. Sci 17, E1375 (2016).This paper demonstrates that patient-derived tumour tissue can be transplanted into zebrafish and that the behaviour of these xenografts reflects the clinical course of the patients’ disease.
- 206.Gaudenzi G et al. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in neuroendocrine tumors. Endocrine 57, 214–219 (2016). [DOI] [PubMed] [Google Scholar]
- 207.Stoletov K, Montel V, Lester RD, Gonias SL & Klemke R High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc. Natl Acad. Sci. USA 104, 17406–17411 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Reymond N et al. RhoC and ROCKs regulate cancer cell interactions with endothelial cells. Mol. Oncol 9, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Weis S, Cui J, Barnes L & Cheresh D Endothelial barrier disruption by VEGF-mediated Src activity potentiates tumor cell extravasation and metastasis. J. Cell Biol 167, 223–229 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Harney AS et al. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Discov. 5, 932–943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Stoletov K et al. Visualizing extravasation dynamics of metastatic tumor cells. J. Cell Sci 123, 2332–2341 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hagedorn EJ et al. Integrin acts upstream of netrin signaling to regulate formation of the anchor cell’s invasive membrane in C. elegans. Dev. Cell 17, 187–198 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chen MB, Lamar JM, Li R, Hynes RO & Kamm RD Elucidation of the roles of tumor integrin β1 in the extravasation stage of the metastasis cascade. Cancer Res. 76, 2513–2524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Reymond N et al. Cdc42 promotes transendothelial migration of cancer cells through β1 integrin. J. Cell Biol 199, 653–668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pagès F et al. Immune infiltration in human tumors: a prognostic factor that should not be ignored. Oncogene 29, 1093–1102 (2010). [DOI] [PubMed] [Google Scholar]
- 216.Condeelis J & Pollard JW Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 124, 263–266 (2006). [DOI] [PubMed] [Google Scholar]
- 217.Roh-Johnson M et al. Macrophage contact induces RhoA GTPase signaling to trigger tumor cell intravasation. Oncogene 33, 4203–4212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wyckoff JB et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 67, 2649–2656 (2007). [DOI] [PubMed] [Google Scholar]
- 219.Goswami S et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 65, 5278–5283 (2005). [DOI] [PubMed] [Google Scholar]
- 220.Robinson BD et al. Tumor microenvironment of metastasis in human breast carcinoma: a potential prognostic marker linked to hematogenous dissemination. Clin. Cancer Res 15, 2433–2441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.He S et al. Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J. Pathol 227, 431–445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang J et al. Novel mechanism of macrophage-mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res. 75, 306–315 (2015). [DOI] [PubMed] [Google Scholar]
- 223.Fior R et al. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc. Natl Acad. Sci. USA 114, E8234–E8243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mezawa Y & Orimo A The roles of tumor- and metastasis-promoting carcinoma-associated fibroblasts in human carcinomas. Cell Tissue Res. 365, 675–689 (2016). [DOI] [PubMed] [Google Scholar]
- 225.Bernards R & Weinberg RA A progression puzzle. Nature 418, 823 (2002).This paper supports the view that genes that drive tumour initiation and progression may overlap substantially.
- 226.Steeg PS Targeting metastasis. Nat. Rev Cancer 16, 201–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kodura MA & Souchelnytskyi S Breast carcinoma metastasis suppressor gene 1 (BRMS1): update on its role as the suppressor of cancer metastases. Cancer Metastasis Rev. 34, 611–618 (2015). [DOI] [PubMed] [Google Scholar]
- 228.Nguyen DX & Massagué J Genetic determinants of cancer metastasis. Nat. Rev. Genet 8, 341–352 (2007). [DOI] [PubMed] [Google Scholar]
- 229.Nguyen DX, Bos PD & Massagué J Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284 (2009).This paper proposes the view that metastasis genes can be identified.
- 230.Meehan WJ et al. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J. Biol. Chem 279, 1562–1569 (2004). [DOI] [PubMed] [Google Scholar]
- 231.Horak CE et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res. 67, 11751–11759 (2007). [DOI] [PubMed] [Google Scholar]
- 232.Sahni S et al. The metastasis suppressor, N-myc downstream-regulated gene 1 (NDRG1), inhibits stress-induced autophagy in cancer cells. J. Biol. Chem 289, 9692–9709 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wessels D, Lusche DF, Kuhl S, Heid P & Soll DR PTEN plays a role in the suppression of lateral pseudopod formation during Dictyostelium motility and chemotaxis. J. Cell Sci. 120, 2517–2531 (2007). [DOI] [PubMed] [Google Scholar]
- 234.Lee T & Montell DJ Multiple Ras signals pattern the Drosophila ovarian follicle cells. Dev. Biol 185, 25–33 (1997). [DOI] [PubMed] [Google Scholar]
- 235.Dang TT, Prechtl AM & Pearson GW Breast cancer subtype-specific interactions with the microenvironment dictate mechanisms of invasion. Cancer Res. 71, 6857–6866 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Westcott JM et al. An epigenetically distinct breast cancer cell subpopulation promotes collective invasion. J. Clin. Invest 125, 1927–1943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Edme N, Downward J, Thiery J-P & Boyer B Ras induces NBT-II epithelial cell scattering through the coordinate activities of Rac and MAPK pathways. J. Cell Sci 115, 2591–2601 (2002). [DOI] [PubMed] [Google Scholar]
- 238.Kalluri R & Weinberg RA The basics of epithelial-mesenchymal transition. J. Clin. Invest 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Tsuji T, Ibaragi S & Hu G Epithelialmesenchymal transition and cell cooperativity in metastasis. Cancer Res. 69, 7135–7139 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Riihimäki M, Hemminki A, Sundquist J & Hemminki K Patterns of metastasis in colon and rectal cancer. Sci. Rep. 6, 29765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Entenberg D et al. In vivo subcellular resolution optical imaging in the lung reveals early metastatic proliferation and motility. Intravital 4, e1086613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ramakrishna R & Rostomily R Seed, soil, and beyond: the basic biology of brain metastasis. Surg. Neurol. Int 4, S256–S264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ren G, Esposito M & Kang Y Bone metastasis and the metastatic niche. J. Mol. Med 93, 1203–1212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Peinado H et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017). [DOI] [PubMed] [Google Scholar]
- 245.Avgustinova A et al. Tumour cell-derived Wnt7a recruits and activates fibroblasts to promote tumour aggressiveness. Nat. Commun 7, 10305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Calvo F et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Melero I, Rouzaut A, Motz GT & Coukos G T-Cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 4, 522–526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Senbabaoglu Y et al. The landscape of T cell infiltration in human cancer and its association with antigen presenting gene expression. bioRxiv 10.1101/025908 (2015). [DOI] [Google Scholar]
- 249.Nakaya Y & Sheng G EMT in developmental morphogenesis. Cancer Lett. 341, 9–15 (2013). [DOI] [PubMed] [Google Scholar]
- 250.Thiery JP, Acloque H, Huang RYJ & Nieto MA Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 (2009). [DOI] [PubMed] [Google Scholar]
- 251.Zheng X et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Fischer KR et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015).References 251 and 252 show that, in contrast to established dogma, EMT is not necessary for metastasis of lung or pancreatic cancers but rather confers chemoresistance.
- 253.Steinestel K, Eder S, Schrader AJ & Steinestel J Clinical significance of epithelialmesenchymal transition. Clin. Transl Med 3, 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Bronsert P et al. Cancer cell invasion and EMT marker expression: a three-dimensional study of the human cancer-host interface. J. Pathol 234, 410–422 (2014). [DOI] [PubMed] [Google Scholar]